Abstract
Objective
The inflammasome complex is a driver of organ damage in systemic lupus erythematosus (SLE). While type I interferons (IFNs) are well established as mediators of SLE pathogenesis, their role in inflammasome activation in SLE has not been assessed. Thus, we examined type I IFNs as regulators of the inflammasome and identified interferon regulatory factor 1 (IRF1) as critical for inflammasome hyperactivity in SLE.
Methods
SLE patients fulfilled ≥4 ACR criteria and were recruited from the University of Michigan Lupus Cohort. Primary monocytes were isolated from SLE patients or healthy controls by negative selection, treated with inflammasome activators in the presence or absence of IFNα, and IL-1β secretion was measured by ELISA. Expression levels of interferon and inflammasome-related molecules were assessed by real-time PCR and Western blotting. IRF1 expression was specifically downregulated by siRNA transfection and a chemical inhibitor.
Results
SLE monocytes exhibited increased expression and enhanced activation of the inflammasome by ATP when compared to control monocytes. Expression of inflammasome and interferon-regulated genes was significantly correlated in lupus, but not control, monocytes. Inflammasome activity was increased after prolonged exposure to IFNα. Reduction of IRF1 expression via siRNA blocked caspase-1 upregulation after treatment with IFNα. Importantly, hyperactivity of the inflammasome in lupus monocytes was significantly reduced after knock-down or inhibition of IRF1.
Conclusion
Prolonged type I IFN exposure, as seen in SLE patients, primes monocytes for robust inflammasome activation in an IRF1-dependent manner. IRF1 inhibition may serve as a novel target for treatment of SLE-associated inflammation and organ damage.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by formation of autoantibodies, deposition of immune complexes, and pleotropic end-organ damage. Dysregulation of both the innate and adaptive immune responses is central to the development of SLE, and recent evidence from human and murine studies identifies the innate signaling complex termed the inflammasome as dysregulated in SLE and a central contributor to development of lupus nephritis and other associated organ damage(1–10).
Inflammasomes are multi-protein complexes which utilize a central scaffold and adaptor molecules to recruit and activate caspase-1. Active caspase-1 then cleaves the pro-inflammatory cytokines IL-1β and IL-18 to their active forms. The best characterized inflammasome is one which contains the nucleotide-binding oligomerization domain (NOD)-like receptor 3 (NLRP3), the adapter molecule apoptosis-associated speck-like protein that contains a CARD (ASC), and caspase-1. This inflammasome complex is hyper-activated in lupus macrophages(2), is activated by lupus-specific autoantibodies(1, 6, 7), and contributes to the development of nephritis(11, 12).
Despite this plethora of research describing the importance of the inflammasome in SLE, the mechanisms which regulate inflammasome activation in this disease have not been well-characterized. Experimental evidence collected in the past decade has identified type I interferons (IFN), and particularly interferon α (IFNα), as important contributors to the pathogenesis of SLE in human and murine models(13–18). Many patients with SLE have elevated levels of IFN-induced gene expression or “interferon signature,” which correlates with the presence of autoantibodies, nephritis, and disease activity(18). The effects of type I IFNs on inflammasome activity are unclear. Studies have identified type I IFN signaling as a negative regulator of inflammasome gene expression and activity in PBMCs and bone marrow-derived macrophages (BMDMs)(19, 20). However, other studies have identified correlations between elevations in IFN-induced gene expression and activation of the inflammasome in SLE(4, 21). Monocytes are sentinels of the IFN signature in autoimmune diseases(22) and are also an important source of inflammasome activation. Thus, in this study we chose to clarify the effects of IFNα on inflammasome function in both control and lupus monocytes in order to better understand how an environment with chronically activated type I IFNs, as seen in SLE patients, changes inflammasome activity. We find increased inflammasome activity in SLE monocytes and identify the chronicity of IFNα exposure as a dichotomous regulator of inflammasome activity. Importantly, we demonstrate that IFNα upregulation of interferon regulatory factor-1 (IRF1) is required for upregulation of caspase-1 and the enhanced IL-1β production from SLE monocytes. These data identify an important intersect between type I IFNs and the inflammasome and may suggest an additional mechanism by which blockade of type I IFN signaling may result in improvement of SLE activity(23).
Materials and Methods
Reagents and antibodies
Ficoll-Paque™ Plus and fetal bovine serum (FBS) were from GE Healthcare (Pittsburgh, PA). LPS was purchased from Sigma–Aldrich (St. Louis, MO). RPMI 1640 was from Lonza (Walkersville, MD). IFNα recombinant was from Schering Corporation (Kenilworth, NJ). 4,5,6,7-Tetrabromobenzotriazole (TBB) was purchased from Tocris (Avonmouth, Bristol, UK). Anti-human IRF1 (D5E4), caspase-1 (D7F10), Phospho-STAT1(Tyr701)(D4A7), Phospho- STAT 2(Tyr690)(D3P2P), STAT 1(D1K9Y), STAT 2, and β-actin were purchased from cell signaling technology (Danvers, MA). HRP-conjugated goat anti-rabbit IgG was from Abcam (Cambridge, MA).
Subjects
All prospectively recruited patients and controls gave written, informed consent and were treated according to the declaration of Helsinki. The study protocol was approved by the Institutional Review Board of the University of Michigan Medical School. SLE patients fulfilled >4 ACR criteria(24) and were recruited from the University of Michigan Lupus Cohort. We recruited 23 SLE patients diagnosed and treated at the University of Michigan hospital clinic and 23 gender-, age- and race-matched healthy controls for studies that compared SLE to control monocytes. An additional 25 controls were recruited for studies on control cells alone. See Table I for patient and control information. For the microarray study, 90 cases of DLE and SCLE biopsies were identified via a SNOMED search of the University of Michigan Pathology Database using the search terms “lupus” and “cutaneous lupus”. Patients who met both clinical and histologic criteria for DLE or SCLE were included in the study.
Table 1.
