Abstract
Primitive round- or spindle-cell EWSR1-negative undifferentiated sarcomas harboring CIC-DUX4 gene fusion are the most common form of Ewing-like sarcomas. These tumors primarily occur in peripheral soft tissues, but examples have been described within viscera and the brain. As far as we are aware, CIC-DUX4 positive primary epidural spinal sarcoma has not been reported. Herein, we describe a T5–T6 epidural tumor in a 15-year-old girl in which many neoplastic cells had moderate and focally abundant cytoplasm, including plasmacytoid or rhabdoid cells, rather than the more common Ewing-like morphology described in the majority of such tumors. The diagnosis was confirmed by fluorescent in situ hybridization after the tumor was found to be WT-I positive, and comprehensive genomic profiling demonstrated breakpoints in exon 20 and exon 1 of the CIC and DUX4 genes, respectively. After treatment with local radiation and systemic chemotherapy, resected recurrent tumor demonstrated more pleomorphic neoplastic cells as well as intracytoplasmic eosinophilic globules and nuclear pseudoinclusions which may reflect therapy-related changes. Unfortunately, there was further progression of tumor including the development of intracranial lesions, and the patient succumbed to her tumor 22 months after the original resection.
Keywords: CIC-DUX4, comprehensive genomic profiling, spinal dura, undifferentiated sarcoma
Introduction
CIC-rearranged sarcomas are a group of EWSR1-negative primitive round- or spindle-cell sarcomas that most commonly harbor translocations between the human homologue of the Drosophila capicua (CIC) gene on chromosome 19q13 and the double-homeobox 4 (DUX4) gene on chromosome 4q35 or 10q26.3.1–10 Occasional cases occurring in peripheral sites harbor variant translocation in which the CIC gene is fused with the forkhead box O4 (FOXO4) gene on chromosome Xq13.11 While the majority of these tumors are primarily described in peripheral soft tissues, visceral, bone, and cerebrum occurrences (Table 1) have been reported.2–14 Sturm et al. recently reclassified a subset of central nervous system-primitive neuroectodermal tumors (CNS-PNETs) based on DNA methylation profiles; one of the four categories included the CNS Ewing sarcoma family of tumors with CIC alteration (CNS EFT-CIC), the majority of which harbored rearrangement of the CIC gene.14 All previously reported primary CNS CIC-rearranged tumors originated in the cerebrum,9,13,14 but herein, we report a case of a primary thoracic spine epidural CIC-DUX4 sarcoma.
Table 1.
Sites of Previously Reported CIC-rearranged Tumors.a
| Primary Site | Number of Cases | Age (years) | References |
|---|---|---|---|
| Trunk wall and head and neck soft tissue | 31 | Median 25 (range 6–73) | 2–5, 7–10 |
| Limbs | 34 | Median 32.5 (range 6–62) | 5, 7–10 |
| Internal trunk | 6 | Median 23.5 (range 19–40) | 5, 7, 9 |
| Primary bone | 3 | Median 44 (range 14–47) | 5, 7, 10 |
| Viscerab and adjacent structures | 6 | Median 25.5 (range 9–37) | 2, 5, 7, 9, 12 |
| Brainc | 11 | Adults are 64 and 24 | 9, 13, 14 |
| Median of 1.5 in EFT-CIC cases (range < 1–8)d |
EFT-CIC, Ewing family of tumors with CIC alteration.
This tally of cases excludes some reports. The overall median presenting age for tumors at peripheral sites from these reports is 29 years (range 6–73 years).
Visceral sites include lung, stomach, and kidney.
The brain case reported in Yoshida et al.9 is also fully presented as a case report by Ito et al. A case report of CIC-rearranged undifferentiated small round cell sarcoma in the cerebrum. Diagn Cytopathol. 2016; 44: 828–832.
Data obtained from supplemental data accessible from the online version of Sturm et at.14
Materials and Methods
The clinical history, evaluation, treatment, and follow-up pathologic findings in resection and postmortem specimens, and molecular studies used to characterize the tumor are reviewed. Review of this case was performed as part of an institutional review board approved study (no. 018813).
