Abstract
Formation of the bivalent interaction between the Notch intracellular domain (NICD) and the transcription factor CSL is a key event in Notch signaling because it switches Notch-responsive genes from a repressed state to an activated state. Interaction of the intrinsically disordered RAM region of NICD with CSL is thought to both disrupt binding of corepressor proteins to CSL and anchor NICD to CSL, promoting interaction of the ankyrin domain of NICD with CSL through an effective concentration mechanism. To quantify the role of disorder in the RAM linker region on the effective concentration enhancement of Notch transcriptional activation, we measured the effects of linker length variation on activation. The resulting activation profile has general features of a worm-like chain model for effective concentration. However, deviations from the model for short sequence deletions suggest that RAM contains sequence-specific structural elements that may be important for activation. Structural characterization of the RAM linker with SV-AUC and NMR spectroscopy reveals that the linker is compact and contains three transient helices and two extended and dynamic regions. To test if these secondary structure elements are important for activation, we made sequence substitutions to change the secondary structure propensities of these elements and measured transcriptional activation of the resulting variants. Substitutions to two of these nonrandom elements (helix 2, extended region 1) have effects on activation, but these effects do not depend on the nature of the substituting residues. Thus, the primary sequences of these elements, but not their secondary structures, are influencing signaling.
Keywords: Intrinsically disordered proteins, protein-protein interactions, reporter gene assays, NMR spectroscopy, analytical ultracentrifugation
Graphical abstract
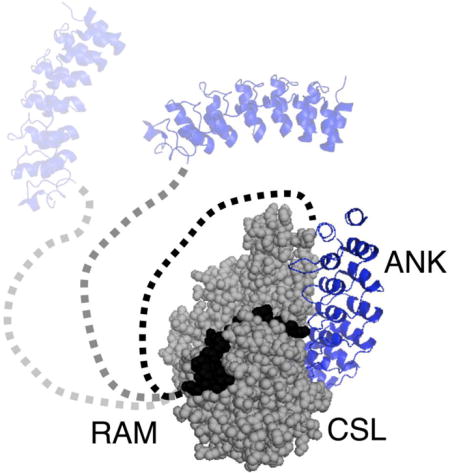
INTRODUCTION
The Notch pathway is a conserved signaling pathway in metazoans that is crucial to cell fate determination.1, 2 Errors in Notch signaling can lead to developmental disorders,3 adult-onset diseases4 and cancer.5, 6 The Notch receptor is a single-pass type I transmembrane receptor.7 In canonical Notch signaling, association of the extracellular region of the Notch receptor with DSL ligands attached to neighboring cells triggers proteolytic cleavage of the receptor, releasing the intracellular domain of the Notch receptor (NICD) from the membrane (Figure 1).2 Upon release, NICD translocates to the nucleus where it interacts with the transcription factor CSL (CBF-1/RBP-J, Su(H), Lag-1) and a coactivator protein from the Mastermind family (MAML) to form ternary complexes at the promoters of Notch target genes, such as HES and HEY in mammals.2 Ternary complex formation activates transcription of Notch target genes by displacing corepressor proteins and by recruiting transcription machinery to the target gene promoters.
Figure 1. Schematic of Notch signaling.
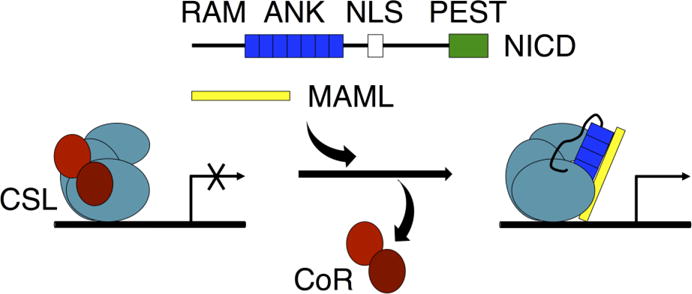
NICD (containing RAM, ANK, NLS, and PEST regions) enters the nucleus after it is cleaved from the Notch receptor upon receptor activation. The RAM region and ANK domain of NICD bind to CSL at the promoter region of Notch target genes. Binding of NICD to CSL disrupts binding of co-repressor (CoR) proteins and promotes binding of the co-activator protein MAML.
NICD contains the N-terminal approximately 135-residue intrinsically disordered region referred to as RAM (RBP-j associated molecule) that is followed by seven ankyrin repeats, nuclear localization sequences and a C-terminal PEST degradation sequence (Figure 1).2 NICD forms a bivalent interaction with CSL through the RAM and ankyrin (ANK) domains. The N-terminus of RAM, centered on the highly conserved ΦWΦP motif, binds with high affinity to the beta-trefoil domain (BTD) of CSL.8 ANK binds to the other two domains of CSL: the N-terminal domain (NTD) and the C-terminal domain (CTD).8–11 The interaction of ANK with CSL produces a cleft that interacts with the N-terminal helix of MAML to stabilize the ANK:CSL interaction.9, 10, 12
Compared to commonly studied intrinsically disordered protein (IDP)-mediated interactions, the RAM:CSL interaction is atypical. For some IDPs involved in cell signaling, interaction with a binding partner is coupled to the folding of secondary structure elements within the IDP.13 In contrast, much of RAM appears to remain disordered when bound to BTD, forming a “fuzzy” complex.10, 14 Additionally, the interactions of IDPs with their binding partners often have modest affinities, with dissociation constants in the micromolar range.15, 16 This allows for short-lived interactions that can easily tune signaling output. In contrast, the RAM:CSL interaction has a very high affinity (KD = 22 nM).17 This high affinity RAM:CSL interaction may be important for displacing corepressor proteins that bind to CSL and suppress transcription.18, 19
Additionally, the bivalent interaction of NICD with CSL may increase the effective concentration of ANK near the CTD and NTD of CSL to promote ANK:CSL interaction and promote active ternary complex formation.20, 21 A simple worm-like chain (WLC) polymer statistics model of the linker between RAM and ANK predicts that when RAM is bound to CSL, the effective concentration of ANK at its binding site is maximized when the linker is 80 to 100 residues in length. This length is approximately equal to the lengths of the linker regions of the Notch receptor paralogs.21 This model also predicts that shortening or lengthening the linker would decrease transcription activity by disrupting ternary complex formation.
