Abstract
Triple-negative breast cancers have unfavorable outcomes due to their inherent aggressive behavior and lack of targeted therapies. Breast cancers occurring in BRCA1 mutation carriers are mostly triple-negative and harbor homologous recombination deficiency, sensitizing them to inhibition of a second DNA damage repair pathway by, e.g., PARP inhibitors. Unfortunately, resistance against PARP inhibitors in BRCA1-deficient cancers is common and sensitivity is limited in BRCA1-proficient breast cancers. RK-33, an inhibitor of the RNA helicase DDX3, was previously demonstrated to impede non-homologous end-joining repair of DNA breaks. Consequently, we evaluated DDX3 as a therapeutic target in BRCA pro- and deficient breast cancers and assessed whether DDX3 inhibition could sensitize cells to PARP inhibition. High DDX3 expression was identified by immunohistochemistry in breast cancer samples of 24% of BRCA1 (p = 0.337) and 21% of BRCA2 mutation carriers (p = 0.624), as compared to 30% of sporadic breast cancer samples. The sensitivity to the DDX3 inhibitor RK-33 was similar in BRCA1 pro- and deficient breast cancer cell lines, with IC50 values in the low micromolar range (2.8–6.6 μM). A synergistic interaction was observed for combination treatment with RK-33 and the PARP inhibitor olaparib in BRCA1-proficient breast cancer, with the mean combination index ranging from 0.59 to 0.62. Overall, we conclude that BRCA pro-and deficient breast cancers have a similar dependency upon DDX3. DDX3 inhibition by RK-33 synergizes with PARP inhibitor treatment, especially in breast cancers with a BRCA1-proficient background.
Keywords: DDX3, DEAD box RNA helicase, PARP inhibitor, DNA damage repair, BRCA, Breast cancer
Introduction
Increased genomic instability is one of the underlying hallmarks of cancer [1]. Cancer cells often acquire a deficiency in one of the DNA damage repair (DDR) pathways, to allow continued proliferation in the presence of genetic aberrations. This leads to a greater dependency on the remaining pathways to deal with endogenous and exogenous DNA damage [2, 3], which is the principle behind synthetic lethality of pharmacologic PARP inhibition in cancers with a BRCA1/BRCA2 mutation.
Women harboring a germline mutation in the BRCA1 or BRCA2 genes are at high risk of developing breast cancer, due to a deficiency in a DNA double-strand break (DSB) repair mechanism, homologous recombination (HR) [4]. BRCA1-related breast cancers are mostly estrogen receptor, progesterone receptor and HER2/neu negative (triple negative; TN) [5]. Patients with TN breast cancer (TNBC) have an unfavorable prognosis, due to the inherent aggressive behavior of this molecular subtype and the lack of targeted therapies [6]. PARP inhibitors, such as olaparib, inhibit base excision repair (BER), a single-strand break (SSB) repair mechanism, and have shown great promise in the treatment of tumors with a BRCA1 or BRCA2 mutation. However, a significant proportion of these patients show primary resistance [7]. Although only 5–10% of TNBC occurs in patients with a germline BRCA1 mutation, BRCA-proficient TNBCs are also characterized by impairments of DDR pathways. However, the effect of PARP inhibitors as a monotherapy in this group of patients is limited [8]. Therefore, development of new treatment strategies—specifically targeting BRCA1-related—and TNBC is urgently required.