Immunohistochemistry
Immunohistochemistry (IHC) was performed using Envision+ Dual Link, Peroxidase (Dakocytomation) or standard streptavidin technique (Jackson) with a 3-3’-diaminobenzadine (DAB) chromogen. The primary anti-bodies utilized for classification of this tumor were as follows: CD99 (M; O13; Signet), desmin (M; 33; Accurate Chemical & Scientific), smooth muscle actin (SMA) (M; IA4; Dako), chromogranin A (R; Dako), epithelial membrane antigen (EMA) (M; E29; Cell Marque), S-100 (P; Dako), vimentin (M; V9; BioGenex), cytokeratin cocktail (M; AE1 and AE3 and CAM 5.2, KA4 and UC2/PR-10-11; Becton-Dickerson and Zymed/ThermoFisher Scientific), CD45 (M; PD7/26 and 2B11; Cell Marque); CD138 (M; MI15; Dako), CD31 (M; JC/70A; Cell Marque), CD34 (M; QBEnd/10; Dako), PLAP (M; 8A9; Dako), CD68 (M; KPI; Cell Marque), CD117 (R; Diagnostics Biosystems), GFAP (M; GA-5 Biogenex), INI-1 (M; BAF47; BD Bioscience), and WT-1 (M;6F-H2; Dako).
Comprehensive Genomic Profiling
Comprehensive genomic profiling (CGP) was performed on DNA/RNA extracted from 10-µm-thick formalin-fixed paraffin-embedded (FFPE) sections using the FoundationOne Heine panel (Foundation Medicine, Inc, Cambridge, MA) for hematologic malignancies and soft tissue tumors.15 The adaptor-ligated sequencing library was captured by solution hybridization using a bait-set targeting. DNA-seq was used to analyze 405 cancer-related genes and selected introns from 31 genes (full details available in the supplemental data accessible from the online version of He et al.15). RNA-seq was used to analyze 265 genes that are frequently rearranged.15 The library was sequenced in a CLIA-certified, CAP-accredited, and NYS-approved laboratory to 859×median coverage for DNA and 4.2 million on-target distinct pairs for RNA on a HiSeq2500 sequencer. Proprietary algorithms were used to accurately detect base substitutions, insertions, deletions, copy number alterations, and rearrangements.15
Results
Initial Patient Presentation
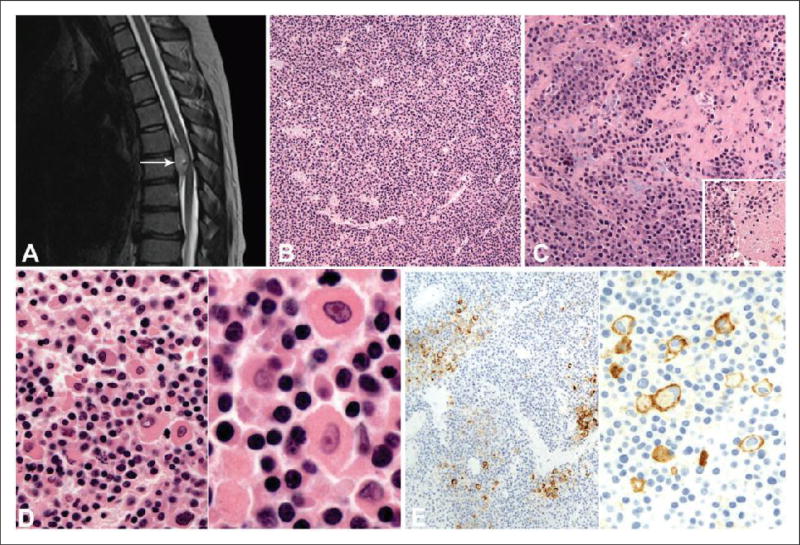
A 15-year-old girl presented with back pain that was exacerbated by an injury during gymnastics training. Magnetic resonance imaging (MRI) demonstrated a 2 cm epidural tumor at T5–T6. T2-weighted imaging with and without contrast revealed regional spinal cord compression with a possible focus of central cystic change (Figure 1(a)). A costo-transversectomy approach via T4–T7 laminectomy was used to resect the tumor.
Figure 1.
