Abstract
Multi-subunit transcription factors (TF) direct RNA polymerase (pol) III to synthesize a variety of essential small transcripts such as tRNAs, 5S rRNA and U6 snRNA. Use by pol III of both TATA-less and TATA-containing promoters, together with progress in the Saccharomyces cerevisiae and human systems towards elucidating the mechanisms of actions of the pol III TFs, provides a paradigm for eukaryotic gene transcription. Human and S.cerevisiae pol III components reveal good general agreement in the arrangement of orthologous TFs that are distributed along tRNA gene control elements, beginning upstream of the transcription initiation site and extending through the 3′ terminator element, although some TF subunits have diverged beyond recognition. For this review we have surveyed the Schizosaccharomyces pombe database and identified 26 subunits of pol III and associated TFs that would appear to represent the complete core set of the pol III machinery. We also compile data that indicate in vivo expression and/or function of 18 of the fission yeast proteins. A high degree of homology occurs in pol III, TFIIIB, TFIIIA and the three initiation-related subunits of TFIIIC that are associated with the proximal promoter element, while markedly less homology is apparent in the downstream TFIIIC subunits. The idea that the divergence in downstream TFIIIC subunits is associated with differences in pol III termination-related mechanisms that have been noted in the yeast and human systems but not reviewed previously is also considered.
INTRODUCTION
RNA polymerase III (pol III) synthesizes a large variety of small RNAs of cellular and viral origin (1). In order to produce sufficient amounts of small RNAs of the correct specific structure, the initiation, termination and recycling phases of transcription by pol III must be accurate and efficient. This requires an elaborate array of cooperating proteins that comprise pol III and its associated transcription factors (TF). The variety of TATA-less and TATA-containing promoters that direct pol III transcription, together with the multi-subunit TFs that recognize them, provide a paradigm for eukaryotic gene transcription (1–5).
Recent studies suggest that the fission yeast Schizosaccharomyces pombe represents an attractive model of eukaryotic transcription because it exhibits similarities to and differences from other pol III systems (6–8). For example, unlike the TATA-less tRNA promoters that have been extensively characterized in human, Xenopus, Drosophila and Saccharomyces cerevisiae (1–3), many tRNA genes in S.pombe contain an upstream TATA sequence (9), which comprises an essential promoter element (M.Hamada and R.Maraia, manuscript submitted) suggesting function similar to the U6 snRNA gene (10). Another example is the differential α-amanitin sensitivity of S.pombe, S.cerevisiae and human pols III, the pattern of which correlates with the sensitivity of these polymerases to a minimal length termination signal (6,8). Moreover, an essential subunit of S.cerevisiae TFIIIC, TFC6p, and the human regulatory factor (h)TFIIICβ were shown to be orthologs only after Sfc6p, a subunit of S.pombe TFIIIC, was characterized (7).
We have been systematically searching through S.pombe genomic sequences for pol III and related components and have previously reported on four subunits of TFIIIC (7). The recent completion of the sequencing of the S.pombe DNA and annotation of a protein database provides an opportunity to compile the S.pombe homologs of all of known pol III components in S.cerevisiae and humans.
OVERVIEW OF POL III TRANSCRIPTION: COMBINATORIAL USE OF PROMOTER ELEMENTS
Although a short summary of pol III transcription is appropriate here, the reader is referred to several excellent reviews for more detailed information (1–5,11). Based on promoter structures and factor requirements, genes transcribed by pol III have been divided into three types. The promoters of 5S rRNA genes (type 1) are composed of a major internal element, the C box, as well as additional elements that vary among species (11). tRNA genes, adenovirus VA genes, Alu sequences and other short interspersed elements constitute type 2 genes. The promoters of these genes are also internal, consisting of two highly conserved sequence elements, a proximal A box and a more distal B box, within the transcribed region. While the A box resides within 10–20 bp of the start site of transcription, the distance between the A and B boxes is variable, including a range of different length introns and variable stem regions found in eukaryotic tRNA genes (11). A separate control element, the pol III terminator, resides 20–25 bp downstream of the B box of tRNA genes. TFIIIC binds along the entire length of a tRNA gene, beginning just upstream of the start site of transcription and extending through the terminator (3). The ability to accommodate the variety of different A box–B box distances usually found in tRNA genes (30–60 bp) is mediated, at least in part, by TFC4p, a large protein with multiple tetratricopeptide repeats (TPR) that appears to provide the flexibility required for such elasticity (12; see below).