DDX3, also known as DDX3X, is DEAD box RNA helicase that has been associated with several cytosolic steps of mRNA processing [9] and plays an oncogenic role in the development of breast [10] and several other types of cancer [11–14]. DDX3 was found to have anti-apoptotic properties [15] and to stimulate cell cycle progression [11, 12], migration [10] and invasion [16]. In addition, DDX3 was shown to be upregulated in TNBC [17]. RK-33 was developed as a small molecule inhibitor of DDX3 and showed promising preclinical activity as a radiosensitizer in models of lung [11] and prostate cancer [14]. Interestingly, the radiosensitizing capacities of RK-33 were attributed to inhibition of non-homologous end joining (NHEJ), a second DNA DSB repair mechanism. Inhibition of NHEJ makes RK-33 an interesting candidate for the treatment of BRCA1-related breast cancer and TNBC. Given their preexisting DDR deficiency, we hypothesized that BRCA-deficient breast cancers might be dependent on DDX3 and therefore sensitive to DDX3 inhibition with RK-33. In addition, DDX3 inhibition could potentially sensitize cells to PARP inhibition. This study therefore focused on evaluating DDX3 as a therapeutic target in BRCA pro- and deficient breast cancer and assesses whether there is a potential synergistic interaction between DDX3 inhibition and PARP inhibition.
Materials and methods
Patient samples
Archived formalin-fixed paraffin-embedded breast cancer samples from 103 germline BRCA1 mutation carriers and 29 germline BRCA2 mutation carriers were previously processed into a tissue microarray (TMA) and compared against a TMA with 265 consecutive breast cancer cases not known to bear mutations in these genes (further denoted “sporadic”) [18]. All patients in the hereditary group had been referred to the clinical genetics department of one of the three academic hospitals in the Netherlands (VUMC, UMC Utrecht and UMC Groningen) and tissue was retrieved from the pathology departments of these hospitals or of local surrounding hospitals. All TMAs included multiple cores per patient. As we used anonymous archival leftover pathology material, no ethical approval or informed consent is required according to Dutch legislation [19], as this use of redundant tissue for research purposes is part of the standard treatment agreement with patients in our hospitals [20].
Immunohistochemistry
Four-μm-thick sections were deparaffinized in xylene and rehydrated in decreasing ethanol dilutions. Endogenous peroxidase activity was blocked with 1.5% hydrogen peroxide buffer for 15 min and was followed by antigen retrieval by boiling for 20 min in 10 mM citrate buffer (pH 6.0). Slides were subsequently incubated for 1 h with anti-DDX3 (1:1000, pAb r647 [21]), followed by poly-HRP-anti-mouse/rabbit/rat IgG (BrightVision, Immunologic, Duiven, the Netherlands) as a secondary antibody for 30 min. Peroxidase activity was developed with diaminobenzidine. The slides were lightly counterstained with hematoxylin and mounted. Appropriate positive and negative controls were used throughout.
Scoring was performed by consensus of two observers. Cytoplasmic DDX3 expression was fairly homogeneous, but the intensity varied and was therefore scored semi-quantitatively as absent (0), low (1), moderate (2) or strong (3). Cases with score 0–2 were classified as having low DDX3 expression and evaluated against cases with strong expression as before [17].
Statistics
DDX3 expression and other clinicopathological characteristics were compared between tumors in patients with a germline mutation in BRCA1 or BRCA2 versus sporadic breast cancers. Discrete variables were compared by Chi-square or Fisher’s exact test. Student’s t test and Mann–Whitney U tests were calculated for normally and non-normally distributed variables, respectively. Multivariate analysis was performed by including all factors significantly associated with both DDX3 expression and BRCA mutation status in a logistic regression model. Effect modifiers were identified by including multiplicative interaction terms into the model. P values smaller than 0.05 were considered statistically significant. All statistical analyses were performed with R version 3.2.0.
Immunoblotting
All cells were harvested at 50–70% confluency. Cells were lysed in SDS extraction buffer and sonicated on ice. 30 μg protein was loaded on SDS-PAGE gels for gel electrophoresis. The blots were probed overnight with primary antibodies against DDX3 (1:1000, mAb AO196) [21], β-actin (1:10000, A5441, Sigma-Aldrich), followed by appropriate secondary antibodies, development with ECL (Bio-Rad, Hercules, CA, USA) and imaging with a G:BOX Chemi XR5 (Syngene, Frederick, MD, USA).