To test the effective concentration model for bivalent NICD:CSL interaction, we measured transcription activities of NICD variants with sequence insertions and deletions in the RAM region. The activation profile is generally consistent with the WLC model of the disordered linker; however, it deviates from the model for short internal deletions. These deviations are likely due to specific sequence and/or structural elements within the RAM linker. To attempt to identify these elements, we used NMR spectroscopy and sedimentation velocity analytical ultracentrifugation (SV-AUC) to structurally characterize the isolated RAM region of NICD. We found several secondary structure elements in the region where short deletions produce variations in transcriptional activation. We tested the importance of these secondary structure elements to Notch signaling and find sensitivity to sequence changes in these regions, but not to secondary structure changes.
RESULTS
The effects of insertions and deletions in the RAM region on NICD transcriptional activation
Covalent connection between the N-terminal RAM binding site and ANK domain via the RAM linker has been shown to be important for transcriptional activation and has been proposed to increase the effective concentration of ANK near CSL.20–22 For this mechanism, linker length should modulate effective concentration. To test the importance of linker length in Notch signaling, we measured transcription activities of NICD variants containing sequence insertions and deletions in the RAM linker region using a dual-luciferase reporter assay in HeLa cells (Figure 2). The inserted and deleted sequences are distant from the ΦWΦP motif, so direct contacts between RAM and the BTD of CSL should be unaffected. For insertions, the duplicated linker sequence is the segment of the RAM linker with the lowest amount of sequence conservation among Notch homologs to minimize sequence-specific effects.
Figure 2. Transcription activities of RAM linker sequence deletions and duplications.
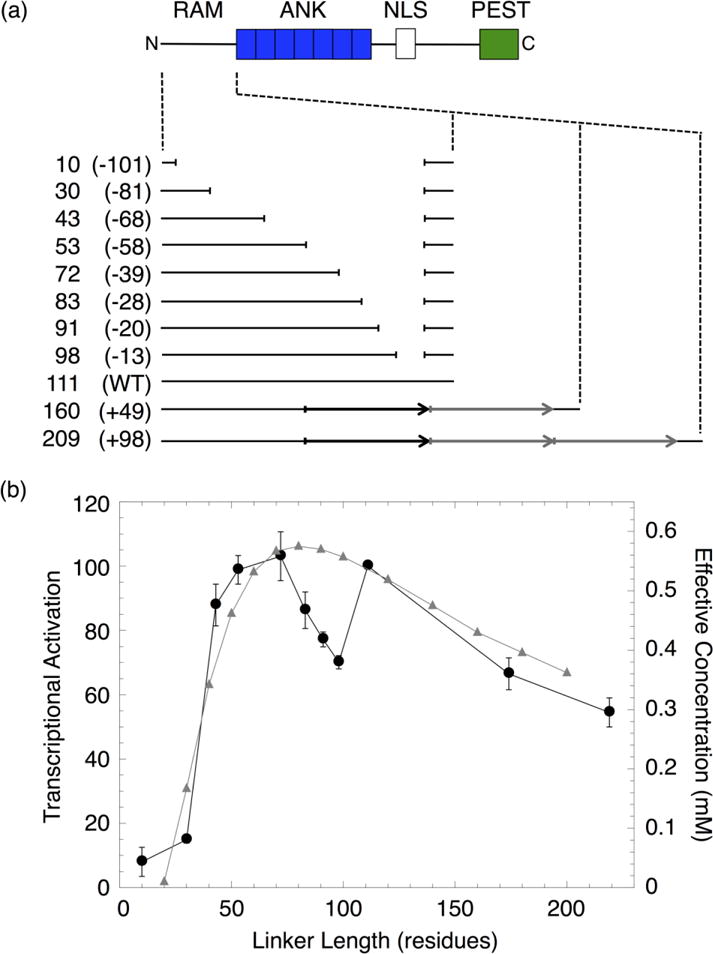
(a) Schematic of NICD and constructs with sequence deletions and duplications in the RAM region. Each construct is labeled with the length, in residues, of the RAM linker region and the number of residues deleted or inserted (in parentheses). The duplicated sequence (black arrow) was replicated once (+49) or twice (+98) (gray arrows). (b) NICD-mediated transcriptional activation (black) and effective concentration (gray) of constructs with different RAM linker lengths. Effective concentrations were calculated from the worm-like chain model.21
The transcription activities of the RAM insertion and deletion series show maximal transcriptional activation around the wild-type linker length. Activation decreases for both large increases and decreases in linker length. The decrease in activation is gradual as linker length increases, but is quite sharp as linker length decreases. Thus, activation is more tolerant of sequence insertions than deletions. The residual levels of activation (~10% of wild-type) obtained after removal of 81 or more residues may result from binding of the RAM and ANK domains of two separate NICD molecules to CSL (Figure S1).
This transcriptional activation profile is generally consistent with the effective concentration profile calculated from a WLC model proposed by Bertagna et. al.21 The maximum effective concentration for the WLC, using persistence and residue lengths of 4 and 3.8 Å respectively, is obtained around the wild-type linker length. Effective concentration for the WLC decreases broadly and sharply at shorter and longer lengths, respectively. Despite this overall agreement, there are significant discrepancies between the activation and effective concentration profiles for short deletions. Decreasing the 111-residue wild-type linker by 13 residues decreases transcriptional activation to 70% of the wild-type level. Activation remains below that of wild-type upon further deletions of 7 and 15 residues (91- and 83-residue linkers). Further deletions of 11 and 30 residues (72- and 53-residue linkers) restore transcriptional activation to that of wild-type. This variation would not be predicted from a simple statistical polymer model. Instead, this variation is likely the result of removing sequence-specific structural elements within the linker region that affect the dimensions of the RAM structural ensemble. Changes to the RAM structural ensemble from disrupting local structural elements may affect its ability to position the ANK near its binding site on CSL and thus decrease ternary complex formation and transcriptional activation. It is also possible that the sequence-specific elements in the linker interact directly with CSL and influence activation; however, interaction between the RAM linker and CSL has not been observed in crystallographic studies of the ternary complex.10
The intrinsically disordered RAM region contains three transient α-helices and two extended and dynamic regions
To identify sequence and structure elements within the RAM region of NICD that may be important for Notch transcriptional activation, we used NMR spectroscopy to resolve local structural and dynamic features of the isolated RAM region of the Notch receptor. The 15N-1H HSQC spectrum of the 133-residue RAM polypeptide shows narrow chemical shift dispersion in the 1H dimension, which is typical of disordered proteins (Figure 3a).23, 24 However, most peaks appear to be adequately resolved at 800 MHz for assignment by triple resonance methods. Using standard triple resonance experiments, we assigned 86% (100 residues) of the RAM non-proline backbone resonances. Repeating sequence motifs prevented assignment of the resonances for the first ten residues.