Summary characteristics of lupus and control patients
| SLE (n=23) | Control (n=48) | p value | |
|---|---|---|---|
| Mean Age (+/−SEM) | 44.9 +/− 2.3 | 41.5 +/−1.5 | 0.21 |
| Gender | 0.84 | ||
| Female (%) | 91 | 90 | |
| Male (%) | 9 | 10 | |
| Medications (%) | |||
| Oral prednisone ≤10 mg daily | 35 | ||
| Oral prednisone >10 mg daily | 0 | ||
| Mycophenolate Mofetil | 30 | ||
| Belimumab | 9 | ||
| Antimalarials | 87 | ||
| Methotrexate | 0 | ||
| Antibody Status | |||
| ANA positive (%) (ever positive) | 78 | ||
| Anti-DNA positive (%) (at sampling) | 52 | ||
| Mean titer (IU/mL) | 63.6 (range 0–337) | ||
| SLEDAI at time of sampling (mean) | 2.7 (range 0–9) |
Human peripheral monocytes preparation
50 ml of heparinized blood was obtained from SLE and controls. The buffy coat was obtained after density centrifugation on 15 ml Ficoll-Paque™ Plus (5:3 blood to Ficoll ratio). Red blood cells were removed via hypotonic lysis. CD14+CD16− monocytes were isolated using negative selection via EasySep™ Human Monocyte Enrichment Kit (Stemcell Technology, Vancouver, BC, Canada). Monocyte population purity was assessed by flow cytometry and deemed to be 87% after negative selection.
Monocytes culture and treatment
Control and SLE monocytes were plated onto 48-well or 96-well tissue culture plates at a density of 0.5 × 106 cells/well in 0.5 ml RPMI1640 plus 10% FBS or 0.2 × 106 cells/well in 0.2 ml RPMI1640 plus 10% FBS respectively. Monocytes were cultured in the presence or absence of 1000 unit/ml human IFNα overnight. In some cases, 50µM TBB was added 1 hr prior to IFN treatment. The next morning, the cells were stimulated with or without 100 ng/ml LPS, or vehicle for 4 hours. This was followed by treatment with or without 5mM of ATP to stimulate inflammasome activation. In some experiments, IFNα was also used concurrently with LPS. To assess effects of control and SLE serum on inflammasome activity, monocytes were cultured in 50% serum overnight in the presence or absence of 50µM TBB prior to inflammasome activation with LPS and ATP.
ELISA
The supernatants from cultured monocytes were assayed for IL-1β using the Human IL1β ELISA Ready-SET-Go kits (eBioscience, San Diego, CA) following the manufactory’s instructions.
Western blot
Monocyte cultures were directly lysed in 2× protein sample buffer after supernatant removal. Protein from each sample (0.8 × 105 cells) was separated on 10% or 12% acrylamide gel and then transferred to the Amersham™ Protran™ 0.2µM NC nitrocellulose membrane (GE Healthcare). The membranes were blocked with 5 % nonfat dry milk and incubated overnight at 4 °C with primary antibodies (1:1,000 dilution) followed by HRP-conjugated anti-rabbit IgG. Protein expression bands were detected by chemiluminescence using WesternBright™ Quantum Western blot detection reagent (Advansta, Menlo Park, CA), and the protein bands were imaged by Omega Lum C (Gel Company, San Francisco, CA). Expression quantification was completed with ImageJ software.
RNA isolation and real-time quantitative PCR
Total RNA were isolated from human monocytes using Direct-Zol RNA miniprep kit (Zymo Research Corporation, Irvine, CA) following the manufacturer’s instruction. 1 µg of total RNA was transcribed into cDNA using M-MLV Reverse Transcriptase and oligo(dT) primer (Invitrogen, Carlsbad, CA). Real time PCR was performed in triplicate using SYBR Green PCR Master Mix from Applied Biosystems (Foster City, CA) with the assistance of the University of Michigan DNA sequencing core. The primer sequences are shown in Supplemental Table I. Gene expression was normalized to β-actin and relative gene expression to normal controls was calculated by the comparative threshold cycle method and fold-change (FC) was expressed as 2−ddCT(25). Interferon scores were calculated as previously described (26, 27) for IFI44, MXA and PRKR. Briefly, the mean fold change of healthy control samples was calculated and the SLE IFN gene score was calculated as (SLE(gene)FC – control(mean)FC)/StDev control. The three gene scores were then summed to provide a total SLE IFN gene score. Control scores were calculated similarly.
siRNA and transfection
siRNA targeting human IRF1(hIRF1-siRNA) or AllStars Negative Control siRNA (SCR) were purchased from Qiagen (Valencia, CA). The siRNA target sequence for IRF1 is CTGGCTAGAGATGCAGATTAA. Electroporation of hIRF1-siRNA or SCR into monocytes was performed with the 4D–Nucleofector™ System (Lonza) using the P3 Primary Cell 4D–Nucleofector® × Kit S. Briefly, 1 × 106 monocytes were resuspended in 20 µL of P3 Primary Cell 4D–Nucleofector™ × Solution containing 250nM hIRF1-siRNA or SRC respectively, transferred into the Nucleocuvette™ Vessels and electroporation were performed using standard program optimized for human monocytes. Immediately after electroporation, 0.5 × 106 cells from each sample were diluted into 500 µl RPMI/5%FBS, plated onto 48 –well plate and cultured overnight. Then the cells were treated with IFNα, LPS and ATP as above.
RNA Isolation and Microarray Procedures
Formalin-fixed paraffin embedded (FFPE) blocks of skin biopsies were obtained and five 10µm sections were cut with a microtome. RNA was extracted using the E.N.Z.A. FFPE RNA Kit (Omega Bio-tek) following manufacturer’s instructions. Complementary DNA (cDNA) was prepared (NuGEN, Ovation PicoSL WTA System V2 Manual, P/N M01226 v4) from approximately 30 ng total RNA. 2.5 µg cDNA was biotinylated using the NuGEN Encore Biotin Module (Encore Biotin Module Manual, P/N M01111 v6). The Poly-A RNA Control Kit was used as the routine procedure at the University of Michigan microarray core. Labeled cDNA was hybridized at 48°C to Affymetrix Human Gene ST 2.1 array plates, which were then washed, stained and scanned using the Affymetrix GeneTitan system (software version 3.2.4.1515) with the assistance of the University of Michigan DNA Sequencing Core. Quality control and RMA (Robust Multi-array Average) (28) normalization of CEL files were performed in R software version 3.1.3 using custom CDF version 19 and modified Affymetrix_1.44.1 package from BrainArray (http://brainarray.mbni.med.umich.edu/brainarray/default.asp). Log2 expression values were batch corrected using Combat implemented into GenePattern (http://www.broadinstitute.org/cancer/software/genepattern/). The baseline expression was defined as minimum plus one standard deviation of the median of all genes. A variance filter of 80% was then applied. Of the 25,582 unique genes represented on the Human ST2.1 chip, a total of 20,410 genes passed the defined criteria. The normalized data file was uploaded to the Gene Expression Omnibus (GEO) Web site (http://www.ncibi.nlm.nih.gov/geo/) under accession number GSE81071 and will be available upon acceptance of this manuscript.
