Abstract
γ-Secretase is a multiprotein complex that catalyzes intramembrane proteolysis associated with Alzheimer’s disease and cancer. Here, we have developed potent sulfonamide clickable photoaffinity probes that target γ-secretase in vitro and in cells by incorporating various photoreactive groups and walking the clickable alkyne handle to different positions around the molecule. We found that benzophenone is preferred over diazirine as a photoreactive group within the sulfonamide scaffold for labeling γ-secretase. Intriguingly, the placement of the alkyne at different positions has little effect on probe potency but has a significant impact on the efficiency of labeling of γ-secretase. Moreover, the optimized clickable photoprobe, 163-BP3, was utilized as a cellular probe to effectively assess the target engagement of inhibitors with γ-secretase in primary neuronal cells. In addition, biotinylated 163-BP3 probes were developed and used to capture the native γ-secretase complex in the 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO) solubilized state. Taken together, these next generation clickable and biotinylated sulfonamide probes offer new tools to study γ-secretase in biochemical and cellular systems. Finally, the data provide insights into structural features of the sulfonamide inhibitor binding site in relation to the active site and into the design of clickable photoaffinity probes.
Keywords: Alzheimer’s disease, presenilin, β-amyloid, clickable photoaffinity probe, benzophenone, diazirine, click walking
Graphical abstract
Photoaffinity labeling (PAL) has been a valuable approach to determine the biological targets of numerous small molecules from natural products to screening hits.1,2 Moreover, a PAL based photophore walking approach, through incorporating a photoreactive moiety at various positions of the probe, has been developed to examine the conformational changes within the active site of endogenous γ-secretase induced by mutations and small molecule binding.3–8 However, design of photoprobes has been a challenging task due to the difficulty of maintaining the potency and specificity of parent compounds while imparting the probes with photo-cross-linking and target visualization/retrieval capabilities. Furthermore, small molecule–protein interactions often depend on the integrity of the native cellular environment, making it advantageous to develop probes that can covalently capture their targets in live cells.9 The advent of cell-permeable clickable PAL probes has provided a major advance toward these goals.10–16 Photo-activatable groups including benzophenone, aryl azide, and diazirine are often used for cross-linking.17 Despite well studied photochemistry and chemical reactivities of these species, selection of photoreactive groups is not straightforward. In some cases, a diazirine- or aryl azide-based photoreactive probe is advantageous, while in other cases a benzophenone-based probe is preferred.1,2,18–25 Benzophenone-based probes are commonly utilized for identification and characterization of γ-secretase,4–6,26–31 a macromolecular complex linked with Alzheimer’s disease (AD).32,33 Fuwa et al.34 compared different photoreactive groups and showed that only benzophenone containing γ-secretase inhibitor probes cross-linked to presenilin (PS), the catalytic subunit of γ-secretase,35 or SPP, but not phenyl-diazirine probes, despite similar inhibitory potencies. However, diazirine γ-secretase modulator probes have successfully cross-linked to PS,31 suggesting diazirine is a viable option for targeting γ-secretase.
We have developed 163-BPyne based on BMS-708163 (BMS-163) (Figure 1a) and identified that it targets PS-1.4 163-BPyne maintained the sub-nanomolar potency of BMS-163, and the photoreactive group could be introduced in the final step of the synthesis (see compound synthesis in Supporting Information). As a result, we felt that this represented an ideal scaffold to explore the molecular determinants for effective photoprobe design. The present study directly compares the efficiency of PAL of a series of BMS-163-based clickable photoaffinity probes that incorporate various photoreactive groups with placement of the clickable alkyne handle at different positions within the photoprobe scaffold. We have found that benzophenone is preferred over diazirine and that electron withdrawing groups such as nitro affect the specificity and photoreactivity of these probes. Importantly, the placement of the alkyne had a big impact on the efficiency of the tandem PAL–click chemistry conjugation of reporter groups and the ability to detect probe-labeled proteins. Ultimately, this work resulted in the γ-secretase inhibitor activity-based probe 163-BP3 that successfully labeled endogenous γ-secretase in live neurons. Furthermore, a biotinylated probe was developed that pulled-down the γ-secretase complex from solubilized membranes.
Figure 1.
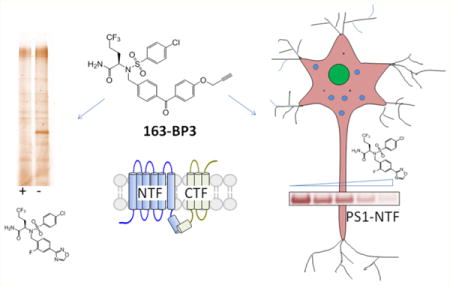
Comparison of photoreactive groups and para substituted benzophenones for labeling PS1–NTF. (a) Chemical structures of probes. (b) IC50 values of these probes against γ-secretase for Aβ40, Aβ42, and NICD1 production. (c) Comparison of PS1–NTF labeling efficiency. Probes (20 nM) were incubated with HeLa membranes with and without 2 μM competitor BMS-163, UV-irradiated to cross-link, and tagged with biotin azide via click chemistry, followed by pull-down with streptavidin and Western blot for PS1–NTF. (d) Probes (20 nM) were incubated with HeLa membranes with and without 2 μM competitor BMS-163, UV-irradiated to cross-link, and tagged with TAMRAZ–azide (see Supplementary Figure 2 for the structure of TAMRA–azide) via click chemistry, followed by in-gel fluorescence (left). Coomassie blue staining (right) shows equal amounts of total protein loaded on the gel. The arrow points to PS1–NTF at approximately 30 kDa.
