Abstract
The malaria parasite Plasmodium falciparum invades human red blood cells via interactions between host and parasite surface proteins. By analyzing genome sequence data from human populations, including 1269 individuals from sub-Saharan Africa, we identify a diverse array of large copy number variants affecting the host invasion receptor genes GYPA and GYPB. We find that a nearby association with severe malaria is explained by a complex structural rearrangement involving the loss of GYPB and gain of two GYPB-A hybrid genes, which encode a serologically distinct blood group antigen known as Dantu. This variant reduces the risk of severe malaria by 40% and has recently risen in frequency in parts of Kenya, yet it appears to be absent from west Africa. These findings link structural variation of red blood cell invasion receptors with natural resistance to severe malaria.
Malaria parasites cause human disease by invading and replicating inside red blood cells, which can lead to life-threatening complications that are a major cause of childhood mortality in Africa (1, 2). The invasion of red blood cells is orchestrated by the specific binding of parasite ligands to erythrocyte receptors (3), a stage at which genetic variation could influence the progression of infection. Indeed, a human genetic variant that prevents erythrocytic expression of the Duffy antigen receptor for chemokines (DARC), which is essential for invasion by Plasmodium vivax, is thought to have undergone a selective sweep resulting in the present-day absence of P. vivax malaria across most of sub-Saharan Africa (4). In contrast the main cause of malaria in Africa, P. falciparum, has an expanded family of erythrocyte binding ligands that target a different set of human receptors, most of which appear not to be required for invasion (5–7). Two such invasion receptors are the glycophorins GYPA and GYPB, which are abundantly expressed on the erythrocyte surface and underlie the MNS blood group system (6, 8–10). The antigenic complexity of this system as well as rates of amino acid substitution and levels of diversity in African populations have led to speculation that this locus is under evolutionary selection due to malaria (8, 11–13).
In a recent genome-wide association study (GWAS), we identified alleles associated with protection from severe malaria on chromosome 4, between FREM3 and the cluster of genes encoding GYPE, GYPB and GYPA (14). Although the association signal did not extend to these genes and a functional variant was not identified, interpretation and further analysis of the association signal was inhibited by several factors. First, the GWAS samples were collected at multiple locations in sub-Saharan Africa, where levels of human genetic diversity are higher than in other parts of the world. This diversity remains underrepresented in genome variation reference panels. Second, the glycophorin genes are in a region of segmental duplication that is difficult to characterize due to high levels of paralogy. Notably, the region is known to harbor structural variation that contributes to the MNS blood group system but has not been characterized by next generation sequence data (15, 16). Here we aim to capture additional variation in sub-Saharan African populations, including structural variation, to determine the underlying architecture of the association signal in this region.
An African-enriched reference panel in the glycophorin region
We constructed a reference panel with improved representation of sub-Saharan African populations from countries where malaria is endemic. We performed genome sequencing of 765 individuals from 10 ethnic groups in the Gambia, Burkina Faso, Cameroon and Tanzania, including 207 family trios (100 bp paired end (PE) reads, mean coverage 10x; Tables S1, S2). We focused on a region surrounding the observed association signal (chr4:140Mb-150Mb; GRCh37 coordinates). Genotypes at single nucleotide polymorphisms (SNPs) and short indels in the region were called and computationally phased (17–19) and combined with Phase 3 of the 1000 Genomes Project (20) to obtain a reference panel of 3,269 individuals, including 1,269 Africans and a further 157 individuals with African ancestry (Fig. S1; Tables S1, S3). We imputed variants from this panel into the published severe malaria GWAS dataset comprising 4,579 cases of severe malaria and 5,310 population controls from the Gambia, Kenya and Malawi and tested for association as described previously (14). The signal of association, formerly identified and replicated at SNPs lying between FREM3 and GYPE, extends over a region of at least 700 kb, and includes linked variants within GYPA and GYPB where association is only apparent with the additional African reference data (Fig. S2).
Identification of copy number variants
We next assessed copy number variation in the glycophorin region (defined here as the segmental duplication within which the three genes lie) for the sequenced reference panel individuals. The high level of sequence identity between the duplicate units presents a challenge for short read sequence analysis due to ambiguous mapping (Fig. 1A) (21). We therefore focused on changes in read depth at sites of high mappability and developed a hidden Markov model (HMM) to infer the underlying copy number state for each individual in 1600 bp windows. We grouped individuals carrying similar copy number paths to define copy number variants (CNVs) and assign individual CNV genotypes.
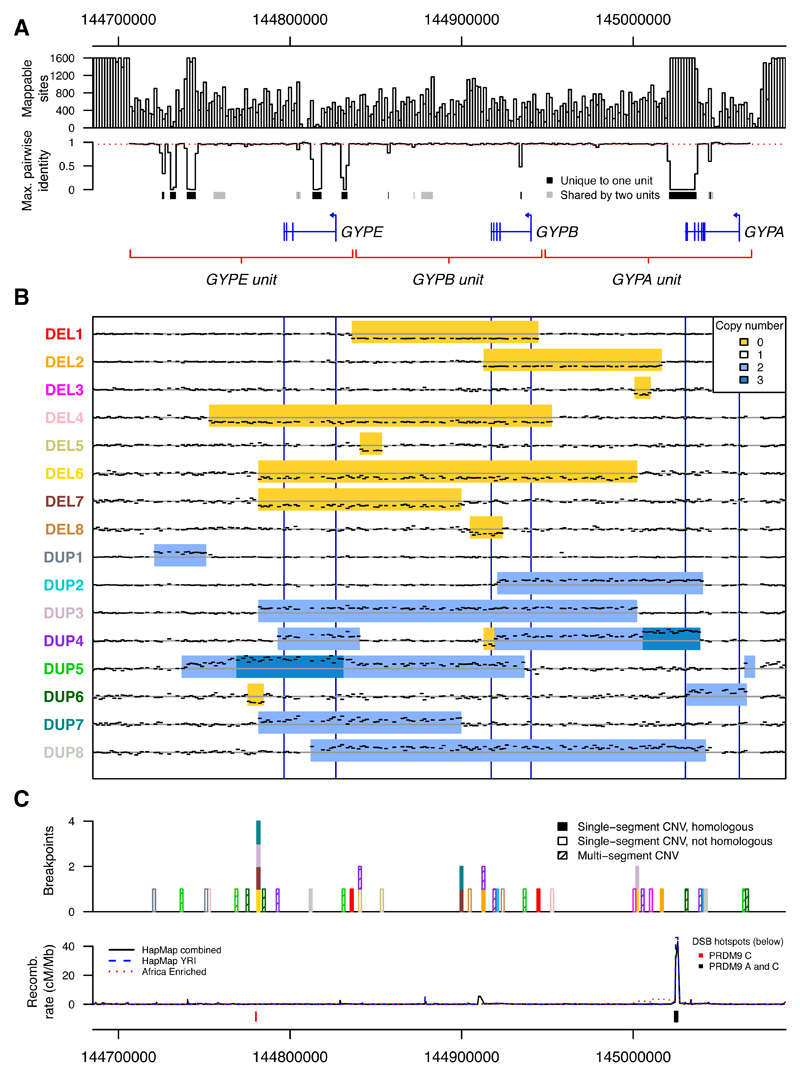
Fig. 1. Copy number variants in the glycophorin region.
(A) Mappability in the glycophorin region. The number of mappable sites and the maximum pairwise identity between homologous locations in the segmental duplication are shown in 1600 bp windows. Pairwise identity is inferred from a multiple sequence alignment, with mean of 0.96 indicated with a red dashed line. The locations of protein-coding genes and the segmentally duplicated units are indicated below, with sequences of at least 100 bp that are unique to one or two out of the three segmentally duplicated units marked in black and gray, respectively. (B) Sequence coverage in 1600 bp windows and copy number for non-singleton CNVs. Black dashes show the mean normalized sequence coverage across heterozygous individuals not carrying another CNV. Only windows input to the HMM are shown. The inferred CNVs are indicated with deletion in yellow, duplication in light blue, and triplication in dark blue. A horizontal gray line indicates the expected coverage without copy number variation and blue vertical lines mark the locations of the three genes. (C) Positions of breakpoints, colored as the variant names in (B) and shaded by whether the variant has a single pair of homologous breakpoints, a single pair of non-homologous breakpoints, or is a multi-segment CNV. The recombination rate from LD-based recombination maps (52, 53) and locations of DSB hotspots (24) are annotated below.
Across the 3,269 samples, we identified eight deletions and eight duplications that were found in ≥2 unrelated individuals (referred to below as non-singleton CNVs), as well as at least 11 singleton variants (Figs. 1B, S3) (22). For reference, we label these variants by copy number type (DEL for deletion, DUP for duplication), and number them in order of frequency. To validate the CNV calls we analyzed transmission in family trios and observed segregation as expected with few exceptions (Table S4) (22). We also compared the CNV calls with the 1000 Genomes Project structural variant analysis (23), and found highly consistent copy number inference (98.8% of individuals have the same overall copy number call) but substantial improvements to individual genotypes in our analysis (Fig. S4) (22). Validation of the breakpoint of the most common variant (DEL1) by Sanger sequencing further confirmed the accuracy of our method (Figs. S5, S6 and Table S5) (22).
