Abstract
The ETS transcriptional repressor ETV6 (or TEL) is autoinhibited by an α-helix that sterically blocks its DNA-binding ETS domain. The inhibitory helix is marginally stable and unfolds when ETV6 binds to either specific or non-specific DNA. Using NMR spectroscopy, we show that folding of the inhibitory helix requires a buried charge–dipole interaction with helix H1 of the ETS domain. This interaction also contributes directly to autoinhibition by precluding a highly conserved dipole-enhanced hydrogen bond between the phosphodiester backbone of bound DNA and the N terminus of helix H1. To probe further the thermodynamic basis of autoinhibition, ETV6 variants were generated with amino acid substitutions introduced along the solvent exposed surface of the inhibitory helix. These changes were designed to increase the intrinsic helical propensity of the inhibitory helix without perturbing its packing interactions with the ETS domain. NMR-monitored amide hydrogen exchange measurements confirmed that the stability of the folded inhibitory helix increases progressively with added helix-promoting substitutions. This also results in progressively reinforced autoinhibition and decreased DNA-binding affinity. Surprisingly, locking the inhibitory helix onto the ETS domain by a disulfide bridge severely impairs, but does not abolish DNA binding. Weak interactions still occur via an interface displaced from the canonical ETS domain DNA-binding surface. Collectively, these studies establish a direct thermodynamic linkage between inhibitory helix stability and ETV6 autoinhibition, and demonstrate that helix unfolding does not strictly precede DNA binding. Modulating inhibitory helix stability provides a potential route for the in vivo regulation of ETV6 activity.
Keywords: ETS family, protein folding, charge–dipole interaction, NMR spectroscopy, hydrogen exchange
Introduction
Normal cellular function requires the tight control of protein activity, loss of which can result in aberrant growth and disease. Autoinhibition is one such important control mechanism, whereby one part of a protein, typically referred to as the inhibitory module or domain, interacts with another part to negatively regulate its function [1,2]. Relief or reinforcement of autoinhibition via protein partnerships, post-translational modifications or proteolysis in response to cellular signaling events enables the control of numerous protein functions, spanning gene expression to metabolism and transport.
Autoinhibition of DNA binding has emerged as a key regulatory mechanism for the ETS family of transcription factors [3]. There are 28 ETS paralogs in humans, all of which contain a conserved ETS domain that mediates binding to DNA sequences containing a 5′GGA(A/T)3′ motif [4,5]. Despite this commonality, several distinct mechanisms of autoinhibition have been discovered across the ETS family. For example, ETS-1 has an inhibitory module consisting of an intrinsically disordered serine-rich region (SRR), as well as four helices flanking its ETS domain. DNA binding is allosterically coupled to helix unfolding and the disruption of the inhibitory module [6,7]. Furthermore, ETS-1 autoinhibition is reinforced by multi-site phosphorylation of the SRR [6,8] and relieved through partnerships with additional transcription factors such as Runx1 [9–11]. In contrast, the ETS transcriptional repressor ETV6 (or TEL) is inhibited by an α-helix that sterically occludes the DNA-binding interface of its ETS domain [12,13]. This inhibitory helix also unfolds upon binding both specific and non-specific DNA [14]. In the case of ERG, weak autoinhibition is reported to involve flexible sequences flanking its ETS domain [15].
Underlying these differences, the autoinhibitory mechanisms exhibited by various ETS family members also share several common features [3]. The inhibitory modules are formed by sequences appended to the ETS domain. These sequences, which are either marginally stable helices or predominantly disordered, readily undergo conformational transitions. Furthermore, the inhibitory elements dampen dynamics in the ETS domain that, by analogy to the well-characterized lac repressor [16], are postulated to be important for binding non-specific and specific DNA [6,7,13,15].
In this study, we have dissected the thermodynamic basis of ETV6 autoinhibition. Initially, we identified a buried charge–dipole interaction that is critical to the folding of the inhibitory helix. This interaction positions the inhibitory helix onto the ETS domain and prevents the formation of a conserved hydrogen bond to the phosphodiester backbone of DNA. Subsequently, we investigated how the stability of the inhibitory helix is linked to autoinhibition. Amino acid substitutions that progressively enhance the stability of the inhibitory helix against unfolding also progressively enhance autoinhibition. Finally, we determined that ETV6 can still weakly bind DNA even when the inhibitory helix is locked onto the ETS domain via a disulfide bridge. Thus, despite blocking the canonical DNA-recognition surface of the ETS domain, unfolding of the inhibitory helix does not necessarily precede DNA binding. Together, these data lead to a detailed thermodynamic model for the autoinhibition of specific and non-specific DNA binding by ETV6. This model also provides a foundation for understanding possible mechanisms for regulating ETV6 activity in a cellular context.
Results
Helix H5 has low intrinsic helical propensity
Autoinhibited ETV6 has two helices, H4 and H5, appended to the C-terminus of its ETS domain (Fig. 1a). The presence of these helices reduces the nanomolar-range affinity of ETV6 for a consensus ETS binding site by ~50-fold [12]. Using NMR spectroscopy and X-ray crystallography, we have demonstrated previously that helix H5 sterically blocks the canonical DNA interaction surface of the ETS domain and unfolds when ETV6 binds either specific (cognate) or non-specific DNA [13,14]. Furthermore, although well formed, helix H5 is only marginally stable and poised to unfold [13]. Thus, the modest energetic penalty of this requisite conformational change leads to a net reduction in DNA binding affinity. Together, these observations provide a straightforward structural and thermodynamic mechanism for ETV6 autoinhibition.
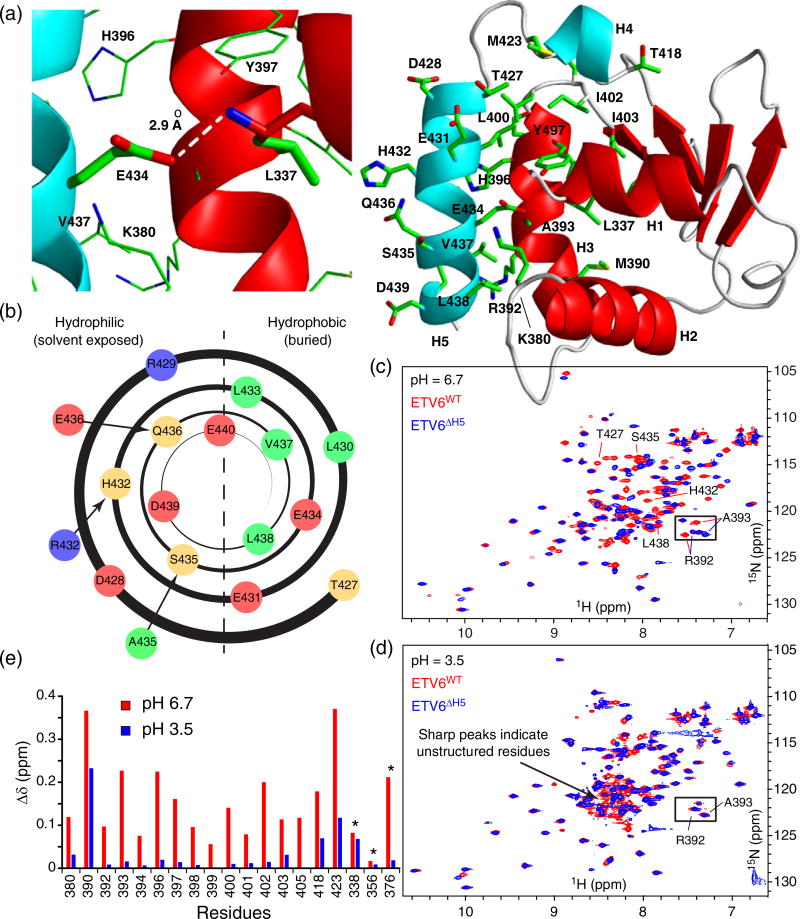
Fig. 1.
Negatively charged Glu434 is required for folding of the inhibitory helix H5. (a) Packing interactions of helix H5 (cyan) with the ETS domain (red; 2LF8.pdb). Right panel: residues in the inhibitory module and its interface with the ETS domain are shown as sticks (oxygen, red; nitrogen, blue; sulfur, yellow; carbon, green). Left panel: expanded view of the buried Glu434 sidechain and its interaction with the amide nitrogen of Leu337 at the N-terminus of helix H1. (b) Helical wheel representation of helix H5 (hydrophobic residues in green, polar and His in light orange, negatively charged in red and positively charged in blue). The helix is amphipathic with the hydrophobic side packing against the ETS domain and the hydrophilic side exposed to solvent. Glu434 interrupts the hydrophobic pattern. Stabilizing amino acid substitutions introduced into helix H5 are also indicated outside the helical wheel (Table 1). Overlaid 15N-HSQC spectra of ETV6WT (red) and ETV6ΔH5 (blue) at pH 6.7 (c) and pH 3.5 (d). Several peaks corresponding to helix H5 residues in ETV6WT (Thr427, His432, Ser435 and Leu438) are resolved at pH 6.7, yet absent at pH 3.5 and most likely cluster in the central “random coil” region of the spectrum. (e) Combined amide 1HN–15N and Trp indole 1Hε1–15Nε1 (*) chemical shift differences (Δδ) between ETV6WT and ETV6ΔH5 at pH 6.7 and 3.5 are presented for residues in the ETS domain that interface with helix H5 (including Arg392 and Ala393, boxed in the spectra). Along with the appearance of sharp signals with random coil chemical shifts, the close similarity of the spectra recorded for the two proteins at pH 3.5, but not at pH 6.7, indicates that helix H5 in ETV6WT unfolds under acidic conditions.