RESULTS
The 163-based Benzophenone Probes Covalently Label PS1-NTF, whereas the Diazirine-Containing Probe Does Not
Initially we prepared phenyl diazirine 163-DZ to compare its photolabeling efficiency with that of 163-BP1 (Figure 1a, synthesis of all probes can be found in the Supporting Information). Photoactivation of diazirine generates a highly reactive carbene species that either cross-links to target proteins or gets quenched by solvent. 163-DZ remains a potent γ-secretase inhibitor with IC50 for the production of Aβ40, Aβ42, and NICD1 of 0.56, 0.46, and 1.51 nM, respectively (Figure 1b). Despite its comparable inhibitory potency to 163-BP1, 163-DZ exhibits no labeling of PS1–NTF (Figure 1c, lane 2 vs 12) after clicking with biotin azide and enrichment. We also attempted UV irradiation for 30 min at 365 nm followed by irradiation for 15 min at 300 nm according to an alternative protocol for diazirine cross-linking31 but were not successful in labeling PS1–NTF (data not shown). Furthermore, in-gel fluorescence showed that 163-BP1 specifically labels a band at ~30 kDa corresponding to the molecular weight of PS1–NTF that is blocked by excess BMS-163 (Figure 1d, lane 1 and 2). No specifically labeled bands were detected with 163-DZ (Figure 1d, lane 11 and 12). Coomassie blue staining demonstrated that equivalent amounts of membrane proteins were loaded on the gel (Figure 1d). One possible explanation for the difference in labeling profiles observed for the two probes is that the reactive carbene could be solvent exposed and get quenched before it is able to label the target protein (vide infra). However, a final proof of this notion requires a high resolution structure of the γ-secretase–163-DZ complex.
Next, we investigated how substituents on the benzophenone affect labeling efficiency and specificity (Figure 1a,b). Probes with substituents ortho to the benzophenone carbonyl were not prepared because of the increased probability that the photoreactive species could undergo an intramolecular reaction or the ortho substituent could destabilize the planar benzophenone conformation and thus reduce protein-target cross-linking.36 Therefore, we focused on the para position and synthesized several probes with para-substituted benzophenones (Figure 1a,b): 163-BP1–OMe contains an electron donating methoxy group, 163-BP1–F contains a fluoride, which has inductive electron withdrawing properties, 163-BP1-NO2 contains a nitro group, which has strong inductive and mesomeric electron withdrawing properties, and 163-BP1–DZ is a hybrid containing a benzophenone with an electron withdrawing p-substituted diazirine. We had hoped that the hybrid photoreactive moiety would have enhanced protein cross-linking due to the dual photoactivatable groups. These substitutions did not significantly affect potencies against γ-secretase (Figure 1b), except for 163-BP1–DZ, which still had single digit nanomolar IC50s for the production of Aβ40, Aβ42, and NICD1 (2.53, 2.29, and 6.61 nM, respectively).
When we examined the efficiency of photo-cross-linking, we found that 163-BP1, 163-BP1-OMe, and 163-BP1–F displayed similar labeling as judged by anti-PS1–NTF Western blot and TAMRA in-gel fluorescence (Figure 1c,d), suggesting that these substitutions have little effect on the photoreactivity of the benzophenone. However, 163-BP1–NO2, despite the potent IC50 value of 1 nM, does not label PS1 (Figure 1c, lanes 7 and 8) and shows strong nonspecific labeling by in-gel fluorescence (Figure 1d, left panel, lanes 7 and 8). This probe labeled many off-target proteins that were not competed by BMS-163, including a very strong band at 45 kDa (Figure 1d). It has been reported that benzophenones preferentially react with methionine residues within a 13 Å radius,37,38 and researchers have taken advantage of this characteristic to perform “methionine proximity assays”37 to determine the location of photoinsertion of a benzophenone.39,40 Moreover, p-nitro-benzoyl-phenylalanine has been shown to preferentially label the γ-CH2 or ε-CH3 on the thioether side chain of methionine residues during protein cross-linking, with very little cross-linking to other amino acid side chains.37 Therefore, it is plausible that 163-BP1-NO2 would only be a functional photoprobe if a methionine residue was near the binding site of the sulfonamide compounds. There are 13 methionine residues in PS1 (Supplementary Figure 1). It is difficult to predict the PS1 binding site without knowing the attachment site of these probes. It appears that the BP1–NO2 probe is capable of crosslinking to proteins but does not cross-link at the BMS-163 binding site. It is likely that the BMS-163 binding site does not contain a methionine; therefore BP1–NO2 does not label PS1–NTF; however, other possibilities should also be considered. For example, it is also possible that the addition of the p-nitro group results in too much nonspecific reactivity, which ultimately makes it difficult to see specifically labeled bands from in-gel fluorescence or to pull-down specifically biotinylated proteins.