The variants ranged in length from 3.2 kb (the minimum possible with our method) to >200 kb and included deletions and duplications of entire genes. Loss of GYPB was a common feature, with five different forms of GYPB deletion among the non-singleton CNVs (Fig. 1B). Hybrid gene structures were another common feature, with two non-singleton CNVs predicted to generate GYPB-A or GYPE-A hybrids (Fig. S7). Some variants are predicted to correspond to known MNS blood group antigens while others have not previously been reported (Table S6) (22). Of the non-singleton CNVs, half (8/16) had a single pair of breakpoints in homologous parts of the segmental duplication, consistent with formation via non-allelic homologous recombination (NAHR; Fig. 1C). Of these, four share a breakpoint position, which coincides with a double-strand break (DSB) hotspot active in a PRDM9 C allele carrier (Figs. 1C, S8) (24).
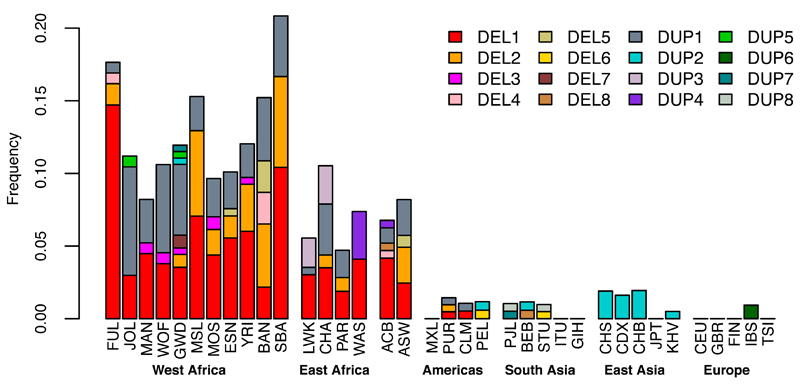
CNVs in the glycophorin region were observed more frequently in Africa than other parts of the world (Fig. 2). In this dataset, the combined allele frequency of glycophorin CNVs in African populations was 11% compared to 1.1% in non-African populations, and most of the non-singleton CNVs (13/16) were identified in individuals of African ancestry. Among fourteen different ethnic groups sampled in Africa, the estimated frequency ranged from 4.7-21% with the highest frequencies in west African populations.
Fig. 2. Frequency of CNVs in the sampled populations.
Populations are grouped on the basis of geographical proximity; abbreviations can be found in Tables S1 and S3.
Association with severe malaria
We sought to incorporate CNVs into the phased reference panel with the aim of imputing into our GWAS dataset. Computational phasing of CNVs is challenging, as published methods do not model CNV mutational mechanisms or non-diploid copy number at smaller variants within CNVs. To work around this, we excluded SNPs and short indels within the glycophorin region and relied on the trio structure of sequenced individuals to resolve haplotype phase between CNVs and flanking SNPs (18, 25). Haplotype clustering and cross-validation predict good imputation performance for the three highest frequency CNVs, DEL1, DEL2, and DUP1 as well as for DUP4 (Figs. S9, S10, S11).
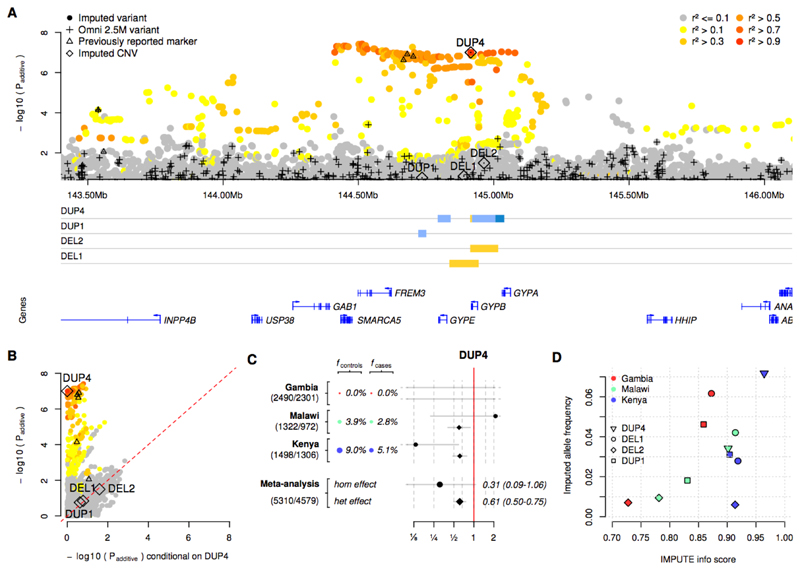
We used this panel to impute CNVs into the severe malaria GWAS samples (Fig. S12) and tested for association as before. One of the imputed CNVs, DUP4, is associated with decreased risk of severe malaria (odds ratio, OR=0.60; 95% CI 0.50-0.72; Padditive = 9.9x10-8; computed under an additive model of association using fixed-effect meta-analysis; Fig. 3A). Across populations, evidence for association at DUP4 is among the strongest of any variant in our data. Moreover, conditioning on the imputed genotypes at DUP4 in the statistical association model removes signal at all other strongly associated variants including the previously reported markers of association (e.g., conditional Padditive=0.32 at rs186873296; Figs. 3B, S13). DUP4 has an estimated heterozygous relative risk of 0.61 (95% CI 0.50-0.75) and its genetic effect appears to be consistent with an additive model, although the low frequency of homozygotes makes it difficult to distinguish the extent of dominance (homozygous relative risk 0.31; 95% CI 0.09-1.06; n=24 homozygotes; Fig. 3C). Analysis of different clinical forms of severe malaria showed that DUP4 reduced the risk of both cerebral malaria and severe malarial anaemia to a similar degree (Table S7). We noted some evidence of additional associations in the region, including a possible protective effect of DEL2 (OR=0.63; 95%CI=0.42-0.94, Padditive=0.02), but no evidence of association with the more common DEL1, or with GYPB deletion status overall (P>0.1 using logistic regression). These results are compatible with a primary signal of association that is well explained by an additive effect of DUP4.
Fig. 3. Evidence of association.
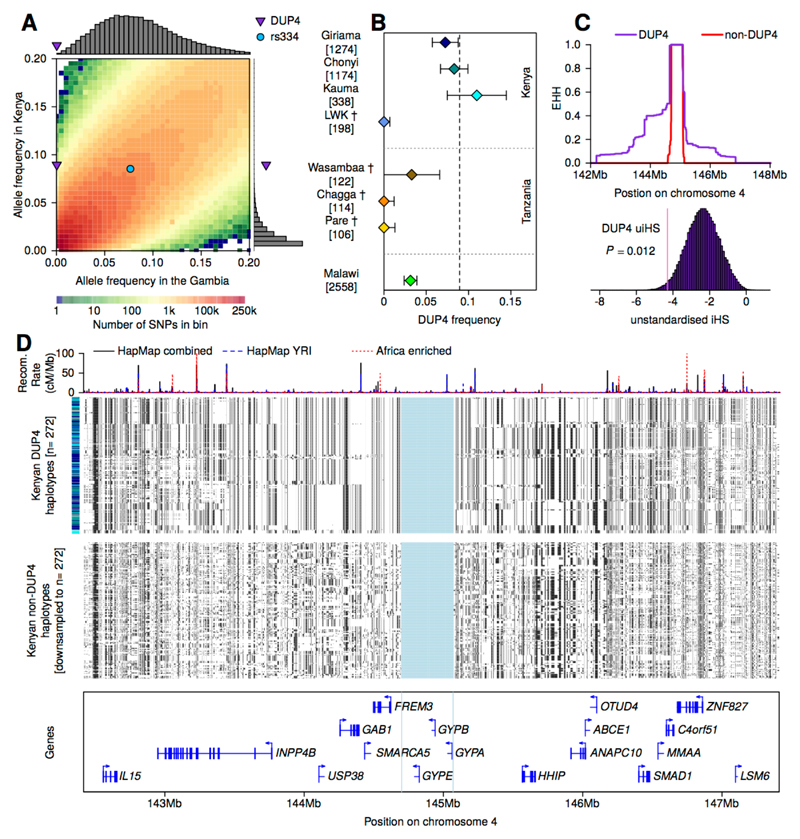
(A) The evidence for association at SNPs, short indels, and CNVs across the glycophorin region. P-values are computed under an additive model of association using meta-analysis across the three African populations included in our study. Points are colored by LD with DUP4 in east African reference panel populations. Directly typed SNPs are denoted with black plusses, and CNVs with diamonds. Black triangles represent SNPs where the association signal was previously reported and replicated in further samples. CNV copy number profiles, as in Fig. 1B, and protein-coding genes are shown below. (B) Comparison of unconditional association test P-values (y axis, as in panel (A)), and after additionally conditioning on genotypes at DUP4 (x axis). (C) The evidence for association at DUP4. Colored circles and text show the estimated allele frequency of DUP4 in population controls and severe malaria cases. To the right is the odds ratio and 95% confidence interval for DUP4 heterozygotes (diamonds) and homozygotes (circles) relative to non-carriers. The bottom two rows represent effect sizes computed by fixed-effect meta-analysis. Sample sizes (number of controls/number of cases) are denoted to the left. (D) Comparison of IMPUTE info score and expected imputed allele frequency for the four annotated CNVs.
