ABSTRACT
Fanconi anemia (FA) is a rare disease, in which homozygous or compound heterozygous inactivating mutations in any of 21 genes lead to genomic instability, early-onset bone marrow failure and increased cancer risk. The FA pathway is essential for DNA damage response (DDR) to DNA interstrand crosslinks. However, proteins of the FA pathway have additional cytoprotective functions that may be independent of DDR. We have shown that many FA proteins participate in the selective autophagy pathway that is required for the destruction of unwanted intracellular constituents. In this Cell Science at a Glance and the accompanying poster, we briefly review the role of the FA pathway in DDR and recent findings that link proteins of the FA pathway to selective autophagy of viruses and mitochondria. Finally, we discuss how perturbations in FA protein-mediated selective autophagy may contribute to inflammatory as well as genotoxic stress.
KEY WORDS: Selective autophagy, Fanconi anemia, Mitophagy, Inflammasome, Virophagy, DNA damage response
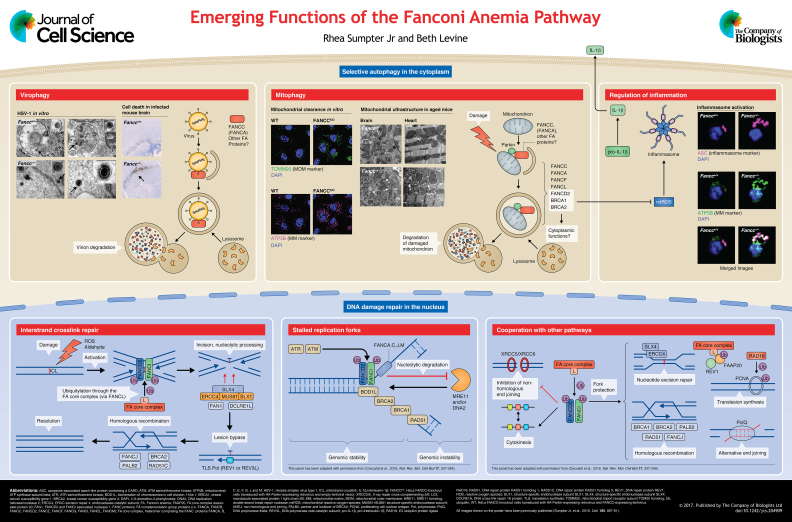
Summary: We discuss recently discovered links between the Fanconi anemia (FA) pathway and selective autophagy of viruses and mitochondria. Potential implications of these findings in FA pathophysiology are examined.
Introduction
Fanconi anemia (FA), first described by Guido Fanconi in 1927, is a rare and potentially devastating disease associated with inactivating homozygous or compound heterozygous mutations − except in the case of the gene for Fanconi anemia complementation group (FANC) B (FANCB), which is X chromosome-linked − in any of 21 FA genes (Garaycoechea and Patel, 2014). Affected patients suffer from bone marrow failure (with a 90% prevalence in the first decade of life), increased cancer susceptibility − particularly to acute myelogenous leukemia and squamous cell carcinomas of the upper digestive and urogenital tracts, and premature gonadal failure. Although some patients are phenotypically normal, congenital defects are often associated with FA, e.g. skeletal and skin abnormalities (Neveling et al., 2009). Although FA is a rare disease, inherited and somatic mutations or epigenetic silencing of FA genes are also found in many cancers. These include breast and ovarian cancers in patients with mutations of the inherited breast cancer susceptibility gene 1 (BRCA1), or breast cancer susceptibility gene 2 (BRCA2), as well as pancreatic, brain and other cancers that are associated with a loss-of-function of FA genes (D'Andrea, 2010; Mamrak et al., 2016) (see Box 1). In this Cell Science at a Glance article and accompanying poster, we will briefly review the role of the FA pathway in DNA damage response (DDR) and then explore a recently described function of FA proteins in selective autophagy and its implications for our understanding of the pathophysiology of FA.
Box 1. FA proteins and proteins associated with FA featuring in this article.