Figure 3. 15N-1H HSQC spectra of uniformly labeled 15N RAM.
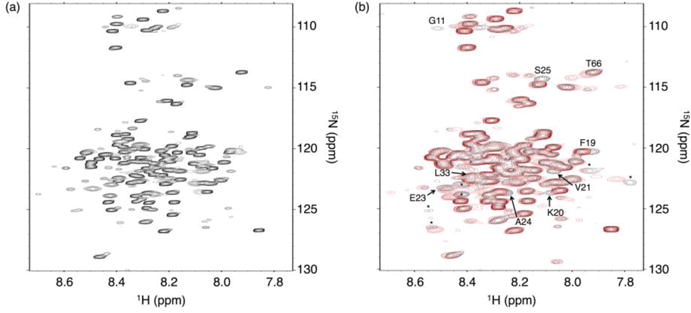
(a) Spectrum of 15N RAM at 800 MHz. Most resonances are reasonably well dispersed for an IDP, facilitating assignment with standard triple resonance experiments. (b) Overlay of 15N RAM spectra with (red) and without (black) a 1.4:1 molar excess of CSL at 600 MHz. Resonances that move upon RAM:CSL binding are labeled and correspond to residues at the RAM N-terminus. Unassigned resonances that move are labeled with an asterisk. Conditions: 25 mM sodium phosphate pH 6.5, 50 mM sodium chloride, 0.1 mM TCEP, and 1 mM EDTA at 20 °C.
To detect the residues of RAM that interact with CSL, we measured 15N-1H spectra of 15N RAM with and without a molar excess of CSL (Figure 3b). For the most part, chemical shift changes are restricted to the ΦWΦP region. Only one residue within the RAM linker (T66) shifts upon CSL binding. Therefore, the RAM:CSL interaction is localized to the RAM N-terminus and, with the possible exception of T66, does not involve highly specific interactions between CSL and the residues in the RAM linker. This observation is consistent with crystallographic studies of the NICD:CSL complex.10
In addition, the resonances of RAM are uniformly broader in the presence of CSL. If RAM was fully disordered and bound to CSL solely through the ΦWΦP region, then broadening would be most pronounced for resonances near the ΦWΦP region. Uniform broadening demonstrates that the linker resonances are affected by the increased molecular weight of the CSL complex. This is consistent with our observations that RAM is compact and has regions of structural bias in the unbound state.
To identify regions of secondary structure bias within RAM that may be important for transcriptional activation, we determined secondary Cα chemical shifts, 15N-1H amide residual dipolar couplings (RDC), and 1H-15N NOEs. These three quantities are commonly used to identify secondary structure bias in IDPs.23,24 Secondary Cα chemical shift values provide information on secondary structure due to differences in local bonding architecture for α-helical and β-strand structures. 15N-1H Amide RDC values report on secondary structure due to local alignment of the 15N-1H spin vector with the magnetic field.25 We used several different methods to analyze secondary chemical shifts. Although there are minor variations among the different prediction methods, the same general trends are observed (Figure 4a, S2). Most of the RAM secondary Cα chemical shifts are close to zero and most of the 15N-1H RDC values are slightly negative, consistent with RAM being largely devoid of secondary structure.25 However, there are three regions of RAM that contain uniformly positive Cα secondary chemical shift and 15N-1H RDC values (identified as H1, H2 and H3, Figure 4). These values indicate bias toward α-helical structure.25, 26 Two additional regions of RAM (identified as E1 and E2, Figure 4) have continuously negative Cα secondary chemical shift and 15N-1H RDC values. These values indicate bias toward extended conformations. Interestingly, some of E2 becomes α-helical upon interaction of NICD with CSL.10
Figure 4. Local structural features of RAM determined by NMR spectroscopy.
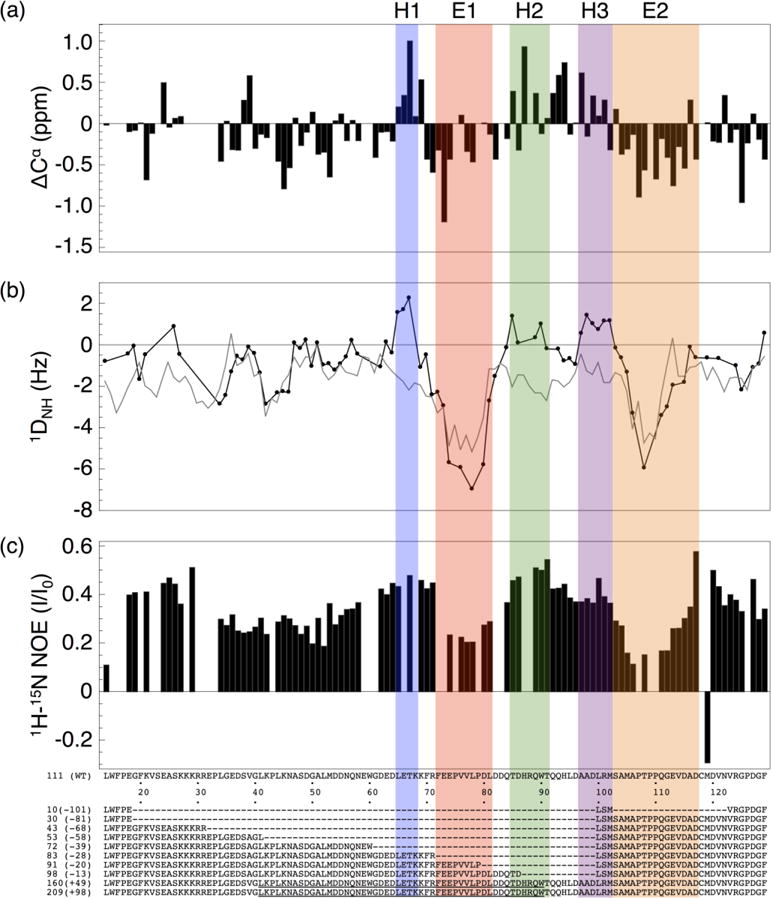
(a) Cα Chemical shift deviations from random coil values.26 (b) 15N-1H Amide residual dipolar coupling values (black circles), and values predicted by Flexible-Meccano27 (gray). (c) 1H-15N Heteronuclear NOE values. Based on these three parameters, we identify three transiently helical segments H1-H3 (blue, green and purple), and two extended and dynamic segments E1 and E2 (green and orange). The first twelve and last five residues of RAM are not shown because of missing assignments.