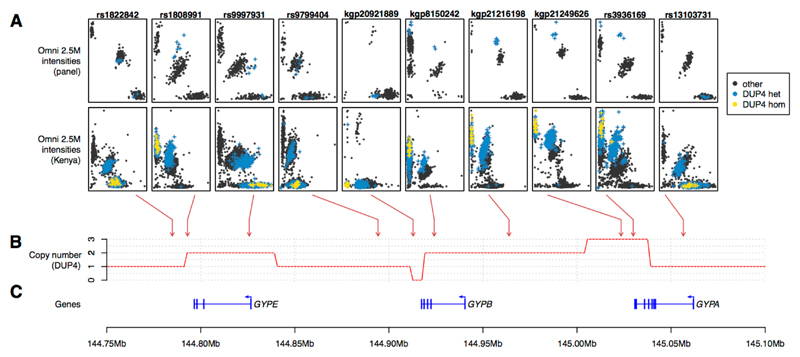
DUP4 is imputed with high confidence in both east African populations (Fig. 3D), where it is at substantially higher frequency than in the reference panel (Fig. S12). To independently confirm the imputed DUP4 genotypes, we analyzed SNP microarray data for intensity patterns indicative of copy number variation (Fig. 4) using a Bayesian clustering model informed by the sequenced DUP4 carriers (Fig. S14, Table S8) (22). Classification of GWAS samples was highly concordant with the imputed DUP4 genotypes in the east African populations (r2=0.96 in Kenya; r2=0.88 in Malawi; Table S9). Surprisingly, both imputation and the microarray intensity analysis suggest there may be no copies of DUP4 present among the 4791 Gambian individuals in the GWAS. This large frequency difference places DUP4 as an outlier compared with imputed variants at a similar frequency in the Gambia or in Kenya genome-wide (P=1.7x10-3 and P=5x10-3, respectively, based on the empirical distribution of frequencies; Fig. 5A). DUP4 also varies in frequency within east Africa (Fig. 5B). Computation of haplotype homozygosity provides evidence that DUP4 is carried on an extended haplotype (P=0.012 for iHS (26, 27) based on the empirical distribution for variants of similar frequency genome-wide; Fig. 5C-D) that may have risen to its current frequency in Kenya relatively recently. We note that DUP4 is also absent from all but two of the reference panel populations (Fig. 2).
Fig. 4. The effect of DUP4 on SNP array intensities.
(A) Normalized Illumina Omni 2.5M intensity values at selected SNP assays across the glycophorin region for reference panel individuals (top row; N=367 individuals from Burkina Faso, Cameroon, and Tanzania) and Kenyan GWAS individuals (second row, N = 3,142). Blue and yellow points represent individuals heterozygous or homozygous for DUP4 respectively, as determined by the HMM in reference panel individuals and by imputation in Kenya (genotypes with posterior probability at least 0.75). Arrows denote the mapping location of these SNPs. (B) The copy number profile of DUP4. (C) Position of the glycophorin genes and exons.
Fig. 5. DUP4 frequency and haplotype homozygosity.
(A) The empirical joint allele-frequency spectrum for population controls in the Gambia and Kenya, in 0.5% frequency bins between 0 and 20%. The frequencies of DUP4 and rs334 are highlighted. Histograms show the frequency in the Gambia of SNPs in the DUP4 frequency bin in Kenya (8.5-9%, top) and the frequency in Kenya of SNPs in the DUP4 frequency bin in the Gambia (0-0.5%, right). (B) The estimated frequency of DUP4 in east African populations, shown with 95% confidence intervals and the number of haplotypes sampled. Estimates are from population controls in the GWAS or from the HMM genotype calls in the reference panel (daggers). The dotted vertical line denotes the overall frequency of DUP4 in Kenyan controls. (C) Extended haplotype homozygosity (EHH) computed outward from the glycophorin region for DUP4 haplotypes and non-DUP4 haplotypes in Kenya, after excluding other variants within the glycophorin region. Below, the empirical distribution of unstandardized iHS for all typed SNPs within 1% frequency of DUP4 in Kenyan controls, with empirical P-value annotated. (D) The 272 haplotypes imputed to carry DUP4 and a random sample of 272 non-DUP4 haplotypes in Kenya. Haplotypes in each panel are clustered on 1 Mb extending in either direction from the glycophorin region, which is shaded in blue. The bar on the left depicts the population for each haplotype with colors as in panel (B). Recombination rate estimates (52, 53) are shown above and protein-coding genes below.
The physical structure of DUP4
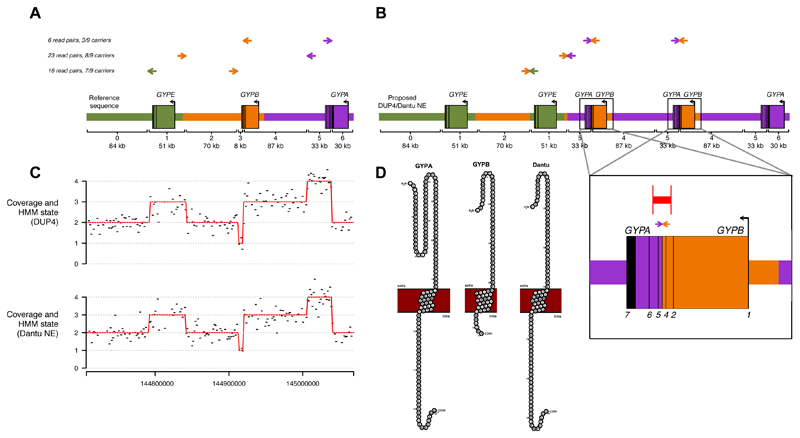
The copy number profile of DUP4 is complex, with a total of six copy number changes that cannot have arisen by a single unequal crossover event from reference-like sequences (Figs. 1B, 4B). At the gene level, this copy number profile corresponds to duplication of GYPE, deletion of the 3’ end of GYPB, duplication of the 5’ end of GYPB and triplication of the 3’ end of GYPA. To begin to understand the functional consequences of DUP4, we sought to reconstruct the physical arrangement of this variant by pooling data across the nine carriers in the sequenced reference panel (eight Wasambaa individuals from Tanzania including three parent-child pairs, and a single African Caribbean individual from Barbados). First, analysis of coverage along a multiple sequence alignment of the segmental duplication corroborated the location of the six copy number changes from the HMM, with two pairs of breakpoints at homologous locations in the alignment (Fig. S15).
Next, we looked for sequenced read pairs spanning CNV breakpoints, which provide direct evidence of the structure of the underlying DNA. We identified read pairs that were mapped near breakpoints but with discordant positions (MQ>=1, absolute insert size > 1000 bp), including longer read data we generated for the 1000 Genomes individual who carries DUP4 (HG02554; 300 bp PE reads on Illumina MiSeq to 13x coverage). Discordant read pairs supported the connection between each pair of homologous breakpoints as well as between the remaining two breakpoints, which lie in non-homologous sequence (Figs. 6A, S16). On the basis of the combined evidence from copy number changes, discordant read pairs, and homology between inferred breakpoints, we generated a model of the DUP4 chromosome that contains five glycophorin genes (Fig. 6B).
Fig. 6. The structure of DUP4.
(A) Discordant read pairs mapped near DUP4 copy number changes. Colored arrows represent read pairs from DUP4 carriers, with paired reads shown on the same horizontal line and the direction of the arrows depicting the strand and position as mapped to the human reference sequence. The number of such read pairs and distinct carriers is given to the left. A schematic of the reference sequence is below with colors indicating the segmentally duplicated units. Brackets delineate segments with different copy number in DUP4, numbered and labeled with their length to the nearest kb. (B) The structure of DUP4, inferred by connecting sequence at breakpoints based on sequence homology and discordant read pairs. Arrows depict the concordant positions of the read pairs in (A) on this structure, and the order of reference segments is shown below. Inset: detail of the inferred GYPB-A hybrid genes, indicating the positions of discordant read pairs (arrows), PCR primers (vertical red lines) and the resulting product (horizontal red line). (C) Normalized coverage in 1600 bp windows (black) and HMM path (red) for a DUP4 carrier (top) and for an individual serotyped as Dantu+ (NE type; bottom), on the same x axis as (A). (D) Protein sequences of GYPA, GYPB, and the Dantu hybrid within the cell membrane depicting the extracellular, transmembrane, and intracellular domains as visualised with protter (54).
A prominent functional change on this structure is the presence of two GYPB-A hybrid genes, supported by several read pairs within intron 4 of GYPA and GYPB and the copy number profile. We confirmed the hybrid sequence by PCR-based Sanger sequencing of a 4.1 kb segment spanning the breakpoint (Figs. 6B, S17, S18 and Table S10) (22). These data localize the breakpoint to a 184 bp section of GYPA and GYPB where the two genes have identical sequence (Fig. S19). If translated, the encoded protein would join the extracellular domain of GYPB to the transmembrane and intracellular domains of GYPA, creating a peptide sequence at their junction that is characteristic of the Dantu antigen in the MNS blood group system (28, 29). Moreover, like DUP4, the most common Dantu variant (termed NE type, here referred to as Dantu NE) is reported to have two such hybrid genes and lack a full GYPB gene (30). We sequenced genomic DNA from an individual serologically determined to be Dantu positive, and of NE type (150 bp PE reads on Illumina HiSeq to 18x coverage) and analyzed it using our HMM. The coverage profile and HMM-inferred copy number path, indistinguishable from those of DUP4 carriers, confirm identification of DUP4 as the molecular basis of Dantu NE (Fig. 6C-D).
In addition to duplicate GYPB-A hybrid genes, these data reveal the full structure of this Dantu variant, including a duplicated copy of GYPE and the precise location of six breakpoints. Either complex mutational events or a series of at least four unequal crossover events are needed to account for the formation of this variant (confirmed by simulation; Fig. S20) (22). However, we find no potential intermediates and no obvious relationship between DUP4 and other structural variant haplotypes in the present dataset (Fig. S9) (22). Further analysis of discordant read pairs identifies a number of shorter discrepancies relative to the reference sequence that are consistent with gene conversion events (Fig. S21) and could be functionally relevant (e.g., Fig. S22).
