Abstract
The HBx protein of hepatitis B virus has been found to co-opt a host-cell enzyme that targets the Smc5/6 protein complex for degradation. The finding identifies Smc5/6 as a cellular antiviral factor.
Like special agents Mulder and Scully in the science-fiction television show The X-Files, the hepatitis B virus research community has for decades been chasing its own unsolved mystery — the truth about the enigmatic ‘X’ protein of this virus. The HBx protein was first shown to be involved in transcriptional activation, and has since been implicated in diverse cellular pathways, including signal transduction, apoptotic cell death, cell-cycle regulation and DNA repair. But how HBx exerts its effects has remained unclear. In this issue, Decorsière et al.1 (page 386) provide intriguing evidence that HBx mediates the degradation of a host antiviral (restriction) factor by interacting with the ubiquitin–proteasome system — the major protein-degradation system in cells.
Hepatitis B virus (HBV) is a small DNA virus that has a partially double-stranded genome and replicates through an RNA intermediate. After entry into a host cell, the genome is converted to covalently closed circular DNA (cccDNA) that exists as a mini-chromosome in the nucleus and serves as the template for viral gene transcription. Viruses that are related to the HBV that infects humans have been discovered in many species, including ducks, woodchucks, squirrels and diverse species of bat. The virus predominantly infects liver cells, and can cause chronic infection even in the presence of an intact immune response.
Much of the uncertainty about the function of HBx has been attributed to the limitations of the experimental models in which it has been studied, because most are based on non-infectious systems. However, it is clear that HBx is required for effective HBV infection in vivo; woodchuck hepatitis virus that harbours defects in the gene encoding this protein is poorly infective2,3.
Many host factors are known to interact with HBx. Among them, damaged DNA binding protein 1 (DDB1) was first identified using a genetic approach4. This interaction was subsequently validated by structural and functional studies. But how this seemingly unrelated DNA-damage-response protein is involved in the function of HBx remained unclear. Like many typical breakthroughs in science, advances in an unrelated field provided the connection.
The ubiquitin–proteasome system has garnered much attention because of its central role in many cellular processes5, and its components and biochemical pathways are well understood. Among them is an E3 ligase enzyme named CRL4. This member of the Cullin-RING ubiquitin ligase family uses DDB1 as an adaptor protein with which to target specific proteins for degradation. Several viral-gene products are known to target the CRL4–DDB1 complex6, suggesting that the ubiquitin–proteasome system may be a common cellular pathway exploited by viruses to ensure productive infection. Decorsière et al. set out to identify proteins targeted for destruction by the complex formed between CRL4, DDB1 and HBx.
Using a clever protein-interaction approach, the authors identified the Smc5/6 protein complex, which is involved in several aspects of chromosome biology, as one such target (Fig. 1). HBx redirects the enzymatic function of CRL4 to target Smc5/6 for ubiquitina-tion — a modification that marks the protein for degradation. They then used genetic and biochemical methods to show that the Smc5/6 complex indeed associates with the HBV genome (probably with the cccDNA, although this is not shown definitively) to inhibit viral transcription. A previous study7 of Cullin-RING ligases did not identify the Smc5/6 complex as a target of CRL4, so it is unclear whether the complex is a natural substrate of CRL4. It is possible that HBx alters the substrate specificity of CRL4 such that the enzyme targets cellular proteins, in addition to the Smc5/6 complex, for degradation.
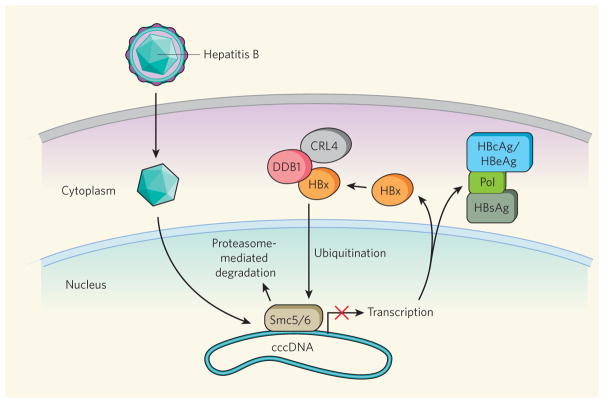
Figure 1. Hepatitis B virus evasion of cellular antiviral function.