Whereas recognition of the type 2 promoter is responsible for direct recruitment of the TFIIIC complex, recognition of the 5S promoter is principally mediated by a single polypeptide, TFIIIA. While TFIIIC also binds along the length of the 5S gene in the presence of TFIIIA and is required for transcription, the exact mechanism of TFIIIC recruitment to the type 1 promoter remains unclear (3). For type 1 and 2 promoters the proximal subunits of TFIIIC direct TFIIIB to bind upstream of the transcription start site (3,13,14). TFIIIB then recruits and positions pol III over the initiation site and remains stably bound to the DNA through multiple rounds of re-initiation by pol III (15).
As mentioned above, accurate and efficient termination as well as rapid re-initiation are important aspects of pol III transcription. The pol III termination signal is comprised of oligo(dT) in the non-template strand and has been reported to stimulate re-initiation by pol III on transcription complexes in the S.cerevisiae and human systems (16,17). TFIIIC or associated components contact the terminator regions of tRNA (and other) genes in S.cerevisiae and humans (3,18–21). In the human system this binding by TFIIIC components is mediated in part by the partially characterized activities referred to as TFIIIC1 and TFIIIC0 and is associated with increased termination and re-initiation (17–20,22). Several factors have been reported to stimulate termination and/or re-initiation by pol III in the human system; these include topoisomerase I, positive factor 4 (PC4), a pol II co-activator, nuclear factor 1 (NF1, see below) and the La antigen, a RNA UUU-OH-terminus-binding protein (17,19,23–26), although this function of La has recently been disputed (27,28). As alluded to above, a commonality of these latter proteins is that they also appear to function in aspects of nucleic acid metabolism apart from pol III transcription. It is important to note for the purposes of this review that, in contrast to the human system, no evidence of TFIIIC involvement in termination and/or re-initiation in S.cerevisiae has been observed and TFIIIB alone can direct multiple round transcription in the budding yeast (15).
Type 3 genes include the vertebrate U6, 7SK, hY4, hY5 and H1 snRNA genes, which utilize an upstream TATA element that functions as one component of an entirely upstream multi-partite promoter (29–32). The well-characterized promoter of the human U6 snRNA gene consists of an upstream TATA box, a proximal sequence element (PSE) and a distal sequence element (DSE). The PSE functions with the TATA element to recruit the TFs SNAPc/PTF and a TFIIIB-like activity (TFIIIB-α, see below) (30,33–35). It is noteworthy that the SNAPc complex is also used by pol II and associated general TFs to promote transcription of U1 (and several other) snRNA genes (36–39), although the latter do not contain TATA elements (40,41). The transcriptional activator Oct-1 is recruited by the upstream DSE (42–45) and functions in part by promoting binding of the SNAPc complex to the PSE (37,46). A stable initiation complex is formed by cooperative interactions between TFIIIB, SNAPc and Oct-1 bound to their respective promoter elements, in part mediated by a nucleosome that is positioned between the DSE and PSE (44,45,47).
In contrast to the vertebrate U6 snRNA gene, the promoter structure of the S.cerevisiae (sc)U6 gene lacks a PSE and DSE but instead includes a downstream B box, the latter of which resides ∼120 bp beyond the terminator (48,49). Thus, although the scU6 snRNA gene (SNR6) uses TATA and B box elements that are external to the transcribed region and are therefore not considered as type 1 or 2, it is to be distinguished from the vertebrate type 3 genes, which use TATA and other promoter elements that are entirely upstream. It is also noteworthy that although the unusually lengthy A box–B box distance in the scU6 gene may be sub-optimal for transcription complex assembly, it appears that scTFIIIC nonetheless participates in recruiting TFIIIB to the TATA element, apparently in conjunction with Nhp6p, a non-histone chromatin protein of the HMG1 class (50,51). While S.cerevisiae TFIIIB (scTFIIIB) can be recruited directly by the upstream TATA of the scU6 gene in vitro (52), scTFIIIC is required to activate U6 transcription in vivo (53).