To highlight the regions of secondary structural bias, the RAM 15N-1H RDC values can be compared to calculated random coil RDC values generated by the program Flexible-Meccano.27 Flexible-Meccano models an excluded volume random coil structural ensemble for a given polypeptide sequence and calculates average residual dipolar coupling values from this ensemble. The random coil RDC values that Flexible-Meccano calculates for RAM are generally similar to the experimental RDC values over much of the RAM region (Figure 4b). However, there are significant deviations in the regions with α-helical bias, emphasizing that these sequences have non-random structures.
To characterize the dynamic properties of RAM, we measured 1H-15N NOE intensities. For backbone amide 1H-15N pairs, NOE intensities are sensitive to the local reorientational dynamics of a specific peptide unit in the picosecond to nanosecond timescale.28 Ordered residues have higher NOE intensity ratios than dynamic residues. In general, the RAM 1H-15N NOE ratios are higher than the values typical for disordered regions, which tend to be negative (Figure 4c).29,30 This suggests that RAM is overall less dynamic than a typical IDP and may also explain the uniform resonance broadening of RAM upon interaction with CSL. Within the RAM sequence, the transiently helical regions have relatively high NOE values. This indicates that the helical regions are relatively rigid on the picosecond to nanosecond timescale. The two extended regions have the lowest 1H-15N NOE values observed for RAM, showing that these regions are the most dynamic on this timescale.
Sequence substitutions to regions of RAM with transient secondary structure have a modest effect on transcription activity
To test the importance of the RAM secondary structure elements on Notch transcriptional activation, we substituted these regions with sequences designed to alter secondary structure composition. We made block sequence substitutions with either alanine residues (to increase helicity), glycine residues (to disrupt helicity), or proline residues (to decrease flexibility and disrupt helicity, Figure 5a).31, 32 Region E2 was not substituted because it is close to ANK and substitutions may directly affect the interaction between NICD and CSL. All of the resulting NICD variants were tested for their ability to activate transcription through CSL.
Figure 5. Transcription activities of block sequence substitutions in regions of RAM containing residual secondary structure.
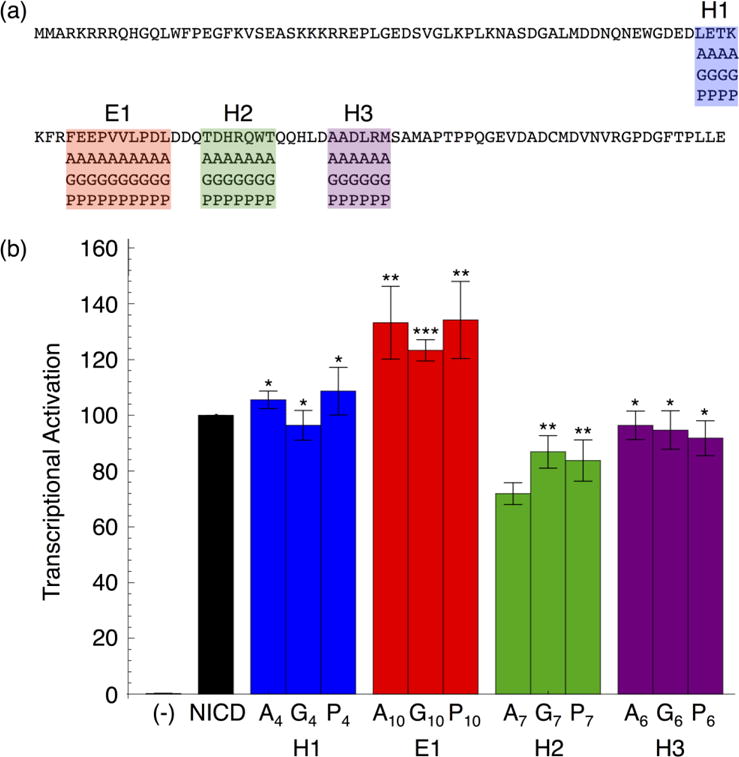
(a) RAM sequence with block substitution boundaries. Each RAM variant contains one helical or extended region substituted with alanine, glycine, or proline residues. (b) Transcriptional activation of RAM sequence substitutions in NICD. Substitutions in the E1 and H2 regions change transcriptional activation, but this change does not depend on secondary structure type. Error bars show the standard error of the mean and statistical significance compared to NICD is labeled as ***P < 0.001, **P < 0.01, and *P < 0.05.
Overall, the transcription activities of these RAM secondary structure variants are similar to wild-type levels (Figure 5b). Substitutions to regions H1 and H3 result in transcription activities that are within error of wild-type levels. Therefore, these regions do not affect activation, even though the resonance for T66, which is in H1, shifts in the 15N-1H HSQC spectrum of 15N RAM bound to CSL. Substitutions to region H2 result in a ~30% decrease in transcription activation. NICD variants with substitutions to region E1 exhibit a ~30% increase in transcription activation. Importantly, these changes are independent of the sequences of the substitutions. Therefore, the sequences but not the secondary structure biases of these two regions appear to modulate signaling.
The effects of these sequence substitutions on transcription activity are consistent with the effects of sequence deletion. Deletion of the H2 region in the 98(−13) RAM linker decreases activation, as does sequence substitutions to H2. Activation is restored when the E1 region is deleted in the 72(−39) linker; likewise, sequence substitution of E1 increases transcriptional activation. These observations suggest that there are positive and negative control regions within the RAM linker.