Discussion
Here we use whole-genome sequence data to identify at least 27 CNVs in the glycophorin region that segregate in global populations. In this study, 14% of sub-Saharan African individuals carry a variant that affects the genic copy number relative to the reference assembly. Our description of these variants complements the existing literature on antigenic variation associated with the MNS blood group system and offers additional insights. For example, the frequency of GYPB deletion is broadly commensurate with previous surveys of the S–s–U– blood group phenotype linked to absence of the GYPB protein, but the GYPB deletions in our data differ from the reported molecular variant (Fig. S23) (16, 22, 31–33).
Of the array of glycophorin CNVs identified, one (DUP4) is associated with resistance to severe malaria and explains the previously reported signal of association (14). While there may be other functional mutations on this haplotype, we propose that the direct consequences of this rearrangement are likely to drive the underlying causal mechanism for resistance to severe malaria. DUP4 was not present in the 1000 Genomes Phase 1 reference panel (used in (14)), and exists as a singleton in the 1000 Genomes Phase 3 reference panel. Thus, as previously observed at the sickle cell locus (34), mapping of the association signal by imputation was only possible with the inclusion of additional individuals in the reference panel.
Through additional sequencing, we have shown that DUP4 corresponds to the variant encoding the Dantu+ (NE type) blood group phenotype, thus linking the predicted hybrid genes to a serologically distinct hybrid protein that is expressed on the red blood cell (29, 35). The few existing studies of Dantu+ (NE type) erythrocytes indicate high levels of the hybrid GYPB-A protein and lower levels of GYPA than wild type cells (35, 36). A single study reports parasite growth to be impaired in Dantu+ cells (37), making Dantu NE one of many glycophorin variants that have been hypothesized to influence malaria susceptibility or shown to have an effect in vitro (12, 37–40). Our results regarding a specific protective effect of DUP4, and the lack of evidence for other protective CNVs, suggest the relevance of these effects in natural populations may be complicated. We caution that many of the other CNVs are rare, such that larger sample sizes and direct typing may be required to test their effect in vivo.
These findings then raise the question of how DUP4 protects against malaria. GYPA and GYPB are exclusively expressed on the erythrocyte surface and are targeted by parasites during invasion (6, 7). P. falciparum EBA175 binds to the extracellular portion of GYPA (41), which is preserved in DUP4. P. falciparum EBL1 binds to the extracellular portion of GYPB (42) which is duplicated in DUP4 but joined to intracellular GYPA. The significance of the extra copy of GYPE or the absence of full GYPB in DUP4 is uncertain, since GYPE is not known to be expressed at the protein level (8, 43), and we do not observe evidence that absence of GYPB alone confers protection (Fig. 3A). GYPA and GYPB are known to form homodimers as well as heterodimers in the red cell membrane (33), so these copy number changes could have complex functional effects. There are physical interactions between GYPA and band 3 (encoded by SLC4A1) at the red cell surface (44) and parasite binding to GYPA appears to initiate a signal leading to increased membrane rigidity (45). Thus the GYPB-A hybrid proteins seen in DUP4 could potentially affect both receptor-ligand interactions and the physical properties of the red cell membrane.
Previous surveys of the Dantu blood group antigen have indicated that it is rare (Table S11) (33, 46–48). We find that DUP4 is absent or at very low frequency outside parts of east Africa, with a frequency difference and extended haplotype consistent with a recent rise in frequency in Kenya. In contrast, the malaria-protective variant causing sickle-cell anaemia (rs334 in HBB), which is thought to be under balancing selection, has a similar frequency in both the Gambia and Kenya (Fig. 5A). One possibility for why DUP4 is not more widespread, given its strong protective effect against malaria, is that it has arisen recently without time for gene flow to facilitate its dispersion. Alternatively, this frequency distribution could be consistent with balancing selection, for example if it protects only against certain strains of P. falciparum that are specific to east Africa. The glycophorin region is near a signal of long-term balancing selection, and measures of polymorphism in both the human glycophorins and P. falciparum EBA175 have been suggestive of diversifying selection (11, 12, 49, 50). Although apparently not directly related to these signals, current selection on DUP4 may represent a snapshot of the long-term evolutionary processes acting at this locus. Mapping the allele frequency of DUP4 across additional populations could help clarify the nature of selection.
Recent GWAS have confirmed three other loci associated with severe malaria (HBB, ABO, ATP2B4), all of which are also related to red blood cell function (14, 51). However, only the association with GYPA and GYPB directly involves variation in invasion receptors. These receptors have been found to be non-essential in experimental models (7, 9), yet this result indicates important functional roles in natural populations. Intriguingly, there is marked variation among P. falciparum strains in preference for different invasion pathways in vitro (7); field studies that account for parasite heterogeneity and tests for genetic interactions may therefore be important in determining how DUP4 affects parasite invasion. The discovery that a specific alteration of these invasion receptors confers substantial protection provides a foundation for experimental studies on the precise functional mechanism, and may lead us towards novel parasite vulnerabilities that can be utilized in future interventions against this deadly disease.
Materials and methods
Sample collection and sequencing
Sequencing of African individuals
Blood samples from a total of 773 healthy individuals from 10 ethnic groups in four countries in sub-Saharan Africa were collected by MalariaGEN partners (www.malariagen.net) and the MRC Unit in the Gambia (http://www.cggh.org/collaborations/mrc-unit-the-gambia) as part of ongoing projects (Fig. S1A and Tables S1, S2). Individuals were from the general population, with most collected in family trios except in Burkina Faso where individuals are unrelated. Genomic DNA was extracted and sequencing was performed on Illumina HiSeq 2000 at the Wellcome Trust Sanger Institute with 100 bp paired end reads to an average of 10x coverage. Reads were mapped to the GRCh37 human reference genome with additional sequences as modified by the 1000 Genomes Project (hs37d5.fa; (20)), using BWA (55) with base quality score recalibration (BQSR) and local realignment around known indels as implemented in GATK (56, 57).
Sequence data curation
We used GATK HaplotypeCaller to compute an initial set of genotype likelihoods across samples at a genome-wide set of variants, including polymorphic sites from 1000 Genomes Phase 3 (58). We computed average coverage across the genome for each individual using BEDTools genomecov (59) and excluded seven Bantu individuals from Cameroon with less than 2x coverage across the genome, and one Wollof individual from the Gambia with less than 6x coverage and greater than 10% missing call rate in the GATK analysis. All further analyses described here are based on the 765 non-excluded individuals.
We inferred the sex of sequenced samples based on the ratio of X chromosome coverage to autosomal coverage. To infer family relationships, we used lcMLkin (60) to compute maximum likelihood pairwise kinship estimates from the GATK-estimated genotype likelihoods at a thinned set of ~26,000 SNPs genome-wide. We then ran PRIMUS (61) to infer pedigrees from the kinship estimates and compared the inferred and reported relationships. Based on this we manually curated the family structure of sequenced samples by removing relationships incompatible with trio structure (IBD1 < 0.9 for parent-child relationship), swapping three individuals between trios with clear sample mixups, and exchanging parental labels in two trios to be consistent with the genetic sex of the parents. The curated dataset contains 207 trios, 16 duos, and 115 individuals without nominal close relationships. All trios and duos are unrelated to each other except for one extended family in the Wollof from the Gambia, which consists of a quad (two parents and two children, here encoded as two trios), where one of the children is a parent in an additional trio.
1000 Genomes sequence data
The 2,504 individuals from 26 populations in the 1000 Genomes Phase 3 release (20) were analyzed. Bam files containing reads mapped to GRCh37 were downloaded from the 1000 Genomes FTP site (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/; Fig. S1B and Table S3).
Overview of the glycophorin region
The glycophorin gene cluster on chromosome 4 results from segmental duplication events in the ancestor to African great apes (62) and is not related in sequence to GYPC on chromosome 2. By aligning segments of the reference sequence against one another, we identified the region of segmental duplication, here referred to as the glycophorin region, as chr4:144,706,830-145,069,066, and the three paralogous units of the segmental duplication as: GYPE unit, chr4:144,706,830-144,837,481; GYPB unit, chr4:144,837,482-144,947,716; GYPA unit, chr4:144,947,717-145,069,066. Each gene occupies ~30 kb toward the end of its ~120 kb repeat unit. We generated a multiple sequence alignment of the reference sequence for the three units by running kalign (63) with default parameters, and calculated pairwise identity in 1600 bp windows along this alignment (Fig. 1A). The glycophorin genes are transcribed on the negative strand, and we adopt the convention of numbering exons and introns as they occur in the GYPA transcript (exon 3 is a pseudoexon in GYPB and exons 3 and 4 are pseudoexons in GYPE). We focus on the three protein-coding genes, but note that a long noncoding RNA is annotated between GYPE and GYPB (LOC101927636). The coordinates here, and throughout the paper, are given with respect to GRCh37.
Construction of a regional reference panel
Identification of polymorphic sites and genotype likelihood computation
To construct a regional reference panel we focused on the 10 Mb region chr4:140-150Mb, including a 500 kb margin at either end. We first assembled a list of previously identified SNPs and short indels from the 1000 Genomes Phase 3 (20), the Illumina Omni 2.5M array, the ExAC project (64) and the European Variation Archive (downloaded on 24th February 2016; www.ebi.ac.uk/eva/), totaling 421,670 variants. We then used freebayes v1.0.2 (19) to calculate genotype likelihoods at these sites as well as at newly identified putatively polymorphic sites, across the 3,269 sequenced individuals. Freebayes was run in 10 kb chunks across the region. We filtered freebayes output to include all previously identified variants and high quality novel variants, i.e., novel variants with quality (QUAL) > 1, presence of supporting reads on both strands (SAF > 0 and SAR > 0) and both sides of the variant (RPL>1 and RPR>1), and high quality per alternate observation (QUAL/AO>10). In total, the filtered output contained 424,909 variants, of which 412,795 were among the previously identified variants.