Position of the Alkyne Tag Has a Strong Impact on Probe Functionality: Click Walking
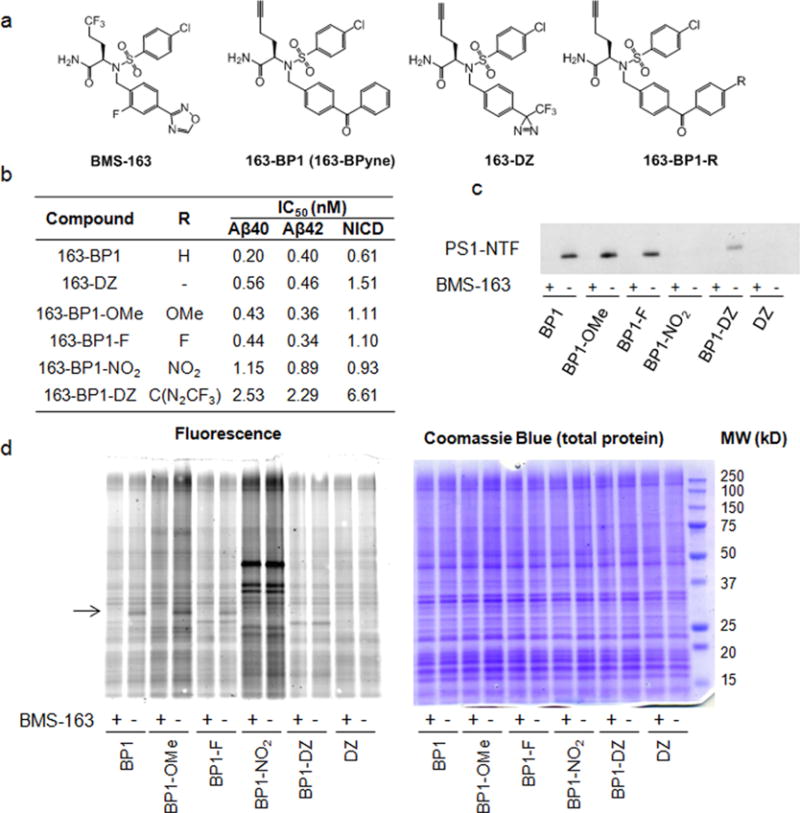
In addition to optimizing our photo-cross-linking moiety, we also wanted to understand how placement of the alkyne would affect the activity and functionality of the BMS-163 probes. Therefore, we performed click walking by designing two additional probes that have different alkyne placements (Figure 2a, see compound synthesis in Supporting Information). Surprisingly, although the placement of the alkyne within the molecule had virtually no effect on the inhibitory activity of the molecule (Figures 2b and 1b), the overall labeling efficiency is significantly impacted (Figure 2c,d). Labeling of PS1–NTF is barely detected with 163-BP2, whereas 163-BP3 provides drastically enhanced tandem PAL–click chemistry relative to 163–BP1 (Figure 2c). No specific bands, as judged by the ability of excess BMS-163 to block labeling, were detected for 163-BP2 by in-gel fluorescence (Figure 2d). A specific band at ~30 kDa was detected for 163-BP1 and 163-BP3 (Figure 2d). However, the intensity of the 163-BP3 labeled band is much stronger than that of 163-BP1, which is consistent with Western blot data for PS1–NTF (Figure 2c). There are several possible explanations for this: (1) each probe binds slightly differently within the pocket, creating a different geometric alignment of the benzophenone with nearby amino acids, thus changing the labeling efficiency of the benzophenone; (2) the steric environment of the alkyne in each probe-labeled adduct may be different, which could influence the efficiency of the click chemistry conjugation reaction; (3) the propargyl substituent on 163-BP3 could enhance reactivity of the benzophenone, but this seems unlikely considering the very modest difference between 163-BP1 and 163-BP1–OMe (Figure 1).
Figure 2.
Location of the alkyne within the BMS probes has a strong impact on functionality. (a) Chemical structures of the probes with varied alkyne placement. (b) Inhibitory potencies of 163-BP2 and 163-BP3. (c) Indicated probes (20 nM) were incubated with HeLa membranes in the presence or absence of 1 μM competitor BMS-163 and UV-irradiated, followed by click chemistry with biotin azide, pull-down with streptavidin resin, and Western blot analysis for PS1-NTF. (d) Indicated probes (20 nM) were incubated with HeLa membranes in the presence or absence of 1 μM competitor BMS-163 and UV-irradiated, followed by click chemistry with TAMRA–azide and in-gel fluorescence (left). Coomassie blue staining shows equal amounts of total protein loaded on the gel (right).
Development of Biotinylated BMS-163-based Probes That Capture the γ-Secretase Complex under Native Conditions
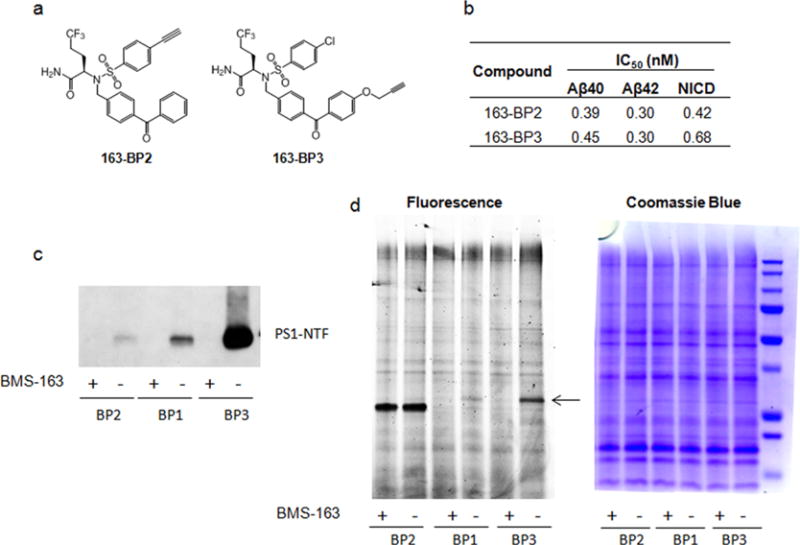
To explore the accessibility of the alkyne group for the in situ click reaction when the probe is bound to γ-secretase, we directly conjugated biotin onto 163-BP1 and 163-BP3 using CuAAC (copper catalyzed azide–alkyne cycloaddition) chemistry. The resulting compounds are 163-BP1–biotin and 163-BP3–biotin (Figure 3a). The compound 163-BP1–biotin had no γ-secretase inhibition for Aβ40 production at concentrations up to 10 μM, nor was the probe able to label and pull-down PS1–NTF compared with 163-BP1 (Figure 3b). This illustrates the advantage of using a small alkyne tag in combination with click chemistry as opposed to direct conjugation of the biotin moiety. On the other hand, 163-BP3–biotin was a potent γ-secretase inhibitor with an IC50 of 5 nM for Aβ40 production. Notably, 163-BP3–biotin exhibited even more efficient labeling of PS1–NTF than 163-BP3 (Figure 3c), which presumably reflects the efficiency of the click reaction needed to attach biotin azide in the case of 163-BP3. The fact that 163-BP1–biotin loses all activity and 163-BP3–biotin maintains good inhibitory and labeling activity suggests that the alkyne in 163-BP1 is in a strictly limited space within the binding pocket, while the alkyne in 163-BP3 is in a solvent exposed area. If the location of the alkyne in 163-BP1 is pointed toward the inside of the binding pocket, it may be less accessible for click chemistry with biotin azide. Similarly, adding a bulky biotin linker at this location would likely abrogate activity. The notion that the benzophenone is solvent exposed is consistent with the lack of labeling observed with 163-DZ as previously mentioned. However, the final proof requires a high resolution costructure of these probes with γ-secretase.