The RAM structural ensemble is compact due to electrostatic interactions
The above studies demonstrate that specific sequence elements that are distant from the CSL binding site affect transcriptional activation, but they do not conform to a simple secondary structure code. To examine higher order structural properties that may mediate these sequence-specific effects, we explored the hydrodynamic properties of RAM using sedimentation velocity analytical ultracentrifugation (SV-AUC, Figure S3). In addition to detecting possible oligomerization, SV-AUC reports on shape and compaction, which are sensitive to intra-chain contact and electrostatic interactions. Sedimentation coefficients of RAM were determined by direct boundary fitting of SV-AUC data to a single-species model with SEDANAL.33 Sedimentation coefficients were used to calculate the hydrodynamic radii (RH) of RAM using the Svedberg equation combined with Stokes’ Law (Figure 6, Table S1).
Figure 6. Hydrodynamic radii (RH) of RAM determined by SV-AUC at various concentrations of NaCl.
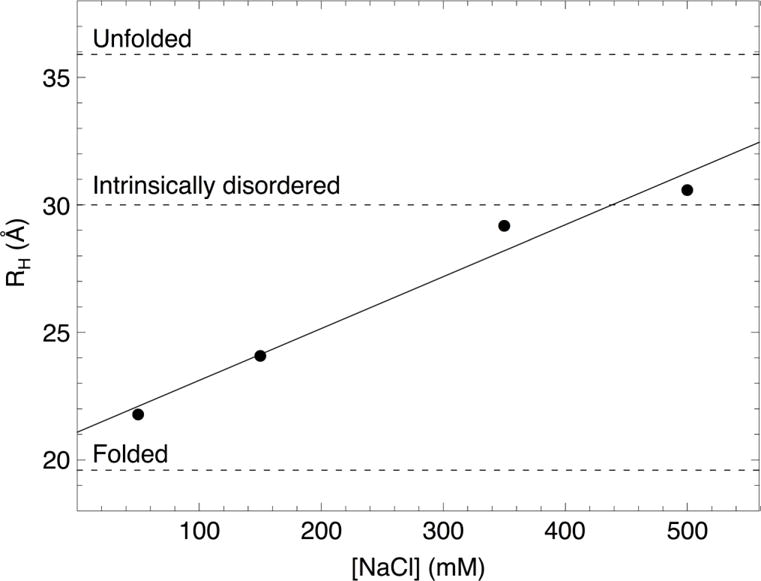
The solid line is a linear fit to the data. The dashed lines are RH values calculated from scaling relationships for folded and unfolded proteins48 and for intrinsically disordered proteins39 of the same molecular weight as RAM at physiological salt concentration.
The RH values for RAM are larger than expected for a folded protein, which is consistent with RAM being largely disordered (Figure 6). However, the RH value for RAM increases significantly with salt concentration. This suggests that stabilizing electrostatic interactions lead to compact configurations at low salt concentrations. At 150 mM NaCl, RAM is significantly more compact than expected for an intrinsically disordered protein of the same molecular weight.
DISCUSSION
Many IDPs, like the RAM region of NICD, bind target proteins to modulate cell signaling pathways. The RAM:CSL interaction is a key component of Notch ternary complex formation that leads to activation of downstream genes. Here we test various mechanisms by which the RAM linker may modulate Notch signaling, ranging from polymer statistical effects to local sequence effects. By combining structural insights at local (NMR) and global (SV-AUC) levels with functional studies in mammalian cells, we find that the RAM linker has a significant amount of non-random structure and that some of these structured regions contribute to signaling activity.
Both polymer statistics and local sequence effects in RAM influence Notch signaling
The overall agreement between transcriptional activation from our linker deletion and insertion series and the effective concentration estimates from the WLC model of Notch transcriptional activation (Figure 2) supports the idea that the RAM linker promotes interaction by optimizing effective concentration. However, we also see sharp changes in activation resulting from short deletions of linker length. The effective concentration model predicts minimal effects for the deletion of short, random coil sequences because the ability of the linker to position ANK near its CSL binding site should not be significantly altered. Rather, this sharp decrease in activation is likely due to the removal of sequence-specific elements within the linker region that impact NICD:CSL interaction. It is also possible that the insertions and deletions influence transcriptional activation because they affect NICD nuclear transport and/or post-translational modifications.
These sequence-specific effects may come from the influence of these segments on the dimensions of the RAM structural ensemble rather than from specific interactions between these segments and CSL. The 15N-1H HSQC spectrum of 15N RAM bound to CSL (Figure 3b) and crystallographic studies by Wilson and Kovall10 show that RAM does not specifically interact with CSL outside of the well-characterized N-terminal ΦWΦP binding site. Instead of disrupting interactions between RAM and CSL, removing sequence-specific elements within the linker may cause the chain to expand, resulting in decreased effective concentration and reduced activation. Deleting additional linker residues may restore wild-type levels of activation by significantly reducing the contour length, so that ANK is tethered closer to its CSL binding site.
The sequence-specific structural elements within the RAM linker region identified by solution NMR spectroscopy overlap the short deletions that modulate Notch transcriptional activation. Our block-substitution studies of these structural elements show that while some of these elements (E1 and H2) do contribute to signaling, these perturbations are independent of changes in secondary structure propensity. This demonstrates that there is not a secondary structure code for activation within the RAM linker.
This behavior differs from several systems where IDP function is affected by stabilization of transient α-helices within the IDP. For the Sgs1:Top3/Rmi1 and p53:mdm2 systems, Daughdrill and coworkers showed that enhancing or disrupting residual helicity correspondingly changes the binding affinities of the IDPs for their partners and alters functional effects in vivo.30, 34 For the PUMA:MCL-1 system, Rogers and Clarke showed that disrupting residual helicity in PUMA decreases its binding affinity for MCL-1.35
Our observation that sequence changes in RAM secondary structure regions affect Notch transcriptional activation is similar to observations from studies of the p27:cyclin A-Cdk2 system by Bienkiewicz et. al.36 and Otieno and Kriwacki.37 These studies showed that either enhancing or disrupting residual helicity in p27 decreases its ability to inhibit cyclin A-cdk2 kinase activity. The two studies described conflicting results of enhancing helicity in p27 on binding affinity, but both showed that disrupting helicity in p27 decreases its binding affinity for cyclin A/cdk2.