Genotype calling and phasing
We next aimed to generate a high-quality set of genotype calls in the 765 individuals collected by MalariaGEN partners. We focused on sites with combined mean coverage between 7.5x and 13.5x, excluding approximately 4% and 1% of variants with depth below or above this range respectively, and restricted to variants with allele count at least 2, leaving a total of 117,531 variants. We produced an initial set of genotype calls at these variants using BEAGLE 4.0 (beagle.r1399; (17)) without specifying family information. Based on the initial calls, we removed variants with more than five mendelian errors in the 207 trios, strong evidence for deviation from Hardy-Weinberg equilibrium (PHWE < 1x10-3 in the 542 individuals without parents in the dataset) or more than one alternate allele. We then re-ran BEAGLE on the remaining set of 111,167 variants, including family information, to produce genotype calls. We phased these genotypes using SHAPEIT2, specifying 400 selected and 200 random conditioning states, an effective population size of 17,469, and 10 burn-in, 8 pruning and 50 main iterations, and including trio information.
Finally, to form a joint reference panel across all individuals, we merged the phased haplotypes with the 1000 Genomes Phase 3 haplotypes (20) at the overlapping set of variants. The merged reference panel, which contains 96,676 variants in the region chr4:139.5-150.5Mb, is available for use in other studies (see https://www.malariagen.net/resource/23).
Imputation and association testing at SNPs and short indels
Imputation and association testing
We used both the 1000 Genomes Phase 3 reference panel (20) and the merged reference panel described above to impute genotypes into the previously published sets of severe malaria cases and population controls from the Gambia, Kenya and Malawi (14). In brief, we ran IMPUTE2 (65, 66) in 2 Mb chunks, using Illumina Omni 2.5M genotype calls as previously described. A total of 5,621 (the Gambia), 5,203 (Malawi), and 5,583 (Kenya) genotyped variants were included in the 11 Mb region considered. We specified 1,000 copying states (-k_hap 1000), an effective population size of 20,000 (-Ne 20000) as recommended for African populations, and a buffer region of 500 kb. We removed reference panel individuals with parents in the panel before imputation.
We tested for association in each population under additive, dominant, recessive and heterozygote models using a logistic regression model as implemented in the program SNPTEST, including five principal components to account for population structure. We restricted analysis here to severe malaria cases with nonzero parasitaemia, as measured by blood slide at the time of admission, as this is likely to be the most accurate phenotype. For downstream meta-analysis, we omitted results for variants at low frequency (MAF < 0.5%) or with low imputation quality (IMPUTE info < 0.75) in each population.
We used fixed-effect meta-analysis to compute an estimate of the odds ratio across populations, along with a standard error and P-value, for each model. For additive model we refer to the resulting P-value as Padditive. Additionally, we computed a model-averaged Bayes factor (BFavg) reflecting the overall evidence for association as previously described (14). Meta-analysis results under the two imputation reference panels, as well as for the previously published 1000 Genomes Phase 1 reference panel imputation, are shown in Fig. S2.
Functional annotation of variants
We used Variant Effect Predictor (VEP; (67)) to annotate the functional consequences of all variants with evidence of association (BFavg>1). We noted one variant with VEP IMPACT rating ‘moderate’ with some evidence of association (rs147343123; nonsynonymous in exon 1 of GYPA, predicted deleterious; association test BFavg=17.34, Padditive=1.4x10-3 for 1000 Genomes imputation; BFavg=121.3, Padditive=1.5x10-4 for full panel imputation) (Fig. S2). We also noted that the nonsynonymous variant in FREM3 exon 1 previously found to have evidence of association (rs181620317; see (14)), is imputed at much lower frequency using both the 1000 Genomes Phase 3 panel and the full reference panel (e.g. frequency = 0.2% in Kenya using the full reference panel), and shows no evidence for association using genotype imputed from these panels.
Identification of copy number variation
Method to call copy number variation
To identify large CNVs across the glycophorin region, we implemented a hidden Markov model (HMM) to infer the underlying copy number state from the observed read counts. The input to the HMM is read depth, averaged over sites in windows of fixed length (here, 1600 bp) for each individual. To reduce the problems with mapping in the region, we included only (i) reads with at least a mapping quality (MQ) of 20; (ii) mappable sites, defined as sites with mappability >0.9, where mappability of a site is the mean value of the CRG mappability track for all 100-mers overlapping that site (21); (iii) windows with ≥25% of sites fulfilling this criterion; windows with fewer mappable sites were considered uninformative.
We modeled the mean depth of coverage for individual i at window j, di,j, as normally distributed with mean and variance dependent on the assumed underlying copy number k (k=2 is the normal diploid state):
For copy number k=0 (homozygous deletion), we used a truncated normal distribution (truncated at 0) and assigned a variance of 0.04 to allow for spurious mapping. To account for systematic variation in coverage along the genome, we estimated a window-specific factor (wj) proportional to how much individuals with no copy number variation (k=2) are above or below their mean in that window. These define the emission probabilities for the HMM. We used a fixed transition matrix that assumes a low rate of switching with approximately 0.999 probability of remaining in the current copy number state; 1x10-4 for leaving the diploid state; 0.001 probability of returning to diploid (k=2) from a non-diploid state; and 1x10-5 probability of switching among non-diploid states.
We estimated μi, σi and wj by starting with an initial guess assuming everyone to be diploid across the region, and then running the Viterbi algorithm separately for each individual. We then recalculated μi and σi for each individual, only including windows in which the copy number is inferred to be 2, and wj for each window only including individuals in which the copy number is inferred to be 2. We iterated this algorithm until no further changes in the inferred underlying copy number paths were observed for any individuals.
CNV calling in the 3,269 sequenced individuals
We applied the HMM method described above to the full set of 3,269 individuals with sequence data in an 850 kb region including the glycophorin genes (chr4:144.35-145.20Mb). After inferring the copy number state paths for each individual, we then considered variants to be the same across individuals if the direction of copy number change was the same and both end points were within three bins of each other. Heterozygous triplications and homozygous duplications were differentiated by looking across individuals; variants that were always found in copy number state 4 were attributed as triplications. We excluded copy number variable segments that covered only a single bin or were outside the segmental duplication. We then considered copy number variable segments that were always found together to be a single CNV, and manually refined the few other copy number variable segments that were not found separately from other CNVs (22). This process identified 16 non-singleton variants and 28 singleton variants (Figs. 1B, S3), although we note several caveats about the singleton CNVs including some that likely correspond to more common variants (22).
To validate the CNVs and genotype calls, we assessed inheritance in the MalariaGEN trios and compared genotype calls for the 1000 Genomes individuals with those released in the 1000 Genomes Phase 3 paper on structural variants (Table S4, Fig. S4) (22, 23). We also designed PCR primers on either side of the DEL1 variant and generated Sanger sequence that confirmed and localized this breakpoint (Figs. S5, S6 and Table S5) (22).
Phasing and imputation of CNVs
Initial phasing of CNVs
We investigated whether CNVs could be accurately phased relative to surrounding SNP variation in the regional reference. Collectively, non-singleton CNVs in our dataset cover a total of 350 kb (Fig. 1B), which extends over most of the region of segmental duplication surrounding the glycophorin genes. In principle, inference of CNV haplotype phase might benefit from copy-number-aware genotype calls at smaller variants within CNVs. However, implementing this is challenging as the state-of-the art phasing methods assume samples are diploid. A further possible issue is that read mapping, and hence the quality of SNP genotype calls, is likely to be impaired in such regions of segmental duplication.
Motivated by these observations, we took the following approach to phasing CNVs which leverages the family trio structure of sequenced individuals to infer accurate phase, using both SHAPEIT2 (18) and MVNCALL (25). First, we focused on the 765 sequenced individuals collected by MalariaGEN partners. We removed variants within the region of segmental duplication (here taken as chr4:144.7-145.07Mb) from the reference panel, and replaced these with CNV genotype calls, to form a single file with genotypes at the CNVs and flanking SNPs and indels. SHAPEIT2 requires each variant to be assigned a single genomic position. For each CNV longer than 10 kb we included the genotypes for that variant at the start-, mid- and endpoints of the CNV; for variants less than 10 kb we used the midpoint. We then ran SHAPEIT2 with parameters as for SNP/indel phasing to produce phased genotype calls, treating each CNV as a separate, biallelic variant. Next, to phase CNVs into the 1000 Genomes Project individuals, we extracted the Omni 2.5M sites with allele frequency > 1% from the 1000 Genomes Project phased haplotypes and removed the region of segmental duplication. We used MVNCALL to phase each non-singleton CNV into this scaffold, again placing each CNV greater than 10 kb in length at its start-, mid-, and end positions.
We assessed accuracy of phasing by considering patterns of LD between CNVs and variants in the left and right flanking regions (Figs. S9, S10). We noted high LD between some CNVs and variants to the left of the region, including for DEL1, DEL2, DUP1 and DUP4. LD estimated from phased haplotypes captured most of the correlation between genotypes across the region, as expected in an outbred population if haplotype phase is accurate. The small deviations observed may be due to the presence of a small number of switch errors, or potentially to population substructure.