Statistical analysis
Data are presented as the mean + SEM. The two-tailed unpaired Student’s t test was used for analysis of differences between simultaneously cultured control and SLE groups. For studies examining effects on samples with multiple treatment conditions, paired two-tailed student’s t test between untreated and treated groups was used. Linear regression was used for comparison of monocyte IFN score with inflammasome and inflammatory genes and for comparison of IRF1 with CASP1 from microarray data. Both data sets were normally distributed. p-values <0.05 were considered as statistically significant.
Results
SLE monocytes demonstrate increased inflammasome gene expression and IFN signature
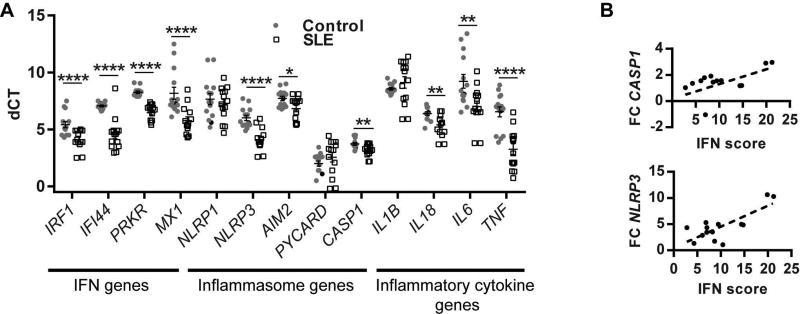
In order to develop an understanding of the relationship between the inflammasome and type I IFN exposure, we first examined inflammasome and interferon-regulated gene expression in monocytes from SLE patients and normal controls. Real-time qPCR on freshly isolated monocytes demonstrates that mRNA levels of inflammasome genes such as NLRP3, AIM2 and CASP1 were elevated in lupus monocytes compared to healthy controls (Figure 1A). mRNA levels of genes downstream of type I IFN signaling (IRF1, PRKR, IFI44 and MX1) as well as the pro-inflammatory cytokines IL6 and TNF were also significantly higher in SLE monocytes (Figure 1A).
Figure 1. Inflammasome genes are increased in SLE monocytes and correlate with type I IFN gene expression.
Total RNA was collected from freshly isolated SLE (n=14) and control monocytes (n=14); A. mRNA levels of inflammasome and Type I IFN genes were measured by real time qPCR and ΔCT calculated vs. β-actin. Graph represents ΔCT for 14 patients and 14 controls (each dot is average of triplicate) +/− SEM for each tested gene. B. Fold change (FC) of indicated genes were plotted vs. the IFN score for each SLE patient. *=p<0.05, **=p<0.01, ****=p<0.0001.
Linear regression to evaluate expression of interferon-regulated genes, expressed as an interferon score(18), compared with inflammasome genes demonstrated a strong positive correlation of CASP1 and NLRP3 in SLE patients, but not in controls (Figure 1B and Supplemental Table II). In contrast to inflammasome genes, the pro-inflammatory gene TNFA was not significantly correlated with the IFN-regulated genes in SLE monocytes (Supplemental Table II). This suggests that the IFN/inflammasome correlation is not a generalized inflammatory effect. These data raise the hypothesis that chronic exposure to type I IFNs may result in upregulation of inflammasome components and facilitate inflammasome activation.
Prolonged, but not short-term, exposure to IFNα enhances inflammasome activity in control and SLE monocytes
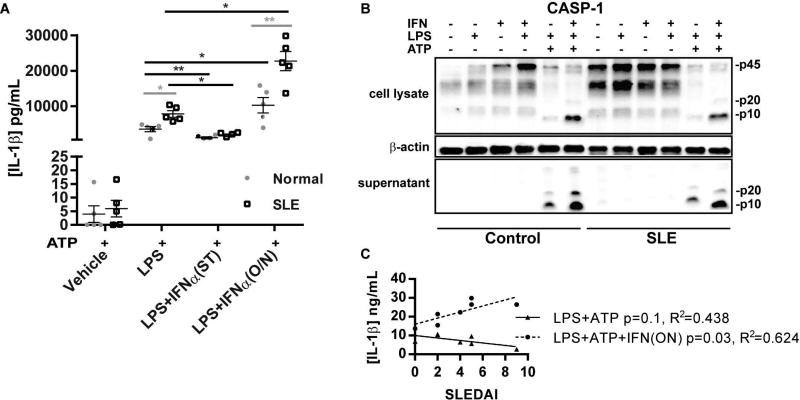
To determine whether IFNα promotes inflammasome activation in monocytes, monocytes from SLE patients and controls were primed with LPS in the presence or absence of simultaneous addition of IFNα followed by activation of the NLRP3 inflammasome with ATP(29, 30). We found that at baseline, SLE monocytes had significantly enhanced IL-1β production when compared to control monocytes (Figure 2). Consistent with previous macrophage studies(20), we detected inhibition of IL-1β release when cells were treated with simultaneous (ST) IFNα (Figure 2). However, knowing that SLE patients are exposed to chronic IFNα signaling, we examined whether there were differential effects of long-term IFNα exposure on inflammasome activation. Thus, SLE and control monocytes were treated with or without IFNα overnight prior to LPS priming and inflammasome activation with ATP treatment. As shown in Figure 2, release of IL-1β was significantly enhanced by overnight IFNα exposure in control monocytes and to a greater extent in SLE monocytes. This effect was seen at doses as low as 100 IU of IFNα, which is estimated to be equivalent to SLE plasma(31) (Supplemental Figure 1). Interestingly, the IL-1β production correlated with SLEDAI disease activity following long-term IFN exposure but not at baseline (Figure 2C). These data suggest that prolonged exposure to type I IFNs, prior to inflammasome priming with LPS, promotes inflammasome activity and that more active patients may be more susceptible to the effect of type I IFNs.