In these examples, helical propensity within the IDP affects binding affinity and function because the transient α-helices are directly involved in protein-protein interactions and become fully helical when bound to their partners. For Notch signaling, transcriptional activation is affected by bias toward both helical and extended structures within RAM that are distant in sequence from the CSL binding site. In addition, these regions do not appear to undergo structural changes upon interaction of RAM and CSL because we find almost no chemical shift changes for the linker region in the 15N-1H HSQC spectrum of 15N RAM bound to CSL (Figure 3b). Because the regions of RAM with structural bias do not specifically interact with CSL or seem to change their structures upon RAM:CSL binding, they may affect NICD:CSL interaction by influencing the overall structure of the disordered polypeptide.
RAM compaction may be important to Notch signaling
To gain insight into other structural properties of RAM that may be important for Notch transcriptional activation, we characterized RAM compaction and electrostatic interactions using SV-AUC. We found that RAM is significantly more compact than a typical IDP of its molecular weight. The high salt dependence of RH from 50–500 mM NaCl suggests that compact conformations are stabilized by favorable electrostatic interactions and that these are partially screened at higher concentrations of salt. RAM has a large number of charged residues (35% D, E, K, R). All-atom simulations and experiments have shown that the compaction of IDPs is strongly influenced by the patterning of these charged residues.38, 39, 40 By manipulating charge patterning, we should be able to test the relationship between sequence, global structure, and transcriptional activation. Local structural features, such as secondary structure elements, may also affect the compaction of RAM. RAM compaction may be important for transcriptional activation because it may influence the ability of RAM to enhance ANK binding through the effective concentration mechanism.
MATERIALS AND METHODS
Expression and purification of unlabeled, 15N-, and 15N, 13C-RAM
The RAM polypeptide used in these experiments spans residues 1758 to 1890 from human Notch1. The polypeptide has an extra Met residue on the N-terminus and Leu and Glu residues on the C-terminus resulting from the cloning procedure used to insert the gene fragment into a modified pET vector. RAM was expressed in BL21(DE3*) Escherichia coli cells. Unlabeled and 15N-RAM were grown in auto-induction media supplemented with 0.05 g/L kanamycin sulfate and 15NH4Cl when appropriate.41 15N, 13C-RAM was grown in M9 minimal media supplemented with 15NH4Cl, 0.05 g/L kanamycin sulfate, 2 mM MgSO4, 100 μM CaCl2, and 2.0 g/L 13C D(+)glucose.42 All cultures were grown at 37 °C to an OD600 of 0.6. For M9 cultures, isopropyl-beta-D-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. Protein expression was induced overnight after lowering the growth temperature to 20 °C for all cultures. Bacteria were collected by centrifugation and were stored at −80 °C.
Cell pellets were resuspended in 50 mL water and 5 cOmplete mini EDTA-free protease inhibitor tablets (Roche) per liter of culture. Resuspended cells were lysed by boiling in a water bath for 20 min with occasional stirring. Cooled cell lysates were clarified by centrifugation. Lysis supernatants were diluted 1:1 with 50 mM sodium phosphate pH 7.0 and 2 mM β-mercaptoethanol.
RAM was purified from the diluted lysis supernatant by applying it to a bench-top column with Q Sepharose™ anion exchange resin (Sigma) equilibrated with 25 mM sodium phosphate pH 7.0, 100 mM NaCl, 0.1 mM TCEP. The protein-bound column was washed with 5 column volumes (CV) of equilibration buffer. RAM was eluted with 3 CV of 25 mM sodium phosphate pH 7.0, 300 mM NaCl, 0.1 mM TCEP. The eluate was concentrated to a volume of 5 mL and exchanged into buffer containing 25 mM sodium phosphate pH 7.0, 150 mM NaCl, 1 mM β-mercaptoethanol. The concentrated elution was further purified by chromatography on a HiPrep™ 26/60 Sephacryl™ S-100 size-exclusion column (GE Healthcare Life Sciences) using an Äkta FPLC (GE Healthcare Life Sciences).
Expression and purification of CSL
The CSL construct spans residues 25 to 449 of human CSL isoform 1 and was inserted into the pMAL-c2x vector modified with a His12 tag on the N-terminus of MBP and a tobacco etch virus (TEV) protease recognition site between MBP and CSL. MBP-CSL fusion protein was expressed in BL21(DE3*) E. coli cells grown in LB media to an O.D. of 0.6 to 0.8. Expression was induced O/N at 20 °C with the addition of 0.8 mM IPTG. Bacteria were collected by centrifugation and lysed by homogenization after resuspension in 25 mM Tris-HCl pH 8.0, 50 mM NaCl, 0.5 mM TCEP. Cell lysates were clarified by centrifugation. Lysis supernatant was treated with DNase I and Benzonase (Sigma) for 1 hr at room temperature. MBP-CSL was purified from the treated lysate by applying it to a bench-top column with Ni-NTA agarose resin equilibrated in 25 mM Tris-HCl pH 8.0, 500 mM NaCl, 0.5 mM TCEP. The protein-bound column was washed with 5 CV of equilibration buffer with 100 mM imidazole. MBP-CSL was eluted with 3 CV of equilibration buffer with 750 mM imidazole. The eluate was dialyzed O/N with His6-tagged TEV protease into cation exchange buffer A (25 mM sodium phosphate pH 7.0, 100 mM NaCl, and 0.5 mM TCEP).
CSL was purified from the dialysate by applying it to a bench-top with Ni-NTA agarose resin equilibrated cation exchange buffer A. The column was washed with 5 CV cation exchange buffer A with 100 mM imidazole. The column flow-through and wash were collected and applied to a HiTrap™ SP HP cation exchange column (GE Healthcare Life Sciences) equilibrated with cation exchange buffer A. CSL was eluted with a linear NaCl gradient (0.1 mM to 1 M).