Haplotype-based curation and re-phasing of CNVs
Some singleton CNVs have similar copy number profiles to more common CNVs in the HMM-based calls (22) and after phasing we observed that haplotypes carrying several of these clustered with the corresponding common variant (DEL9 with DEL1; DEL11, DEL12 and DEL14 with DEL2; DUP15 and DUP18 with DUP1). We reasoned that these are likely to represent the same variant, with differences in calling potentially due to noise in coverage profiles or variation in the other chromosome. We merged these singletons with the corresponding non-singleton CNV and repeated the procedure described above, using SHAPEIT2 and MVNCALL to re-phase CNVs and flanking short variants then merging the two reference panels. We also noted that haplotypes carrying DEL4 cluster with DUP1, which shares a similar breakpoint (Fig. 1B-C). A plausible explanation for this is that DUP1 arose by NAHR on a DEL4 background (Fig. S24) (22).
For subsequent imputation steps we restricted the combined panel to the set of haplotypes from unrelated individuals (i.e. removing the children of family trios and duos). Three individuals in the 1000 Genomes data carry CNVs that are not singletons in the overall dataset but are private to that individual within the 1000 Genomes data (HG01986, carrying DEL4; HG02554, carrying DUP4, and HG02585, carrying DUP5). Of these, we noted in particular that HG02554 appears to have a switch error in the 1000 Genomes (explaining clustering of opposite haplotypes with other DUP4 haplotypes on either side of the region; Fig. S9). Because our approach phases variants in the 1000 Genomes separately and these are singletons in that dataset, we also excluded these three individuals from the panel used for imputation.
Cross-validation of CNV imputation in the reference panel
To evaluate how well CNVs are likely to be imputed in our GWAS dataset, we performed a cross-validation experiment using the African reference panel individuals as follows. For each individual, we removed the individual and his/her family members (if present) from the phased reference panel haplotypes to form a subsetted panel. We also extracted genotype calls for that individual at variants on the Illumina Omni 2.5M array from the reference panel genotypes, excluding variants within the glycophorin region. We used these genotypes and the subsetted panel to re-impute CNVs for that individual.
We evaluated CNV re-imputation by computing the correlation between HMM-based genotype calls and genotype dosages from the re-imputation (Fig. S11) and by direct comparison of HMM and re-imputed calls. We note that DEL4 carriers were imputed with some confidence as carrying DUP1, consistent with the shared haplotype for these variants and the higher frequency of DUP1. Given the functionally distinct nature of these variants, this affects interpretation of imputed DUP1 genotypes.
Among CNVs >10 kb in length, we noted little variation between the three imputation locations (leftmost breakpoint, midpoint, and rightmost breakpoint of the CNV), except for DUP4, where the right endpoint had slightly higher imputation performance (Fig. S11). For all analyses presented in this paper we refer to the midpoint imputation of each CNV.
Imputation of CNVs
We used the combined panel to impute CNVs into the three GWAS datasets. Imputation settings were as described above. To ensure imputation was based on flanking SNPs, we removed SNPs within the glycophorin region from the genotype data before imputation. We evaluated imputation performance in the GWAS data by comparing the overall expected allele frequency against the IMPUTE info metric and another metric of confidence in imputed CNV call probabilities, the proportion of expected frequency of CNV heterozygote or homozygote that is due to genotypes with at least 90% probability (Figs. 3D, S12A-B). We also compared the expected frequency in control samples with the frequency in the geographically nearest reference panel population (Fig. S12C).
Analysis of association at CNVs
Association with severe malaria
We tested for association with each CNV in each population using logistic regression including five principal components as covariates, and computed both fixed-effect and Bayesian meta-analyses as described above for SNPs and indels. To directly estimate the effect of heterozygote and homozygote genotypes, we modified SNPTEST to fit the logistic regression model with a separate parameter for heterozygote and homozygote genotypes in a missing data likelihood framework that integrates over imputation uncertainty. To assess evidence for association after accounting for DUP4, we used QCTOOL v2 (http://www.well.ox.ac.uk/~gav/qctool_v2/) to extract the additive and heterozygote imputed dosages of DUP4 for each individual, and repeated the association test and meta-analysis at SNPs, indels and CNVs conditioning on these dosages (Figs. 3B, S13).
Association with clinical subtypes
Severe malaria-affected children in our data are recorded as either having cerebral malaria (CM), severe malarial anaemia (SMA), or other nonspecific severe malaria phenotype (OTHER). To assess the association of DUP4 with these subphenotypes, we fit a multinomial logistic regression model with these outcome levels using population controls as a baseline (Table S7). A small number of individuals are annotated as both CM and SMA and were excluded from this analysis.
Association with gene content
To test for association with overall copy number of each glycophorin gene, we used the imputed genotype probabilities and the copy number profile of non-singleton CNVs (Fig. 1B) to compute the expected number of copies of each gene in each GWAS sample. Since overlap of some CNVs with genes is partial, we measured copy number separately at the transcription start and end sites. We also computed the expected number of hybrid genes as formed by DUP2, DUP4 and DUP8. We tested for association with each copy number measure by fitting a logistic regression model across the three populations, including the genic dosage and five principal components in each population. Overall genic copy number was significantly associated with severe malaria status (e.g. P=3x10-7 for dosage at transcription start sites), as was the expected number of hybrid genes. However, these effects were well explained by the effect of DUP4 (P > 0.14 after conditioning on the expected dosage of DUP4). We note that several of these predictors are strongly correlated with DUP4 dosage; this analysis could not distinguish between an effect of DUP4 and that of copy number of GYPE, copy number of the GYPA transcription end site, or the total copy number of hybrid genes (P > 0.13 for either effect in a joint model).
We also specifically tested whether the number of deleted copies of GYPB, or the presence of one or two deleted copies, was associated with severe malaria status, but saw no evidence of association (P>0.48).
Population genetic analysis
Frequency differentiation
We used estimated minor allele frequencies (MAF) at both typed and imputed variants from (14) for the 2,490 population controls from the Gambia and 1,498 population controls from Kenya to investigate the extent to which the observed frequency difference of DUP4 is extreme relative to other variants genome-wide. For this comparison, we included all autosomal variants having IMPUTE info >0.75 and estimated frequency ≥0.5% in at least one of the populations (14,973,426 variants in total). We binned variants into 0.5% MAF bins and noted the frequency estimates for DUP4 (fKenya=0.0895 and fGambia=0.0003) and, for comparison, the sickle cell anaemia-causing allele (rs334:T; MAF=0.0853 in Kenya and 0.0766 in the Gambia based on direct Sequenom typing of this SNP in these samples (68)).
To quantify the extent to which DUP4 is an outlier, we computed two empirical P-values based on the marginal distributions. Specifically, we computed PGambia|Kenya as the proportion of variants with MAF less than or equal to fGambia in the Gambia, among all variants with MAF within 1% of fKenya in Kenya (empirical PGambia|Kenya<1.2x10-6; 0 of 831,956 variants in this bin). Similarly, we computed PKenya|Gambia as the proportion of variants with MAF equal to or greater than fKenya in Kenya, among all variants with MAF within 1% of fGambia in the Gambia (empirical PKenya|Gambia=1.7x10-3; 2,851 of 1,710,922 variants in this bin).
We note that the computation of PGambia|Kenya is sensitive to the frequency of DUP4 in the Gambia, which may be underestimated due to poor imputation performance. To account for this we recomputed PGambia|Kenya assuming 1% frequency in the Gambia (adjusted empirical PGambia|Kenya=5x10-3) and refer to this value in the main text.
Haplotype homozygosity
To assess haplotype homozygosity, we first used SHAPEIT2 to phase imputed CNV genotypes onto haplotypes defined by directly-typed SNPs in the region chr4:139.5-150.5Mb, excluding SNPs in the glycophorin region. We specified 400 selected and 200 random copying states (--states 400, --states-random 200), an effective population size of 17,469 as recommended for African populations, and included reference panel haplotypes to inform phasing. EHH (27) was then computed around DUP4 using the rehh R package. We used a custom script to compute an unstandardized integrated haplotype score (uiHS) (26), using recombination rate estimates from the HapMap combined recombination map (52).
To generate a distribution of uiHS at SNPs with a similar frequency to DUP4, we computed uiHS at those genotyped SNPs (14) where the human ancestral allele could be inferred using the six primate EPO alignments (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/supporting/ancestral_alignments/), and the derived allele frequency was in the 2% frequency bin centered around fKenya (0.0795-0.0995). We computed an empirical P-value as the proportion of SNPs with uiHS less than or equal to that observed at DUP4 (P=0.0119; 996 of the 83,419 variants in this bin). We note that the exclusion of the glycophorin region from the estimate at DUP4 is likely to be conservative, since in effect it adds a constant term equal to the recombination length of the removed interval (approximately 0.15 cM) to both the numerator and denominator of the statistic.
Resolution of the structure of DUP4
Discordant read pair analysis
We began by remapping reads from each of the nine heterozygous DUP4 carriers genome-wide using bwa mem, which has better performance for longer reads (69). Because even longer reads should facilitate more confident mapping around the breakpoints, we also obtained DNA from Coriell for HG02554, the 1000 Genomes sample carrying DUP4, and generated 300 bp PE sequence data on Illumina MiSeq at the High Throughput Genomics core at the Wellcome Trust Centre for Human Genetics at the University of Oxford. We mapped these reads to the same GRCh37 human reference (hs37d5.fa) with bwa mem, yielding 13x genomic coverage. For all re-mapped bam files, we marked duplicates with Picard MarkDuplicates and excluded duplicate reads from further analysis.