Figure 3.
Biotinylated photoaffinity probes. (a) Chemical structure of the biotinylated probes. (b, c) Biotinylated probes and clickable probes (20 nM) were incubated with HeLa membranes in the presence or absence of 1 μM competitor BMS-163 and UV-irradiated, followed by click chemistry with biotin azide for clickable probes only, pull-down with streptavidin resin, and Western blot analysis for PS1–NTF. (d) The solubilized γ-secretase was captured under native conditions (0.25% CHAPSO) by 163-BP3–biotin and 163-BP3–L-biotin. The bound complex was eluted and analyzed by Western blot analysis.
Next, we examined whether biotinylated BMS-163-based probes can be used to isolate the γ-secretase complex. We have previously demonstrated that a longer linker between biotin and the active-site-directed L685,458-based inhibitors is needed for interaction of the probe with both γ-secretase and streptavidin.41 We thus synthesized 163-BP3–L-biotin (linker ~41 Å) that contains a longer linker than 163-BP3–biotin (linker ~21 Å) (Figure 3a). HeLa cell membrane was solubilized with 1% 3-[(3-cholamidopropyl)-dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO); the ensuing fraction was diluted to 0.25% CHAPSO and captured with 163-BP3–biotin or 163-BP3–L-biotin under native conditions. The isolated γ-secretase complex was analyzed by Western blot (Figure 3d). Both probes are capable of capturing the complex that contains essential subunits nicastrin (NCT), PS1–NTF, PS1–CTF, Aph-1, and Pen2,42 which is in contrast to L685,458-based probes that need a linker length of at least 34 Å.41 These findings suggest that the BMS-163 binding site is closer to the surface of the γ-secretase complex than the L685,458 binding pocket, which further supports the hypothesis that the location of the photoreactive group of these BMS-163-based probes is near the interface of PS and the aqueous environment.
163-BP3 Functions As a Cellular Probe for Determining the Engagement of γ-Secretase in Primary Neuronal Culture
Understanding target engagement and selectivity is an important facet of drug discovery.43 IC50 values are often measured in vitro using purified protein or cellular lysates, which may not recapitulate the real effect in vivo. The design of clickable photoaffinity probes can provide membrane penetrant probes capable of determining the target occupancy in live cells, which presents a more physiologically relevant environment to address a compound’s IC50. The cellular potency of 163-BP3 was determined in a whole-cell assay using Chinese hamster ovary cells overexpressing wtAPP (CHO-APP) and the IC50 for inhibition of Aβ42 production was 2.7 nM (n = 5). Therefore, the probe demonstrated excellent cellular potency and permeability and could be employed to determine target occupancy in live cells. We used 163-BP3 to assess the occupancy of BMS-163 on γ-secretase in primary cortical neurons. Cultured primary cortical neurons were treated with BMS-163 at the indicated concentration, followed by incubation with 163-BP3. Neurons were photolabeled and lysed, followed by click chemistry with biotin azide and enrichment of labeled proteins on streptavidin. Occupancy of γ-secretase was analyzed with immunoblot using PS1–NTF antibody (Figure 4a). We observed a dose-dependent decrease in 163-BP3 photolabeling of PS1–NTF with increasing concentrations of BMS-163 (EC50 = 16 nM) (Figure 4b). This finding strongly indicates that 163-BP3 is a valuable probe to assess the binding of BMS-163 and related γ-secretase inhibitors to neuronal γ-secretase.
Figure 4.
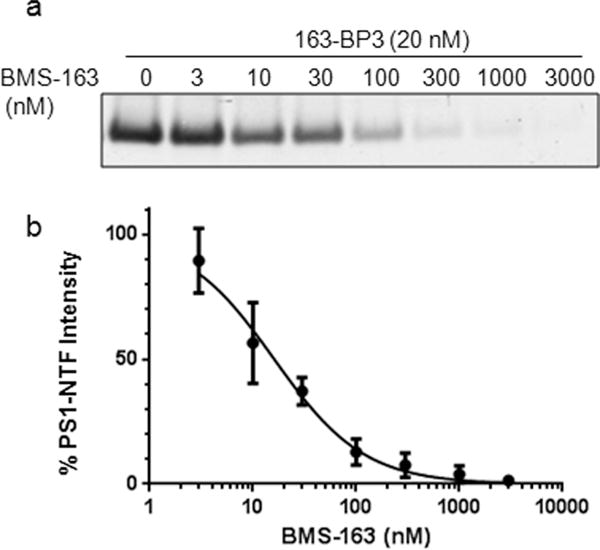
Labeling of PS1–NTF in live cortical neurons. (a) Western blot of PS1–NTF showing dose dependent competition of 163-BP3 labeling with BMS-163. (b) Inhibitory curve for competition with BMS-163.