Cell culture
The NICD construct used in these experiments spans residues 1758–2555 of human Notch1 in a pcDNA3.1(+) vector. The NICD internal deletion constructs were generated by removing the RAM region from the NICD vector and inserting PCR products with the desired sequences. These constructs have the following boundaries: wild-type NICD (1758–2555), NICD−13 (1758–1839, 1853–2555), NICD−20 (1758–1832, 1853–2555), NICD−28 (1758–1824, 1853–2555), NICD−39 (1758–1813, 1853–2555), NICD−58 (1758–1794, 1853–2555), NICD−68 (1758–1784, 1853–2555), NICD−81 (1758–1771, 185–2555), NICD−101 (1758–1772, 1874–2555), and NICD−RAM (1853–2555). NICD internal duplication constructs were generated by duplicating and triplicating residues 1794–1843 to generate the +49 and +98 linker. Deletion constructs contain point substitutions at deletion boundaries. NICD secondary structure sequence substitutions were produced by substituting residues 1822–1825 (H1), 1829–1838 (E1), 1842–1848 (H2) and 1854–1859 (H3) with alanine, glycine, or proline residues through QuikChange mutagenesis (Agilent) or synthetic gene fragments.
HeLa cells were maintained in Dulbecco’s Modified Eagle Medium with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin at 37 °C. Cells were seeded 24 hours prior to transfection in 24 well-plates at a density of 2×105 cells per well. For insertion and deletion experiments, cells were transfected with 225 ng TP1-luc reporter plasmid (firefly luciferase under the control of a 10x CSL binding site promoter), 75 ng Renilla transfection control plasmid (Promega), 60 ng NICD-expressing plasmid, and 140 ng empty pcDNA3.1(+) vector. For secondary structure sequence substitution experiments, cells were transfected with 225 ng TP1-luc reporter plasmid, 75 ng Renilla transfection control plasmid, and 100 ng NICD-expressing plasmid. An empty plasmid was used as a negative control for all experiments. Transfections were performed using Lipofectamine LTX (Life Technologies) according to the manufacturer’s instructions. Cells were harvested 40–44 hrs after transfection by lysing in Passive Lysis Buffer and were assayed using the Dual-luciferase Reagent on a GloMax Multi microplate luminometer following the manufacturer’s directions (Promega). For each transfection, luciferase activities were measured in quadruplicate. For each construct, transfections were repeated at least three times. The reported activities are relative to wild-type NICD and are reported as the mean ± standard error of the mean. For block sequence substitutions, Student’s t-test was used to compare mean values with NICD and one-way ANOVA was applied with a significance value of p < 0.001.
NMR spectroscopy
Double- (15N-1H HSQC) and triple-resonance spectra (HNCACB, HNCO, HN(CA)CO, HN(CA)N, HN(COCA)N, 15N-edited HMQC-NOESY-HSQC) for backbone assignments were collected on samples of uniformly 15N- or 15N, 13C-labeled RAM at concentrations of 1–2 mM in 25 mM sodium phosphate pH 6.5, 50 mM NaCl, 0.1 mM TCEP, 1 mM EDTA, 5% D2O. All spectra were collected at 20 °C on a Varian Inova 800 MHz spectrometer equipped with a cryoprobe. Data were processed with NMRpipe43 and resonance assignments were made with the aid of CARA.44 Resonance assignments are deposited in the BMRB (accession number 11593).
15N-1H RDC values were determined by recording two sets of IPAP spectra in liquid crystalline alignment media45 and isotropic solution. RDC values that differ by more than 1.1 Hz between the two experiments are not reported. Predicted random coil 15N-1H RDC values were calculated in Flexible-Meccano v1.1 from 133,000 conformers of the RAM sequence.27
1H-15N Heteronuclear NOE values were determined as the ratios of the intensities of resonances in the presence and absence of proton saturation.28 The reported ratios are an average of two experiments. Ratios that differ by more than 0.1 between the two experiments are not reported.
Sedimentation Velocity Analytical Ultracentrifugation
RAM was dialyzed extensively in 25 mM sodium phosphate pH 7.0, 50–500 mM NaCl, 0.1 mM TCEP, 1 mM EDTA. Dialyzed RAM was diluted with dialysis buffer to 100 μM. This sample was serially diluted with dialysis buffer to produce additional samples with 75 μM, 50 μM and 25 μM RAM. The four samples and dialysis buffer references were loaded into AUC cells containing SedVel60K 1.2 cm meniscus-matching centerpieces (SpinAnalytical) and sapphire windows. The rotor containing the AUC cells was spun at 50,000 rpm to match the sample and reference menisci and to adjust the interference optics settings of the Beckman XL-I centrifuge. The rotor was removed to mix the samples and was returned to the centrifuge to equilibrate to 20 °C for at least 1.5 hr. Sedimentation velocity experiments were run at 50,000 rpm collecting interference scans every 30 s for 8 hr.
Sedimentation g(s*) distributions were generated in SEDANAL33 from a set of scans captured immediately after the sedimentation boundary had cleared the meniscus. Boundaries captured later in the experiment were very broad due to diffusion. The selected scans were used for direct boundary fitting with the Lamm equation in SEDANAL to determine the sedimentation coefficient and protein concentrations. The RAM molecular weight and partial specific volume were calculated from the RAM sequence in SEDNTERP46 and were held constant in the fits, along with the refractive index increment. The Δc/Δt curves for all four protein concentrations were fit simultaneously.
Fitted sedimentation coefficients were converted to hydrodynamic radii (RH) values by substituting Stoke’s law into the Svedberg equation47 to obtain the following equation:
RH values can be calculated by substituting the measured sedimentation coefficients (s), molecular weight (M), partial specific volume , solution density (ρ), Avogadro’s number (NA), and solution viscosity (η). The RAM molecular weight and partial specific volume along with solvent densities and viscosities were determined in SEDNTERP.
Supplementary Material
HIGHLIGHTS.
The disordered RAM region of NICD interacts with CSL during Notch activation
RAM linker length variation shows features consistent with chain statistics
Additional sequence elements in RAM have specific effects on Notch activation
RAM transient secondary structure has modest effects on Notch activation
IDP structural elements distant from interaction sites can affect IDP function
Acknowledgments
We thank The Johns Hopkins University Biomolecular NMR Center and the Center for Molecular Biophysics for providing facilities and resources. We thank Dr. Jacob Marold for help with AUC experiments. This work was supported by NIH grant GM060001 to D.B.