Official protein names are given in bold
BRAC1: breast cancer susceptibility gene 1 (also known as FANCS)
BRAC2: breast cancer susceptibility gene 2 (also known as FACD, FANCD1)
BRIP1: BRCA1 interacting protein C-terminal helicase 1 (also known as FANCJ)
ERCC4: excision repair 4, endonuclease catalytic subunit (also known as FANCQ)
FAAP20: FA core complex associated protein 20
FAN1: FANCD2 and FANCI-associated nuclease 1
FANCA: Fanconi anemia complementation group (FA comp group) A protein
FANCB: FA comp group B protein
FANCC: FA comp group C protein
FANCD2: FA comp group D2 protein (also known as FACD, FANCD)
FANCE: FA comp groupE
FANCF: FA comp group F
FANCG: FA comp group G
FANCI: FA comp group I protein
FANCL: FA comp group L protein (also known as E3 ubiquitin-protein ligase FANCL)
FANCM: FA comp group M protein
MAD2L2: mitotic arrest-deficient-2-like 2 (also known as REV7, FANCV)
PALB2: partner and localizer of BRCA2 (also known as FANCN)
RAD51: RAD51 recombinase (also known as FANCR)
RAD51C: RAD51 paralog C (also known as FANCO)
SLX4: structure-specific endonuclease subunit SLX4 (also known as FANCP)
UBE2T: ubiquitin-conjugating enzyme 2 T (also known as FANCT)
XRCC2: X-ray repair cross-complementing 2 (also known as FANCU)
Role of the FA pathway in DDR to of interstrand crosslinks
The FA pathway is required for the DDR to interstrand crosslinks (ICLs), highly toxic lesions in which adjacent bases on opposite DNA strands are covalently linked (Mamrak et al., 2016; Michl et al., 2016). It has been estimated that each human cell has to repair ∼10 ICLs/day (Grillari et al., 2007) and that as few as 20-40 unresolved ICL lesions can lead to cell death (Clauson et al., 2013). ICLs are generated during treatment with certain chemotherapeutic agents (e.g. nitrogen mustards, Mitomycin C, or Cisplatin) (Lopez-Martinez et al., 2016). However, even in the absence of exogenous insults, endogenous sources, such as reactive oxygen species (ROS) and their peroxidated intermediates, as well as reactive aldehydes, induce- ICLs (Clauson et al., 2013; Lopez-Martinez et al., 2016), although their relative contributions to the burden of ICLs in a given cell population are difficult to quantify. Unrepaired ICLs, ultimately, lead to DNA breakage and chromosomal rearrangements, and promote the development and/or progression of cancer (see below) (Ceccaldi et al., 2016).
Detection of ICLs by DDR surveillance proteins results in the activation of an eight-member FA core complex (consisting of FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FANCM) that directs the E3 ubiquitin ligase activity of FANCL to monoubiquitylate and thereby activate the DNA-binding heterocomplex between Fanconi anemia group D2 protein (FANCD2) and FANCI (see poster). The active FANCD2−FANCI complex, in turn, orchestrates the concerted downstream actions of the FA pathway and accessory proteins to resolve the ICL lesion (Boisvert and Howlett, 2014; Kim and D'Andrea, 2012; Mamrak et al., 2016; Renaudin et al., 2016) (see poster). Thus, FA proteins are crucial factors in the maintenance of genomic integrity; in fact, the FA pathway is the only known mechanism of repair for ICLs that, if left unrepaired, promote genotoxic stress, genomic instability and tumorigenesis.
Other nuclear functions of the FA pathway in genome maintenance and DDR
In addition to its canonical role in ICL repair, accumulating evidence indicates that subsets of FA proteins also participate in additional pathways that are crucial to maintain genomic integrity (see poster) (reviewed in detail by Ceccaldi et al., 2016). For example, heterodimers of ubiquitylated FANCD2 and FANCI (and, additionally, the FA core complex and accessory proteins) localize to stalled replication forks (Lossaint et al., 2013), where they protect nascent (single-stranded) DNA from degradation by nucleases (Schlacher et al., 2011, 2012) and suppress inappropriate new origin firing and entry into mitosis (Chaudhury et al., 2013; Chen et al., 2015). These functions prevent a destabilization or collapse of the replication fork and, thus, any resulting genomic instability (Ceccaldi et al., 2016). Members of the FA pathway are also required for the suppression of non-homologous end-joining (Renaud et al., 2016), the correct segregation of chromosomes during cytokinesis (Chan et al., 2009; Naim and Rosselli, 2009), as well as for numerous other forms of non-ICL-directed DDR, including nucleotide excision repair (Kelsall et al., 2012), translesion synthesis (Haynes et al., 2015), homologous recombination (Haynes et al., 2015) and alternative end joining (Nguyen et al., 2014). Thus, FA pathway proteins participate in diverse non-canonical (i.e. non-ICL repair) roles in concert with other pathways that are involved in genomic stability and DDR to ameliorate many forms of genotoxic stress.
Clues to a DDR-independent role for FANCC in cytoprotection
Although hypersensitivity to DNA damage is the best known phenotype of cells that are deficient in FA-gene function (Ceccaldi et al., 2016), it has long been appreciated that, at least some, genes of the FA pathway play additional cytoprotective roles, such as protection from cell death induced by ROS (Schindler and Hoehn, 1988), proinflammatory cytokines (Haneline et al., 1998; Whitney et al., 1996) or growth factor withdrawal (Cumming et al., 1996). Previous studies have offered some insight into possible mechanisms underlying the cytoprotective effects of the FA protein FANCC. For example, FANCC-mediated protection against proinflammatory cytokine-induced cell death is correlated with its biochemical interactions with signal transducer and activator of transcription 1 (STAT1) (Pang et al., 2000), protein kinase R (PKR; officially known as EIF2AK2) (Pang et al., 2002) and stress-inducible heat shock protein 70 (HSPA1A) (Pang et al., 2002, 2001b). FANCC-mediated protection against growth factor withdrawal-induced cell death is associated with its interaction and activation of the xenobiotic- and ROS-detoxifying enzyme glutathione S-transferase P1 (GSTP1) (Cumming et al., 2001).