(a), MRI without contrast shows the T5–T6 ventral epidural tumor (arrow) that measured 2.4 cm and is T2 hyperintense, but not isointense with cerebrospinal fluid. A small (9 mm) fluid-containing focus within the tumor is radiologically consistent with cystic change. (b), Hematoxylin and eosin (H&E) section shows a diffuse arrangement of neoplastic cells with prominent vasculature. (c), The neoplastic cells are focally noted in a fibrous and myxoid stroma. Inset: Focal necrosis is noted within the tumor. (d), On high-power magnification, a large proportion of cells demonstrate moderate cytoplasm and nuclei with coarse chromatin (left and right panels). A smaller proportion of neoplastic cells demonstrate more abundant eosinophilic cytoplasm including cells with rhabdoid or plasmacytoid morphology (left panel). Occasional cells have nuclei with vesicular chromatin and prominent nucleoli (right panel). (e), An immunohistochemical stain with the CD99 antibody. Low-power magnification demonstrates membranous staining of a small proportion of neoplastic cells (left panel). On high-power magnification, membranous staining of rhabdoid/plasmacytoid cells is noted (right panel).
Histopathologic and Ancillary Findings in Original Resection
The tumor was predominantly composed of a diffuse arrangement of neoplastic cells with prominent vasculature (Figure 1(b)). Focally, fibrous and myxoid stroma and necrosis of neoplastic cells were noted (Figure 1(c)). A large proportion of neoplastic cells contained round or oval nuclei with coarse chromatin and moderate eosinophilic cytoplasm (Figure 1(d)). However, a subset of neoplastic cells had larger nuclei, with vesicular chromatin, prominent nucleoli, and abundant eosinophilic cytoplasm imparting a rhabdoid or plasmacytoid morphology (Figure 1(d)). Adjacent sections were used for IHC studies. Virtually all of the neoplastic cells were positive for vimentin, retained positive nuclear staining with the antibody to INI-1, and approximately 5% had membranous CD99 immunoreactivity (Figure 1(e)). Focally, CD68 was positive and scattered cells were immunoreactive for pan-cytokeratin. The tumor cells exhibited no immunoreactivity to GFAP, EMA, desmin, SMA, CD138, CD45, PLAP, CD117, chromogranin A, CD31, CD34, and S-100. RT-PCR and fluorescent in situ hybridization (FISH) performed on FFPE tissue at a reference center were negative for EWSR1 rearrangement, and FISH was negative for SYT and FUS rearrangements. Therefore, a diagnosis of high-grade malignant neoplasm without further classification was rendered.
Follow-up Clinical Course and Diagnostic Studies
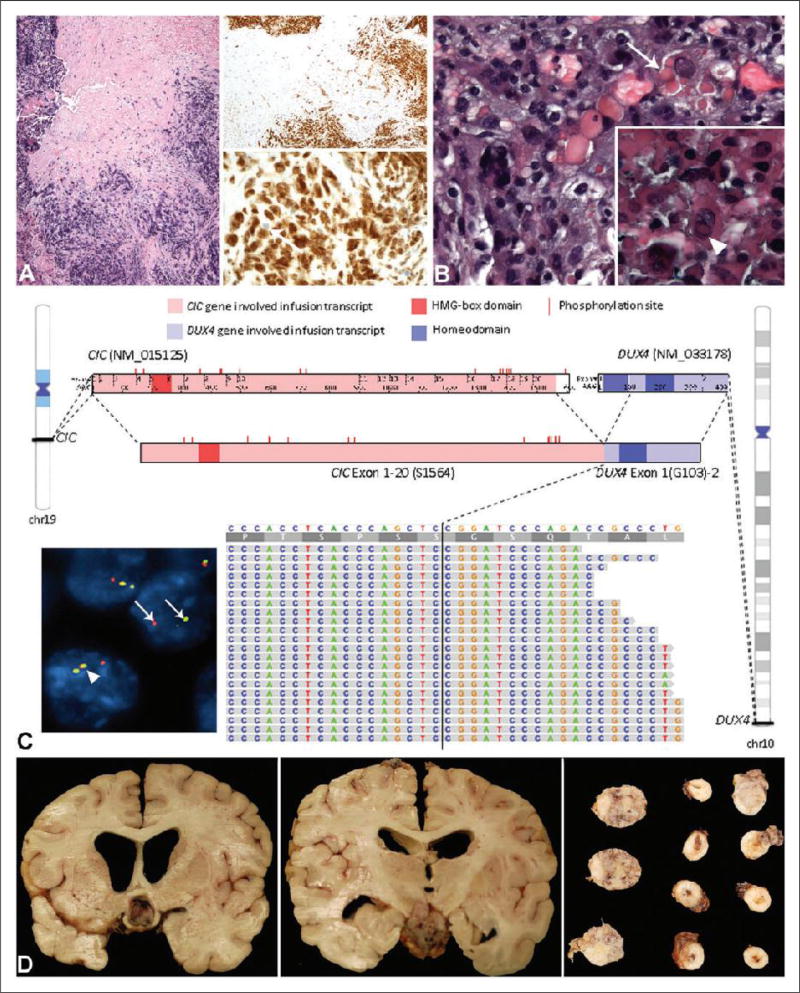
One month after the initial gross total resection, the tumor recurred at the operative site. Despite 6 months of radiation and steroid therapy, the tumor continued to enlarge. The patient underwent endoscopic biopsy and resection of the recurrent tumor. The histopathological features were similar to the original tumor (Figure 2(a)). The same patterns of immunoreactivity were observed as noted in the initial specimen. In addition, diffuse nuclear WT-1 immunoreactivity was demonstrated with the antibody to the N-terminal of WT-1 (Figure 2(a)). As a result, the case was submitted for external consultation and testing for CIC-rearrangement on FFPE tissue which was positive by FISH. Additionally, the tumor was tested for and found to be negative for BCOR-CCNB3 fusion, EWSR1, FUS, and ERG rearrangement, all by FISH performed on FFPE tissue.