We then used samtools to pull out read pairs where both reads had a primary alignment to the glycophorin region with MQ≥1 and an absolute insert size ≥1 kb. For the 300 bp data, we allowed one of the reads in the pair to be mapped to the glycophorin region with MQ=0. Across samples, there were 434 such read pairs. We grouped read pairs where both ends were mapped within 1 kb of each other and identified those near the HMM-inferred breakpoints.
To view how uniquely the discordant read pairs matched each of the three possible homologous positions in the segmental duplication, we used the mapped position and the cigar string to place each read into the multiple sequence alignment and then identified positions in the read with a match or mismatch to each of the three aligned reference sequences using custom scripts in R (e.g., Fig. S16).
Molecular assays and Sanger sequencing
Briefly, a 4.1 kb fragment spanning the GYPB-A hybrid breakpoint (located between exons 4 and 5) was amplified by PCR using primers designed against GYPB and GYPA sequences (Fig. S17 and Table S10). In practice, it is difficult to design specific primers due to the high homology and the assay amplifies both GYPA and GYPB-A hybrid sequences. To separate these, we identified a restriction enzyme site (BlpI [5'…GC/TNAGC…3']) that cleaves the GYPA sequence but not that of the hybrid. PCR products were digested and then separated on an agarose gel. We excised the 4.1 kb band and obtained Sanger sequence of the region around the putative breakpoint (Figs. S18, S19). A full description of the design and protocols is given in (22).
Sequencing and copy number analysis of a serologically-typed Dantu+ individual
We obtained DNA from the International Blood Group Reference Laboratory in Bristol, UK from an archived reference sample that had been serologically typed as Dantu+ (NE type). This sample was collected in 1992 and the individual was originally from Natal, South Africa. The DNA was sequenced with 150 bp PE reads on Illumina HiSeq 4000 by the High Throughput Genomics core at the Wellcome Trust Centre for Human Genetics at the University of Oxford. Reads were mapped to the same GRCh37 human reference genome (hs37d5.fa) with bwa mem, yielding 18x coverage. To generate CNV calls, we ran our HMM method on this individual alone, without using the window-specific factors.
Simulations of DUP4 formation
To determine possible routes to formation of DUP4, we implemented a computer program in C++ to iteratively generate structural rearrangements via unequal crossing over, allowing breakpoints to occur at any of the six locations observed for DUP4 with no constraint based on homology. In brief, we encode the reference haplotype as a string of seven segments delineated by the coverage breakpoints (i.e., as the string 0123456; Fig. 6A). We ran the program through three generations of events, where the first generation produced all possible events between two reference haplotypes and the second and third generations allowed unequal crossing over between any two haplotypes from previous generations. This brute force search allows us to place a lower limit of four on the number of events required to form DUP4. For additional details on the program and search, see (22).
Supplementary Material
One sentence summary.
Resolution of copy number changes in African populations reveals a complex structural variant that encodes hybrid genes and confers resistance to severe malaria.
Acknowledgements
We thank all the study participants and the members of the MalariaGEN consortium. A list of researchers involved at each study site can be found at https://www.malariagen.net/projects/consortial-project-1/malariagen-consortium-members.
The MalariaGEN Project is supported by the Wellcome Trust (WT077383/Z/05/Z) and the Bill & Melinda Gates Foundation through the Foundations of the National Institutes of Health (566) as part of the Grand Challenges in Global Health Initiative. The Resource Centre for Genomic Epidemiology of Malaria is supported by the Wellcome Trust (090770/Z/09/Z). This research was supported by the Medical Research Council (G0600718; G0600230; MR/M006212/1). Chris C.A. Spencer was supported by a Wellcome Trust Career Development Fellowship (097364/Z/11/Z). The Wellcome Trust also provides core awards to The Wellcome Trust Centre for Human Genetics (090532/Z/09/Z) and the Wellcome Trust Sanger Institute (098051).
Eric Achidi received partial funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement N° 242095 – EVIMalaR and the Central African Network for Tuberculosis, HIV/AIDS and Malaria (CANTAM) funded by the European and Developing Countries Clinical Trials Partnership (EDCTP). Thomas N. Williams is funded by Senior Fellowships from the Wellcome Trust (076934/Z/05/Z and 091758/Z/10/Z) and through the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement N° 242095 – EVIMalaR. The KEMRI-Wellcome Trust Programme is funded through core support from the Wellcome Trust. Carolyne Ndila is supported through a strategic award to the KEMRI-Wellcome Trust Programme by the Wellcome Trust (084538). Tanzania/KCMC/JMP received funding from MRC grant number (G9901439). The Malawi-Liverpool-Wellcome Trust Clinical Research Programme (MLW) is a Major Overseas Programme of the Wellcome Trust. Malcolm Molyneux was funded by a Wellcome Trust Research Leave Fellowship. Vysaul Nyirongo was supported on the MLW core grant. V.D.M. was funded by Istituto Pasteur-Fondazione Cenci Bolognetti, BioMalPar and Evimalar (European Community FP6,FP7).
We thank the staff of the WTSI Sample Logistics, Genotyping, Sequencing and Informatics facilities and the WTCHG High-Throughput Genomics core for their contributions to sample handling and generation and processing of sequence data.
Footnotes
Author contributions: Writing group: E.M.L., G.B., G.B.J.B., K.A.R., C.C.A.S., D.P.K.; Data analysis: E.M.L., G.B., G.B.J.B., Q.S.L., D.P.K., G.M.C., K.A.R., C.C.A.S.; Study site lead investigators: K.A.B., D.J.C., D.M., S.B.S., E.A., K.M., T.N.W., C.D., H.R., E.R., M.M., T.T.; Sample collection and curation: K.A.B., D.J.C., M.J., F.S-J., E.C.B., V.D.M., D.M., S.B.S., E.A., T.O.A., K.M., C.M.N., N.P., T.N.W., C.D., A.M., H.R., E.R., D.K., M.M., V.N., T.T.; Serotyped sample curation and handling: N.T., L.T., S.G.; Sample processing, sequencing, data management and project coordination: G.B., K.K, E.D, J.S, V.C., C.H., A.E.J., K.R., K.A.R.; Experimental design and targeted assay development: E.M.L., G.B., C.H., A.E.J., K.R., K.A.R., C.C.A.S., D.P.K.
The regional combined reference panel and association summary statistics, multiple sequence alignment of the glycophorin segmental duplication, and Sanger sequences are available at https://www.malariagen.net/resource/23. The sequence data generated for HG02554 and the Dantu+ (NE type) individual have been deposited in the European Nucleotide Archive under study accession number PRJEB20081. Accompanying scripts are available at https://github.com/malariagen/glycophorin_cnvs.
References
- 1.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. World Malaria Report. 2015 [Google Scholar]
- 3.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Langhi DM, Jr, Bordin JO. Duffy blood group and malaria. Hematology. 2006;11:389–398. doi: 10.1080/10245330500469841. [DOI] [PubMed] [Google Scholar]
- 5.Gaur D, Mayer DC, Miller LH. Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int J Parasitol. 2004;34:1413–1429. doi: 10.1016/j.ijpara.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Satchwell TJ. Erythrocyte invasion receptors for Plasmodium falciparum: new and old. Transfus Med. 2016;26:77–88. doi: 10.1111/tme.12280. [DOI] [PubMed] [Google Scholar]
- 7.Wright GJ, Rayner JC. Plasmodium falciparum erythrocyte invasion: combining function with immune evasion. PLoS Pathog. 2014;10:e1003943. doi: 10.1371/journal.ppat.1003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartron J-P, Rouger P. Blood cell biochemistry. Plenum Press; New York: 1995. Molecular basis of human blood group antigens; p. xx. 492 p. [Google Scholar]
- 9.Hadley TJ, et al. Falciparum malaria parasites invade erythrocytes that lack glycophorin A and B (MkMk). Strain differences indicate receptor heterogeneity and two pathways for invasion. J Clin Invest. 1987;80:1190–1193. doi: 10.1172/JCI113178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasvol G, et al. Glycophorin as a possible receptor for Plasmodium falciparum. Lancet (London, England) 1982;2:947–950. doi: 10.1016/s0140-6736(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 11.Baum J, Ward RH, Conway DJ. Natural selection on the erythrocyte surface. Mol Biol Evol. 2002;19:223–229. doi: 10.1093/oxfordjournals.molbev.a004075. [DOI] [PubMed] [Google Scholar]
- 12.Ko WY, et al. Effects of natural selection and gene conversion on the evolution of human glycophorins coding for MNS blood polymorphisms in malaria-endemic African populations. Am J Hum Genet. 2011;88:741–754. doi: 10.1016/j.ajhg.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang HY, Tang H, Shen CK, Wu CI. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol Biol Evol. 2003;20:1795–1804. doi: 10.1093/molbev/msg185. [DOI] [PubMed] [Google Scholar]
- 14.Malaria Genomic Epidemiology Network. Band G, Rockett KA, Spencer CC, Kwiatkowski DP. A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature. 2015;526:253–257. doi: 10.1038/nature15390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patnaik SK, Helmberg W, Blumenfeld OO. BGMUT Database of Allelic Variants of Genes Encoding Human Blood Group Antigens. Transfus Med Hemother. 2014;41:346–351. doi: 10.1159/000366108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumenfeld OO, Huang CH. Molecular genetics of glycophorin MNS variants. Transfus Clin Biol. 1997;4:357–365. doi: 10.1016/s1246-7820(97)80041-9. [DOI] [PubMed] [Google Scholar]