Some of the earliest work on FANCC indicated that at least some of its cytoprotective roles might be independent of its role in DDR. For example, already in 1996, Yamashita et al. demonstrated that cells of FA patients that endogenously express the naturally occurring FANCC mutant c.67delG (which results in the use of an alternative start codon and the deletion of 54 N-terminal amino acids) were indistinguishable from FA patient cells that harbor a null mutation in FANCC with respect to their hypersensitivity to mitomycin C (MMC)-induced DNA damage (Yamashita et al., 1996). However, patients that carry the c.67delG mutant have a milder clinical course than patients with null mutations in FANCC (i.e. mutations that abrogate both its DDR and cytoprotective roles), suggesting that FANCC has other disease-modulating functions that are independent of its role in DDR (Neveling et al., 2009). Indeed, in a previous study, the FANCC c.67delG mutant was shown to rescue interferon-γ (IFNγ)-induced STAT1 activation, which is correlated with resistance to cell death induced by IFNγ and/or tumor necrosis factor-α (TNFα), to the same extent as wild-type FANCC (Pang et al., 2001a). Taken together, these in vitro results and clinical observations hint at the existence of a clinically important additional function or functions of FANCC beyond its role in DDR.
Identification of FA proteins as putative selective autophagy factors
Autophagy is a phylogenetically conserved cellular housekeeping pathway that plays pleiotropic roles in cellular and organismal homeostasis (Levine and Kroemer, 2008). In contrast to general (e.g. starvation-induced) autophagy, which is thought to be nonspecific, a specialized form of autophagy that has been termed selective autophagy, specifically targets unwanted cytoplasmic contents (e.g. viruses, intracellular bacteria, damaged mitochondria and endoplasmic reticulum, lipid droplets, peroxisomes) for engulfment by double-membraned vesicles (the so-called autophagosomes) and then delivered to lysosomes for destruction (Khaminets et al., 2016). Although our understanding of the mechanism(s) of selective autophagy has progressed significantly in recent years, much remains to be learned about this process before it can be harnessed for clinical use, for instance, to treat infectious diseases, cancer and conditions related to aging.
To this end, we performed a high-content, image-based, genome-wide screen to identify host factors involved the selective autophagy of Sindbis virus (virophagy) (Orvedahl et al., 2011). This screen identified genes that blocked the colocalization of a red fluorescent Sindbis virus capsid protein with microtubule-associated protein 1 light chain 3 alpha (MAP1LC3A, also known as LC3) conjugated to green fluorescent protein (GFP-LC3), a marker of autophagosomes. To our surprise, three FA genes, FANCC, FANCF and FANCL that, like the other FA genes, had no known association with autophagy, were confirmed positives in the virophagy screen. We also found a high degree of overlap between the positives from the virophagy screen and the genes that are required for another type of selective autophagy – Parkin-mediated autophagy of damaged mitochondria (a form of mitophagy) – suggesting a conservation of cellular factors required for the selective autophagy of seemingly unrelated cytoplasmic cargoes.
FANCC is required for virophagy
We first confirmed that FANCC is not required for non-selective starvation-induced autophagy but is required for Sindbis virophagy in murine embryonic fibroblasts (MEFs) because the colocalization of mCherry-labeled Sindbis virus capsids and autophagosomes is reduced in Fancc−/− MEFs (Sumpter et al., 2016). We also found that virophagy of a herpes simplex virus type 1 (HSV-1) mutant that lacks a 20 amino acid region of the infected cell protein 34.5 (ICP34.5) neurovirulence gene product − rendering it incapable of binding to the essential autophagy protein Beclin 1 and inhibiting autophagy − is impaired in Fancc−/− MEFs. Ultrastructural analysis of the cytoplasm of wild-type MEFs revealed that HSV-1 nucleocapsids are primarily located within autolysosomes and in the process of being degraded. In contrast, in Fancc−/− MEFs, intact HSV-1 nucleocapsids and enveloped virions can be found free in the cytoplasm, suggesting a failure of their targeting to autophagosomes/autolysosomes (see poster). Similar to other previously identified virophagy factors, such as sequestosome 1 (SQSTM1) (Orvedahl et al., 2010) and SMAD-specific E3 ubiquitin protein ligase 1 (SMURF1) (Orvedahl et al., 2011), FANCC (as well as FANCA) interacted biochemically with the Sindbis virus capsid protein. Additionally, both FANCC and Sindbis virus capsid protein can be immunoprecipiated within LC3-positive membrane structures, which are indicative of autophagosomes.
Importantly, Fancc−/− mice are more susceptible to death after intracerebral inoculation with both the neuronotropic RNA virus (Sindbis) and the neuronotropic DNA virus (HSV-1) (Sumpter et al., 2016). This increased susceptibility to lethal CNS infection is associated with increased neuronal cell death (as measured by TUNEL staining; see poster) and increased viral antigen loads, but not with raised levels of infectious virus titers. These phenotypes in mice with deletion of a selective autophagy factor (FANCC) are consistent with those observed in previous studies of mice with neuronal deficiency of Atg5 − a core component of the autophagy machinery (Orvedahl et al., 2010; Yordy et al., 2012). These results suggest that the autophagy pathway exerts its pro-survival role during viral infections of the CNS by removing excess viral protein products and preventing cell death due to proteotoxicity, i.e. damage of cellular functions due to protein misfolding.