Figure 2. SLE monocytes demonstrate increased inflammasome activation and this is enhanced by pre-exposure to type I IFNs.
Monocytes from 5 different SLE and 5 different control subjects in duplicate were treated with and without IFNα either one night before (O/N) or simultaneously (ST) with LPS treatment. 5 mM ATP was added 2 hours before harvesting to activate the NLRP3 inflammasome and release mature IL-1β. Supernatants were collected and IL-1β production was measured by ELISA. Each collected sample is shown as a dot +/− SEM. Comparisons between control and SLE were made via unpaired Student’s t-test (in gray), and comparisons within control or SLE samples with and without IFN treatment were made via paired Student’s t-test (in black). *=p<0.05, **=p<0.01. B. Representative Western blot for caspase-1 of cell lysate and extracellular media (supernatant) from samples from Figure 2A. Pro (inactive) 45kDa form and active (p20 and p10 forms) are denoted. C. Comparison of SLEDAI scores vs. IL-1β production via linear regression for SLE samples treated with and without IFNα overnight prior to inflammasome activation.
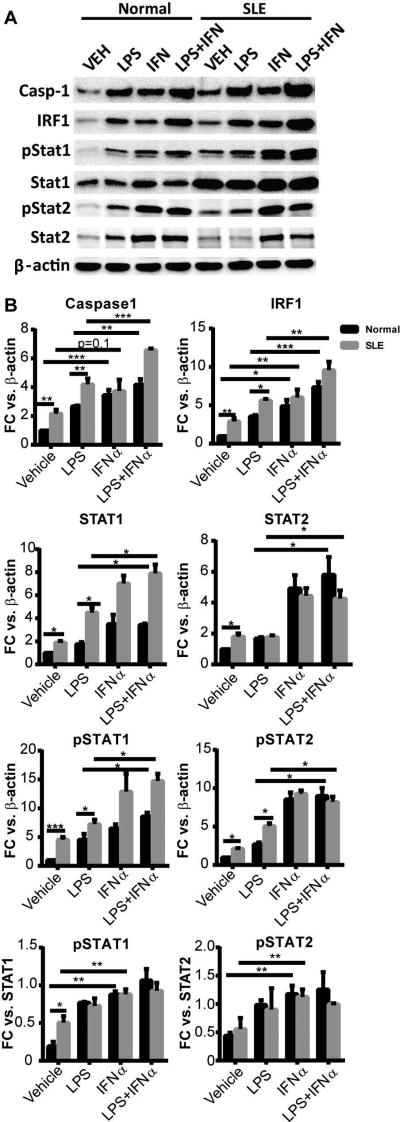
In order to examine the effects of prolonged type I IFN exposure on expression of involved proteins, Western blot analysis of control and SLE monocytes was completed. Evidence of chronic type I IFN activation of SLE monocytes at baseline was noted via increased phospho-STAT1 and IRF1 expression. Importantly, p45 caspase-1 expression was also increased in SLE vs. control monocytes at baseline (Figure 3). Overnight exposure to IFNα alone significantly upregulated phosphorylation of STAT1 and STAT2 and upregulated IRF1 and caspase-1 in control monocytes. IFNα induced moderate, but not significant, increases of caspase-1 over baseline elevated levels in SLE monocytes. Addition of LPS after overnight IFNα exposure resulted in even greater expression of caspase-1 and IRF1 in SLE and control monocytes (Figure 3), implying a synergistic effect of TLR and IFN signaling for caspase-1 expression. Taken together, these data suggest that exposure to type I IFNs prior to priming and activation of the inflammasome results in enhanced caspase-1 expression and increased inflammasome activity whereas if TLR4 and IFN signals are received simultaneously, inflammasome activity is inhibited.
Figure 3. Type I IFN enhances caspase-1 expression.
A. Representative Western blot of LPS- and/or IFNα-treated monocytes. Cell lysates were collected, separated by SDS-PAGE and blotted with indicated antibodies. P45 pro-form of Caspase-1 is shown. B. Protein band density of caspase-1, IRF1, pSTAT1, pSTAT2, STAT1 and STAT2 was digitally quantified by ImageJ. The results are expressed as mean+SEM fold change vs. vehicle-treated control monocytes normalized to β-actin or STAT1/2 as indicated; n = 5 control and SLE samples. *=p<0.05, **=p<0.01, ***=p<0.001 via Student’s paired t test.
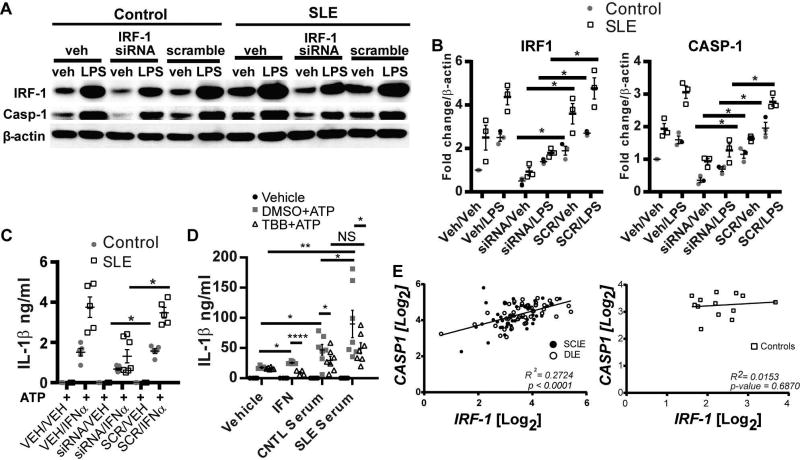
Knockdown of IRF1 effectively attenuates IFNα-mediated caspase-1 expression and inflammasome hyper-activation in SLE monocytes
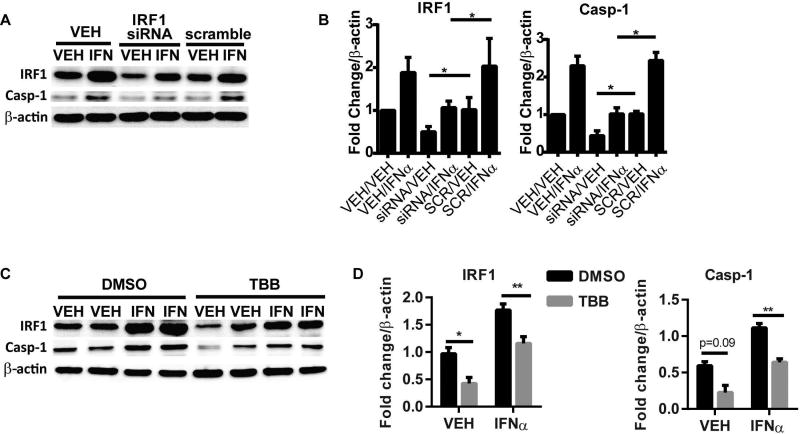
IRF1 is a transcription factor that is activated by type I IFN signaling and is important for development of (32), and prediction of treatment response in lupus nephritis(33). Importantly, IRF1 has been identified as a central regulator of target genes in lupus monocytes(34) and has been reported as a transcription factor of caspase-1(35, 36). Our data support a parallel regulation of IRF1 and caspase-1 downstream of IFNα. To determine whether IFNα-mediated increase in caspase-1 expression required IRF1, we knocked down the expression of IRF1 by hIRF1-siRNA in control monocytes followed by treatment with or without IFNα. As shown in Figure 4A and B, we were able to achieve a 50% reduction in IRF1 through this method. Importantly, knockdown of IRF1 significantly inhibited the basal and IFNα-induced expression of caspase-1 (Figure 4 A and B). Inhibition of IRF1 transcriptional activity with TBB, an inhibitor of casein kinase II, an upstream positive regulator of IRF1, (37) also gave similar results (Figure 4C and D).