In this study, we sought to understand the physicochemical basis of helix H5 stability and its association with the ETS domain. Inspection of the sequence of the inhibitory helix reveals that its constituent residues, collectively, have very low helical propensities (Table 1). That is, as an isolated peptide, this sequence is predicted to be predominantly disordered and not to adopt a helical conformation [17]. This is due to the presence of several residues, such as Asp, Thr, Val and His, that disfavor helix formation [18]. Also, when formed, a positively charged Arg close to the N terminus and negatively charged Asp and Glu close to the C terminus of helix H5 will be unfavorably positioned with respect to its electrostatic macro-dipole. Hence, the inhibitory helix is intrinsically unstable and requires additional interactions with the ETS domain for its conformational integrity.
Table 1.
Characterization of ETV6 variants
| Helix H5 sequencea | AGADIRb (%) | L337 PFc |
H5 Avg. PFd |
ΔΔGHX (kcal/mol)e | KD (nM)f | Fold-increase inhibitiong | |
|---|---|---|---|---|---|---|---|
| ETV6ΔH5 | No helix H5 | ndh | 0.1i | ||||
| ETV6WT | 427TDRLEHLESQVLDE440 | 0.4 | ndh | 13 | 6 ± 1 | 1 | |
| ETV6E434Q | TDRLEHLQSQVLDE | 0.6 | ndh | Unfolded H5 | |||
| ETV6E434D | TDRLEHLDSQVLDE | 0.4 | ndh | Unfolded H5 | |||
| ETV6H432R | TDRLERLESQVLDE | 1.6 | 11 | 17 | 0.1 | 30 ± 10 | 5 |
| ETV6RAE | TDRLERLEAEVLDE | 3.2 | 20 | 23 | 0.3 | 90 ± 10 | 15 |
| ETV62C-ox | TDRLEHLESQCLDE | 0.4 | 116 | 68 | 1.0 | 20,000 ± 9000j | 3000 |
The amino acid substitutions are bold underlined.
AGADIR predicted helical propensity for each given sequence as an isolated polypeptide [17].
Measured at 35 °C and 11.7 T (500 MHz) NMR field strength.
Measured at 30 °C and pH values of 6.4 and 7.2. PFs of residues 430 to 440 were averaged.
, where ΔGHX = RTln(PF) for unfolding to an exchange competent state was calculated from the average PFs for the helix H5 residues 430 to 440.
Measured by EMSA with a 23-bp DNA duplex containing the ETS recognition sequence 5′−GGAA−3′. The data are the average ± standard deviation of at least four measurements.
Fold increase in autoinhibition with respect to inhibited ETV6WT.
Not detected.
Value reported [13].
Measured by NMR-based titrations using a 15-bp DNA containing a consensus 5′−GGAA−3′ sequence.
Inhibitory helix H5 is stabilized by a charge–dipole interaction with the ETS domain
Helix H5 is generally amphipathic and packs against the ETS domain via its hydrophobic surface (Fig. 1a and b). However, it is striking that Glu434 interrupts this amphipathic pattern. In the NMR spectroscopically derived structural ensemble of an inhibited ETV6 fragment, Glu434 in helix H5 is fully buried and positioned to interact with the N-terminus of helix H1 of the ETS domain (Fig. 1a). Depending on its pKa value in this buried state, the Glu434 sidechain carboxyl group could be neutral or negatively charged. In both forms, it can hydrogen bond with the amide NH of Leu337 (the first residue of helix H1), whereas when present as carboxylate, it can also partake in an electrostatic interaction with the macro-dipole of helix H1 [19]. Either way, the positioning of Glu434 that accompanies helix H5 folding directly prevents the formation of a conserved dipole-enhanced hydrogen bond between Leu337 and a phosphodiester oxygen in the backbone of DNA [14,20]. The importance of these interactions is consistent with the loss of autoinhibition when Glu434 is replaced with an alanine [12].
To probe the role of Glu434 in helix H5 stability, we monitored the conformation of ETV6WT (Table 1) over a range of pH values from 6.7 to 3.5 using 15N heteronuclear single quantum correlation (HSQC) spectra. At pH values below ~4.0, signals from amides in helix H5 disappeared while new peaks appeared in the central region of the 15N-HSQC spectra (Fig. 1c and d). Although not assigned to specific residues, these sharp peaks with random coil chemical shifts are attributed to residues in an unfolded H5 that result from acidification of the protein. To verify this conclusion, we note that the chemical shifts of the residues in the ETS domain that pack against helix H5 are highly sensitive to the presence of this helix [14]. We reasoned that the chemical shifts for these interfacial residues should be similar when helix H5 is unfolded or deleted. Hence, we compared their amide 15N and 1HN chemical shifts in the context of ETV6WT versus ETV6ΔH5 (lacking H5) at both pH 6.7 and 3.5 as reporters for the conformational state of helix H5. As expected, the chemical shifts of many amides in these two protein constructs were very different at pH 6.7 due to the presence or absence of a folded helix H5. In contrast at pH 3.5, these chemical shift differences decreased significantly, indicating that helix H5 is indeed unfolded and no longer packed against the ETS domain (Fig. 1e). Hence, the negatively charged form of Glu434 appears to be required to maintain the structural integrity of the inhibitory helix H5.
To confirm this conclusion, Glu434 was replaced with glutamine and aspartate. Based on an analysis of amide 15N and 1HN chemical shifts, helix H5 is also predominantly unstructured in ETV6E434Q at pH 6.7 (Supplementary Fig. S1). Gln434 can potentially form a hydrogen bond with Leu337 in helix H1, but cannot partake in a charge–dipole interaction. Thus, the charged state of Glu434 indeed appears critical for its interaction with helix H1. Changing Glu434 to aspartic acid, which shortens the sidechain while maintaining the ionizable carboxyl group, yielded chemical shifts for ETV6E434D that are intermediate between those of ETV6WT and ETV6ΔH5 (Supplementary Fig. S1). Therefore, helix H5 is destabilized by the E434D substitution, although not to the same extent as with E434Q or, even more so, acidification of the WT protein. This highlights the strict distance dependence of the Glu434–helix H1 interaction as expected for a buried electrostatic contact. The analysis of these ETV6 variants also demonstrates that the pH-dependent folding of helix H5 does not arise from additional Glu, Asp and His residues along the helix H5–ETS domain interface that likely titrate in the pH 6.7 to 3.5 range.
Collectively, these experiments show that a negatively charged Glu434 sidechain is required for the proper folding of helix H5 and its packing against the ETS domain. Unfortunately, we were unable to extract the pKa values of Glu434 in the context of folded and unfolded helix H5 from the NMR-monitored titrations due to complex patterns of chemical shift perturbations (CSPs) arising from ionization and conformation equilibria. However, by thermodynamic linkage, when buried due to the favorable folding of helix H5, the pKa value of Glu434 must be less than when it is solvent exposed with the helix unfolded. The latter is likely near the pKa ~4.4 of a glutamic acid in a random coil polypeptide [21].
Amino acid substitutions designed to stabilize inhibitory helix H5 and strengthen its interactions with the ETS domain
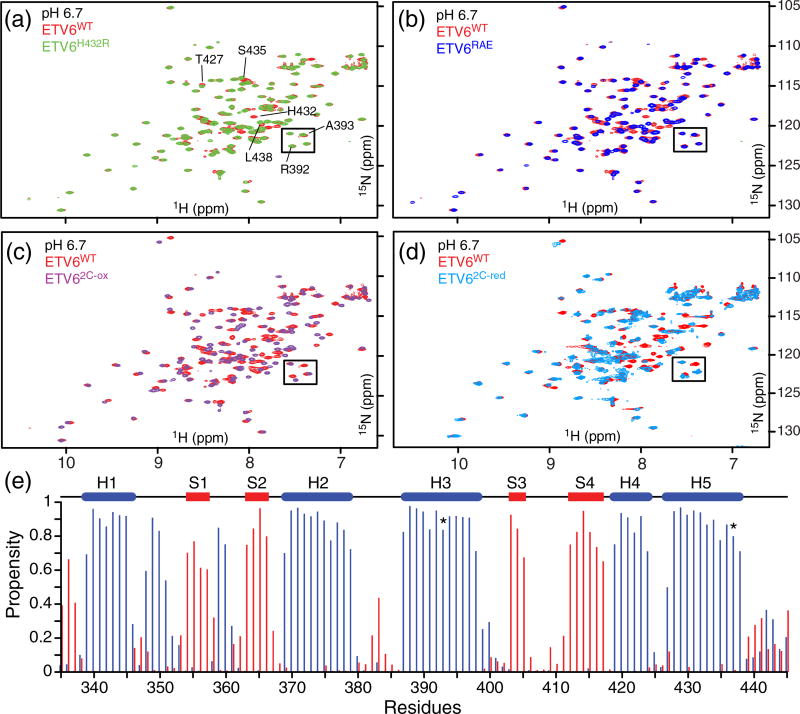
We hypothesized that ETV6 DNA-binding autoinhibition is directly dependent upon the stability of the inhibitory module. To test this hypothesis, amino acid substitutions were introduced into helix H5 in the attempt to increase its helical propensity (Table 1). Importantly, these changes were restricted to the solvent-exposed surface of the helix so that its packing interactions with the ETS domain would not be perturbed. The 15N-HSQC spectra of these ETV6 variants confirmed that each contained a well-folded ETS domain (Fig. 2a and b). A properly folded helix H5 in ETV6H432R and ETV6RAE was also inferred from the close similarity of the amide chemical shifts of residues along the DNA-binding interface with those of the corresponding residues in ETV6WT.