Figure 2.
(a), Hematoxylin and eosin (H&E) and WT-1 stains of the recurrent tumor. The H&E section (left panel) shows neoplastic cells with associated crush artifact. On low-power magnification, there is diffuse staining of the tumor with the antibody to WT-1 (right upper panel). The high-power magnification of the WT-1 stain shows primarily nuclear staining of the neoplastic cells (right lower panel). An occasional intermixed nucleus without staining is noted (arrow). (b), H&E section of the tumor from the third resection after radiation and chemotherapy. There are more pleomorphic nuclei including multilobated nuclei and eosinophilic cytoplasmic globules are noted within a subset of cells (arrow). Inset: Occasional neoplastic cells also demonstrate nuclear pseudoinclusions (arrowhead). (c), Comprehensive genomic profiling (CPG) image with fluorescent in-situ hybridization (FISH) as inset on the lower left. CPG identifies a CIC-DUX4 fusion transcript, in which exons 1–20 of CIC [NM_015125, chr19:42,788,817-42,799,949 (hg19); breakpoint at S1564] is fused with exon 1 of DUX4 [NM_033178, chr10:135,480,558-135,485,241 (hg19); breakpoint at G103]. The fused transcript maintains the HMG-box domain of CIC as well as most of the MAPK phosphorylation sites. One complete homeodomain in DUX4 is maintained. The IGV screenshot at the bottom shows the alignment of RNA reads that cross the CIC-DUX4 breakpoint to the CIC-DUX4 fusion sequence. Exact breakpoint locations are indicated by vertical black lines. CIC phosphorylation sites are annotated based on UniProtKB entry Q96RK0 (CIC_HUMAN). HMG-box: high-mobility group box. Inset: FISH performed on a section from the autopsy specimen demonstrates rearrangement of the centromeric probe (red) and telomeric probe (green) of the CIC gene (arrows). This contrasts with a nucleus without rearrangement in which the overlapping red, yellow, and green signals are noted (arrowhead). (d), Collage of sections of the intracranial and spinal tumor from the autopsy. The left panel shows tumor compression of the optic chiasm, while the middle panel shows tumor adjacent to the medial temporal cortex. The right panel demonstrates tumor around the spinal cord (right two columns) and from the cerebello-pontine angle (left three sections).
Whole body positron emission tomography-computed tomography (PET-CT) and MRI scans of the brain performed after the second resection were negative for intracranial tumor. Treatment with systemic chemotherapy protocol for the Ewing sarcoma family of tumors caused a transient 70% reduction in volume but later it progressed. Tumor recurrence prompted an additional resection just 18 months after her original surgery. The histopathological features of the third resected tumor were remarkable for the presence of a subset of cells with pleomorphic multilobated nuclei, cells with eosinophilic cytoplasmic globules, and occasional cells demonstrated nuclear pseudoinclusions (Figure 2(b)).