Figure 4. Inhibition of IRF1 expression prevents IFNα upregulation of caspase-1.
A and B. Control monocytes were transfected with hIRF1-siRNA or SCR by electroporation and then treated with vehicle or IFNα for 6 hours. Caspase-1 and IRF1 expression was analyzed by Western Blot. P45 pro-form of Caspase-1 is shown. Densitometry analysis of IRF1 and caspase-1 protein expression are shown in B. C and D. Control monocytes were incubated with 50 µM TBB, an inhibitor of IRF1 regulator casein kinase II, prior to 6 hr stimulation with IFNα. Caspase-1 and IRF1 expression were analyzed by Western Blot. Densitometry analysis of IRF1 and caspase-1 protein expression are shown in D. Data are expressed as mean+SEM fold change vs. vehicle-treated control; n= six experiments for A/B and n=4 experiments for C/D. *=p<0.05, **=p<0.01.
In order to determine whether inhibition of IRF1 was sufficient to normalize caspase-1 overexpression and inflammasome hyperactivation in SLE monocytes, which are chronically exposed to type I IFNs, SLE and control monocytes were then transfected with SCR or hIRF1-siRNA followed by priming of the inflammasome with LPS and activation with ATP. Reduction in IRF1 expression significantly down-regulated baseline and LPS-induced caspase-1 expression in control and SLE monocytes (Figure 5A,B). Importantly, IRF1 inhibition normalized IL-1β activation and secretion following LPS priming and ATP-mediated inflammasome activation in SLE monocytes to levels seen in control monocytes, and had further inhibitory effects on IL-1β production in control cells (Figure 5C). Inhibition of IRF1 with TBB also inhibited IFNα-mediated enhancement of IL-1β release after inflammasome activation (Figure 5D). These data suggest that type I IFN-mediated enhanced IRF-1 activity in SLE monocytes is driving amplified inflammasome activity.
Figure 5. IRF1 regulates Caspase-1 expression and IL-1β production in SLE monocytes.
A. Lupus and control monocytes were transfected with hIRF1-siRNA or SCR and treated with vehicle or LPS for 4 hours. Cell lysates were analyzed by Western blot for indicated proteins. P45 pro-form of Caspase-1 is shown. B. Densitometry analysis of A. Data are expressed as mean +SEM fold change vs. β-actin; n=3. C. Measurement of IL-1β by ELISA for monocytes treated as in A followed by stimulation of the inflammasome with 5 mM ATP. Points represent the average of duplicates for 5 different SLE patients and controls. D. Monocytes were stimulated overnight with IFNα or 50% control or SLE serum in the presence or absence of DMSO (vehicle) or 50µM TBB followed by LPS priming and inflammasome activation with 5mM ATP. IL-1β production was measured by ELISA. Dots represent average of duplicates +SEM for seven unique control and SLE serums on two separate experiments. E. Linear regression of CASP1 and IRF1 Log2 mRNA expression values (as assessed by microarrays) in DLE and SCLE lesional biopsies (left) or control skin (right). Comparisons made in B-D by paired Student’s t-test. *=p<0.05, **=p<0.01, ****=p<0.0001.
In order to see whether IFN activity in SLE serum can enhance inflammasome activation in control monocytes, we incubated control monocytes with IFNα or serum from control or SLE patients overnight prior to inflammasome activation with LPS and ATP. We were able to detect a robust increase in IL-1β release from monocytes that were pre-incubated overnight with control serum prior to inflammasome stimulation and this effect was amplified with the use of SLE serum (Figure 5D). Importantly, we were able to significantly diminish the enhanced effect of SLE serum on inflammasome activation via IRF-1 inhibition with TBB, returning IL-1β release to levels obtained with control serum. Taken together, these data suggest that interferons present in lupus serum can enhance inflammasome activation in an IRF-1-dependent manner.
Cutaneous lupus lesions are marked by strong type I IFN signatures(38, 39) and monocyte infiltration of lesions(40, 41). In order to confirm relevance of caspase-1 and IRF1 dysregulation in an SLE disease phenotype, expression of both genes were assessed in lesional skin biopsies from 47 discoid lupus erythematosus (DLE) and 43 subacute cutaneous lupus erythematosus (SCLE) patients and from 13 normal control biopsies. As shown in Figure 5E, there was a strongly significant correlation between IRF1 and CASP1 expression in both cutaneous lupus subtypes. This correlation also held when each disease subtype was considered separately: DLE CASP1 vs. IRF1 R2=0.3315 and p=<0.0001; SCLE CASP1 vs. IRF1 R2=0.2129 and p=0.002. No correlation between IRF1 and CASP1 expression was noted in control skin. Taken together, these data suggest that chronic type I IFN exposure drives enhanced inflammasome activity in SLE in an IRF1-dependent manner.