Fig. 2.
Stabilizing amino acid substitutions do not markedly perturb the structure of the ETS domain or inhibitory helix H5. Overlaid 15N-HSQC spectra of the variants ETV6H432R (a), ETV6RAE (b), ETV62C-ox (c) and ETV62C-red (d) with ETV6WT at pH 6.7. Well-resolved peaks for several helix H5 residues are labeled in panel a. Also identified are signals from Arg392 and Ala393 (boxed), two residues in the ETS domain with chemical shifts diagnostic of a folded helix H5. The close similarity of the spectra of ETV6H432R and ETV6RAE to that of ETV6WT confirms that the changes do not alter packing of helix H5 on the ETS domain. In the case of ETV62C-red, some spectral differences arise from the A393C and V437C substitutions at the ETS domain–helix H5 interface. (e) Although spectral perturbations are more pronounced with ETV62C-ox, secondary structure propensities predicted from its 1HN, 15N, 13Cα and 13Cβ chemical shifts with the algorithm MICS [22] confirm that helix H5 remains folded (blue: α-helix; red β-strand; coil: not shown; all three propensities sum to 1). In the cartoon, blue cylinders and red bars indicate α-helices and β-strands, respectively. The positions of Cys393 and Cys437 are indicated (*).
Based on the steric mechanism of autoinhibition, we also hypothesized that locking the inhibitory helix onto the DNA-binding interface would severely impair DNA binding. A variant was therefore designed to provide a disulfide bond between helix H5 and the DNA-binding interface (ETV62C-ox, Table 1). This was accomplished by changing Ala393 in the DNA-recognition helix H3 and Val437 in the inhibitory helix H5 to cysteines. The formation of disulfide bond in the ETV62C-ox variant was confirmed from SDS-PAGE assays (not shown), as well as by the diagnostic 13Cβ chemical shifts of its cysteine residues [23]. Although the presence of the disulfide bond perturbed the spectrum of the ETV62C-ox ETS domain (Fig. 2c and Supplementary Fig. S2), an MICS analysis [22] of its 1HN, 15N, 13Cα and 13Cβ chemical shifts confirmed that all strands and helices, including helix H5, remained intact (Fig. 2e). Reducing the disulfide bond restored ETV6WT-like chemical shifts for ETV62C-red, albeit with some perturbations due to the A393C and V437C changes at the ETS domain–helix H5 interface (Fig. 2d and Supplementary Fig. S2).
Amide hydrogen exchange demonstrates that designed amino acid substitutions stabilize helix H5
NMR-based hydrogen exchange (HX) experiments were used to determine the stability of the wild-type and variant ETV6 ETS domains with residue-level resolution. The protection factors (PFs) for backbone amides were determined by complementary protium–deuterium and protium–protium HX experiments. A PF is given by the ratio kint/kex, where kex is the experimentally measured HX rate constant and kint is the intrinsic rate constant predicted for the corresponding unstructured sequence under the same conditions of pH, temperature and solvent [24,25]. In the commonly observed EX2 regime, the PF is the inverse of equilibrium constant for fluctuations between a “closed” state, where the exchange of the amide proton with the bulk solvent is prevented, and a transient “open” exchange-competent state [26]. Thus, PFs provide a per residue measure of the local free energy of stability [ΔG°HX = RTln(PF)] of a protein for conformational fluctuations detectable by HX.
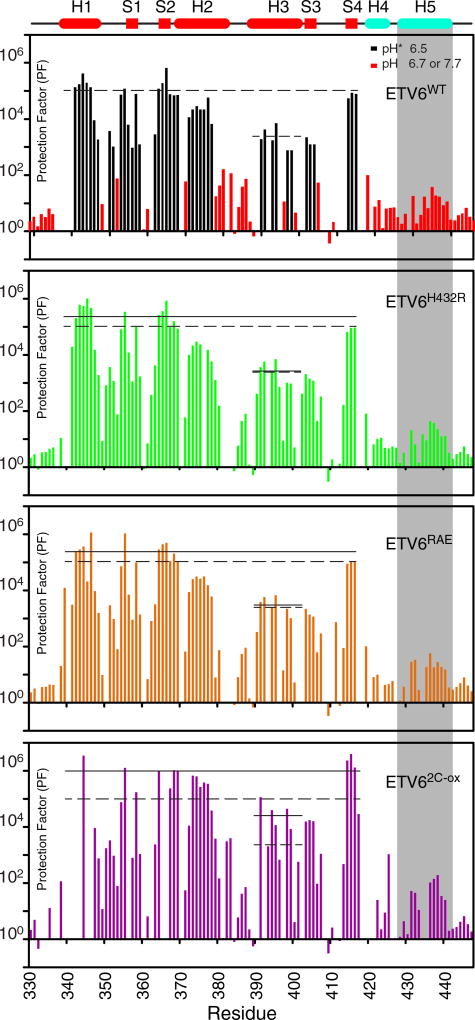
The HX measurements for ETV6WT (Fig. 3) agree well with those reported previously [13]. Amides in helix H1 and strands S1, S2 and S4 in the core ETS domain have the highest PFs (~105) and likely exchange only through global fluctuations [27]. Although integral to the ETS domain fold, the DNA-recognition helix H3 has intermediate PFs (~103), indicating that it undergoes sub-global fluctuations [13]. Helix H3 in ETS-1 also shows moderated PFs, and we have postulated that these dynamic properties are a conserved feature of ETS domains that are important for binding non-specific and specific DNA sequences [6,7,28]. In contrast, helix H5 is marginally stable in ETV6WT with PFs of only ~ 10 (Figs. 3 and 4). Thus, although well defined in the NMR-derived structural ensemble of this protein, helix H5 readily samples unfolded states detectable via HX.
Fig. 3.
HX measurements provide a measure of local and global protein stability. HX PFs for ETV6WT and its variants were merged from a combination of experiments (Supplementary Table S1). In the case of ETV6WT, black and red bars indicate data collected by protium–deuterium HX (uncorrected pH* 6.5) and protium–protium CLEANEX (pH 6.7 or 7.7) experiments, respectively. This is not differentiated for the three variants. The estimated PF errors are 5% to 15%, and in the upper cartoon, cylinders and rectangles indicate α-helices and β-strands, respectively. The average PFs for the ETS domain (residues in helices H1 and H2 and strands S1, S2 and S4), and for the recognition helix H3 are shown as solid lines for each variant species in its respective panel; the corresponding values for ETV6WT are shown as dashed lines in all panels. For better visualization, the data for residues in helix H5 (gray shading) are overlaid in Fig. 4. A comparison of these average PFs shows that the designed substitutions stabilize the ETS domain against HX.
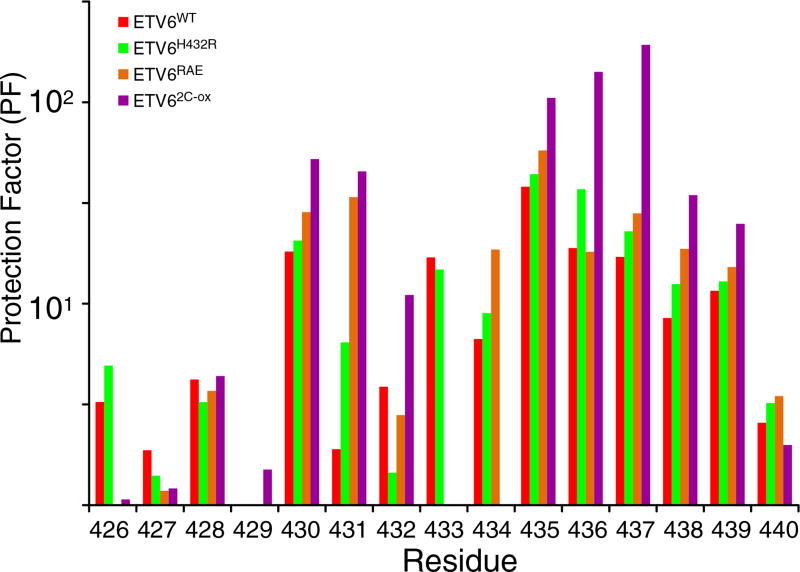
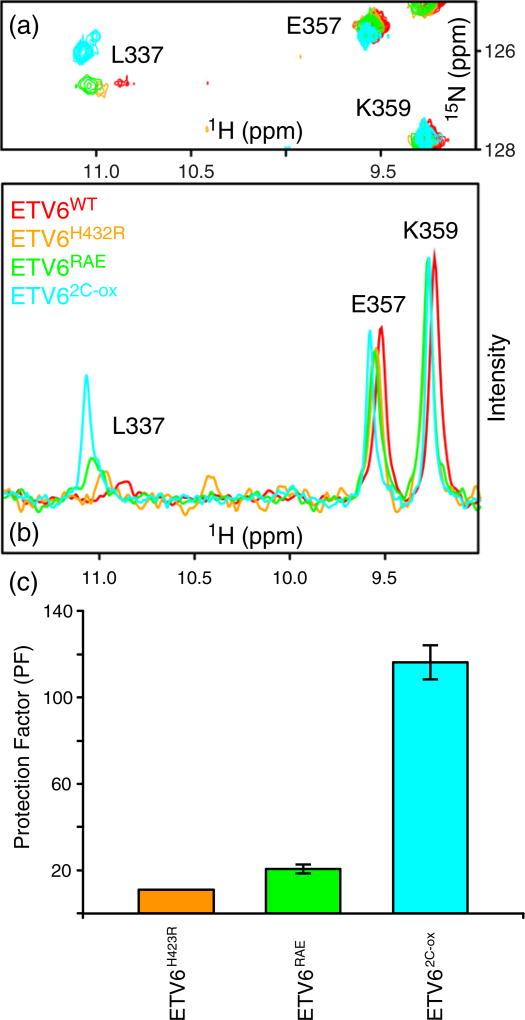
Fig. 4.