Some pol III-transcribed genes do not fit into types 1, 2 or 3 because they use combinations of promoter elements different from those described above (1). However, in many if not all of the above cases an important function of the various promoter elements and their associated TFs (e.g. PSE/SNAPc versus B box/TFIIIC) is to bring TFIIIB to the start site of transcription and stabilize it there; in some cases this involves interactions between the TATA-binding protein (TBP) of TFIIIB and a TATA promoter element, while in other cases TBP, with associated TFIIIB subunits, is induced to bind the upstream DNA in the absence of a TATA motif (54).
TRANSCRIPTION FACTOR IIIC
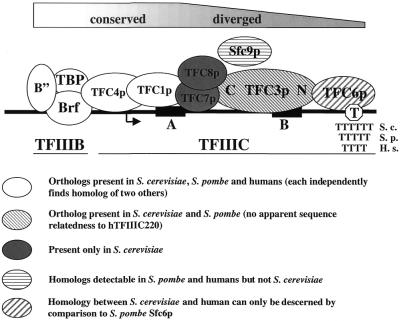
scTFIIIC, historically referred to as τ (55), is composed of six essential subunits, TFC1p/τ95, TFC3p/τ138, TFC4p/τ131/PCF1, TFC6p/τ90, TFC7p/τ55 and TFC8p/τ60 (21,56–62). The spatial organization of scTFIIIC subunits on an intron-containing tRNATyr gene and a 5S rRNA gene has been determined by photocrosslinking (63–65). Comprehensive results obtained using genetic methods (4,66) and results for human TFIIIC (14,67) are in general agreement with this model, which is schematically represented in Figure 1.
Figure 1.
Highly schematized cartoon of TFs assembled onto a tRNA gene (3) that also reflects the relative evolutionary conservation of individual TFIIIB and TFIIIC subunits, as described in the text. For most of the subunits the nomenclature follows that for S.cerevisiae (4). The positions of the A box and B box promoter elements are indicated. The arrow indicates the direction and site of initiation of transcription. T indicates the site of transcription termination. The minimal numbers of T sites required for efficient termination by S.cerevisiae (6, S. c.), S.pombe (5, S. p.) and human (4, H. s.) pols III are indicated schematically (8).
Human TFIIIC (hTFIIIC) has been chromatographically separated into two sub-complexes, hTFIIIC2, which exhibits B box-specific binding, and hTFIIIC1, which stimulates binding by hTFIIIC2 and is required for transcription activity (20,22,68). hTFIIIC2 is similar to scTFIIIC, composed of five subunits that together recognize the type 2 promoter (hTFIIIC220/TFIIICα, hTFIIIC110/TFIIICβ, hTFIIIC102/TFIIICγ, hTFIIIC90/TFIIICδ and hTFIIIC63/TFIIICɛ) (14,67,69–72). Although the molecular composition of hTFIIIC1 activity has not been defined, recent studies suggest that NF1 may be a component of hTFIIIC1 and TFIIIC0 (19). NF1 binds near the terminator, interacts with the two TFIIIC downstream subunits, TFIIICα and TFIIICβ, and stimulates transcription of the VA1 RNA gene at levels stoichiometrically similar to TFIIIC (19).
Five subunits of S.pombe TFIIIC (spTFIIIC) were identified by BLAST, four of which have been characterized biochemically and/or genetically (7; Table 1). Sfc1p is homologous to TFC1p and hTFIIIC63, orthologs that bind near the A box (3,14,65). In S.cerevisiae TFC1p forms a sub-complex with another subunit that appears to be involved in responding to carbon nutrients (56), suggesting a role for TFC1 (and, by association, hTFIIIC102) in the integration of environmental stimuli.