Cell viability assay
MCF7 and MDA-MB-231 were purchased from ATCC (ATCC, Manassas, VA, USA). MDA-MB-468, MDA-MB-435, SUM149-PT and HCC1937 were a kind gift of Shyam Sharan (NCI, Frederick, MD, USA). For cell viability assays 1 × 103–3 × 103 cells were plated per well in a 96-well plate. The following day RK-33 or DMSO (vehicle control) was added. The number of viable cells was estimated after 72 hours of drug exposure with an MTS assay. For this, the cells were incubated with MTS reagent (CellTiter 96 Aqueous One Solution, Promega, Madison, WI, USA) for 2 h, after which absorbance was measured at 490 nm with a Victor3V plate reader (PerkinElmer, Waltham, MA, USA).
Colony-forming assay
Synergy between RK-33 and olaparib was evaluated by colony-forming assays, as this is the most used assay to evaluate PARP inhibitor efficacy [22]. For HCC1937, 2500 cells were plated in 60-mm dishes. For all other cell lines 200–600 cells were plated in 6-well plates and allowed to attach overnight. Cells were treated with RK-33, olaparib or a combination of both, in the IC50 ratio for 24 h, followed by either fresh media or fresh media containing olaparib addition every 4 days. When colonies reached a size of more than 50 cells, they were fixed in methanol with 0.5% crystal violet. Colonies were counted and survival fractions were calculated.
Synergy analysis
Monotherapy and combination therapy curves of multiple independent experiments were modeled with nonlinear mixed-effects modeling, using the mixlow R package [23]. To evaluate dose–response interactions combination indices with 95% confidence intervals were calculated for every fraction affected (Fa) according to the Loewe additivity principle [24], as formulated in Eq. 1.
| (1) |
CA,x and CB,x are the concentrations of olaparib and RK-33 in combination to achieve fraction affected x. ICx,A and ICx,B are the concentrations of the olaparib and RK-33 alone to achieve the same effect. Synergy was defined as a combination index significantly lower than one.
Results
DDX3 expression in BRCA1-deficient breast cancer patient samples
To assess the dependence on DDX3 in cancer with a DDR deficiency, we evaluated DDX3 expression by immunohistochemistry in breast cancer samples of 103 germline BRCA1 mutation carriers, 29 germline BRCA2 mutation carriers and 265 women with sporadic breast cancer (Fig. 1 and Table 1). Strong cytoplasmic DDX3 expression was observed in 30% of sporadic cases, as compared to 24% of BRCA1 (p = 0.337) and 21% of BRCA2-related cases (p = 0.624), indicating that these mutations do not cause an increase in DDX3 expression levels. We did observe the usual known differences between our sporadic and BRCA-deficient study populations, such as lower age (p < 0.001) and higher grade (p = 0.014) in both BRCA1- and BRCA2-related cases and higher MAI (p < 0.001), negative ER status (p < 0.001), negative PR status (p < 0.001) and more frequent basal-like molecular classification (p < 0.001) in BRCA1 mutation carriers.
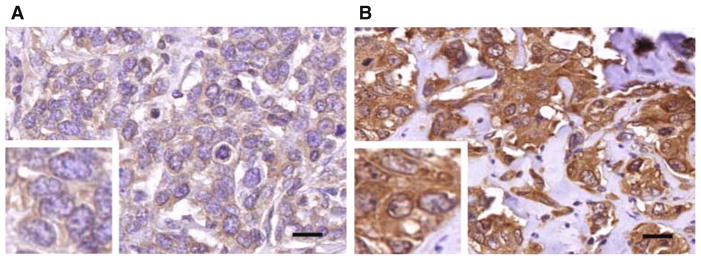
Fig. 1.
DDX3 expression in BRCA1-related breast cancer. Example of low (a) and high (b) immunohistochemical DDX3 expression in breast cancer occurring in patients with a germline BRCA1 mutation. Scale bar is 20 μm
Table 1.