HX measurements demonstrate the stabilization of helix H5. PFs for helix H5 residues in ETV6WT and its variants are shown. The data were derived from protium– protium CLEANEX experiments recorded at several temperature and pH values (Supplementary Table S1). The estimated PF error is 5% to 15%. Most residues exhibit a progressive increase in protection against HX in the order of ETVWT < ETV6H432R < ETV6RAE < ETV62-C-ox indicating progressive stabilization of helix H5 by the designed substitutions. Residue-specific variations within this trend could arise for many reasons, including exchange from partially, rather than fully, unfolded states of the inhibitory helix, as well as errors in the measured (kex) and predicted (kint) rate constants for each residue and the assumption that PFs are independent of sample pH and temperature over the range of conditions used for these measurements. Accordingly, the averaged PFs for the inhibitory helix amides are summarized in Table 1 and Fig. 6.
Relative to ETV6WT, the variants ETV6H432R and ETV6RAE have a ~ 2-fold increase in PFs for the core secondary structure elements of the ETS domain, whereas ETV62C-ox has a ~5-fold increase (Fig. 3). Thus, these amino acid substitutions modestly stabilize the protein on a global level. The disulfide bond to Cys393 in helix H3 also increases its protection against HX. Most importantly, the substitutions generally retard the HX of amides throughout helix H5 (Fig. 4). Recognizing that the first three amides in a regular α-helix do not form intra-helical hydrogen bonds, we averaged the PFs for residues 430 to 440 to obtain a measure of H5 stability for each protein. As summarized in Table 1, the variants ETV6H432R, ETV6RAE and ETV62C-ox displayed small but progressively increasing average PFs relative to the ETV6WT. Thus, the designed substitutions indeed stabilize helix H5.
Designed amino acid substitutions also stabilize the interaction of helix H5 with helix H1 of ETS domain
As described above, the charge–dipole interaction between Glu434 and helix H1 is crucial for helix H5 stability. Leu337 is the first residue of helix H1 and is very sensitive to this interaction. In particular, this amide of Leu337 has highly downfield shifted 1HN signal (~11 ppm) that is consistent with a hydrogen-bonding interaction with the Glu434 sidechain. Also, the 1HN–15N peak corresponding to the amide of Leu337 is almost at the noise level in the 15N-HSQC spectrum of ETV6WT at 25 °C and pH 6.7 (Fig. 5). However, its signal sharpens with increasing temperature (20 °C to 35 °C) and decreasing magnetic field strength (not shown). This diagnostic behavior indicates that the Leu337 amide signal is broadened by fast-intermediate regime conformational exchange [29]. It is plausible that such exchange broadening originates from an msec–µsec timescale conformational equilibrium of helix H5 between its folded and unstructured states, which in turn results in different chemical shifts for the amide of Leu337 due to the presence or absence, respectively, of an interacting Glu434.
Fig. 5.
Stabilization of helix H5 leads to increased HX protection and reduced conformational exchange broadening for Leu337. An expanded region (a) and corresponding 1H skyline projections (b) of the overlaid 15N-HSQC spectra of ETV6WT and three variants (25 °C and pH 6.5). These spectra show that the 1HN–15N signal of Leu337 sharpens and increases in intensity as helix H5 is stabilized by amino acid substitutions. (c) The substitutions also increase the HX PFs for the Leu337 amide, as measured by a CLEANEX experiment at 35 °C using a 500-MHz spectrometer. These experimental conditions of higher temperature and lower magnetic field strength favored signal detection. Due to its very weak signal, we could not measure the PF of Leu337 in ETV6WT.
Relative to ETV6WT, the amide signal of Leu337 signal sharpened in the 15N-HSQC spectra of ETV6H432R, ETV6RAE and ETV62C-ox (Fig. 5). Therefore, stabilizing helix H5 reduces its exchange broadening. With a sharper signal, we were also able to measure the PFs for Leu337 in all three variants. These PFs increased progressively with the increasing stability of helix H5 across this series (Table 1). Collectively, these results confirm that Leu337 is sensitive to the folding–unfolding of helix H5 and further validates the presence of charge–dipole interaction between Glu434 and this N-terminal residue of helix H1.
Stabilization of inhibitory helix reduces DNA binding
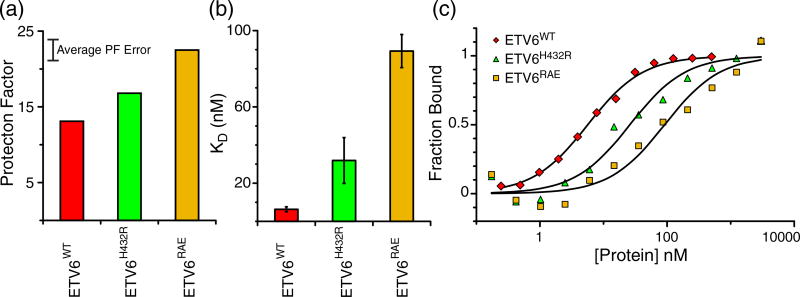
To determine the effect of inhibitory helix stabilization on DNA binding, equilibrium dissociation constants (KDvalues) were measured for wild-type and variant ETV6 fragments. These data were obtained via electrophoretic mobility shift assays (EMSAs) with DNA duplexes bearing the ETS recognition sequence 5′−GGAA−3′ (Table 1 and Fig. 6). The KD value of 6 nM for ETV6WT is in good agreement with our previous DNA binding studies [13]. Also, although only an estimated KD < 1 nM could be obtained for ETV6ΔH, this is consistent with a value of 0.1 nM reported for the uninhibited fragment of ETV6WT [13]. Most importantly, progressively stabilizing the inhibitory helix H5 resulted in progressively weaker DNA-binding affinity for the ETV6H432R and ETV6RAE proteins. This supports our hypothesis that ETV6 autoinhibition scales with the stability of the inhibitory module.
Fig. 6.
Helix H5 stabilization weakens ETV6 DNA binding. (a) Average PFs of helix H5 residues of ETV6WT and the stabilized variants ETV6H432R and ETV6RAE. An approximate 10% PF error is indicated. (b) Equilibrium dissociation constants (KD values) for ETV6WT, ETV6H432R and ETV6RAE. These KD values increase as the PFs and hence stability of helix H5 increase. (c) Representative EMSA DNA binding data. Symbols and solid lines represent fraction DNA bound as determined from band intensities of free DNA and data fitting to a 1:1 binding model, respectively.
Disulfide-blocked ETV62C-ox binds DNA weakly via a displaced interface
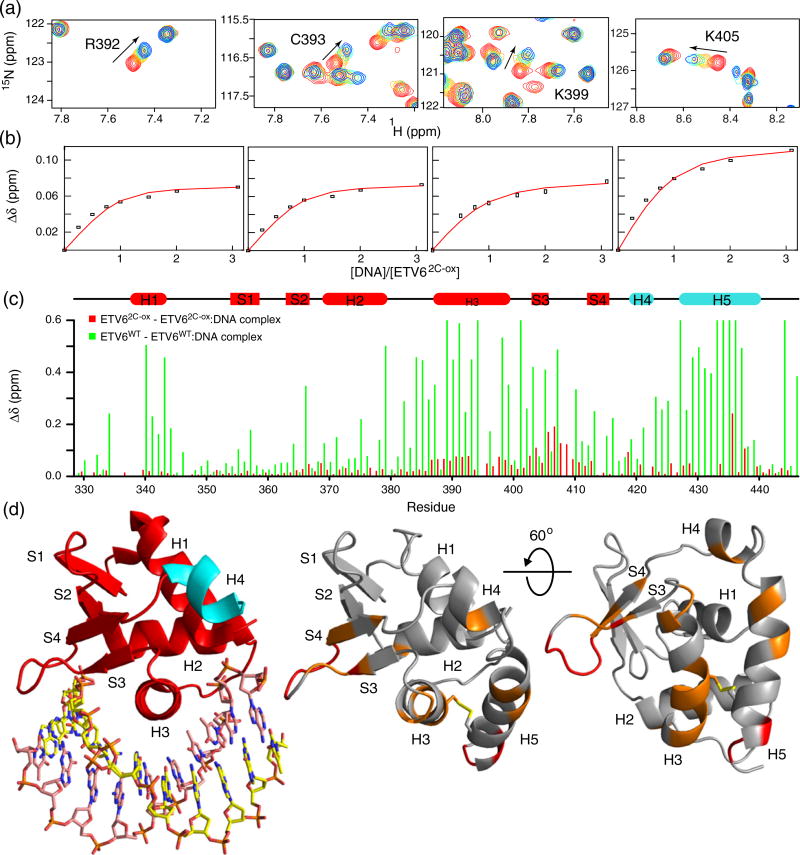
The interaction of ETV62C-ox with DNA was investigated by NMR methods. In particular, the titration of 15N-labeled ETV62C-ox with a 15-bp DNA duplex containing the ETS recognition sequence 5′−GGAA−3′ was monitored via 15N-HSQC spectroscopy (Fig. 7a). Surprisingly, despite the covalent attachment of helix H5 along the ETS domain recognition helix H3, the protein still bound DNA, albeit weakly. Fitting of the titration data yielded a KD value of 20 ± 9 µM (Fig. 7b and Table 1). This corresponds to an ~3000-fold reduction in affinity relative to the inhibited wild-type protein.