Taken together, the in vitro and in vivo studies with Sindbis virus and HSV-1 (see poster) indicate that FANCC is an adaptor protein during virophagy that functions in antiviral host defense in the CNS (Sumpter et al., 2016). The precise mechanism by which FANCC and, potentially, additional FA proteins function to deliver viral cargoes for autophagic destruction remains to be elucidated, as do the precise mechanisms by which FANCC protects mice in vivo against lethal neuronotropic viral disease.
FANCC is required for mitophagy in a DDR-independent manner
While by definition selective autophagy is associated with exquisite cargo selectivity, there is also extensive evidence for shared mechanisms that are involved in the selective targeting of diverse intracellular cargoes (Khaminets et al., 2016). For example, the E3 ligase Parkin plays an important role in both the removal of damaged mitochondria (mitophagy) (Pickrell and Youle, 2015) and the autophagic targeting of the intracellular bacterium Mycobacterium tuberculosis (Manzanillo et al., 2013). Moreover, as noted above, there is extensive overlap between genes (including FANCC, FANCF and FANCL) that are required for selective virophagy and mitophagy − another form of selective autophagy (Orvedahl et al., 2011).
The concept that FA proteins can function in mitophagy is attractive as a potential mechanism that contributes to the cytoprotective functions of FANCC (discussed above) and a previously described role for FA proteins in mitochondrial quality control. Kumari et al. found that FA-deficient cells have a number of mitochondrial defects, including elevated levels of mitochondrial ROS, decreased mitochondrial membrane potential, decreased ATP production, impaired oxygen uptake and abnormal mitochondrial morphology (Kumari et al., 2014), although the molecular basis of the latter phenotype has not been described yet.
We confirmed a role for FANCC in mitophagy by using CRISPR/Cas9-mediated deletion in HeLa cells (see poster), FANCC mutant fibroblasts of FA patients and bone marrow-derived macrophages (BMDMs) from Fancc-deficient mice (Sumpter et al., 2016). Furthermore, we observed the accumulation of damaged mitochondria in post-mitotic tissues (i.e. brain and heart) of aged Fancc−/− mice (see poster) (Sumpter et al., 2016), a finding that is consistent with a defect in mitochondrial quality control in vivo. We could also show that FANCC (as well as FANCA) interacted biochemically with the E3 ubiquitin ligase Parkin and translocated to mitochondria (see poster) in a Parkin- and mitochondrial damage-dependent manner (Sumpter et al., 2016).
The role of FANCC in Parkin-mediated mitophagy appears to be genetically distinct from its role in DDR. Specifically, the c.67delG FANCC mutant, which − as mentioned above − is not functional in DDR but fully functional in protecting cells against cytokine-induced death, can completely restore Parkin-mediated mitophagy in FANCC knockout cells (Sumpter et al., 2016). Although these results strongly suggest that the DDR function of FANCC is dispensable for Parkin-mediated mitophagy, further studies are required to rule out other potential nuclear functions of FANCC in mitophagy.
Defective FANCC-mediated mitophagy and suppression of mtROS results in aberrant inflammasome signaling
Damaged mitochondria produce mitochondrial reactive oxygen species (mtROS) that activate the NACHT, LRR and PYD domain-containing protein 3 (NLRP3) inflammasome pathway, resulting in the caspase-1-mediated cleavage of pro-IL-1β (pro-interleukin-1β) followed by secretion of the potent proinflammatory cytokine interleukin-1β (IL-1β) (Zhou et al., 2011), and mitophagy downregulates inflammasome signaling by removing mtROS (Zhong et al., 2016) (see poster). Given the emerging interrelationships between mitophagy, mtROS and inflammasome activation, coupled with previous reports that link FANCC deficiency and increased mitochondrial ROS production (Kumari et al., 2014) or hyperactivation of inflammasome signaling (Garbati et al., 2013), we sought to determine whether a failure to clear mtROS-producing damaged mitochondria is responsible for enhanced inflammasome activation in FANCC-deficient cells. Indeed, primary BMDMs from Fancc−/− mice, primed with the pathogen-associated molecular pattern (PAMP) lipopolysaccharide (a component of the cell wall of Gram-negative bacteria) and then treated with extracellular ATP (to mimic the ‘danger’ signal from nearby dying cells) (de Zoete et al., 2014), results in increased mtROS generation, hypersecretion of IL-1β, and generation of more and larger apotosis-associated speck-like protein containing a containing a caspase recruitment domain (PYCARD, hereafter referred to as ASC) specks (i.e. markers of inflammasome assembly, overlying damaged mitochondria; see poster) (Sumpter, 2016). In agreement, addition of MitoTEMPO, a mtROS-specific free radical scavenger, dramatically reduces IL-1β secretion and ASC speck formation, which is consistent with a model in which failure of FANCC-mediated mitophagy during inflammasome activation results in mtROS-driven hyperactivation of the inflammasome and associated hypersecretion of IL-1β (see poster).