CGP was performed and found to be negative for targetable genomic alterations, but a CIC-DUX4 fusion transcript was demonstrated in which an in-frame fusion of exon 20 of the CIC gene at the S1564 break-point to exon 1 of the DUX4 gene at the G103 breakpoint (Figure 2(c)). The predicted fusion protein retained most of the functional regions of the CIC gene, including the DNA-binding high-mobility group (HMG) box and most of the putative MAPK phosphorylation sites. One complete homeodomain in DUX4 is maintained in the fusion transcript. Realigning RNA-seq reads to this fusion transcript identified over 140 reads that cross the breakpoint (Figure 2(c)).
Despite continued management with systemic chemotherapy, the patient’s disease progressed and caused her death. Multiple repeat MRI studies of the brain, with and without contrast, were consistently negative for tumor until two months prior to her death when intracranial tumor was demonstrated for the first time.
At autopsy, a large contiguous mass was present in the cerebellum, hypothalamus, infundibulum, optic chiasm, and throughout the brainstem and spinal cord, with extension into the lumbosacral region and cauda equina. Intramedullary spinal cord tumor was most pronounced at upper cervical and thoracic levels (Figure 2(d)). The morphology of the tumor in the autopsy specimens was similar to the original morphology.
Discussion
Undifferentiated small round- or spindle-cell sarcomas are highly aggressive tumors arising primarily in soft tissues of children and young adults. CIC-rearranged sarcomas represent the majority of primitive undifferentiated sarcomas. Due to morphologic and immunohistochemical overlap with Ewing sarcoma, CIC-rearranged tumors have been referred to as Ewing-like sarcoma; however, they lack the translocations involving EWSR1 gene on chromosome 22 or FUS gene fused to the ETS transcription factor family.1
As reviewed by Specht et al.,7 individual cases of these undifferentiated sarcomas were first recognized to have the characteristic karyotypic translocation.7 In 2006, Kawamura-Saito et al.2 reported that the karyo-typic translocation corresponded to fusion of the CIC and DUX4 genes. This finding was confirmed by Yoshimotoa et al.3 in their group of pediatric undifferentiated sarcomas. Subsequently, other authors reported on larger series of CIC-DUX4 sarcomas.4–9 Italiano et al. described one of the largest series to date and reported that over 60% of their EWSR1-negative primitive or undifferentiated sarcomas belonged to the group of CIC-DUX4 sarcomas. Specht et al.7 reported WT-1 nuclear positivity using the antibody to the N-terminal in the majority of their CIC-rearranged sarcomas 6 months after the original resection of this tumor. This discovery facilitated the submission of this case for CIC testing when the second resection was performed and diffuse nuclear WT-1 positivity was demonstrated.
CIC-rearranged sarcomas are more typically described as small round- and spindle-cell tumors that have foci of myxoid stroma, microcyst formation, and typically vesicular nuclear chromatin with prominent nucleoli.1–13 While foci with larger cells may be seen focally in CIC-rearranged sarcomas,10 this case demonstrated a large proportion of cells with moderate cytoplasm and focally cells with abundant cytoplasm accompanied by limited nuclear pleomorphism in the original resection. It may be that some of the features encountered in the third resection, including the more pleomorphic nuclei, eosinophilic cytoplasmic globules, and nuclear pseudoinclusions are related to treatment. Morphologic changes in CIC-rearranged sarcomas due to local radiation and systemic chemotherapy have not been addressed in the available literature.
As indicated above, in addition to CIC-DUX4 fusion, CIC-FOXO4 fusion has also been described in tumors occurring in peripheral soft tissue.11 Furthermore, CIC-NUTM1 fusion was reported in a subset of CNS-PNETs by Sturm et al.14 DNA methylation profiles performed by these authors demonstrated that the majority of 323 CNS PNETs were indistinguishable from that of 211 well-known CNS reference tumors,14 but 77 cases (~24%) formed 4 separate clusters: CNS neuroblastoma with FOXR2 activation (44 cases), CNS EFT-CIC tumors (12 cases, all in children), CNS high-grade neuroepithelial tumors with MN1 alteration (11 cases), and CNS high-grade neuroepithelial tumors with BCOR alteration (HGNET-BCOR; 10 cases, predominantly in children).