Fig. 7.
Disulfide-bridged ETV62C-ox binds DNA weakly via a displaced interface. (a) Expanded regions of the overlaid 15N-HSQC spectra of 15N-labeled ETV62C-ox recorded with increasing concentrations of a 15-bp DNA duplex containing the ETS recognition sequence 5′−GGAA−3′. The DNA/protein molar ratios were 0, 0.25, 0.5, 1.0, 1.5, 2.0 and 3.1, and the initial protein concentration was 0.2 mM. Red peaks: free ETV62C-ox; blue peaks: ETV62C-ox with 3.1-fold DNA. For clarity, not all titration points are included. (b) Weak binding was observed in the fast exchange limit, and fitting the CSPs (Δδ) for six residues (R392, C393, K399 and K405 shown; G408 and Q441 not shown) to a 1:1 binding isotherm yielded an average KD value of 20 ± 9 µM. (c) Histogram of CSPs for the free versus DNA-bound forms of ETV62C-ox (3.1:1 DNA/protein in red) and ETV6WT (1:1.1:1 in green). The data for the latter protein are based on De et al. [14] and are clipped at an upper limit of 0.6 ppm. (d) When mapped onto a model of ETV62C-ox (right), the small CSPs for this disulfide-bridged protein localize to the S3–S4 wing and helices H3 and H5 (>0.04 ppm, orange; >0.1 ppm, red). This binding interface is displaced from the canonical ETS domain DNA-recognition surface seen with ETV6ΔH5 lacking H5 (4MHG.pdb; left). Also in contrast to ETV6WT, the lack of significant CSPs for helix H5 indicates that it remains folded in the DNA-bound state of ETV62C-ox.
The amide CSPs of ETV62C-ox accompanying DNA binding were also substantially smaller than those exhibited by ETV6WT (Fig. 7c). As discussed in detail previously [14], upon titration with specific DNA, many amides near the canonical ETS domain DNA-binding interface of ETV6WT exhibit large amide 1HN and 15N chemical changes. Additional amides are also perturbed due to the unfolding of helix H5. In contrast, residues throughout helix H5 of ETV62C-ox yielded small CSPs upon the addition of DNA, demonstrating that the disulfide-bridged helix remained intact. DNA-induced CSPs in ETV62C-ox map to residues along helix H5, the N-terminal portion of helix H3 and the “wing” between strands S3 and S4. These broadly cluster to same positively charged region of the ETS domain used for DNA binding by the wild-type protein. However, involvement of helix H5 and the lack of CSPs for amides in helices H1 and the turn of the helix (H2)–turn–helix (H3) illustrates that the interaction surface ETV62C-ox only partially overlaps the conserved ETS domain interface of the wild-type protein (Fig. 7). Thus, locking the inhibitory helix with a disulfide bond prevents full access to the canonical DNA-binding site and uncovers a weak interaction via a displaced interface.
Control experiments with ETV62C in the reduced state (ETV62C-red) resulted in more ETV6WT-like chemical shifts with respect to ETV62C-ox (Supplementary Fig. S2A). Moreover, the chemical shifts of a 1:1 complex of ETV62C-red with a 15-bp DNA duplex containing the ETS recognition sequence 5′−GGAA−3′ were also very similar to those of ETV6WT in complex with this DNA (Supplementary Fig. S2B). Thus, reducing the disulfide bond restores DNA binding to the canonical interface and also results in a tight binding complex. This is evidenced further by large CSPs similar to those exhibited by ETV6WT.
Discussion
Charge–dipole interaction plays critical role in ETV6 autoinhibition
The current model of ETV6 autoinhibition centers on the unfolding of an appended inhibitory helix H5 that otherwise sterically blocks the DNA-binding surface of its ETS domain. Consistent with the ~50-fold attenuation of affinity for specific DNA sites, helix H5 is marginally stable and poised to unfold [13]. Indeed, based on its sequence, the inhibitory residues have very low helical propensities and, in isolation, will not adopt a helical conformation. In this study, we investigated the factors contributing to the folding of helix H5 and its interaction with the adjacent ETS domain. In particular, we identified and characterized a charge–dipole interaction that is necessary for the stability of the ETV6 inhibitory helix. This interaction, which cannot be fully recapitulated by a glutamine or an aspartic acid, involves a buried negatively charged Glu434 sidechain in helix H5 that interacts with the amide of Leu337 at the positive end of the helix H1 macro-dipole.
The helix macro-dipole plays important roles in stabilization of protein structure, in ligand binding and in enzyme catalysis [19,30,31]. This macro-dipole can be approximated by a half-positive charge at the N-terminal end of a helix and a half-negative charge at its C-terminus [30]. These effective charges can interact in a distance- and orientation-dependent manner with adjacent charged or dipolar moieties. For example, theoretical calculations estimate that the interaction of an anionic group, such as that of a glutamate sidechain, at a 5-Å distance from the positive dipole of a 10-Å long helix can contribute up to 12 kcal/mol of attractive energy [32]. However, in the case of the ETV6 Glu434–helix H1 pairing, the net interaction is substantially weaker and may be offset by the thermodynamically linked energetic cost of helix H5 folding and desolvating the glutamate sidechain into a low dielectric environment at the ETS domain interface.
The macro-dipole of helix H1 plays an important role in recognition and binding of DNA sites by the ETS family of transcription factors. In all ETS domain–DNA complexes characterized to date, the amide NH of the N-terminal residue in helix H1 forms a highly conserved hydrogen bond with a backbone phosphodiester oxygen [33]. Thus, by interacting with the N-terminus of helix H1, Glu434 both provides stability to the inhibitory helix H5 and blocks a critical ETS domain–DNA contact. These linked processes contribute directly to the steric mechanism of ETV6 DNA-binding autoinhibition.
In contrast to ETV6, ETS-1 autoinhibition arises from the allosteric interactions of its ETS domain with an appended helical inhibitory module, combined with steric contributions from the adjacent intrinsically disordered SRR [6–8]). The inhibitory module in ETS-1 is offset from the DNA-binding interface and is formed predominantly by the hydrophobic packing of helices from the flanking N-terminal (HI-1, HI-2) and C-terminal (H4, H5) inhibitory sequences, along with helix H1 of the intervening ETS domain [28,34]. Previous studies have shown that eliminating or perturbing the helix H1–DNA interaction not only weakens DNA binding but also alters autoinhibition in ETS-1 [20]. Furthermore, the N-terminus of helix H1 is a focal point for the cooperative partnership of ETS-1 and Runx1 that relieves autoinhibition [11]. These results implicate helix H1 as a key link for the allosteric communication between the ETS-1 inhibitory module and its DNA-binding site. Thus, despite their distinct mechanisms, the interactions of helix H1 with their respective inhibitory sequences play central roles in DNA-binding autoinhibition for both ETV6 and ETS-1.
Inhibitory helix stability drives ETV6 autoinhibition
To experimentally investigate the dependence of ETV6 autoinhibition on helix H5 stability, we introduced a series of amino acid substitutions that were predicted to increase the helical propensity of its inhibitory sequence. Only solvent exposed sidechains were changed so that packing interactions with the core ETS domain were not perturbed. Amide HX measurements revealed a small increase in the average PFs of helix H5 amides due to the replacement of His432 with arginine (ETV6H432R), and a slightly larger increase due to the additional changes of Ser435 to alanine and Gln436 to glutamate (ETV6RAE). Similar trends were observed for the HX of Leu337, which is at the N-terminus of helix H1 and in a hydrogen bond with Glu434, as discussed above. Consistent with our predictions, these HX data confirmed that the surface substitutions modestly stabilize the folding of helix H5 and reinforce its packing against the ETS domain. In parallel, EMSA experiments revealed small decreases in the DNA-binding affinity of ETV6H432Rand, more so, ETV6RAE relative to the wild-type protein. Thus, ETV6 autoinhibition is directly linked to the stability of helix H5 and its packing onto the DNA-binding interface of the ETS domain.