Additional FA proteins are required for mitophagy
The role for FA proteins in Parkin-mediated mitophagy seems to extend to multiple family members. At least in siRNA knockdown studies, the core complex proteins FANCF, FANCL and FANCA (the most commonly mutated protein in FA patients) are required, along with FANCD2, BRCA1 and BRCA2, which have no known direct functions outside the nucleus (see poster) (Sumpter et al., 2016). Moreover, another group recently found that cells from patients that carry FANCA (or FANCC) mutations have impaired levels of both basal and mitochondrial uncoupling agent-induced mitophagy (Shyamsundar et al., 2016).
Currently, it is impossible to conclude whether the different FA proteins function directly in mitophagy or indirectly, i.e. as a result of other defined cellular functions, including the nuclear roles discussed above. Although it conceptually more straightforward to postulate direct roles for FA proteins that are already known to have cytoplasmic functions, the possibility of direct roles for other FA proteins without any known cytoplasmic functions warrants further investigation. A mitochondrial localization of some of these proteins has been described, although, except for a role in mitophagy, no mitochondrial function is currently known. For example, FANCD2 that is not monoubiquitylated (and, therefore, not targeted by the FA core complex) is constitutively present in immunoprecipitated mitochondria (Sumpter et al., 2016), and a fraction of BRCA1 is located at the mitochondrial matrix (Coene et al., 2005).
A general role for DDR pathways in mitochondrial quality control and mitophagy?
In recent years, evidence has mounted that defects in multiple non-FA DDR pathways result in defective mitophagy, leading to accumulation of damaged mitochondria and concomitant increased intracellular oxidative stress (Drake et al., 2017). These defects include mutations in the ataxia-telangiectasia (A-T) double-strand break repair pathway (Valentin-Vega et al., 2012), the Cockayne syndrome B transcription-coupled repair pathway (Scheibye-Knudsen et al., 2012) and the xeroderma pigmentosum group A nucleotide excision repair pathway (Fang et al., 2014). Interestingly, the A-T serine/threonine kinase (ATM) has also been shown to be required for pexophagy, the removal of peroxisomes (which are also a source of intracellular ROS) by selective autophagy (Zhang et al., 2015). This suggests that, like FANCC, ATM is also required for the removal of diverse substrates by the selective autophagy pathway. As with FA proteins, more studies are needed to determine the precise molecular mechanisms by which other non-FA, DDR pathway proteins function in selective autophagy.
Physiological consequences of defects in mitophagy
A causal connection between mitophagy and tumor suppression is strongly supported by the observations that mitophagy-associated proteins, including the E3 ubiquitin-protein ligase parkin (PARK2), Bcl-2/adenovirus E1B 19-kDa-interacting protein 3 (BNIP3) and Bcl-2/adenovirus E1B 19-kDa-interacting protein 3-like (BNIP3L) have been linked with cancer (Chourasia et al., 2015). Pathogenic alterations in mitochondrial homeostasis are thought to contribute to tumorigenesis through a variety of mechanisms. These include changes in mitochondrial metabolic pathways to support tumor cell metabolism (metabolic reprogramming), as well as pro-tumor changes in cell signaling due to oxidative post-translational protein modifications (Vyas et al., 2016). Mitochondria-derived ROS also directly or indirectly oxidize cellular macromolecules (e.g. proteins, lipids, DNA), ultimately leading to increased genotoxic stress (Drake et al., 2017; Vyas et al., 2016). Thus, a potential role for mitophagy in the pathogenesis of cancers in FA and other DDR-associated diseases associated with the accumulation of damaged mitochondria warrants further exploration.
Aberrant IL-1β signaling, which is another consequence of defective mitophagy (Zhou et al., 2011), has been implicated in a wide range of pathophysiologies, including autoinflammatory diseases (Zhong et al., 2016), cancer (Zitvogel et al., 2012), metabolic disorders (de Zoete et al., 2014) and senescence (Salama et al., 2014). Drugs targeting the inflammasome pathway are already used to treat some of these diseases (Shao et al., 2015). A crucial unanswered question is to what extent aberrant inflammasome activation, in addition to genotoxic stress, contributes to bone marrow failure, cancer susceptibility and developmental abnormalities − all of which are observed in patients that carry mutations of FANCC and other FA genes (Auerbach, 2009; Ceccaldi et al., 2016; Mamrak et al., 2016; Neveling et al., 2009).
Conclusions
The FA pathway is crucial not only in DDR of ICLs but it also has emerging roles in other nuclear functions (stabilization of replication forks, cytokinesis and other types of DNA repair), as well as in the cytoplasmic process of selective autophagy. Although there are many open questions regarding the mechanism of action of FA proteins in their ‘non-canonical’ roles, the exciting emerging links between the FA pathway and selective autophagy might provide a roadmap to new and unexpected therapeutic targets in order to treat FA and diseases associated with FA gene mutations.