DISCUSSION AND CONCLUSION
We have examined the structure–activity relationship (SAR) of BMS-163-derived clickable photoaffinity probes and developed a series of potent probes with different photoreactive groups and alkyne positions. By directly comparing the tandem photoaffinity labeling–click chemistry conjugation efficiency, we gained valuable insights into the design of clickable photoaffinity probes. We found that in our system a benzophenone is preferred over a phenyl-diazirine for photocross-linking studies. The fact that multiple groups are tolerated at the para position of the benzophenone, including the large linker plus biotin, suggests that the photoreactive moieties are near the solvent-exposed membrane surface of the PS binding site. This would explain why the benzophenone is preferred in our case. It is likely that carbene reactive species get irreversibly quenched whereas the benzophenone ketyl radical reactive species can be regenerated under continued UV irradiation until a productive cross-link is formed. In the absence of binding site information, it is prudent to not limit photoprobe design to one type of photoreactive group but rather include at least one carbene-based and one benzophenone-based photoprobe to maximize chances of success.18
Fluoro or methoxy substitutions at the para position of the distal benzophenone phenyl ring were well tolerated for photolabeling of PS1–NTF. In contrast, we could not detect any labeling of PS1–NTF by the p-nitro-benzophenone photoprobe, despite the fact that nonspecific photolabeling was observed as detected by in-gel fluorescence. Nitro-benzophenone has been suggested to selectively cross-link with methionines that are within a distance of approximately 13 Å, and this has been used as a methionine proximity assay.37 The lack of PS1–NTF labeling by 163-BP1–NO2 suggests that there may not be a methionine residue in the binding site that is in proximity to the benzophenone photoreactive group.
Importantly, we found that different positioning of the alkyne tag within the molecule had little effect on activity but had very significant effect on probe functionality. Therefore, when designing clickable PAL probes, multiple areas of incorporation of the alkyne tag should be considered. In our studies, the propargyl ether substituted benzophenone provided the best tandem photoaffinity labeling–click chemistry conjugation efficiency, and 163-BP3 was used to label γ-secretase and assess target engagement in neurons. Propargyl ether benzophenone represents a minimalist alkyne-containing benzophenone photo-cross-linker for the design of clickable photoaffinity probes. This benzophenone can be prepared with multiple functional groups at the para position of the other phenyl ring (CH2Br, CHO, CH2OH, CO2H, CH2NH2), which constitute versatile intermediates for synthesis of clickable photoaffinity probes.1
Taking advantage of the hypothesized solvent-exposed nature of the alkyne moiety of 163-BP3, we designed potent probes incorporating biotin, which were used to pull-down the γ-secretase complex. Moreover, these findings offer insights into the binding sites of two classes of inhibitors, BMS-163 and L685,458, within the γ-secretase complex. Despite availability of high resolution cryo-EM structures of the γ-secretase complex,44,45 the binding sites of these inhibitors remain unknown. We have demonstrated that two classes of inhibitors bind to distinct sites of γ-secretase.4,33 The present study suggests that the binding site of BMS-163 within γ-secretase is shallower than the active site, to which L685,458 binds. Mapping the BMS-163-based photoprobe binding and attachment site on γ-secretase represents an important step toward elucidating the molecular mechanism of this class of compounds and would offer structural insights into γ-secretase including the interplay with the active site.
METHODS
In Vitro γ-Secretase Activity Assay
Cell-free γ-secretase activity assays were performed similarly to those previously described.4,6,7 Biotinylated recombinant APP substrate, Sb4 (1 μM), or Notch1 substrate, N1–Sb1 (0.4 μM), were incubated with 40 μg/mL HeLa membranes in 0.25% CHAPSO for 2.5 h in the presence or absence of γ-secretase inhibitor compounds. The amount of cleavage product generated was then determined using a detection mixture with cleavage specific antibodies for Aβ42 (10-G3), Aβ40 (G2-10), or Notchl intracelluar domain (SM320) in combination with AlphaLISA protein A (for Aβ42 and NICD) or AlphaLISA anti-mouse (for Aβ40) acceptor beads and streptavidin coated donor beads (PerkinElmer). Equal 20 μL volumes of reaction mixtures and detection mixtures were combined in a 384 well plate and incubated at room temperature overnight, and the AlphaLISA signal was read using the EnVision multilabel plate reader.
Photoaffinity Labeling with Clickable Probes, Followed by Streptavidin Pull-down
Clickable photoprobes were incubated with 800 μg of HeLa cell membranes at 37 °C for 1 h in the presence or absence of indicated γ-secretase inhibitors in 1 mL of PBS and then UV irradiated at 350 nm for 30 min to cross-link the photoreactive probe to nearby proteins.4,5,16 The samples were then ultracentrifuged at 90 000g, and the pellets were resuspended with 200 μL of PBS buffer by homogenization with the TissueLyser at 25 rps for 2 min (Qiagen). Labeled proteins were conjugated with biotin by using Cu-catalyzed azide–alkyne cycloaddition (CuAAC) with 1 mM CuSO4, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 0.1 mM tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA), and 100 μM biotin azide in PBS with 5% t-butanol and 2% DMSO and shaking for 1 h at 25 °C. The samples were then ultracentrifuged at 90 000g to remove click chemistry reagents, and the pellets were again resuspended by homogenization and solubilized in 500 μL of RIPA buffer (50 mM Tris, pH 8, 150 mM NaCl, 0.1% SDS, 1% NP40, 0.5% deoxycholate), followed by centrifugation at 15 000g to remove particulate matter. The supernatant was added to 20 μL of streptavidin ultralink resin slurry and incubated rotating overnight at 4 °C. The streptavidin resin was washed 2 times by centrifugation at 100g with 500 μL of RIPA buffer and then washed another 2 times with Tris-buffered saline with 0.1% Tween-20. Biotinylated proteins were eluted by heating with 30 μL of 2 mM biotin + SDS Laemmli sample buffer for 10 min at 70 °C. Then 25 μL of the eluent was loaded on to an SDS-PAGE gel for protein band separation and then transferred to PVDF membrane and blotted for target proteins with indicated primary antibodies.