Table 1. Comparison of TFIIIC subunits of S.pombe, S.cerevisiae and humana.
| S.pombe TFIIIC | S.cerevisiae homolog | Human homolog | S.pombe cosmid no. | ||||||||||
| Subunit |
Mass (kDa) |
Length (amino acids) |
Subunit |
Length |
Identity (%) |
Similarity (%) |
Alignmentb |
Subunit |
Length |
Identity (%) |
Similarity (%) |
Alignmentb |
EMBL locusc |
| Sfc1p | 53 | 456 | TFC1p | 649 | 23 | 41 | 366 | TFIIIC63 | 519 | 28 | 44 | 319 | SPAC6F12.11c |
| Sfc3p | 154 | 1339 | TFC3p | 1160 | 20 | 41 | 891 | TFIIIC220 | 2140 | d | d | d | SPBC336.07 |
| Sfc4p | 106 | 1006 | TFC4p | 1025 | 26 | 45 | 1017 | TFIIIC102 | 886 | 21 | 37 | 892 | SPCC16C4.14c |
| Sfc6p | 66 | 582 | TFC6p | 672 | 25 | 41 | 411 | TFIIIC110 | 911 | 26 | 46 | 133 | SPBC21H7.05 |
| Sfc9p | 75 | 673 | Nonee | TFIIIC90 | 822 | 22 | 39 | 144 | SPBC8D2 | ||||
aComparisons were conducted using the BLAST 2 Sequences program at NCBI with default parameters. The percent identity and percent similarity over the region of greatest match was recorded.
bThe number of positions in the alignment, including gaps.
cCosmid no./EMBL locus is accessible at http://www.sanger.ac.uk/Projects/S_pombe/FUNCAT/trna_syn.shtml.
dNo significant sequence similarity (see Note Added in Proof).
eNo homolog in S.cerevisiae is apparent by the sequence homology search perfomed here.
Sfc4p is homologous to TFC4p/PCF1 and hTFIIIC102 (7). These proteins are characterized by N-terminal acidic regions, multiple copies of a TPR and a basic helix–loop–helix motif (14,59; Y.Huang and R.Maraia, unpublished observation). TFC4p/hTFIIIC102 are large proteins that interact with various other subunits of the pol III machinery, including at least one component each of TFIIIC, TFIIIB and pol III (14,66). Part of TFC4p/PCF1 lies upstream of the transcription start site and recruits TFIIIB (65,73–75) by interacting principally with scBrf and also with scB″ (75,76). TFC4p/τ131 has also been reported to interact with the pol III subunit scRPC53 (66). The evolutionary conservation of this factor appears to be the highest of all the TFIIIC subunits, as its sequence homology extends over the entire length of all three species (Table 1). An archaeal homolog of TFC4p has also been noted in Methanococcus jannaschii, although the significance of this homology, if any, and the function of the putative polypeptide is unknown (accession no. E64417, PID g2129310).
As suggested by the network of interactions mentioned above, TFC4p/τ131/PCF1p appears to be of central importance to the initiation factor activity of TFIIIC. Intriguingly, several mutations of TFC4p stimulate pol III transcription in vivo and in cell-free extracts (77–79). The PCF1-2 dominant mutation does not affect TFIIIC DNA-binding affinity but leads to increased recruitment of TFIIIB70/Brf1p by a non-equilibrium binding mechanism (79). It is noteworthy for the purpose of this review that the mutated residue in PCF1-2, Thr167, is also found in hTFIIIC102 (corresponding to Thr188) but not in Sfc4p (corresponding to Met172). Accordingly, it is tempting to speculate that since mutation of this residue in scTFC4p leads to increased association of TFIIIC and TFIIIB, the lack of conservation at this position in Sfc4p may reflect a relatively unique aspect of TFIIIC–TFIIIB interactions in fission yeast although this remains to be determined.