An intriguing question is why the FA pathway would be involved in selective autophagy in addition to its role in DDR. We speculate this pathway is involved because ROS can inflict the type of DNA lesion (ICL) the FA pathway is required to repair; moreover, it acts at two crucial stages to protect the cell against genotoxic stress. By helping to remove the main endogenous source of ROS (mtROS from damaged mitochondria), members of the FA pathway minimize the generation of nuclear ICLs. As well as deficiencies in the direct FA DNA repair pathway, the failure to suppress extensive genotoxic stress emanating from the cytoplasm by FA proteins might be important in the pathophysiologies associated with FA, such as cancer and accelerated aging (Brosh et al., 2017; Drake et al., 2017). More broadly, it is possible that other types of DNA damage linked to ROS are also modulated by mitophagy, thus providing a common selective pressure for different DNA damage pathways to function together in selective autophagy (Drake et al., 2017).
Acknowledgements
We are grateful to Angela Diehl for assistance with the graphic design and illustrations of the poster.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Work in B.L.’s laboratory was supported by the National Institutes of Health (grant numbers: K08 AI099150 to R.S., U19 AI109725 to B.L., RO1 CA109618 to B.L.), the Burroughs Wellcome Fund (Career Award for Medical Scientists to R.S.), the University of Texas Southwestern Medical Center President's Research Council (Distinguished Researcher Award to R.S.), and the Cancer Prevention and Research Institute of Texas (grant number: RP120718 to B.L.). Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.204909.supplemental
References
- Auerbach A. D. (2009). Fanconi anemia and its diagnosis. Mutat. Res. 668, 4-10. 10.1016/j.mrfmmm.2009.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert R. A. and Howlett N. G. (2014). The Fanconi anemia ID2 complex: dueling saxes at the crossroads. Cell Cycle 13, 2999-3015. 10.4161/15384101.2014.956475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R. M. Jr, Bellani M., Liu Y. and Seidman M. M. (2017). Fanconi Anemia: a DNA repair disorder characterized by accelerated decline of the hematopoietic stem cell compartment and other features of aging. Ageing Res. Rev. 33, 67-75. 10.1016/j.arr.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Sarangi P. and D'Andrea A. D. (2016). The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 17, 337-349. 10.1038/nrm.2016.48 [DOI] [PubMed] [Google Scholar]
- Chan K. L., Palmai-Pallag T., Ying S. and Hickson I. D. (2009). Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 11, 753-760. 10.1038/ncb1882 [DOI] [PubMed] [Google Scholar]
- Chaudhury I., Sareen A., Raghunandan M. and Sobeck A. (2013). FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 41, 6444-6459. 10.1093/nar/gkt348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-H., Jones M. J. K., Yin Y., Crist S. B., Colnaghi L., Sims R. J. III, Rothenberg E., Jallepalli P. V. and Huang T. T. (2015). ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol. Cell 58, 323-338. 10.1016/j.molcel.2015.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourasia A. H., Boland M. L. and Macleod K. F. (2015). Mitophagy and cancer. Cancer Metab. 3, 4 10.1186/s40170-015-0130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauson C., Scharer O. D. and Niedernhofer L. (2013). Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb. Perspect. Biol. 5, a012732 10.1101/cshperspect.a012732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coene E. D., Hollinshead M. S., Waeytens A. A., Schelfhout V. R., Eechaute W. P., Shaw M. K., Van Oostveldt P. M. and Vaux D. J. (2005). Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol. Biol. Cell 16, 997-1010. 10.1091/mbc.E04-10-0895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming R. C., Lightfoot J., Beard K., Youssoufian H., O'Brien P. J. and Buchwald M. (2001). Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat. Med. 7, 814-820. 10.1038/89937 [DOI] [PubMed] [Google Scholar]
- Cumming R. C., Liu J. M., Youssoufian H. and Buchwald M. (1996). Suppression of apoptosis in hematopoietic factor-dependent progenitor cell lines by expression of the FAC gene. Blood 88, 4558-4567. [PubMed] [Google Scholar]
- D'Andrea A. D. (2010). Susceptibility pathways in Fanconi's anemia and breast cancer. N. Engl. J. Med. 362, 1909-1919. 10.1056/NEJMra0809889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoete M. R., Palm N. W., Zhu S. and Flavell R. A. (2014). Inflammasomes. Cold Spring Harb. Perspect. Biol. 6, a016287 10.1101/cshperspect.a016287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake L. E., Springer M. Z., Poole L. P., Kim C. J. and Macleod K. F. (2017). Expanding perspectives on the significance of mitophagy in cancer. Semin. Cancer Biol. [Epub ahead of print] 10.1016/j.semcancer.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang E. F., Scheibye-Knudsen M., Brace L. E., Kassahun H., SenGupta T., Nilsen H., Mitchell J. R., Croteau D. L. and Bohr V. A. (2014). Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 157, 882-896. 10.1016/j.cell.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea J. I. and Patel K. J. (2014). Why does the bone marrow fail in Fanconi anemia? Blood 123, 26-34. 10.1182/blood-2013-09-427740 [DOI] [PubMed] [Google Scholar]