Photoaffinity Labeling with Clickable Probes, Followed by Fluorescence Gel Scanning
Clickable photoprobes were incubated with 300 μg of HeLa cell membranes for 1 h at 37 °C in the presence of parent compound or vehicle control (DMSO) in 1 mL volume of PBS followed by UV irradiation at 350 nm for 45 min to cross-link the probe to nearby proteins.4,5,16 The samples were then ultracentrifuged at 90 000g, and the pellets were resuspended with 200 μL of PBS. Proteins were labeled with tetramethyl rhodamine (TAMRA) using CuAAC with 1 mM CuSO4, 1 mM TCEP, 0.1 mM TBTA, 45 μM TAMRA–azide in PBS with 5% t-butanol and 2% DMSO and shaking for 1 h at 25 °C in the dark. Labeled proteins were then precipitated with 1 mL of cold acetone at −20 °C for 30 min. Precipitated proteins were centrifuged at 15 000g for 10 min, and the pellet was washed once with 500 μL of cold acetone. After the second centrifugation, the protein pellet was air-dried for 10 min. The protein pellets were then resuspended in 50 μL of PBS buffer using the TissueLyser (Qiagen), followed by solubilization with dye-free Laemmli SDS sample buffer. Samples were centrifugued at 15 000g for 5 min to remove debris, and the resulting supernatants were measured for protein quantification using the Lowry assay. Sample (20 μg) was then loaded on to a 12% SDS-PAGE gel for protein band separation; the gel was washed and scanned for fluorescent bands using the Typhoon Trio Imager. The same gel was then stained with Coomassie blue to compare the total amount of protein loaded for each sample.
Capture of the γ-Secretase Complex
Solubilization and capture of the γ-secretase complex was performed as described.46–48 Briefly, solubilized γ-secretase was incubated with 163-BP–biotin (20 nM) or 163-BP–L-biotin (20 nM) in the presence or absence of excess BMS-163 at 0.25% CHAPSO and then isolated with streptavidin beads. γ-Secretase bound beads were washed with 0.25% CHAPSO three times and eluted with SDS-sample buffer. The ensuing fraction was analyzed by SDS-PAGE and Western blot against γ-secretase components.
Neuronal Cell Labeling
Cortical neurons were cultured and treated as described previously.16 Briefly, 13 DIV cortical neurons were pretreated with competitor (BMS-163) or DMSO control at the indicated concentrations for 30 min at 37 °C/5% CO2. Neurons were then treated with 20 nM 163-BP3 for 1 h at 37 °C/5% CO2. Cells were exposed to UV light for 15 min at 4 °C, washed 3 times with ice-cold PBS, and lysed by sonication in PBS plus Halt protease inhibitor cocktail (ThermoFisher). Membrane fractions were collected by ultracentrifugation at 100 000g and processed as described previously.16 Densitometry analysis was performed using Odyssey application software, v. 2.1.12 (LiCor). Values were normalized to 163-BP3 labeled control without BMS-163, plotted using GraphPad Prism 5 and the curve was fit using the equation (GraphPad Software Inc., San Diego, CA) to determine the EC50. Values were determined from three independent experiments and are reported as mean ± standard error of mean (SEM).
Supplementary Material
Acknowledgments
Funding
This work is supported by NIH Grants R01AG026660 (Y.M.L.) and R01NS076117 (Y.M.L.), Alzheimer Association Grant IIRG-12-242137 (Y.M.L.), and the JPB Foundation (Y.M.L.). Authors also acknowledge the MSK Cancer Center Support Grant/Core Grant (Grant P30 CA008748), Mr. William H. Goodwin and Mrs. Alice Goodwin, the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of MSKCC, and the William Randolph Hearst Fund in Experimental Therapeutics.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneur-o.6b00127.