DISCUSSION
Recent data has supported the role of the inflammasome in contributing to the pathogenesis of SLE(1, 4–7, 9, 10, 42). Here we provide mechanistic data on the intersection of inflammasome activation with another SLE-relevant innate immune pathway: chronic type I IFN exposure. Indeed, we have shown that prolonged exposure to IFNα promotes inflammasome activation in SLE monocytes in an IRF1-dependent manner.
The mechanisms by which the inflammasome impacts SLE etiopathogenesis are being defined. Upregulation of inflammasome gene expression and inflammasome activity has been identified in both human and murine lupus(1, 2, 4, 6, 7, 43). Inhibition of caspase-1 or NLRP3-mediated inflammasome signaling in murine models has demonstrated that inflammasome pathways contribute to disease pathogenesis via promotion of autoantibody production, endothelial dysfunction, and nephritis(5, 9, 10, 12). Activation of the inflammasome occurs downstream of SLE-specific factors, including autoantibodies and dysregulated neutrophil NETs(1, 2, 6, 7), which may implicate the inflammasome as a regulatory switch between autoimmunity and inflammatory-mediated organ damage.
Links between type I IFN and the inflammasome in SLE have been less studied. Bisphenol A, a chemical which upregulates both type I IFN production and inflammasome activity in monocytes(21) may be linked with autoimmunity, but whether the two pathways intersected was not examined. One paper, which examined human PMBC populations, described decreased expression of NLRP3 in SLE vs. control samples(19). However, our data, which utilized purified monocyte populations, clearly finds increased expression of NLRP3 and NLRP3-mediated inflammasome activity in SLE vs. control monocytes. It is possible that analysis of a mixed cell population diluted the expression changes in SLE monocytes in that study. Another paper suggested that IFNβ is repressive to inflammasome activation through production of IL-10 in bone marrow derived murine macrophages(20). In agreement with that paper, we have identified that short-term exposure to IFNα represses inflammasome activation, but prolonged exposure, as seen in SLE, promotes inflammasome activation. This concept is supported by the strong correlation between NLRP3 and type I IFN-regulated genes in SLE but not control monocytes and in cutaneous lupus lesions. Surprisingly, we did not see a correlation between AIM2, an inflammasome scaffold activated by dsDNA and known to be upregulated by type I IFNs(44), and monocyte IFN scores. This may reflect increased levels of circulating BAFF in SLE patients, which is known to repress Aim2 expression(45). The role of Aim2 in SLE is multifaceted and may have inhibitory and promoting effects on autoimmunity (reviewed in (46)). No correlation of another inflammasome scaffold, NLRP1, with IFN scores was noted. NLRP1 has genetic links to SLE(47), and similar to NLRP3, transient type I IFN exposure has been reported to downregulate NLRP1 inflammasomes(20); however the role for NLRP1 in SLE remains undefined.
Our data show that inhibition of IRF1 in SLE monocytes normalizes inflammasome hyperactivity. IRF1 is increasingly becoming a target of investigation in SLE pathogenesis. Deletion of IRF1 is protective in the MRL/lpr model of lupus(48). Expression of IRF1-regulated genes is increased in SLE by an average of 7.8% secondary to increased IRF1 binding to H3K4me3 sites (34). Further, elevated IRF1 expression is found in human lupus nephritis biopsies(49), DLE and SCLE (Figure 5) and in SLE monocytes(34). Increased expression of IRF1 facilitates type I IFN-accelerated lupus nephritis in NZB/NZWF1 mice and its downregulation correlates with reduction in lupus nephritis activity(32). Importantly, IRF1 expression in the kidney may also be predictive of treatment response for lupus nephritis(33).
Addition of serum to monocytes has been shown to facilitate inflammasome activation in part via immune complexes and stimulation of Fcγ receptors in the context of TLR signaling (1, 6, 7, 50). We detected similar enhancement of inflammasome activation when control monocytes were exposed to SLE and control serum, although the effect was greater for SLE serum. Interestingly, the enhanced effects of SLE serum on IL-1β release were returned to the level of those seen in control serum following inhibition of IRF-1 via TBB. This suggests that the increased IFN activity in the SLE serum is a significant contributor to inflammasome activation in an IRF-1 dependent manner. The residual enhancement of inflammasome activity in the presence of TBB is likely secondary to immunoglobulin effects.
Interest in the inflammasome as a promoter of SLE organ damage is a recent and expanding area of research. Our work supports a role for chronic type I IFN exposure as a mechanism by which the inflammasome is hyperactivated in SLE patients and identifies IRF1 as an important regulator of this process. These data will inform development of novel therapeutic targets for SLE.
Supplementary Material
Control monocytes were treated with indicated doses of IFNα overnight followed by priming with LPS and activation of the inflammasome with ATP. IL-1β secretion was measured by ELISA. *=p<0.05, ****=p<0.0001. Experiment was repeated 3 times; each point represents one well of one experiment.
Acknowledgments
Funding for this project was through a generous gift from the Mary Catherine Piazza Lupus Research Fund and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health under Award Number R03AR066337. JL was supported by a grant from the Arthritis National Research Foundation (to JMK) and JMK was also partially supported through NIAMS under Award Number K08AR063668.