- Garbati M. R., Hays L. E., Keeble W., Yates J. E., Rathbun R. K. and Bagby G. C. (2013). FANCA and FANCC modulate TLR and p38 MAPK-dependent expression of IL-1beta in macrophages. Blood 122, 3197-3205. 10.1182/blood-2013-02-484816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillari J., Katinger H. and Voglauer R. (2007). Contributions of DNA interstrand cross-links to aging of cells and organisms. Nucleic Acids Res. 35, 7566-7576. 10.1093/nar/gkm1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneline L. S., Broxmeyer H. E., Cooper S., Hangoc G., Carreau M., Buchwald M. and Clapp D. W. (1998). Multiple inhibitory cytokines induce deregulated progenitor growth and apoptosis in hematopoietic cells from Fac-/- mice. Blood 91, 4092-4098. 10.1203/00006450-199804001-00786 [DOI] [PubMed] [Google Scholar]
- Haynes B., Saadat N., Myung B. and Shekhar M. P. V. (2015). Crosstalk between translesion synthesis, Fanconi anemia network, and homologous recombination repair pathways in interstrand DNA crosslink repair and development of chemoresistance. Mutat. Res. Rev. Mutat. Res. 763, 258-266. 10.1016/j.mrrev.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall I. R., Langenick J., MacKay C., Patel K. J. and Alpi A. F. (2012). The Fanconi anaemia components UBE2T and FANCM are functionally linked to nucleotide excision repair. PLoS ONE 7, e36970 10.1371/journal.pone.0036970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A., Behl C. and Dikic I. (2016). Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6-16. 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Kim H. and D'Andrea A. D. (2012). Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 26, 1393-1408. 10.1101/gad.195248.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari U., Ya Jun W., Huat Bay B. and Lyakhovich A. (2014). Evidence of mitochondrial dysfunction and impaired ROS detoxifying machinery in Fanconi anemia cells. Oncogene 33, 165-172. 10.1038/onc.2012.583 [DOI] [PubMed] [Google Scholar]
- Levine B. and Kroemer G. (2008). Autophagy in the pathogenesis of disease. Cell 132, 27-42. 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Martinez D., Liang C.-C. and Cohn M. A. (2016). Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell. Mol. Life Sci. 73, 3097-3114. 10.1007/s00018-016-2218-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossaint G., Larroque M., Ribeyre C., Bec N., Larroque C., Décaillet C., Gari K. and Constantinou A. (2013). FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol. Cell 51, 678-690. 10.1016/j.molcel.2013.07.023 [DOI] [PubMed] [Google Scholar]
- Mamrak N. E., Shimamura A. and Howlett N. G. (2016). Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 3, 93-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo P. S., Ayres J. S., Watson R. O., Collins A. C., Souza G., Rae C. S., Schneider D. S., Nakamura K., Shiloh M. U. and Cox J. S. (2013). The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501, 512-516. 10.1038/nature12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl J., Zimmer J. and Tarsounas M. (2016). Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 35, 909-923. 10.15252/embj.201693860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V. and Rosselli F. (2009). The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 11, 761-768. 10.1038/ncb1883 [DOI] [PubMed] [Google Scholar]
- Neveling K., Endt D., Hoehn H. and Schindler D. (2009). Genotype-phenotype correlations in Fanconi anemia. Mutat. Res. 668, 73-91. 10.1016/j.mrfmmm.2009.05.006 [DOI] [PubMed] [Google Scholar]
- Nguyen T. V., Riou L., Aoufouchi S. and Rosselli F. (2014). Fanca deficiency reduces A/T transitions in somatic hypermutation and alters class switch recombination junctions in mouse B cells. J. Exp. Med. 211, 1011-1018. 10.1084/jem.20131637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., MacPherson S., Sumpter R. Jr, Tallóczy Z., Zou Z. and Levine B. (2010). Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7, 115-127. 10.1016/j.chom.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Sumpter R. Jr, Xiao G., Ng A., Zou Z., Tang Y., Narimatsu M., Gilpin C., Sun Q., Roth M. et al. (2011). Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480, 113-117. 10.1038/nature10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q., Fagerlie S., Christianson T. A., Keeble W., Faulkner G., Diaz J., Rathbun R. K. and Bagby G. C. (2000). The Fanconi anemia protein FANCC binds to and facilitates the activation of STAT1 by gamma interferon and hematopoietic growth factors. Mol. Cell. Biol. 20, 4724-4735. 10.1128/MCB.20.13.4724-4735.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q., Christianson T. A., Keeble W., Diaz J., Faulkner G. R., Reifsteck C., Olson S. and Bagby G. C. (2001a). The Fanconi anemia complementation group C gene product: structural evidence of multifunctionality. Blood 98, 1392-1401. 10.1182/blood.V98.5.1392 [DOI] [PubMed] [Google Scholar]