Schematic representation of PS1, structure of TAMRA–azide, General Information, and chemical synthesis (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Lapinsky DJ, Johnson DS. Recent developments and applications of clickable photoprobes in medicinal chemistry and chemical biology. Future Med Chem. 2015;7:2143–2171. doi: 10.4155/fmc.15.136. [DOI] [PubMed] [Google Scholar]
- 2.Smith E, Collins I. Photoaffinity labeling in target- and binding-site identification. Future Med Chem. 2015;7:159–183. doi: 10.4155/fmc.14.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shelton CC, Zhu L, Chau D, Yang L, Wang R, Djaballah H, Zheng H, Li YM. Modulation of gamma-secretase specificity using small molecule allosteric inhibitors. Proc Natl Acad Sci U S A. 2009;106:20228–20233. doi: 10.1073/pnas.0910757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crump CJ, Castro SV, Wang F, Pozdnyakov N, Ballard TE, Sisodia SS, Bales KR, Johnson DS, Li YM. BMS-708,163 Targets Presenilin and Lacks Notch-Sparing Activity. Biochemistry. 2012;51:7209–7211. doi: 10.1021/bi301137h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump CJ, Fish BA, Castro SV, Chau DM, Gertsik N, Ahn K, Stiff C, Pozdnyakov N, Bales KR, Johnson DS, Li YM. Piperidine acetic acid based gamma-secretase modulators directly bind to Presenilin-1. ACS Chem Neurosci. 2011;2:705–710. doi: 10.1021/cn200098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian Y, Bassit B, Chau D, Li YM. An APP inhibitory domain containing the Flemish mutation residue modulates gamma-secretase activity for Abeta production. Nat Struct Mol Biol. 2010;17:151–158. doi: 10.1038/nsmb.1743. [DOI] [PubMed] [Google Scholar]
- 7.Chau DM, Crump CJ, Villa JC, Scheinberg DA, Li YM. Familial Alzheimer Disease Presenilin-1 Mutations Alter the Active Site Conformation of gamma-secretase. J Biol Chem. 2012;287:17288–17296. doi: 10.1074/jbc.M111.300483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gertsik N, Chau DM, Li YM. gamma-Secretase Inhibitors and Modulators Induce Distinct Conformational Changes in the Active Sites of gamma-Secretase and Signal Peptide Peptidase. ACS Chem Biol. 2015;10:1925. doi: 10.1021/acschembio.5b00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci U S A. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lapinsky DJ. Tandem photoaffinity labeling-bioorthogonal conjugation in medicinal chemistry. Bioorg Med Chem. 2012;20:6237–6247. doi: 10.1016/j.bmc.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Salisbury CM, Cravatt BF. Optimization of activity-based probes for proteomic profiling of histone deacetylase complexes. J Am Chem Soc. 2008;130:2184–2194. doi: 10.1021/ja074138u. [DOI] [PubMed] [Google Scholar]
- 12.Cisar JS, Cravatt BF. Fully functionalized small-molecule probes for integrated phenotypic screening and target identification. J Am Chem Soc. 2012;134:10385–10388. doi: 10.1021/ja304213w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eirich J, Orth R, Sieber SA. Unraveling the protein targets of vancomycin in living S. aureus and E. faecalis cells. J Am Chem Soc. 2011;133:12144–12153. doi: 10.1021/ja2039979. [DOI] [PubMed] [Google Scholar]
- 14.Ranjitkar P, Perera BG, Swaney DL, Hari SB, Larson ET, Krishnamurty R, Merritt EA, Villen J, Maly DJ. Affinity-based probes based on type II kinase inhibitors. J Am Chem Soc. 2012;134:19017–19025. doi: 10.1021/ja306035v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Hao P, Li L, Tan CY, Cheng X, Chen GY, Sze SK, Shen HM, Yao SQ. Design and synthesis of minimalist terminal alkyne-containing diazirine photo-crosslinkers and their incorporation into kinase inhibitors for cell- and tissue-based proteome profiling. Angew Chem Int Ed. 2013;52:8551–8556. doi: 10.1002/anie.201300683. [DOI] [PubMed] [Google Scholar]
- 16.Pozdnyakov N, Murrey HE, Crump CJ, Pettersson M, Ballard TE, Am Ende CW, Ahn K, Li YM, Bales KR, Johnson DS. gamma-Secretase modulator (GSM) photoaffinity probes reveal distinct allosteric binding sites on presenilin. J Biol Chem. 2013;288:9710–9720. doi: 10.1074/jbc.M112.398602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunner J. New photolabeling and crosslinking methods. Annu Rev Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 18.Park J, Koh M, Koo JY, Lee S, Park SB. Investigation of Specific Binding Proteins to Photoaffinity Linkers for Efficient Deconvolution of Target Protein. ACS Chem Biol. 2016;11:44–52. doi: 10.1021/acschembio.5b00671. [DOI] [PubMed] [Google Scholar]
- 19.Preston GW, Radford SE, Ashcroft AE, Wilson AJ. Analysis of Amyloid Nanostructures Using Photo-crosslinking: In Situ Comparison of Three Widely Used Photo-crosslinkers. ACS Chem Biol. 2014;9:761–768. doi: 10.1021/cb400731s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geurink PP, Florea BI, Van der Marel GA, Kessler BM, Overkleeft HS. Probing the proteasome cavity in three steps: bio-orthogonal photo-reactive suicide substrates. Chem Commun (Cambridge, UK) 2010;46:9052–9054. doi: 10.1039/c0cc03322g. [DOI] [PubMed] [Google Scholar]
- 21.Albertoni B, Hannam JS, Ackermann D, Schmitz A, Famulok M. A trifluoromethylphenyl diazirine-based SecinH3 photoaffinity probe. Chem Commun (Cambridge, UK) 2012;48:1272–1274. doi: 10.1039/c2cc16477a. [DOI] [PubMed] [Google Scholar]
- 22.Chan EW, Chattopadhaya S, Panicker RC, Huang X, Yao SQ. Developing photoactive affinity probes for proteomic profiling: hydroxamate-based probes for metalloproteases. J Am Chem Soc. 2004;126:14435–14446. doi: 10.1021/ja047044i. [DOI] [PubMed] [Google Scholar]
- 23.Tate JJ, Persinger J, Bartholomew B. Survey of four different photoreactive moieties for DNA photoaffinity labeling of yeast RNA polymerase III transcription complexes. Nucleic Acids Res. 1998;26:1421–1426. doi: 10.1093/nar/26.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber PJ, Beck-Sickinger AG. Comparison of the photochemical behavior of four different photoactivatable probes. J Pept Res. 1997;49:375–383. doi: 10.1111/j.1399-3011.1997.tb00889.x. [DOI] [PubMed] [Google Scholar]