DDX3 expression and other clinicopathological characteristics in breast cancer in BRCA1 or BRCA2 germline mutation carriers as compared to sporadic breast cancers
| Sporadic | BRCA1 | P value | BRCA2 | P value | |
|---|---|---|---|---|---|
| DDX3 expression [n (%)] | 0.337a | 0.624a | |||
| Absent | 3 (1) | 0 | 0 | ||
| Weak | 34 (13) | 19 (18) | 5 (17) | ||
| Moderate | 148 (56) | 59 (57) | 18 (62) | ||
| Strong | 80 (30) | 25 (24) | 6 (21) | ||
| Age [median (IQR)] | 58 (18) | 40 (11.5) | <0.001b | 50 (11) | <0.001b |
| B&R grade [n (%)] | <0.001 | 0.014a | |||
| 1 | 45 (17) | 2 (2) | 0 | ||
| 2 | 99 (37) | 15 (16) | 8 (31) | ||
| 3 | 121 (46) | 78 (82) | 18 (69) | ||
| Missing | 0 | 0 | 1 | ||
| MAI [median (IQR)] | 12 (18) | 25 (25) | <0.001b | 17 (13) | 0.110a |
| Missing | 0 | 13 | 3 | ||
| Histological type [n (%)] | 0.064 | 0.667a | |||
| Invasive ductal carcinoma | 225 (85) | 80 (84) | 25 (93) | ||
| Invasive lobular carcinoma | 24 (9) | 4 (4) | 2 (7) | ||
| Other | 15 (6) | 11 (12) | 0 | ||
| Missing | 1 | 8 | 2 | ||
| Tumor size (cm) [median (IQR)] | 2 (2) | 2 (2) | 0.924b | 1 (1) | 0.029b |
| Missing | 22 | 26 | 6 | ||
| ER [n (%)] | <0.001 | 1 | |||
| Negative | 50 (19) | 76 (77) | 5 (19) | ||
| Positive | 215 (81) | 23 (23) | 22 (81) | ||
| Missing | 0 | 4 | 2 | ||
| PR [n (%)] | <0.001 | 0.171 | |||
| Negative | 87 (33) | 81 (83) | 13 (48) | ||
| Positive | 177 (67) | 17 (17) | 14 (52) | ||
| Missing | 1 | 5 | 2 | ||
| HER2 [n (%)] | 0.828 | 1a | |||
| Negative | 243 (92) | 94 (93) | 26 (93) | ||
| Positive | 22 (8) | 7 (7) | 2 (7) | ||
| Missing | 0 | 2 | 1 | ||
| Molecular classification [n (%)] | <0.001a | 0.897a | |||
| Luminal A | 208 (78) | 20 (21) | 21 (81) | ||
| Luminal B | 12 (5) | 4 (4) | 1 (4) | ||
| HER2 overexpressing | 10 (4) | 4 (4) | 0 | ||
| Basal-like | 35 (13) | 67 (71) | 4 (15) | ||
| Unclassified | 0 | 0 | 0 | ||
| Missing |
P value calculated by Chi-square test unless otherwise indicated
n number, B&R Bloom and Richardson, MAI mitotic activity index, IQR interquartile range
Fisher exact test
Mann–Whitney U test
To exclude that we were not observing a correlation between BRCA1/2 mutation status and DDX3 expression levels due to incidental cancelation bias by another confounding factor, we performed logistic regression with all covariates that were associated with both mutation status and DDX3 expression. The presence of a germline BRCA1 mutation became a borderline significant predictor of DDX3 expression (ORadjusted 0.53, p = 0.053) after correction for MAI, histological type and PR status, implying that DDX3 expression may even be lower in BRCA1-related breast cancers of equal MAI, histological type and PR status. No factors were significantly associated with both BRCA2 mutation status and DDX3 expression. No effect modifiers were identified.