A closer inspection of the results summarized in Table 1 and Fig. 6 reveals that the changes in helix H5 PFs and the KD values of ETV6WT, ETV6H432R and ETV6RAE for a cognate DNA site follow parallel trends, yet differ somewhat in relative magnitudes. In addition to the experimental uncertainty associated with measuring small changes in these values, the lack of a close linear correlation between the two parameters could arise due to numerous reasons. Most notably, the two sets of experiments were carried out under different experimental conditions (temperature, sample pH and ionic strength). Also, the PFs are residue specific and their values were averaged to obtain an estimation of the stability of helix H5. This neglects the possibility that partially unfolded states of helix H5, which allow HX, may still inhibit DNA binding. Similar to the SRR of ETS-1 [7], unstructured H5 residues could still transiently interact with the ETS domain. All of these effects may lead to slightly greater changes in autoinhibition than expected based solely on the increased protection of helix H5 amides from exchange with water.
The inhibitory helices of ETS-1 are also marginally stable and readily unfold upon DNA binding [28]. Although the factors leading to this behavior have not been examined in detail, the N-terminal helices (HI-1, HI-2) of ETS-1 also have low predicted helical propensities due, in part, to unfavorable juxtapositioning of charged sidechains. Importantly, the inhibitory module is stabilized through transient interactions with the adjacent SRR. These interactions increase with progressive levels of SRR phosphorylation, thus providing a “rheostatic” control of ETS-1 at the level of DNA binding [6,7]. Conversely, displacement of the inhibitory module by the ETS-interacting domain of Runx1 contributes to the relief of inhibition [9–11]. Additional partnerships, including with Pax5 [35] and with ETS-1 itself [36], also counteract autoinhibition.
By analogy with ETS-1, we postulate that ETV6 could also be regulated through post-translational modifications and heterotypic protein partnerships that alter the stability of its inhibitory helix. Although such processes remain to be identified, it is noteworthy that a dramatic effect on ETV6 binding to tandem DNA sites does come from self-association mediated by its PNT domain [12]. However, the PNT and ETS domains of ETV6 are joined by long intervening sequence that is predicted to be intrinsically disordered. To date, there is no direct link between the self-association of the PNT domain and the structure or function of the inhibited ETS domain.
Thermodynamic model for ETV6 DNA-binding autoinhibition
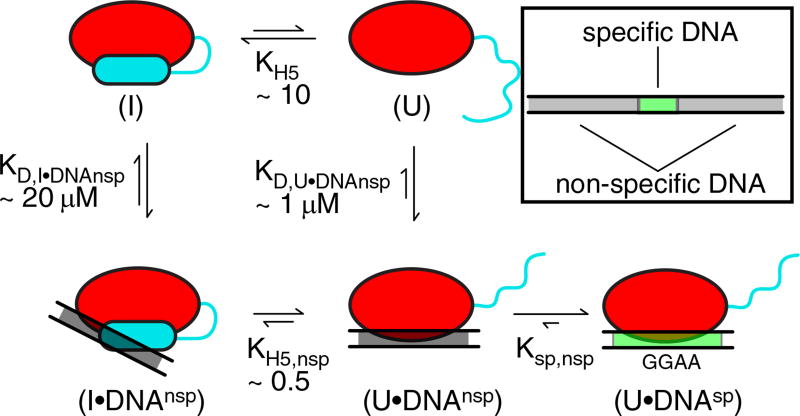
The interaction of ETV6 ETS domain with DNA and its autoinhibition can be represented by a multi-state thermodynamic model (Fig. 8). In the absence of DNA, ETV6 exchanges between an inhibited state (I), where helix H5 blocks the canonical ETS domain DNA-binding interface, and an uninhibited state (U), where residues corresponding to helix H5 are disordered. Based on the average PFs of helix H5, the equilibrium constant (KH5 = [I]/[U]) between these two states is estimated to be ~ 10.
Fig. 8.
Thermodynamic model of ETV6 autoinhibition. The interaction of the ETV6 ETS domain with specific DNA in a background of excess non-specific DNA can be represented by at least a five-state model (red oval, core ETS domain; cyan cylinder or line, folded or unfolded inhibitory helix H5, respectively; gray rectangle, nonspecific DNAnsp; and green rectangle, specific DNAsp). Additional states are possible, but not shown for simplicity. Free ETV6 can adopt inhibited (I) and uninhibited (U) states with helix H5 folded or unfolded, respectively. Inhibited ETV6 interacts with DNAnsp via a displaced DNA-binding interface, whereas the uninhibited ETV6 interacts via the canonical binding interface. Once ETV6 encounters DNAsp, a high-affinity complex is formed. The measured or calculated equilibrium constants for H5 folding in the absence (KH5) or presence (KH5,nsp) of DNAnsp and macroscopic dissociation constants for the protein• DNAnsp complexes with H5 folded (KD,I•DNAnsp) or unfolded (KD,U•DNAnsp) are shown. The macroscopic KD values include binding to multiple sub-sites within the 15-bp duplex DNA used as DNAnsp, of which only one schematic binding mode is shown. Also indicated is the intramolecular dissociation constant from the specific to nonspecific complexes (Ksp,nsp). The clockwise pathway from (I) to (U•DNAnsp) corresponds to a conformational selection model, whereas the counterclockwise pathway can be viewed as an induced fit model.
Because specific promoter sites constitute a small fraction of the genome, the initial encounter of ETV6 most likely occurs with non-specific DNA sequences lacking the ETS recognition sequences 5′−GGAA−3′. Previously, we demonstrated that ETV6 binds nonspecific DNA via its canonical ETS domain interface [14]. Non-specific binding is also autoinhibited by ~5-fold and accompanied by the unfolding of helix H5. However, in contrast to high-affinity specific complexes that are formed through basepair–sidechain hydrogen bonding with recognition sequences, non-specific complexes arise from weaker and less defined electrostatic interactions with the phosphodiester backbone [37,38]. Using isothermal titration calorimetry, the macroscopic KD values of ETV6WT and ETV6ΔH5 for a 15-bp non-specific DNA duplex were measured to be ~5 and ~1 µM, respectively [14]. These are macroscopic values because the 15-bp DNA can be bound at multiple subsites and in either orientation by ETV6. Microscopic KD values for the subsites can be obtained from the isothermal titration calorimetry data by fitting to complex binding isotherms that include neighbor exclusion along a finite lattice [39]. Assuming that the unstructured H5 residues in the uninhibited state (U) do not interfere with DNA binding, the macroscopic KD,U•DNAnsp of this state for a 15-bp non-specific DNA duplex should be similar to the value of 1 µM that was measured for ETV6ΔH5.
Given that helix H5 sterically blocks the ETS domain, it is intuitively reasonable that ETV6 could follow a conformational selection pathway in which helix unfolding transiently yields the uninhibited state (U), which then binds DNA via its canonical interface. However, ETV62C-ox also bound DNA weakly via a displaced interface encompassing the “wing,” part of the recognition helix H3, and the inhibitory helix H5. Surprisingly, the presence of folded helix H5 does not abrogate association with DNA, and the disulphide-locked ETV62C-ox variant reveals an alternative pathway in the model of Fig. 8. The ETV62C-ox–DNA interaction studies were also carried out with a 15-bp DNA duplex and the macroscopic KD value was determined by NMR-monitored titrations to be 20 µM. Although this DNA contained the recognition sequences 5′−GGAA−3′, the covalently linked helix H5 prevented the formation of base-specific hydrogen bonds that would otherwise lead to dramatic and diagnostic NMR spectral changes. We assume that the ETV62C-ox–DNA complex is driven primarily by electrostatic forces and is independent of the DNA sequence. Accordingly, the macroscopic KD,I•DNAnsp of the inhibited state (I) for a 15-bp non-specific DNA duplex estimated to be 20 µM. By thermodynamic linkage, this implies that the equilibrium constant for the unfolding of helix H5 is reduced from ~10 for the free protein to ~0.5 for the bound protein. This demonstrates that ETV6 can also follow an induced fit pathway, whereby the inhibited protein first binds DNA and then undergoes a favorable conformational change with helix H5 unfolding to allow a canonical mode of binding. The presence of both pathways, with the inhibited protein being binding competent, could explain how the inhibitory helix H5 causes ~5-fold autoinhibition rather than the ~10-fold expected from its PFs [14].
Once ETV6 binds non-specific DNA in either its inhibited or uninhibited states, searching, modeled as one-dimensional diffusion, for cognate sites ensues [40,41]. Upon encountering a specific cognate site, the protein and DNA undergo conformational changes in order to make base-specific contacts and thereby form a high-affinity complex [14]. Given that the KD value of ETV6 for specific DNA is three- to four-orders of magnitude lower than for non-specific DNA, it is certain that the helix H5 will remain unfolded in this complex. This could also account for the observation that ETV6 is autoinhibited by ~50-fold for specific DNA binding and only ~5-fold for non-specific binding [14].
In conclusion, using a combination of site-directed mutagenesis, DNA-binding assays and NMR-based structural studies, we have determined the thermodynamic basis of DNA-binding and its autoinhibition in ETV6. These analyses predict potential mechanisms for ETV6 regulatory pathways involving cellular signaling events, such as post-translational modifications and protein partnerships that reinforce or relieve ETV6 autoinhibition by modulating the inhibitory helix stability.