According to Sturm et al., the histology of four cases of CNS EFT-CIC tumors reviewed were characterized by a small-cell morphology, like the other reported cases,9,13 with variable alveolar and fascicular architectural patterns. Two of three of the CNS EFT-CIC tumors analyzed with RNA sequencing demonstrated interchromosomal gene fusion between the CIC gene and the NUT midline carcinoma, family member 1 (NUTM1, located on chromosome 15q14). Both fusions involved exon 16 of the CIC in-frame to exon 4 of NUTM1, such that the DNA-binding high mobility group (HMG) box domain of CIC was retained. This correlates with that finding described for peripheral CIC-rearranged sarcomas. The third case harbored a frameshift deletion in CIC (exon6:c.902delC:p.S301fs). Eight of nine tumors analyzed by a FISH breakapart probes demonstrated rearrangement of the CIC gene, including one of the cases that harbored the CIC-NUTM1 fusion. All tumors in this category were restricted to the cerebrum and none involved the spinal cord or the spinal canal. Parenthetically, intraspinal extraosseus EWS/PNET cases have been reported in the current literature, the majority of which have been as single case reports.16
The median age of the CNS EFT-CIC tumors with CIC-rearrangement was 1.5 years (range < 1–8 years)14 (Table 1), but the two other known CIC-rearranged sarcomas of the brain were reported in 64- and 24 year-old women.9,13 This contrasts with peripheral CIC-rearranged sarcomas in which over 75% of cases have been reported in patients over 18 years of age (median 29 years, range 6–73 years) (Table 1).
It should be noted that none of the cases of CNS HGNET-BCOR tumors reported by Sturm et al. revealed BCOR-CCNB3 fusion as recognized in another of the large group of EWSR1-negative primitive sarcomas of bone and soft tissue.1 Instead DNA and RNA sequencing revealed in-frame tandem duplications of the BCL6 corepressor (BCOR) gene.14 The duplicated region in exon 15 of BCOR is reported to be similar to the BCOR internal tandem duplication described in a subset of clear-cell sarcomas of the kidney (CCSK), infantile undifferentiated round-cell sarcomas and primitive myxoid mesenchymal tumor of infancy.17,18 Interestingly, there were 6 cerebellar, 1 pontine, and 1 spinal HGNET-BCOR tumors, unlike the EFT-CIC tumors that were restricted to the cerebrum. The authors reported that the HGNET-BCOR cases reviewed were composed of spindle and oval cells and often exhibited perivascular pseudorosettes that reportedly gave the tumors an ependymoma-like appearance. An embryonal morphology was only rarely found.
In summary, this CIC-DUX4 positive sarcoma of the spinal dura had a morphology that is less commonly seen in EWSR1-negative undifferentiated sarcomas and demonstrated progressive pleomorphic changes probably attributable to treatment. Therefore, if faced with a neoplasm that is difficult to classify and that does not necessarily adhere to the more typical histology described in EWSR1-negative undifferentiated sarcomas, testing for CIC-rearrangement should be considered. Since different breakpoints in CIC and DUX4 genes and occasional CIC fusion partners rather than DUX4 have been reported, FISH should be the molecular test of choice unless more comprehensive genomic profiling is to be performed.
Acknowledgments
The authors thank Dr Cristina Antonescu, Memorial Sloan-Kettering Cancer Center, New York, NY, who reviewed the case in consultation and performed molecular testing for CIC-rearrangement, BCOR-CCNB3 fusion, EWSR1, FUS, and ERG rearrangement. The contributions of many of our own colleagues who acted as intradepartmental consultants (particularly Dr. Jesse Hart and Dr. Nimesh Patel) in the work up of this case are acknowledged as are our clinical colleagues who cared for this patient. We are also grateful to Karla Alvarez, BS, MB (ASCP), CM, Technical Specialist, Molecular Oncology Texas Children’s Hospital Department of Pathology, for images of the fluorescent in-situ hybridization performed on a sample from the autopsy.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Footnotes
Declaration of Conflicting Interests
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Shan Zhong and Siraj M Ali are employees of and have equity interests in Foundation Medicine, Inc.