- Pang Q., Keeble W., Christianson T. A., Faulkner G. R. and Bagby G. C. (2001b). FANCC interacts with Hsp70 to protect hematopoietic cells from IFN-gamma/TNF-alpha-mediated cytotoxicity. EMBO J. 20, 4478-4489. 10.1093/emboj/20.16.4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q., Christianson T. A., Keeble W., Koretsky T. and Bagby G. C. (2002). The anti-apoptotic function of Hsp70 in the interferon-inducible double-stranded RNA-dependent protein kinase-mediated death signaling pathway requires the Fanconi anemia protein, FANCC. J. Biol. Chem. 277, 49638-49643. 10.1074/jbc.M209386200 [DOI] [PubMed] [Google Scholar]
- Pickrell A. M. and Youle R. J. (2015). The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron 85, 257-273. 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud E., Barascu A. and Rosselli F. (2016). Impaired TIP60-mediated H4K16 acetylation accounts for the aberrant chromatin accumulation of 53BP1 and RAP80 in Fanconi anemia pathway-deficient cells. Nucleic Acids Res. 44, 648-656. 10.1093/nar/gkv1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin X., Koch Lerner L., Menck C. F. M. and Rosselli F. (2016). The ubiquitin family meets the Fanconi anemia proteins. Mutat. Res. Rev. Mutat. Res. 769, 36-46. 10.1016/j.mrrev.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Salama R., Sadaie M., Hoare M. and Narita M. (2014). Cellular senescence and its effector programs. Genes Dev. 28, 99-114. 10.1101/gad.235184.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibye-Knudsen M., Ramamoorthy M., Sykora P., Maynard S., Lin P.-C., Minor R. K., Wilson D. M. III, Cooper M., Spencer R., de Cabo R. et al. (2012). Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J. Exp. Med. 209, 855-869. 10.1084/jem.20111721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler D. and Hoehn H. (1988). Fanconi anemia mutation causes cellular susceptibility to ambient oxygen. Am. J. Hum. Genet. 43, 429-435. [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H. and Jasin M. (2011). Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529-542. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H. and Jasin M. (2012). A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106-116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao B. Z., Xu Z. Q., Han B. Z., Su D. F. and Liu C. (2015). NLRP3 inflammasome and its inhibitors: a review. Front. Pharmacol. 6, 262 10.3389/fphar.2015.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamsunder P., Esner M., Barvalia M., Wu Y. J., Loja T., Boon H. B., Lleonart M. E., Verma R. S., Krejci L. and Lyakhovich A. (2016). Impaired mitophagy in Fanconi anemia is dependent on mitochondrial fission. Oncotarget 7, 58065-58074. 10.18632/oncotarget.11161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R. Jr, Sirasanagandla S., Fernández A. F., Wei Y., Dong X., Franco L., Zou Z., Marchal C., Lee M. Y., Clapp D. W. et al. (2016). Fanconi anemia proteins function in mitophagy and immunity. Cell 165, 867-881. 10.1016/j.cell.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega Y. A., Maclean K. H., Tait-Mulder J., Milasta S., Steeves M., Dorsey F. C., Cleveland J. L., Green D. R. and Kastan M. B. (2012). Mitochondrial dysfunction in ataxia-telangiectasia. Blood 119, 1490-1500. 10.1182/blood-2011-08-373639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S., Zaganjor E. and Haigis M. C. (2016). Mitochondria and cancer. Cell 166, 555-566. 10.1016/j.cell.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney M. A., Royle G., Low M. J., Kelly M. A., Axthelm M. K., Reifsteck C., Olson S., Braun R. E., Heinrich M. C., Rathbun R. K. et al. (1996). Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood 88, 49-58. [PubMed] [Google Scholar]
- Yamashita T., Wu N., Kupfer G., Corless C., Joenje H., Grompe M. and D'Andrea A. D. (1996). Clinical variability of Fanconi anemia (type C) results from expression of an amino terminal truncated Fanconi anemia complementation group C polypeptide with partial activity. Blood 87, 4424-4432. [PubMed] [Google Scholar]
- Yordy B., Iijima N., Huttner A., Leib D. and Iwasaki A. (2012). A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe 12, 334-345. 10.1016/j.chom.2012.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Tripathi D. N., Jing J., Alexander A., Kim J., Powell R. T., Dere R., Tait-Mulder J., Lee J.-H., Paull T. T. et al. (2015). ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 17, 1259-1269. 10.1038/ncb3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Sanchez-Lopez E. and Karin M. (2016). Autophagy, NLRP3 inflammasome and auto-inflammatory/immune diseases. Clin. Exp. Rheumatol. 34, 12-16. [PubMed] [Google Scholar]
- Zhou R., Yazdi A. S., Menu P. and Tschopp J. (2011). A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221-225. 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Kepp O., Galluzzi L. and Kroemer G. (2012). Inflammasomes in carcinogenesis and anticancer immune responses. Nat. Immunol. 13, 343-351. 10.1038/ni.2224 [DOI] [PubMed] [Google Scholar]