Materials and Methods
Design of helix stabilizing amino acid substitutions
The disulfide containing ETV62C-ox was designed using a combination of visual inspection of the ETV6 structure (PDB code: 1FL8) and energy minimization with ROSET-TA [42]. Pairs of residues on helix H5 and the core ETS domain were selected based on the proximity of their Cβ atoms in the ETV6 structure and modeled as cysteines using Pymol [43]. The structural models of these proteins were energy minimized by ROSETTA and then inspected for disulfide bond formation. Out of five pairs (K380C– L438C, A393C–E434C, A393C–V437C, L400C–R426C and L336C–E434C), only A393C–V437C and L336C– E434C formed acceptable disulfide bonds in silico. Both the A393C–V437C and L336C–E434C doubly substituted proteins were expressed and purified from Escherichia coli. The L336C–E434C variant most likely formed both intramolecular and intermolecular disulfide bonds as we observed higher molecular weight bands in SDS-PAGE gels (data not shown). In contrast, the A393C–V437C (ETV62C-ox) variant behaved well in solution and was used for further studies.
Amino acid substitutions to stabilize the inhibitory helix H5 were designed by changing only surface exposed residues such that its packing interactions with the core ETS domain would not be disrupted. The program AGADIR [17] was used to guide this process. ETV6 variants, referred to as ETV6H423R and ETV6RAE, were designed to progressively enhance helix H5 stability with the addition of successive changes summarized in Table 1. These substitutions stabilize the helix by introducing favorable electrostatic interactions between Glu and Arg sidechains at positions “i” and “i + 3”or”i + 4” [17]. Alanine was chosen due to its highest helical propensity among all amino acids [18].
Protein expression and purification
Two fragments of murine ETV6, denoted as ETV6ΔH5 (residues Gly329 to Arg426) and ETV6WT (Gly329 to Asp446), represent the uninhibited (helix H5 deleted) and autoinhibited ETS domains, respectively. Both constructs have the sole wild-type cysteine (Cys334) changed to serine. Additional ETV6 variants (Table 1) were made by site-directed mutagenesis of the ETV6WT construct. Each ETV6 fragment was expressed from pET28b + vectors in E. coli BL21 (λDE3) cells and contained an N-terminal His6-affinity tag followed by a thrombin cleavage site. For 15N/13C (or only 15N) labeling of the protein, cells were grown in M9 minimal media supplemented with 1 g/L 15NH4Cl and 3 g/L 13C6-glucose (or 15 g of 12C6-glucose) as the sole nitrogen and carbon sources, respectively. Using previously described protocols [13,14], the protein samples were purified by Ni+2-affinity chromatography, followed by thrombin cleavage to remove the His6-tag. Gel-filtration chromatography was used as an additional purification and buffer exchange step. Four non-native N-terminal residues (Gly-Ser-His-Met) remained in the final samples. Final protein concentrations were determined by UV absorption using predicted molar absorptivity (ε280) values [44].
NMR spectroscopy
NMR experiments were performed using TCI-cryoprobe equipped Bruker Avance III 500, 600 or 850 MHz spectrometers. Unless stated otherwise, the proteins were 0.15–0.6 mM in sample buffer (20 mM phosphate, 50 mM NaCl, 5% lock D2O) at pH 6.7 and 25 °C. The disulfide bond was reduced in ETV62C-red by the addition of 2 mM TCEP. The collected spectra were processed and analyzed using NMRPipe [45] and Sparky [46], respectively.
The 15N-HSQC spectra of most protein samples were initially assigned at pH 6.7 and 25 °C by comparison with previously published data for ETV6WT and ETV6ΔH5 [14]. Assignments for ETV6H432R and ETV6RAE were then confirmed using 15N-TOCSY-HSQC (τmix = 60 ms) and 15N-NOESY-HSQC (τmix = 120 ms) experiments. In the case of ETV62C-ox, signals from the backbone 1HN, 15N, 13Cα and 13Cβ nuclei of the 15N/13C-labeled protein were assigned using standard heteronuclear scalar correlation experiments [47]. Subsequent 15N-HSQC spectra recorded under different experimental conditions were assigned by tracking the pH- and temperature-dependent amide chemical shifts of each protein.
NMR-monitored pH titrations
Samples of 15N-labeled ETV6WT (0.4 mM) and ETV6ΔH5 (0.6 mM) in NMR buffer (20 mM phosphate, 50 mM NaCl, 5% lock D2O) at 25 °C were used for detailed 15N-HSQC-monitored pH titration studies. The pH value of each sample was adjusted by the addition of small aliquots of 1 M or 0.1 M NaOH and measured using a Thermal Scientific Orion* 3-Star pH meter with an Orion ROSS micro pH electrode. Sample pH values for ETV6WT were 6.7, 6.2, 5.4, 5.1, 4.7, 4.4, 4.0, 3.7 and 3.5, and those for ETV6ΔH5 were 6.7, 6.3, 5.8, 5.3, 4.7, 4.2, 3.9 and 3.6.
Measurement of amide HX by NMR
The temperatures (20 °C to 35 °C) and pH (3.5 to 8.6) conditions for the amide HX studies are summarized in Supplementary Table S1. The slow (minutes to days) exchange rate constants for ETV6WT and its variants were measured at 20 °C from a series of 15N-HSQC spectra collected immediately after dissolving the lyophilized proteins in 99.9% D2O. Following a dead time of ~6 min, four 5-min (acquisition time), four 10-min, six 20-min, four 30-min, five 1-h, five 2-h, six 4-h and five 6-h 15N-HSQC spectra were collected. Only the number of transients per t1 increment was increased for longer acquisition times and improved signal-to-noise ratios. After 3 days, each sample was removed from the spectrometer and stored at 20 °C. Over the next ~2 weeks, 15N-HSQC spectra were recorded intermittently until all amides had exchanged by at least 70%. The reported pH* value (uncorrected for isotope effects) of the sample was then measured. The pseudo-first-order exchange rate constants (kex) were determined by nonlinear least-squares fitting of the peak intensities, It (scaled by the number of transients) to the equation It = I0exp(−kext), where t is the midpoint time of each spectrum and I0 is the initial intensity.
The fast (seconds) exchange rate constants were measured by the CLEANEX-PM method [48] at several temperature and pH values (Supplementary Table S1). At each condition, a series of spectra with 10-, 20-, 30-, 40-, 50-, 60- and 80-ms transfer periods and a reference spectrum with a long (12-s) recycle delay to ensure complete water relaxation were collected. The kex values were obtained as described previously [13].
The PFs for each amide were determined as the ratio of its experimentally measured exchange rate constant (kex) versus the intrinsic exchange rate constant (kint) of the same amino acid sequence in an unstructured polypeptide. The kint values were determined from the program Sphere [49], which uses reference data based on poly-dl-alanine and corrected for amino acid type, temperature, pH and isotope effects [24,25]. The PFs reported in Figs. 3, 4 and 5 and in Table 1 correspond to the kex values measured most reliably by either protium– deuterium or protium–protium exchange. Note that the comparison of PFs assumes an EX2 mechanism with a first-order dependence of kex on pH and an independence of protein stability on sample pH and temperature over the range of conditions used for these measurements [26].
Measurement of ETV62C-ox–DNA binding
DNA oligonucleotides containing the 15-bp consensus sequence 5′−CAAGCCGGAAGTGAG−3′ and its complement were purchased from Integrated DNA Technologies (IDT). The DNA duplex was generated by mixing the single strands at an equimolar ratio (as determined by UV absorbance using IDT-supplied predicted ε260 values), heating to 100 °C and slowly cooling to room temperature. The duplex DNA was purified and exchanged into NMR buffer by gel filtration chromatography. Concentrated DNA was titrated into 0.2 mM 15N-labeled ETV62C-ox and 15N-HSQC spectra collected at each point. The molar ratios of DNA to ETV62C-ox in the titration set were 0, 0.25, 0.5, 0.75, 1.0, 1.5, 2.0 and 3.1. Weak binding occurred in fast exchange limit, thus enabling the tracking of chemical shifts relative to the assigned spectrum of the free protein. Amide CSP values (Δδ =((ΔδH)2+(0.154ΔδN)2)1/2) for each titration point were determined with respect to the free ETV62C-ox. The equilibrium dissociation constant (KD) was determined by averaging the results of non-linear least-squares fitting of the Δδ values of six amides (R392, C393, K399, K405, G408 and Q441) showing the largest spectral perturbations to a 1:1 binding model.
where Δδmax is the maximum, extrapolated change in chemical shift for each residue upon saturation, and DT and PT are total dilution-corrected DNA and protein concentrations, respectively, at each titration point.
To generate ETV62C-red, the disulfide bond was reduced by adding TCEP to a final concentration of 2 mM. Since the disulfide bond is buried in the protein, complete reduction, as monitored by 15N-HSQC spectra, took almost a day. The 15-bp duplexed DNA was added to make a 1:1 ETV62C-red−DNA complex (final concentration of 0.1 mM). In contrast to ETV62C-ox, the reduced ETV62C-red bound this DNA tightly and in the slow exchange regime. Hence, the 15N-HSQC spectrum of DNA-bound ETV62C-red was assigned by comparison to the published spectrum of DNA-bound ETV6WT [14].