- 17.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 19.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. ArXiv e-prints. 2012 [Google Scholar]
- 20.The 1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derrien T, et al. Fast computation and applications of genome mappability. PLoS One. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supplementary text is available as supplementary materials at the Science website
- 23.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratto F, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menelaou A, Marchini J. Genotype calling and phasing using next-generation sequencing reads and a haplotype scaffold. Bioinformatics. 2013;29:84–91. doi: 10.1093/bioinformatics/bts632. [DOI] [PubMed] [Google Scholar]
- 26.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 28.Dahr W, Beyreuther K, Moulds J, Unger P. Hybrid glycophorins from human erythrocyte membranes. I. Isolation and complete structural analysis of the hybrid sialoglycoprotein from Dantu-positive red cells of the N.E. variety. Eur J Biochem. 1987;166:31–36. doi: 10.1111/j.1432-1033.1987.tb13479.x. [DOI] [PubMed] [Google Scholar]
- 29.Blumenfeld OO, Smith AJ, Moulds JJ. Membrane glycophorins of Dantu blood group erythrocytes. J Biol Chem. 1987;262:11864–11870. [PubMed] [Google Scholar]
- 30.Huang CH, Blumenfeld OO. Characterization of a genomic hybrid specifying the human erythrocyte antigen Dantu: Dantu gene is duplicated and linked to a delta glycophorin gene deletion. Proc Natl Acad Sci U S A. 1988;85:9640–9644. doi: 10.1073/pnas.85.24.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahuel C, London J, Vignal A, Ballas SK, Cartron JP. Erythrocyte glycophorin B deficiency may occur by two distinct gene alterations. Am J Hematol. 1991;37:57–58. doi: 10.1002/ajh.2830370115. [DOI] [PubMed] [Google Scholar]
- 32.Lowe RF, Moores PP. S-s-U-red cell factor in Africans of Rhodesia, Malawi, Mozambique and Natal. Hum Hered. 1972;22:344–350. doi: 10.1159/000152509. [DOI] [PubMed] [Google Scholar]
- 33.Daniels G. Human blood groups : Geoff Daniels ; foreword to first edition by Ruth Sanger. 3rd. John Wiley & Sons; Chichester, West Sussex: 2013. p. ix. 544 p. [Google Scholar]
- 34.Jallow M, et al. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009;41:657–665. doi: 10.1038/ng.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahr W, Moulds J, Unger P, Kordowicz M. The Dantu erythrocyte phenotype of the NE variety. I. Dodecylsulfate polyacrylamide gel electrophoretic studies. Blut. 1987;55:19–31. doi: 10.1007/BF00319637. [DOI] [PubMed] [Google Scholar]
- 36.Merry AH, Hodson C, Thomson E, Mallinson G, Anstee DJ. The use of monoclonal antibodies to quantify the levels of sialoglycoproteins alpha and delta and variant sialoglycoproteins in human erythrocyte membranes. Biochem J. 1986;233:93–98. doi: 10.1042/bj2330093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Field SP, Hempelmann E, Mendelow BV, Fleming AF. Glycophorin variants and Plasmodium falciparum: protective effect of the Dantu phenotype in vitro. Hum Genet. 1994;93:148–150. doi: 10.1007/BF00210600. [DOI] [PubMed] [Google Scholar]
- 38.Heathcote DJ, Carroll TE, Flower RL. Sixty years of antibodies to MNS system hybrid glycophorins: what have we learned? Transfus Med Rev. 2011;25:111–124. doi: 10.1016/j.tmrv.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Pasvol G, Jungery M. Glycophorins and red cell invasion by Plasmodium falciparum. Ciba Found Symp. 1983;94:174–195. doi: 10.1002/9780470715444.ch11. [DOI] [PubMed] [Google Scholar]
- 40.Pasvol G, Wainscoat JS, Weatherall DJ. Erythrocytes deficienct in glycophorin resist invasion by the malarial parasite Plasmodium falciparum. Nature. 1982;297:64–66. doi: 10.1038/297064a0. [DOI] [PubMed] [Google Scholar]
- 41.Orlandi PA, Klotz FW, Haynes JD. A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac(alpha 2-3)Gal- sequences of glycophorin A. J Cell Biol. 1992;116:901–909. doi: 10.1083/jcb.116.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer DC, et al. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci U S A. 2009;106:5348–5352. doi: 10.1073/pnas.0900878106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahuel C, Elouet JF, Cartron JP. Post-transcriptional regulation of the cell surface expression of glycophorins A, B, and E. J Biol Chem. 1994;269:32752–32758. [PubMed] [Google Scholar]
- 44.Huang CH, Reid ME, Xie SS, Blumenfeld OO. Human red blood cell Wright antigens: a genetic and evolutionary perspective on glycophorin A-band 3 interaction. Blood. 1996;87:3942–3947. [PubMed] [Google Scholar]
- 45.Chasis JA, Reid ME, Jensen RH, Mohandas N. Signal transduction by glycophorin A: role of extracellular and cytoplasmic domains in a modulatable process. J Cell Biol. 1988;107:1351–1357. doi: 10.1083/jcb.107.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Contreras M, et al. Serology and genetics of an MNSs-associated antigen Dantu. Vox Sang. 1984;46:377–386. doi: 10.1111/j.1423-0410.1984.tb00102.x. [DOI] [PubMed] [Google Scholar]
- 47.Moores P, Smart E, Marais I. The Dantu Phenotype in Southern Africa. Transfus Med. 1992;2:68. [Google Scholar]
- 48.Unger P, et al. The Dantu erythrocyte phenotype of the NE variety. II. Serology, immunochemistry, genetics, and frequency. Blut. 1987;55:33–43. doi: 10.1007/BF00319639. [DOI] [PubMed] [Google Scholar]
- 49.Leffler EM, et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013;339:1578–1582. doi: 10.1126/science.1234070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verra F, et al. Contrasting signatures of selection on the Plasmodium falciparum erythrocyte binding antigen gene family. Mol Biochem Parasitol. 2006;149:182–190. doi: 10.1016/j.molbiopara.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 51.Timmann C, et al. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature. 2012;489:443–446. doi: 10.1038/nature11334. [DOI] [PubMed] [Google Scholar]
- 52.International HapMap Consortium et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinch AG, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Omasits U, Ahrens CH, Muller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11 10 11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lipatov M, Sanjeev K, Patro R, Veeramah K. Maximum Likelihood Estimation of Biological Relatedness from Low Coverage Sequencing Data. 2015 [Google Scholar]
- 61.Staples J, et al. PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am J Hum Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rearden A, Magnet A, Kudo S, Fukuda M. Glycophorin B and glycophorin E genes arose from the glycophorin A ancestral gene via two duplications during primate evolution. J Biol Chem. 1993;268:2260–2267. [PubMed] [Google Scholar]
- 63.Lassmann T, Frings O, Sonnhammer EL. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009;37:858–865. doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malaria Genomic Epidemiology Network. Reappraisal of known malaria resistance loci in a large multicenter study. Nat Genet. 2014;46:1197–1204. doi: 10.1038/ng.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv e-prints. 2013 [Google Scholar]
- 70.Tate CG, Tanner MJ, Judson PA, Anstee DJ. Studies on human red-cell membrane glycophorin A and glycophorin B genes in glycophorin-deficient individuals. Biochem J. 1989;263:993–996. doi: 10.1042/bj2630993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vignal A, et al. A novel gene member of the human glycophorin A and B gene family. Molecular cloning and expression. Eur J Biochem. 1990;191:619–625. doi: 10.1111/j.1432-1033.1990.tb19166.x. [DOI] [PubMed] [Google Scholar]
- 72.Reid ME, Lomas-Francis C, Olsson ML. The blood group antigen factsbook. ed. Third edition. Elsevier/AP; Amsterdam: 2012. p. xii. 745 pages. [Google Scholar]
- 73.Chen TD, Chen DP, Wang WT, Sun CF. MNSs blood group glycophorin variants in Taiwan: a genotype-serotype correlation study of 'Mi(a)' and St(a) with report of two new alleles for St(a) PLoS One. 2014;9:e98166. doi: 10.1371/journal.pone.0098166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Broadberry RE, Chang FC, Jan YS, Lin M. The distribution of the red-cell Sta (Stones) antigen among the population of Taiwan. Transfus Med. 1998;8:57–58. doi: 10.1046/j.1365-3148.1998.00126.x. [DOI] [PubMed] [Google Scholar]
- 75.Dahr W, Pilkington PM, Reinke H, Blanchard D, Beyreuther K. A novel variety of the Dantu gene complex (DantuMD) detected in a Caucasian. Blut. 1989;58:247–253. doi: 10.1007/BF00320913. [DOI] [PubMed] [Google Scholar]
- 76.Tanner MJ, Anstee DJ, Mawby WJ. A new human erythrocyte variant (Ph) containing an abnormal membrane sialoglycoprotein. Biochem J. 1980;187:493–500. doi: 10.1042/bj1870493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fraser GR, Giblett ER, Motulsky AG. Population genetic studies in the Congo. 3. Blood groups (ABO, MNSs, Rh, Jsa) Am J Hum Genet. 1966;18:546–552. [PMC free article] [PubMed] [Google Scholar]
- 78.Wiener AS, Unger LJ, Cohen L. Distribution and heredity of blood factor U. Science. 1954;119:734–735. doi: 10.1126/science.119.3099.734. [DOI] [PubMed] [Google Scholar]
- 79.Mourant AE, Kopeć AC, Domaniewska-Sobczak K. Oxford monographs on medical genetics. ed. 2d. Oxford University Press; London: 1976. The distribution of the human blood groups, and other polymorphisms; p. xiv. 1055 p., 1013 leaves of plates. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.