- 25.Sakurai K, Ozawa S, Yamada R, Yasui T, Mizuno S. Comparison of the reactivity of carbohydrate photoaffinity probes with different photoreactive groups. ChemBioChem. 2014;15:1399–1403. doi: 10.1002/cbic.201402051. [DOI] [PubMed] [Google Scholar]
- 26.Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, Harrison T, Lellis C, Nadin A, Neduvelil JG, Register RB, Sardana MK, Shearman MS, Smith AL, Shi XP, Yin KC, Shafer JA, Gardell SJ. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- 27.Kornilova AY, Bihel F, Das C, Wolfe MS. The initial substrate-binding site of gamma-secretase is located on presenilin near the active site. Proc Natl Acad Sci U S A. 2005;102:3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kukar TL, Ladd TB, Bann MA, Fraering PC, Narlawar R, Maharvi GM, Healy B, Chapman R, Welzel AT, Price RW, Moore B, Rangachari V, Cusack B, Eriksen J, Jansen-West K, Verbeeck C, Yager D, Eckman C, Ye W, Sagi S, Cottrell BA, Torpey J, Rosenberry TL, Fauq A, Wolfe MS, Schmidt B, Walsh DM, Koo EH, Golde TE. Substrate-targeting gamma-secretase modulators. Nature. 2008;453:925–929. doi: 10.1038/nature07055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebke A, Luebbers T, Fukumori A, Shirotani K, Haass C, Baumann K, Steiner H. Novel gamma-secretase enzyme modulators directly target presenilin protein. J Biol Chem. 2011;286:37181–37186. doi: 10.1074/jbc.C111.276972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohki Y, Higo T, Uemura K, Shimada N, Osawa S, Berezovska O, Yokoshima S, Fukuyama T, Tomita T, Iwatsubo T. Phenylpiperidine-type gamma-secretase modulators target the transmembrane domain 1 of presenilin 1. EMBO J. 2011;30:4815–4824. doi: 10.1038/emboj.2011.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumpertz T, Rennhack A, Ness J, Baches S, Pietrzik CU, Bulic B, Weggen S. Presenilin Is the Molecular Target of Acidic gamma-Secretase Modulators in Living Cells. PLoS One. 2012;7:e30484. doi: 10.1371/journal.pone.0030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marjaux E, Hartmann D, De Strooper B. Presenilins in memory, Alzheimer’s disease, and therapy. Neuron. 2004;42:189–192. doi: 10.1016/s0896-6273(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 33.Crump CJ, Johnson DS, Li YM. Development and Mechanism of γ-Secretase Modulators for Alzheimer’s Disease. Biochemistry. 2013;52:3197–3216. doi: 10.1021/bi400377p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuwa H, Takahashi Y, Konno Y, Watanabe N, Miyashita H, Sasaki M, Natsugari H, Kan T, Fukuyama T, Tomita T, Iwatsubo T. Divergent synthesis of multifunctional molecular probes to elucidate the enzyme specificity of dipeptidic gamma-secretase inhibitors. ACS Chem Biol. 2007;2:408–418. doi: 10.1021/cb700073y. [DOI] [PubMed] [Google Scholar]
- 35.Ahn K, Shelton CC, Tian Y, Zhang X, Gilchrist ML, Sisodia SS, Li YM. Activation and intrinsic γ-secretase activity of presenilin 1. Proc Natl Acad Sci U S A. 2010;107:21435–21440. doi: 10.1073/pnas.1013246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorman G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 37.Rihakova L, Deraet M, Auger-Messier M, Perodin J, Boucard AA, Guillemette G, Leduc R, Lavigne P, Escher E. Methionine proximity assay, a novel method for exploring peptide ligand-receptor interaction. J Recept Signal Transduction Res. 2002;22:297–313. doi: 10.1081/rrs-120014603. [DOI] [PubMed] [Google Scholar]
- 38.Wittelsberger A, Thomas BE, Mierke DF, Rosenblatt M. Methionine acts as a “magnet” in photoaffinity crosslinking experiments. FEBS Lett. 2006;580:1872–1876. doi: 10.1016/j.febslet.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 39.Kage R, Leeman SE, Krause JE, Costello CE, Boyd ND. Identification of methionine as the site of covalent attachment of a p-benzoyl-phenylalanine-containing analogue of substance P on the substance P (NK-1) receptor. J Biol Chem. 1996;271:25797–25800. doi: 10.1074/jbc.271.42.25797. [DOI] [PubMed] [Google Scholar]
- 40.Clement M, Cabana J, Holleran BJ, Leduc R, Guillemette G, Lavigne P, Escher E. Activation induces structural changes in the liganded angiotensin II type 1 receptor. J Biol Chem. 2009;284:26603–26612. doi: 10.1074/jbc.M109.012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Placanica L, Tarassishin L, Yang G, Peethumnongsin E, Kim SH, Zheng H, Sisodia SS, Li YM. Pen2 and presenilin-1 modulate the dynamic equilibrium of presenilin-1 and presenilin-2 gamma-secretase complexes. J Biol Chem. 2009;284:2967–2977. doi: 10.1074/jbc.M807269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 43.Simon GM, Niphakis MJ, Cravatt BF. Determining target engagement in living systems. Nat Chem Biol. 2013;9:200–205. doi: 10.1038/nchembio.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, Zhou R, Scheres SH, Shi Y. An atomic structure of human gamma-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human gamma-secretase. eLife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li YM, Lai MT, Xu M, Huang Q, DiMuzio-Mower J, Sardana MK, Shi XP, Yin KC, Shafer JA, Gardell SJ. Presenilin 1 is linked with gamma-secretase activity in the detergent solubilized state. Proc Natl Acad Sci U S A. 2000;97:6138–6143. doi: 10.1073/pnas.110126897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Placanica L, Tarassishin L, Yang G, Peethumnongsin E, Kim SH, Zheng H, Sisodia SS, Li YM. Pen2 and Presenilin-1 Modulate the Dynamic Equilibrium of Presenilin-1 and Presenilin-2 {gamma}-Secretase Complexes. J Biol Chem. 2009;284:2967–2977. doi: 10.1074/jbc.M807269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Placanica L, Chien JW, Li YM. Characterization of an atypical gamma-secretase complex from hematopoietic origin. Biochemistry. 2010;49:2796–2804. doi: 10.1021/bi901388t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