Electrophoretic mobility shift assays
The equilibrium dissociation constants (KD) of ETV6WT and its variants for a 23-bp double-stranded DNA containing the sequence 5′−CGGCCAAGCCGGAAGTGAGTGCC-−3′ and its complement were measured by EMSA [20]. Both oligonucleotides in the duplex had an AlexaFluor-488 fluorescent tag attached to their 5′ base via a flexible polyethylene glycol linker. The HPLC-purified oligonucleotides were purchased from IDT and annealed by heating to 100 °C, followed by slowly cooling to room temperature. Binding reactions were carried out in 25 mM Tris (pH 7.9), 60 mM KCl, 6 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 1 µg/ml poly(dIdC) and 0.1 mg/ml BSA. The reactions, containing 0.5 nM DNA and 0.01 nM to 1 µM protein (ETV6WT, ETV6H432R or ETV6RAE), were incubated at room temperature for 20 min and then on ice for another 20 min. For the higher-affinity uninhibited ETV6ΔH5, 0.04 nM DNA was used. The gel electrophoresis was performed under ice-cold conditions using a BioRad mini-gel system. Gels were scanned with a Typhoon 9200 imager. Band intensities (I) corresponding to the free DNA were quantified by ImageJ [50] and fit to the following equation to determine KD values:
Here, Ibase is the baseline intensity with excess protein, Imax is the maximum intensity without protein and PT is the total protein concentration for each reaction. In the cases of ETV6WT, ETV6H432R and ETV6RAE, the fit KD values were at least 10-fold higher than the DNA concentration (0.5 nM), and thus, the free protein concentrations could be assumed to be equal to that of total protein.
Supplementary Material
Acknowledgments
Soumya De was supported by a Canadian Institutes for Health Research (CIHR) Post-doctoral Fellowship (MFE-135420). This study was funded by the Canadian Cancer Society Research Institute (CCSRI 2011-700772 to L.P.M.), CIHR (MOP-136834 to L.P.M.) and the National Institutes of Health (R01GM38663 to B.J.G.). Instrument support was provided by CIHR, the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund, the UBC Blusson Fund and the Michael Smith Foundation for Health Research. Funding to B.J.G. from Huntsman Cancer Institute/Huntsman Cancer Foundation and Howard Hughes Medical Institute is also acknowledged.
Abbreviations used
- NMR
nuclear magnetic resonance
- CSP
chemical shift perturbation
- HSQC
heteronuclear single quantum correlation
- HX
hydrogen exchange
- EMSA
electrophoretic mobility shift assay
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2016.02.020.
References
- 1.Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 2.Trudeau T, Nassar R, Cumberworth A, Wong ET, Woollard G, Gsponer J. Structure and intrinsic disorder in protein autoinhibition. Structure. 2013;21:332–341. doi: 10.1016/j.str.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharrocks AD. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 5.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, et al. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 7.Lee GM, Pufall MA, Meeker CA, Kang HS, Graves BJ, McIntosh LP. The affinity of ets-1 for DNA is modulated by phosphorylation through transient interactions of an unstructured region. J. Mol. Biol. 2008;382:1014–1030. doi: 10.1016/j.jmb.2008.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desjardins G, Meeker CA, Bhachech N, Currie SL, Okon M, Graves BJ, et al. Synergy of aromatic residues and phosphoserines within the intrinsically disordered DNA-binding inhibitory elements of the Ets-1 transcription factor. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11019–11024. doi: 10.1073/pnas.1401891111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol. Cell. Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrivastava T, Mino K, Babayeva ND, Baranovskaya OI, Rizzino A, Tahirov TH. Structural basis of Ets1 activation by Runx1. Leukemia. 2014;28:2040–2048. doi: 10.1038/leu.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiina M, Hamada K, Inoue-Bungo T, Shimamura M, Uchiyama A, Baba S, et al. A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions. J. Mol. Biol. 2015;427:1655–1669. doi: 10.1016/j.jmb.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Green SM, Coyne HJ, 3rd, McIntosh LP, Graves BJ. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J. Biol. Chem. 2010;285:18496–18504. doi: 10.1074/jbc.M109.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coyne HJ, 3rd, De S, Okon M, Green SM, Bhachech N, Graves BJ, et al. Autoinhibition of ETV6 (TEL) DNA binding: appended helices sterically block the ETS domain. J. Mol. Biol. 2012;421:67–84. doi: 10.1016/j.jmb.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De S, Chan AC, Coyne HJ, 3rd, Bhachech N, Hermsdorf U, Okon M, et al. Steric mechanism of auto-inhibitory regulation of specific and non-specific DNA binding by the ETS transcriptional repressor ETV6. J. Mol. Biol. 2014;426:1390–1406. doi: 10.1016/j.jmb.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regan MC, Horanyi PS, Pryor EE, Jr, Sarver JL, Cafiso DS, Bushweller JH. Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13374–13379. doi: 10.1073/pnas.1301726110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein-DNA recognition as inferred from NMR studies on the Lac repressor system. Chem. Rev. 2004;104:3567–3586. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- 17.Lacroix E, Viguera AR, Serrano L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 18.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson H, Becktel WJ, Matthews BW. Enhanced protein thermostability from designed mutations that interact with alpha-helix dipoles. Nature. 1988;336:651–656. doi: 10.1038/336651a0. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, McIntosh LP, Graves BJ. Inhibitory module of Ets-1 allosterically regulates DNA binding through a dipole-facilitated phosphate contact. J. Biol. Chem. 2002;277:2225–2233. doi: 10.1074/jbc.M109430200. [DOI] [PubMed] [Google Scholar]
- 21.Platzer G, Okon M, McIntosh LP. pH-dependent random coil (1)H, (13)C, and (15)N chemical shifts of the ionizable amino acids: a guide for protein pK a measurements. J. Biomol. NMR. 2014;60:109–129. doi: 10.1007/s10858-014-9862-y. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y, Bax A. Identification of helix capping and b-turn motifs from NMR chemical shifts. J. Biomol. NMR. 2012;52:211–232. doi: 10.1007/s10858-012-9602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma D, Rajarathnam K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- 24.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 26.Englander SW, Kallenbach NR. Hydrogen-exchange and structural dynamics of proteins and nucleic-acids. Q. Rev. Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 27.Li R, Woodward C. The hydrogen exchange core and protein folding. Protein Sci. 1999;8:1571–1590. doi: 10.1110/ps.8.8.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, et al. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J. Biol. Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 29.Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hol WG. The role of the alpha-helix dipole in protein function and structure. Prog. Biophys. Mol. Biol. 1985;45:149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 31.Sali D, Bycroft M, Fersht AR. Stabilization of protein structure by interaction of alpha-helix dipole with a charged side chain. Nature. 1988;335:740–743. doi: 10.1038/335740a0. [DOI] [PubMed] [Google Scholar]
- 32.Hol WG, van Duijnen PT, Berendsen HJ. The alpha-helix dipole and the properties of proteins. Nature. 1978;273:443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- 33.Grishin AV, Alexeevsky AV, Spirin SA, Karyagina AS. Conserved structural features of ETS domain-DNA complexes. Mol. Biol. 2009;43:612–619. [PubMed] [Google Scholar]
- 34.Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J. Biol. Chem. 2002;277:45529–45536. doi: 10.1074/jbc.M206327200. [DOI] [PubMed] [Google Scholar]
- 35.Fitzsimmons D, Lukin K, Lutz R, Garvie CW, Wolberger C, Hagman J. Highly cooperative recruitment of Ets-1 and release of autoinhibition by Pax5. J. Mol. Biol. 2009;392:452–464. doi: 10.1016/j.jmb.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamber EP, Vanhille L, Textor LC, Kachalova GS, Sieweke MH, Wilmanns M. Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization. EMBO J. 2008;27:2006–2017. doi: 10.1038/emboj.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, et al. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 38.Viadiu H, Aggarwal AK. Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol. Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 39.Tsodikov OV, Holbrook JA, Shkel IA, Record MT., Jr Analytic binding isotherms describing competitive interactions of a protein ligand with specific and nonspecific sites on the same DNA oligomer. Biophys. J. 2001;81:1960–1969. doi: 10.1016/S0006-3495(01)75847-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Hippel PH, Berg OG. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 41.Givaty O, Levy Y. Protein sliding along DNA: dynamics and structural characterization. J. Mol. Biol. 2009;385:1087–1097. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Simons KT, Bonneau R, Ruczinski I, Baker D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins Suppl. 1999;3:171–176. doi: 10.1002/(sici)1097-0134(1999)37:3+<171::aid-prot21>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- 43.Schrodinger LLC. The PyMOL Molecular Graphics System. Version 1.3r1. 2010 [Google Scholar]
- 44.Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. The Proteomics Protocols Handbook, 2005. Springer; New York: 2005. Protein Identification and Analysis Tools on the ExPASy Server; pp. 571–607. [Google Scholar]
- 45.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe—a multidimensional spectral processing system based on Unix Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Goddard TD, Kneeler DG. Sparky. third. University of California; San Francisco: 1999. [Google Scholar]
- 47.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spec. 1999;34:93–158. [Google Scholar]
- 48.Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 49.Zhang YZ. Protein and peptide structure and interactions studied by hydrogen exchange and NMRPhD Thesis Structural Biology and Molecular Biophysics. University of Pennsylvania; Pennsylvania: 1995. [Google Scholar]
- 50.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.