Equal sensitivity to the DDX3 inhibitor RK-33 in BRCA1 pro- and deficient breast cancer cell lines
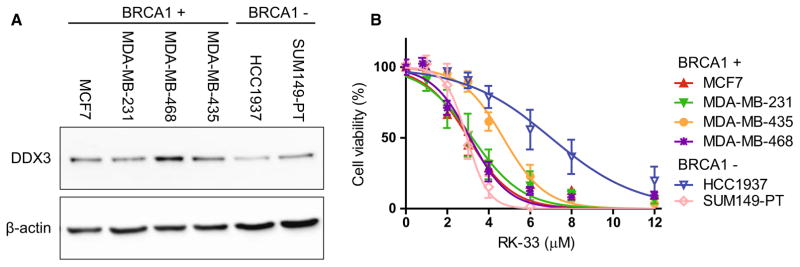
In addition, DDX3 levels were evaluated in two cell lines with a BRCA1 mutation (HCC1937 and SUM149-PT) and four BRCA1-proficient cell lines (MCF7, MDA-MB-231, MDA-MB-435 and MDA-MB-468, Fig. 2a). DDX3 expression was highest in the BRCA1-proficient cell line MDA-MB-468 and lowest in the BRCA1-deficient cell line HCC1937. All other cell lines had similar DDX3 expression levels. Sensitivity to DDX3 inhibition with the small molecule inhibitor RK-33 was evaluated with MTS assays (Fig. 2b). All IC50 values were in the low micromolar range (2.8–6.6 μM). Of the cell lines with a BRCA1 mutation, an IC50 on the lower end of the spectrum (2.9 μM) was observed for SUM149-PT, whereas HCC1937 had the highest IC50 of all cell lines (6.6 μM). Overall, the DDX3 inhibitor RK-33 had a similar in vitro efficacy in BRCA1 pro- and deficient cell lines.
Fig. 2.
Sensitivity of BRCA1 pro- and deficient cell lines to the DDX3 inhibitor RK-33. a Immunoblot showing DDX3 and β-actin expression in BRCA1-proficient cell lines (BRCA1+) and cell lines with mutated BRCA1 (BRCA1−). b MTS assay showing RK-33 cytotoxicity in BRCA1-proficient cell lines and cell lines with mutated BRCA1. Graphs represent mean of independent experiments ± SD
Synergy between DDX3 inhibition with RK-33 and PARP inhibition with olaparib
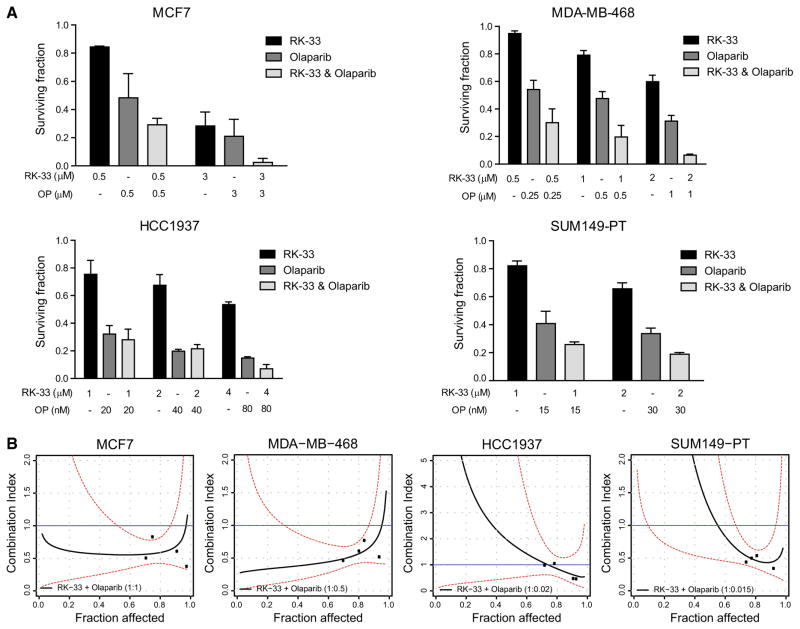
Given the effect of RK-33 on DNA repair, we explored whether any synergy could be observed between the DDX3 inhibitor RK-33 and the PARP inhibitor olaparib. Figure 3 shows the effect of combined RK-33 and olaparib treatment as measured by colony-forming assays. The fraction of cells surviving combination therapy was lower than the surviving fraction of monotherapy in all cell lines, except HCC1937 (Fig. 3a). In order to evaluate whether the cytotoxicity of combined RK-33 and olaparib was more than additive, combination indices were calculated (Fig. 3b). The mean combination index over the 20–95% fraction affected interval was lower than 1 for MCF7 (CI20–95 0.59) and MDA-MB-468 (CI20–95 0.62), indicating synergy in the BRCA1-proficient cell lines. In SUM149-PT, synergy was observed only in case of a high Fa. Although the mean CI20–95 was 1.42, the CI of the actual measured data points (Fa 60–100%) was significantly lower than 1, indicative of synergy in this area. No synergy was observed for HCC1937 (CI20–95 1.71).