References
- 1.Fletcher CDM, Chibon F, Mertens F. Undifferentiated/unclassified sarcomas. In: Fletcher CDM, Bridge J, Hogendoorn PCW, Mertens F, editors. WHO Classification of Tumors of Soft Tissue and Bone. Lyon: International Agency for Research on Cancer; 2013. pp. 236–238. [Google Scholar]
- 2.Kawamura-Saito M, Yamazaki Y, Kaneko K, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimotoa M, Graham C, Chilton-MacNeill S, et al. Detailed cytogenetic and array analysis of pediatric primitive sarcomas reveals a recurrent CIC-DUX4 fusion gene event. Cancer Genet Cytogenet. 2009;195:1–11. doi: 10.1016/j.cancergencyto.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Graham C, Chilton-MacNeill S, Zielenska M, Somers GR. The CIC-DUX4 fusion transcript is present in a subgroup group of pediatric primitive round cell sarcomas. Hum Pathnl. 2012;83:180–189. doi: 10.1016/j.humpath.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 5.Italiano A, Sung YS, Zhang L, et al. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small round cell sarcomas. Genes Chromosomes Cancer. 2012;51:207–218. doi: 10.1002/gcc.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi E-YK, Thomas DG, McHugh JB, et al. Undifferentiated small round cell sarcoma with t(4;19)(q35;q31.1) CIC-DUX4 fusion. A novel highly aggressive soft tissue tumor with distinctive histolopathology. Am J Surg Pathol. 2013;37:1379–1386. doi: 10.1097/PAS.0b013e318297a57d. [DOI] [PubMed] [Google Scholar]
- 7.Specht K, Sung Y-S, Zhang L, et al. Distinct transcriptional signature and immunoprofile of CIC-DUX4 positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas – further evidence towards distinct pathologic entities. Genes Chromosomes Cancer. 2014;53:622–633. doi: 10.1002/gcc.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SC, Buehler D, Choi E-YK, et al. CIC-DUX sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression. Mod Pathol. 2015;28:58–76. doi: 10.1038/modpathol.2014.83. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida A, Goto K, Kodaira M, et al. CIC-rearranged sarcomas. Am J Surg Pathol. 2016;40:313–323. doi: 10.1097/PAS.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 10.Gambarotti M, Benini S, Gamberi G, et al. CIC-DUX4 fusion-positive round-cell sarcomas of soft tissue and bone: a single-institution morphological and molecular analysis of seven cases. Histopathology. 2016;69:624–634. doi: 10.1111/his.12985. [DOI] [PubMed] [Google Scholar]
- 11.Sugita S, Arai Y, Tonooka A, et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma. A genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol. 2014;38:1571–1576. doi: 10.1097/PAS.0000000000000286. [DOI] [PubMed] [Google Scholar]
- 12.Mangray S, Somers GR, He J, et al. Primary undifferentiated sarcoma of kidney harboring novel variant of CIC-DUX4 gene fusion. Am J Surg Pathol. 2016;40:1298–1301. doi: 10.1097/PAS.0000000000000688. [DOI] [PubMed] [Google Scholar]
- 13.Bielle F, Zanello M, Guillemot D, et al. Unusual primary cerebral localization of a CIC-DUX4 translocation tumor of the Ewing sarcoma family. Acta Neuropathol. 2014;128:309–311. doi: 10.1007/s00401-014-1312-0. [DOI] [PubMed] [Google Scholar]
- 14.Sturm D, Orr BA, Toprak UH, et al. New brain entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Abdel-Wahab O, Nahas MK, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127(24):3004–3014. doi: 10.1182/blood-2015-08-664649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Y, Xu T, Chen J, Hu G, Lu Y. Intraspinal Ewing’s sarcoma/primitive neuroectodermal tumors. J Clin Neurosci. 2011;18:601–606. doi: 10.1016/j.jocn.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Argani P, Dehner L, Leuschner I. Mesenchymal tumors occurring mainly children. In: Moch H, Humphrey PA, Ulbright TM, Reuter VE, editors. WHO Classification of Tumors of the Urinary System and Male Genital Organs. Lyon: International Agency for Research on Cancer; 2016. pp. 54–58. [Google Scholar]
- 18.Kao Y-C, Sung Y-Z, Zhang L, et al. Recurrent BCOR internal tandem duplication and YHWAE-NUTM2B/E fusion in undifferentiated round cell sarcoma of infancy. Overlapping genetic features with clear cell sarcoma of kidney. Am J Surg Pathol. 2016;40:1009–1020. doi: 10.1097/PAS.0000000000000629. [DOI] [PMC free article] [PubMed] [Google Scholar]