Footnotes
Conflict of Interest: JMK has received consulting fees from Idera Pharmaceuticals
References
- 1.Zhang H, Fu R, Guo C, Huang Y, Wang H, Wang S, et al. Anti-dsDNA antibodies bind to TLR4 and activate NLRP3 inflammasome in lupus monocytes/macrophages. Journal of translational medicine. 2016;14(1):156. doi: 10.1186/s12967-016-0911-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ. Neutrophil Extracellular Trap-Associated Protein Activation of the NLRP3 Inflammasome Is Enhanced in Lupus Macrophages. The journal of immunology. 2013;190(3):1217–26. doi: 10.4049/jimmunol.1202388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahlenberg JM, Kaplan MJ. The inflammasome and lupus: another innate immune mechanism contributing to disease pathogenesis? Curr Opin Rheumatol. 2014;26(5):475–81. doi: 10.1097/BOR.0000000000000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahlenberg JM, Thacker SG, Berthier CC, Cohen CD, Kretzler M, Kaplan MJ. Inflammasome activation of IL-18 results in endothelial progenitor cell dysfunction in systemic lupus erythematosus. J Immunol. 2011;187(11):6143–56. doi: 10.4049/jimmunol.1101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahlenberg JM, Yalavarthi S, Zhao W, Hodgin JB, Reed TJ, Tsuji NM, et al. An essential role of caspase 1 in the induction of murine lupus and its associated vascular damage. Arthritis & rheumatology (Hoboken, NJ) 2014;66(1):152–62. doi: 10.1002/art.38225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin MS, Kang Y, Lee N, Kim SH, Kang KS, Lazova R, et al. U1-Small Nuclear Ribonucleoprotein Activates the NLRP3 Inflammasome in Human Monocytes. The journal of immunology. 2012;188(10):4769–75. doi: 10.4049/jimmunol.1103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin MS, Kang Y, Lee N, Wahl ER, Kim SH, Kang KS, et al. Self Double-Stranded (ds)DNA Induces IL-1β Production from Human Monocytes by Activating NLRP3 Inflammasome in the Presence of Anti-dsDNA Antibodies. The journal of immunology. 2013 doi: 10.4049/jimmunol.1201195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilaysane A, Chun J, Seamone ME, Wang W, Chin R, Hirota S, et al. The NLRP3 Inflammasome Promotes Renal Inflammation and Contributes to CKD. Journal of the American Society of Nephrology. 2010;21(10):1732–44. doi: 10.1681/ASN.2010020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Wang H, Dai C, Wang H, Zhang H, Huang Y, et al. P2×7 blockade attenuates lupus nephritis by inhibiting NLRP3/ASC/caspase-1 activation. Arthritis & Rheumatism. 2013;65(12):3176–85. doi: 10.1002/art.38174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Zhang H, Huang Y, Wang H, Wang S, Zhao C, et al. Bay11-7082 attenuates murine lupus nephritis via inhibiting NLRP3 inflammasome and NF-κB activation. International Immunopharmacology. 2013;17(1):116–22. doi: 10.1016/j.intimp.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 11.Ka SM, Lin JC, Lin TJ, Liu FC, Chao LK, Ho CL, et al. Citral alleviates an accelerated and severe lupus nephritis model by inhibiting the activation signal of NLRP3 inflammasome and enhancing Nrf2 activation. Arthritis Res Ther. 2015;17:331. doi: 10.1186/s13075-015-0844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Shi X, Qian T, Li J, Tian Z, Ni B, et al. A20 overexpression alleviates pristine-induced lupus nephritis by inhibiting the NF-kappaB and NLRP3 inflammasome activation in macrophages of mice. International journal of clinical and experimental medicine. 2015;8(10):17430–40. [PMC free article] [PubMed] [Google Scholar]
- 13.Eloranta ML, Ronnblom L. Cause and consequences of the activated type I interferon system in SLE. J Mol Med (Berl) 2016 doi: 10.1007/s00109-016-1421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crow MK. Type I interferon in the pathogenesis of lupus. J Immunol. 2014;192(12):5459–68. doi: 10.4049/jimmunol.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal H, Jacob N, Carreras E, Bajana S, Putterman C, Turner S, et al. Deficiency of type I IFN receptor in lupus-prone New Zealand mixed 2328 mice decreases dendritic cell numbers and activation and protects from disease. J Immunol. 2009;183(9):6021–9. doi: 10.4049/jimmunol.0803872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Bethunaickan R, Huang W, Lodhi U, Solano I, Madaio MP, et al. Interferon-alpha accelerates murine systemic lupus erythematosus in a T cell-dependent manner. Arthritis Rheum. 2011;63(1):219–29. doi: 10.1002/art.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thacker SG, Zhao W, Smith CK, Luo W, Wang H, Vivekanandan-Giri A, et al. Type I interferons modulate vascular function, repair, thrombosis and plaque progression in murine models of lupus and atherosclerosis. Arthritis & Rheumatism. 2012 doi: 10.1002/art.34504. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirou K, Lee C, George S, Louca K, Peterson MGE, Crow M. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis and rheumatism. 2005;52(5):1491–503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, Yu C, Yang Z, Wei Q, Mu K, Zhang Y, et al. Deregulated NLRP3 and NLRP1 Inflammasomes and Their Correlations with Disease Activity in Systemic Lupus Erythematosus. The Journal of Rheumatology. 2014;41(3) doi: 10.3899/jrheum.130310. [DOI] [PubMed] [Google Scholar]
- 20.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, et al. Type I Interferon Inhibits Interleukin-1 Production and Inflammasome Activation. Immunity. 2011;34(2):213–23. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Panchanathan R, Liu H, Leung YK, Ho SM, Choubey D. Bisphenol A (BPA) stimulates the interferon signaling and activates the inflammasome activity in myeloid cells. Molecular and cellular endocrinology. 2015;415:45–55. doi: 10.1016/j.mce.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyogoku C, Smiljanovic B, Grun JR, Biesen R, Schulte-Wrede U, Haupl T, et al. Cell-specific type I IFN signatures in autoimmunity and viral infection: what makes the difference? PLoS One. 2013;8(12):e83776. doi: 10.1371/journal.pone.0083776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Furie JM, Werth VP, Khamashta M, Kalunian K, Brohawn P, Illei G, Drappa J, Wang L, Yoo S. Anifrolumab, an Anti-Interferon Alpha Receptor Monoclonal Antibody, in Moderate to Severe Systemic Lupus Erythematosus (SLE). American College of Rheumatology Annual Meeting; San Franscisco, CA. 2015. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Kirou KA, Lee C, George S, Louca K, Papagiannis IG, Peterson MG, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50(12):3958–67. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 27.Ekholm L, Kahlenberg JM, Barbasso Helmers S, Tjarnlund A, Yalavarthi S, Zhao W, et al. Dysfunction of endothelial progenitor cells is associated with the type I IFN pathway in patients with polymyositis and dermatomyositis. Rheumatology (Oxford, England) 2016;55(11):1987–92. doi: 10.1093/rheumatology/kew288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Asgari E, Le Friec G, Yamamoto H, Perucha E, Sacks SS, Kohl J, et al. C3a modulates IL-1beta secretion in human monocytes by regulating ATP efflux and subsequent NLRP3 inflammasome activation. Blood. 2013;122(20):3473–81. doi: 10.1182/blood-2013-05-502229. [DOI] [PubMed] [Google Scholar]