Fig. 3.
Synergy between RK-33 and the PARP inhibitor olaparib. a Bar graphs representing the surviving fraction in a colony formation assay after RK-33, olaparib or combination treatment in BRCA1-proficient (MCF7 and MDA-MB-468) and BRCA1-deficient breast cancer cell lines (HCC1937 and SUM149-PT). b CI-Fa plots showing the combination index (CI) and 95% confidence intervals of RK-33 olaparib combination therapy for different fractions affected (Fa) in BRCA1 pro- and deficient cell lines. Lines represent modeled CI curves. Points represent CI values calculated from measured data points. Dashed red lines represent 95% confidence intervals. Blue lines represent additivity reference line. Graphs represent mean of independent experiments ± SD
Discussion
This study evaluated the efficacy of DDX3 inhibition, by the small molecule inhibitor RK-33, in BRCA-deficient breast cancer, with inherent impaired HR DNA repair pathway, in comparison with BRCA-proficient breast cancer. RK-33 was previously found to inhibit NHEJ, an additional DSB repair mechanism [11]. Therefore, we hypothesized that BRCA-deficient breast cancer might have an increased dependency on DDX3. However, DDX3 expression levels were similar in breast cancers in BRCA1/BRCA2 germline mutation carriers and sporadic breast cancers and BRCA1 pro- and deficient breast cancer cell lines were equally sensitive to RK-33 treatment. We therefore concluded that high DDX3 expression is present in BRCA-mutated breast cancers and that they are sensitive to DDX3 inhibition. However, there was no clear indication of increased DDX3 dependency in BRCA deficient, when compared to BRCA-proficient breast cancers. In addition, we evaluated whether DDX3 inhibition with RK-33 could sensitize BRCA pro- and deficient breast cancer cells to PARP inhibition with olaparib. We found a synergistic interaction mainly in BRCA1-proficient cell lines and to a certain extent in BRCA1-deficient cancers.