After a hepatitis B virus (HBV) particle has entered a host cell (during which it becomes de-coated), its genome is converted to covalently closed circular DNA (cccDNA) that exists as a mini-chromosome in the nucleus and serves as the template for viral gene transcription. Three HBV proteins have well-defined functions: a core protein (named HBcAg, or HBeAg when secreted), a reverse transcriptase enzyme (Pol) and an envelope protein (HBsAg). Decorsière et al.1 reveal that another viral protein, HBx, acts to degrade a cellular antiviral factor, the Smc5/6 protein complex. The authors show that Smc5/6 probably binds to the HBV cccDNA and thus inhibits viral transcription. But HBx interacts with DDB1, an adaptor protein for the cell’s CRL4 E3 ubiquitin ligase enzyme complex, and this results in the Smc5/6 complex being targeted for ubiquitination — a modification that designates the complex for degradation by the cell’s proteasome machinery.
The discovery of Smc5/6 as a viral restriction factor adds to a growing list of intrinsic mechanisms in the cellular defence arsenal against DNA-viral pathogens8,9. It seems that the Smc5/6 complex binds to and suppresses only extrachromosomal (episomal), not chromosomally integrated, HBV DNA. This episome-specific function is reminiscent of another class of antiviral factor, the APOBEC family, which specifically targets episomal foreign DNA for modification and degradation10. It has been suggested that the APOBEC3A protein, whose expression is induced by interferon signalling proteins, binds to and edits HBV cccDNA, leading to cccDNA degradation11. Such pathways reveal that diverse cellular mechanisms have evolved to defend against HBV infection.
The Smc5/6 complex has been implicated in cell-cycle progression, chromosome organization and DNA repair12, but little is known about its involvement in transcriptional regulation. Decorsière et al. have shown that this complex may have a key antiviral role by binding to viral genomes and silencing their transcription. How Smc5/6 targets episomal DNA to exert this silencing effect, and whether it has a similar activity against other DNA viruses, remain to be investigated. There is evidence13 that HBV transcription is tightly regulated by epigenetic modifications (those that alter gene expression without modifying the nucleic-acid sequence), and it seems that HBx epigenetically modifies the HBV mini-chromosome. It is possible that the Smc5/6 complex silences transcription by affecting the epigenetic status of the viral mini-chromosome.
It has been suggested that HBx is present on the HBV mini-chromosome10, although it is not known whether its association with the genome is necessary for the targeted degradation of the Smc5/6 complex. It is also unclear whether HBx interacts directly with Smc5/6; because the complex contains many proteins, it could be that a yet-unidentified factor in Smc5/6 is the direct target of the CRL4–DDB1–HBx complex.
Another remaining puzzle is why the gene that encodes HBx exists in the mammalian but not in avian hepatitis viruses. Did the interaction of this protein with Smc5/6 emerge as mammalian HBV diverged from its avian counterparts? It is intriguing to speculate that the HBx gene might have been acquired from the host as the virus entered mammals more than 10,000 years ago; acquisition of host genes is a typical evolutionary event for many viruses. It seems likely that Decorsière and colleagues’ discovery is not the final episode of the ‘HBx files’.
References
- 1.Decorsière A, et al. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 2.Zoulim F, Saputelli J, Seeger C. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Torii N, Hu Z, Jacob J, Liang TJ. J Clin Invest. 2001;108:1523–1531. doi: 10.1172/JCI13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee TH, Elledge SJ, Butel JS. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosu DR, Kipreos ET. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randow F, Lehner PJ. Nature Cell Biol. 2009;11:527–534. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 7.Emanuele MJ, et al. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavalai N, Stamminger T. Virus Res. 2011;157:128–133. doi: 10.1016/j.virusres.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Wiebe MS, Jamin A. J Virol. 2016 doi: 10.1128/JVI.00178-16. http://dx.doi.org/10.1128/JVI.00178-16. [DOI] [PMC free article] [PubMed]
- 10.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. Nature Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucifora J, et al. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kegel A, Sjögren C. Cold Spring Harb Symp Quant Biol. 2010;75:179–187. doi: 10.1101/sqb.2010.75.047. [DOI] [PubMed] [Google Scholar]
- 13.Levrero M, et al. J Hepatol. 2009;51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]