ABBREVIATIONS
- RAM
RBP-j-associated molecule
- NICD
Notch intracellular domain
- CSL
CBF-1/RBP-j, Su(H), Lag-1
- WLC
worm-like chain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12:9R–13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 4.Louvi A, Arboleda-Velasquez J, Artavanis-Tsakonas S. CADASIL: a critical look at a Notch disease. Dev Neurosci. 2006;28:5–12. doi: 10.1159/000090748. [DOI] [PubMed] [Google Scholar]
- 5.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 6.Ridgway J, Zhang G, Wu Y, Stawicki S, Liang W, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I, Singh M, Chien M, Tan C, Hongo JS, de Sauvage F, Plowman G, Yan M. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–7. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 7.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 8.Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL/intracellular Notch1/Mastermind-like 1 transcriptional activation complex. J Biol Chem. 2003;278:21232–9. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- 9.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–83. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 10.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–96. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 11.Ilagan MXG, Lim S, Fulbright M, Piwnica-Worms D, Kopan R. Real-time imaging of notch activation with a luciferase complementation-based reporter. Sci Signal. 2011;4:rs7. doi: 10.1126/scisignal.2001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SH, Wales TE, Nam Y, O’Donovan DJ, Sliz P, Engen JR, Blacklow SC. Conformational locking upon cooperative assembly of notch transcription complexes. Structure. 2012;20:340–9. doi: 10.1016/j.str.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Shammas SL, Rogers JM, Hill SA, Clarke J. Slow, reversible, coupled folding and binding of the spectrin tetramerization domain. Biophys J. 2012;103:2203–14. doi: 10.1016/j.bpj.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Bio. 2014;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008;283:14781–91. doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, Honjo T. Regulation of Marginal Zone B Cell Development by MINT, a Suppressor of Notch/RBP-J Signaling Pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- 19.Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–77. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch 1 transcription complexes. J Mol Biol. 2008;376:131–40. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertagna A, Toptygin D, Brand L, Barrick D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochem Soc Trans. 2008;36:157–66. doi: 10.1042/BST0360157. [DOI] [PubMed] [Google Scholar]
- 22.Johnson SE, Barrick D. Dissecting and circumventing the requirement for RAM in CSL-dependent Notch signaling. PloS One. 2012;7:e39093–e39093. doi: 10.1371/journal.pone.0039093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyson HJ, Wright PE. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv Protein Chem. 2002;62:311–40. doi: 10.1016/s0065-3233(02)62012-1. [DOI] [PubMed] [Google Scholar]
- 24.Kosol S, Contreras-Martos S, Cedeño C, Tompa P. Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules. 2013;18:10802–28. doi: 10.3390/molecules180910802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen MR, Markwick PRL, Meier S, Griesinger C, Zweckstetter M, Grzesiek S, Bernadó P, Blackledge M. Quantitative determination of the conformational properties of partially folded and intrinsically disordered proteins using NMR dipolar couplings. Structure. 2009;17:1169–85. doi: 10.1016/j.str.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Wishart D, Sykes B. The 13C Chemical-Shift Index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:10. doi: 10.1007/BF00175245. 1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 27.Bernadó P, Blanchard L, Timmins P, Marion D, Ruigrok RWH, Blackledge M. A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc Natl Acad Sci USA. 2005;102:17002–7. doi: 10.1073/pnas.0506202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay J, Kay LE. Backbone Dynamics of a Free and a Phosphopeptide-Complexed Src Homology 2 Domain Studied by 15N NMR Relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 29.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol. 2004;11:358–64. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy JA, Daughdrill GW, Schmidt KH. A transient α-helical molecular recognition element in the disordered N-terminus of the Sgs1 helicase is critical for chromosome stability and binding of Top3/Rmi1. Nucleic Acids Res. 2013;41:10215–27. doi: 10.1093/nar/gkt817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–7. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan L, Tong Y, Xie T, Wang M, Wang J. Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease. Biochemistry. 2007;46:11504–13. doi: 10.1021/bi7009794. [DOI] [PubMed] [Google Scholar]
- 33.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 34.Borcherds W, Theillet F, Katzer A, Finzel A, Mishall KM, Powell AT, Wu H, Manieri W, Dieterich C, Selenko P, Loewer A, Daughdrill GW. Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat Chem Biol. 2014;10:1000–2. doi: 10.1038/nchembio.1668. [DOI] [PubMed] [Google Scholar]
- 35.Rogers JM, Wong CT, Clarke J. Coupled folding and binding of the disordered protein PUMA does not require particular residual structure. J Am Chem Soc. 2014;136:5197–200. doi: 10.1021/ja4125065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bienkiewicz EA, Adkins JN, Lumb KJ. Functional Consequences of Preorganized Helical Structure in the Intrinsically Disordered Cell-Cycle Inhibitor p27Kip1. Biochemistry. 2002;41:752–759. doi: 10.1021/bi015763t. [DOI] [PubMed] [Google Scholar]
- 37.Otieno S, Kriwacki R. Probing the Role of Nascent Helicity in p27 Function as a Cell Cycle Regulator. PLoS ONE. 2012;7:e47177–e47177. doi: 10.1371/journal.pone.0047177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc Natl Acad Sci USA. 2013;110:13392–7. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh JA, Forman-Kay J. Sequence determinants of compaction in intrinsically disordered proteins. Biophys J. 2010;98:2383–90. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller-Späth S, Sorrano A, Hirschfeld V, Hofmann H, Rüegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci U S A. 2010;107:2203–14. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Russell D. Molecular cloning: A laboratory manual. 3rd. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Masse JE, Keller R. AutoLink: automated sequential resonance assignment of biopolymers from NMR data by relative-hypothesis-prioritization-based simulated logic. J Mag Res. 2005;174:133–51. doi: 10.1016/j.jmr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 45.Rückert M, Otting G. Alignment of Biological Macromolecules in Novel Nonionic Liquid Crystalline Media for NMR Experiments. J Am Chem Soc. 2000;122:7793–97. [Google Scholar]
- 46.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. In: Analytical ultracentrifugation in biochemistry and polymer science. Harding SE, Rowe AJ, Horton JC, editors. The Royal Society of Chemistry; Cambridge, U.K.: 1992. pp. 90–125. [Google Scholar]
- 47.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation: A tutorial review. Prot Sci. 2002;11:2067–79. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–31. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