The mechanism behind the increased sensitivity of BRCA-deficient cancers to PARP inhibition is an area of active investigation and several explanations have been proposed [25]. The most accepted mechanism is that PARP inhibitors impair BER, an SSB repair mechanism. Persistent SSBs are converted to DSBs, which are normally repaired by HR, but remain unrepaired in HR-deficient BRCA1/BRCA2 mutant cells [25]. Our observation that PARP inhibitors synergize with NHEJ inhibition by RK-33 fits a variant of this model. Since HR is restricted to the S and G2 phase of the cell cycle [26], accumulation of DSBs is likely to also require NHEJ for repair. This could explain the synthetic lethality observed after combination therapy with the PARP inhibitor olaparib and the DDX3 inhibitor RK-33 and previously reported radiosensitizing capacities of RK-33 [11], since ionizing radiation causes DSB formation as well. Also, it may explain why the dependency on DDX3 is not increased in cells with a BRCA1 mutation. In the absence of an agent that stimulates persistence of SSBs, the DSB production rate may not be high enough to increase the demand for NHEJ as an alternative repair strategy. In addition, the sensitivity of BRCA1-deficient cell lines to PARP inhibition is already very high, making it hard to observe synergy with RK-33 in these cells. A second explanation for PARP inhibitor efficacy in BRCA-deficient tumors is that PARP inhibitors have an activating effect on NHEJ, which is a more error-prone repair pathway than HR [27]. More research on the exact role of DDX3 in different DNA repair pathways is necessary to fully understand the mechanism behind the observed interactions between PARP inhibition and DDX3 inhibition in a BRCA1 pro- or deficient background.
Sensitization of primary resistant BRCA-proficient breast cancers to PARP inhibition by RK-33 could be of specific use in the treatment of TNBC, given their aggressive biology and the lack of specific therapeutic targets [6]. Secondary resistance in BRCA1 mutant cancers is mainly due to restoration of HR defects, often by partially restoring BRCA1 functionality [28]. Since RK-33 sensitizes BRCA1-proficient breast cancer cells to PARP inhibition, DDX3 inhibition could also have an application overcoming secondary resistance against PARP inhibitors. There is a risk of causing normal cell toxicity by enhancing the sensitivity of cancer cells to PARP inhibition [29]. Because cancer cells have higher endogenous DNA damage rates and greater DDR deficiency compared to normal cells, there is a potential therapeutic window for combination therapy with RK-33 and a PARP inhibitor. In previous studies, no toxicity was observed after RK-33 used as a monotherapy or in combination with radiotherapy [11]. Future studies are needed to further evaluate the safety and efficacy of this combination regimen.
Overall, DDX3 expression levels are similar in breast cancers in BRCA1/BRCA2 germline mutation carriers and sporadic breast cancers. BRCA1 pro- and deficient breast cancer cell lines were equally sensitive to RK-33 treatment and therefore show similar DDX3 dependency. Interestingly, DDX3 inhibition with RK-33 synergizes with PARP inhibitor treatment in breast cancer cells, especially in a BRCA1-proficient background.
Acknowledgments
We would like to thank Petra van der Groep, Yvonne Smolders and Joost Bart for selection of patients and assembling TMAs.
Funding This work was financially supported by Utrecht University Alexandre Suerman Stipend (MHVV), the Dutch Cancer Foundation (UU2013-5851; MHVV), the Saal van Zwanenberg foundation (MHVV), the JoKolk foundation (MHVV) and Safeway (VR).
Footnotes
Compliance with ethical standards
Conflict of interest Venu Raman received a patent for the use of RK-33 as a radiosensitizer (US8,518,901). Venu Raman, Guus Bol and Paul van Diest have received a patent for the use of DDX3 as a cancer biomarker (US9,322,831). Paul van Diest and Venu Raman are on the advisory board of Natsar Pharmaceuticals. Authors Marise R Heerma van Voss, Justin D Brilliant, Farhad Vesuna and Elsken van der Wall have no conflict of interest.
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. For this type of study, formal consent is not required. This article does not contain any studies with animals performed by any of the authors.
Informed consent As we used anonymous archival leftover pathology material, no informed consent is required according to Dutch legislation [19], as this use of redundant tissue for research purposes is part of the standard treatment agreement with patients in our hospitals [20].