- 30.Qu Y, Ramachandra L, Mohr S, Franchi L, Harding CV, Nunez G, et al. P2×7 receptor-stimulated secretion of MHC class II-containing exosomes requires the ASC/NLRP3 inflammasome but is independent of caspase-1. J Immunol. 2009;182(8):5052–62. doi: 10.4049/jimmunol.0802968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis & Rheumatism. 2006;54(6):1906–16. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 32.Han X, Wang Y, Zhang X, Qin Y, Qu B, Wu L, et al. MiR-130b ameliorates murine lupus nephritis through targeting type I interferon pathway on resident renal cells. Arthritis & rheumatology (Hoboken, NJ) 2016 doi: 10.1002/art.39725. [DOI] [PubMed] [Google Scholar]
- 33.Parikh SV, Malvar A, Song H, Alberton V, Lococo B, Vance J, et al. Characterising the immune profile of the kidney biopsy at lupus nephritis flare differentiates early treatment responders from non-responders. Lupus science & medicine. 2015;2(1):e000112. doi: 10.1136/lupus-2015-000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Shi L, Song L, Ephrem E, Petri M, Sullivan KE. Interferon regulatory factor 1 marks activated genes and can induce target gene expression in systemic lupus erythematosus. Arthritis & rheumatology (Hoboken, NJ) 2015;67(3):785–96. doi: 10.1002/art.38964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fantuzzi G, Reed D, Qi M, Scully S, Dinarello CA, Senaldi G. Role of interferon regulatory factor-1 in the regulation of IL-18 production and activity. Eur J Immunol. 2001;31(2):369–75. doi: 10.1002/1521-4141(200102)31:2<369::aid-immu369>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 36.Karlsen AE, Pavlovic D, Nielsen K, Jensen J, Andersen HU, Pociot F, et al. Interferon-gamma induces interleukin-1 converting enzyme expression in pancreatic islets by an interferon regulatory factor-1-dependent mechanism. J Clin Endocrinol Metab. 2000;85(2):830–6. doi: 10.1210/jcem.85.2.6366. [DOI] [PubMed] [Google Scholar]
- 37.Lin R, Hiscott J. A role for casein kinase II phosphorylation in the regulation of IRF-1 transcriptional activity. Mol Cell Biochem. 1999;191(1–2):169–80. [PubMed] [Google Scholar]
- 38.Meller S, Winterberg F, Gilliet M, Muller A, Lauceviciute I, Rieker J, et al. Ultraviolet radiation-induced injury, chemokines, and leukocyte recruitment: An amplification cycle triggering cutaneous lupus erythematosus. Arthritis Rheum. 2005;52(5):1504–16. doi: 10.1002/art.21034. [DOI] [PubMed] [Google Scholar]
- 39.Stannard JN, Reed TJ, Myers E, Lowe L, Sarkar MK, Xing X, et al. Lupus skin is primed for IL-6 inflammatory responses through a keratinocyte-mediated autocrine type I interferon loop. The Journal of investigative dermatology. 2016 doi: 10.1016/j.jid.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy Crispin M, Fuentes-Duculan J, Gulati N, Johnson-Huang LM, Lentini T, Sullivan-Whalen M, et al. Gene profiling of narrowband UVB-induced skin injury defines cellular and molecular innate immune responses. The Journal of investigative dermatology. 2013;133(3):692–701. doi: 10.1038/jid.2012.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng G-M, Liu L, Kyttaris VC, Tsokos GC. Lupus Serum IgG Induces Skin Inflammation through the TNFR1 Signaling Pathway. The Journal of Immunology. 2010;184(12):7154–61. doi: 10.4049/jimmunol.0902514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahlenberg JM. Activation of Caspase-1 Signaling Complexes by the P2×7 Receptor Requires Intracellular K <sup>+</sup> Efflux and Protein Synthesis Induced by Priming with Toll-Like Receptor Ligands. Case Western Reserve University; 2004. [Google Scholar]
- 43.Clark KL, Reed TJ, Wolf SJ, Lowe L, Hodgin JB, Kahlenberg JM. Epidermal injury promotes nephritis flare in lupus-prone mice. J Autoimmun. 2015 Dec;65:38–48. doi: 10.1016/j.jaut.2015.08.005. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15(4):453–7. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 45.Panchanathan R, Choubey D. Murine BAFF expression is up-regulated by estrogen and interferons: Implications for sex bias in the development of autoimmunity. Molecular Immunology. 2013;53(1–2):15–23. doi: 10.1016/j.molimm.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choubey D, Panchanathan R. Absent in Melanoma 2 proteins in SLE. Clinical Immunology. 2017;176:42–8. doi: 10.1016/j.clim.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pontillo A, Girardelli M, Kamada A, Pancotto JA, Donadi EA, Crovella S, et al. Polimorphisms in inflammasome genes are involved in the predisposition to systemic lupus erythematosus. Autoimmunity. 2012;45(4):271–8. doi: 10.3109/08916934.2011.637532. [DOI] [PubMed] [Google Scholar]
- 48.Reilly CM, Olgun S, Goodwin D, Gogal RM, Jr, Santo A, Romesburg JW, et al. Interferon regulatory factor-1 gene deletion decreases glomerulonephritis in MRL/lpr mice. Eur J Immunol. 2006;36(5):1296–308. doi: 10.1002/eji.200535245. [DOI] [PubMed] [Google Scholar]
- 49.Han X, Wang Y, Zhang X, Qin Y, Qu B, Wu L, et al. MicroRNA-130b Ameliorates Murine Lupus Nephritis Through Targeting the Type I Interferon Pathway on Renal Mesangial Cells. Arthritis & rheumatology (Hoboken, NJ) 2016;68(9):2232–43. doi: 10.1002/art.39725. [DOI] [PubMed] [Google Scholar]
- 50.Vogelpoel LT, Hansen IS, Rispens T, Muller FJ, van Capel TM, Turina MC, et al. Fc gamma receptor-TLR cross-talk elicits pro-inflammatory cytokine production by human M2 macrophages. Nature communications. 2014;5:5444. doi: 10.1038/ncomms6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Control monocytes were treated with indicated doses of IFNα overnight followed by priming with LPS and activation of the inflammasome with ATP. IL-1β secretion was measured by ELISA. *=p<0.05, ****=p<0.0001. Experiment was repeated 3 times; each point represents one well of one experiment.