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell. 2015;60(4):547–60. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 4.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4(4):511–8. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 5.Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20(9):2310–8. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016 doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–44. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 8.Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12(9):852–61. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 9.Linder P, Fuller-Pace FV. Looking back on the birth of DEAD-box RNA helicases. Biochim Biophys Acta. 2013;1829(8):750–5. doi: 10.1016/j.bbagrm.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P, Jr, et al. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27(28):3912–22. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO mol med. 2015;7(5):648–69. doi: 10.15252/emmm.201404368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heerma van Voss MR, Vesuna F, Trumpi K, Brilliant J, Berlinicke C, de Leng W, et al. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget. 2015;6(29):28312–26. doi: 10.18632/oncotarget.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilky BA, Kim C, McCarty G, Montgomery EA, Kammers K, DeVine LR, et al. RNA helicase DDX3: a novel therapeutic target in Ewing sarcoma. Oncogene. 2016;35(20):2574–83. doi: 10.1038/onc.2015.336. [DOI] [PubMed] [Google Scholar]
- 14.Xie M, Vesuna F, Tantravedi S, Bol GM, van Voss MRH, Nugent K, et al. RK-33 radiosensitizes prostate cancer cells by blocking the RNA helicase DDX3. Cancer Res. 2016;76(21):6340–50. doi: 10.1158/0008-5472.CAN-16-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Wang H, Wang Z, Makhija S, Buchsbaum D, LoBuglio A, et al. Inducible resistance of tumor cells to tumor necrosis factor-related apoptosis-inducing ligand receptor 2-mediated apoptosis by generation of a blockade at the death domain function. Cancer Res. 2006;66(17):8520–8. doi: 10.1158/0008-5472.CAN-05-4364. [DOI] [PubMed] [Google Scholar]
- 16.Chen HH, Yu HI, Cho WC, Tarn WY. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene. 2015;34(21):2790–800. doi: 10.1038/onc.2014.190. [DOI] [PubMed] [Google Scholar]
- 17.Bol GM, Raman V, van der Groep P, Vermeulen JF, Patel AH, van der Wall E, et al. Expression of the RNA helicase DDX3 and the hypoxia response in breast cancer. PLoS ONE. 2013;8(5):e63548. doi: 10.1371/journal.pone.0063548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moelans CB, de Weger RA, van Blokland MT, Ezendam C, Elshof S, Tilanus MG, et al. HER-2/neu amplification testing in breast cancer by multiplex ligation-dependent probe amplification in comparison with immunohistochemistry and in situ hybridization. Cell Oncol. 2009;31(1):1–10. doi: 10.3233/CLO-2009-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Medical Research Involving Human Subjects Act [In Dutch: Wet medisch-wetenschappelijk onderzoek met mensen, WMO]; 1998.
- 20.van Diest PJ. No consent should be needed for using leftover body material for scientific purposes. For BMJ. 2002;325(7365):648–51. [PubMed] [Google Scholar]
- 21.Angus AG, Dalrymple D, Boulant S, McGivern DR, Clayton RF, Scott MJ, et al. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J Gen Virol. 2010;91(Pt 1):122–32. doi: 10.1099/vir.0.015909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 23.Boik JC, Newman RA, Boik RJ. Quantifying synergism/antagonism using nonlinear mixed-effects modeling: a simulation study. Stat Med. 2008;27(7):1040–61. doi: 10.1002/sim.3005. [DOI] [PubMed] [Google Scholar]
- 24.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 25.De Lorenzo SB, Patel AG, Hurley RM, Kaufmann SH. The Elephant and the blind men: making sense of PARP inhibitors in homologous recombination deficient tumor cells. Front Oncol. 2013;3:228. doi: 10.3389/fonc.2013.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17(1):11–6. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci USA. 2011;108(8):3406–11. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouwman P, Jonkers J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clin Cancer Res. 2014;20(3):540–7. doi: 10.1158/1078-0432.CCR-13-0225. [DOI] [PubMed] [Google Scholar]
- 29.Ito S, Murphy CG, Doubrovina E, Jasin M, Moynahan ME. PARP inhibitors in clinical use induce genomic instability in normal human cells. PLoS ONE. 2016;11(7):e0159341. doi: 10.1371/journal.pone.0159341. [DOI] [PMC free article] [PubMed] [Google Scholar]