Sfc3p is a sequence homolog of TFC3p, the B box-binding subunit. While it is clear that Sfc3p and TFC3p are orthologs, neither exhibits recognizable sequence homology to the B box-binding subunit, hTFIIIC220/hTFIIICα, even after multiple reiterations using PSI-BLAST (7). Nonetheless, there may be similarities in the overall architecture of these sequence-specific DNA-binding proteins that hint at a related ancestry. The position of a mutation that creates a conditional allele of TFC3p suggests interaction with TFC6p through the N-terminal third of TFC3p (21). Somewhat similarly, physical evidence indicates that the N-terminal fragment of hTFIIIC220 remains bound to hTFIIICβ while the C-terminal fragment associates with the A box-related hTFIIIC subunits (67). Thus, in a broad general sense, the B box-binding subunits of yeast and human TFIIIC would appear to be orientated such that their C-termini face towards the initiation end of the template while their N-termini face towards the termination end. Moreover, the two domain ‘dumb-bell’ structure noted for yeast TFIIIC may also to be true for human TFIIIC, as reflected in the sensitivity of the B box-binding subunit to proteolysis, somewhat similar to the protease sensitivity of τ138 (67,80,81). Interestingly, this feature is apparently used by poliovirus protease 3C to inactivate TFIIIC during infection by poliovirus (67). Therefore, from these comparisons it would seem as if the apparently similar architectures, interactions with the homologous subunits TFC6p and hTFIIICβ in an ostensible orientation-specific manner and ability to recognize the highly conserved tRNA B box promoter element indicate analogous functions and suggest a common ancestry for TFC3p/τ138/Sfc3p and hTFIIIC220, even though no detectable homology is at present apparent for the yeast and human sequences (Table 1, see below and Note Added in Proof).
Sfc6p was identified as a homolog of TFC6p, the S.cerevisiae protein that cooperates with TFC3p in tRNA gene binding and is crosslinked to the terminator regions of tRNA and 5S rRNA genes (3,21). PSI-BLAST revealed that Sfc6p and TFC6p exhibit sequence homology to hTFIIICβ (7). A temperature-sensitive substitution in TFC3p, Gly349→Glu, which decreases scTFIIIC binding to the tRNA promoter, is suppressed by a compensating substitution, Glu330→Lys, in TFC6p (21,82). It is noteworthy here that while Gly349 in TFC3p is aligned with Gly361 in Sfc3p, suggesting conservation, the immediately surrounding region is also moderately conserved (Y.Huang and R.Maraia, unpublished observation). Likewise, although Glu330 of TFC6p is not exactly aligned with Sfc6p, the surrounding regions are moderately conserved (EHLEMFDK in TFC6p and ETTESVFMR in Sfc6p), suggesting that similar surfaces may interact in both yeasts. In summary, while four subunits of scTFIIIC have homologs in S.pombe, only three of these share sequence homology with hTFIIIC subunits (5; Fig. 1 and Table 1).
Two scTFIIIC subunits, TFC7p and TFC8p, have no apparent homologs in S.pombe or human TFIIIC subunits. One of these, TFC7p, forms a sub-complex with TFC1p in vivo that exists in stoichiometric excess over other TFIIIC subunits (56). It was suggested that TFC7p functions as a nutritional response factor since a deletion of its N-terminal region impairs growth in glycerol- or ethanol-containing media (56). It is therefore conceivable that this subunit may be related to a unique metabolic adaptability of S.cerevisiae. The other scTFIIIC subunit with no apparent counterpart in S.pombe or human, TFC8p, has been shown to interact with scTBP (57). Moreover, a mutant form of scTFIIIC containing functionally impaired TFC8p exhibited a deficiency in scTBP recruitment, suggesting that TFC8p may function with TFC4p to recruit scTFIIIB (57).
A putative fifth S.pombe TFIIIC subunit, tentatively designated Sfc9p, was identified by BLAST using hTFIIIC90 as the query (71). Although the sequence relatedness is among the lowest of the TFIIIC homologs, limited to 144 residues, the significance of the homology is strengthened by the similar lengths of the proteins and more so by the similar position of the homologous regions at the C-termini of both proteins (Table 1 and unpublished observations). Interestingly, hTFIIIC90 shows in vitro direct interactions with the pol III subunit hRPC39, the hTFIIIB subunit hBRF/hTFIIIB90 and the hTFIIIC subunits hTFIIIC220, hTFIIIC110 and hTFIIIC63 (71). In this regard, hTFIIIB90 is reminiscent of scTFC8p in its TFIIIB recruitment function, although no significant sequence homology is apparent at the present time.
INITIATION FACTOR IIIB SUBUNITS AND THEIR INTERACTIONS WITH TFIIIC AND POL III ARE HIGHLY CONSERVED
scTFIIIB has been well characterized, consisting of three components, scTBP (13), TFIIB-related factor (scBrf/PCF4/TFIIIB70) (83–85) and scB″/TFIIIB90/TFC5 (86–89). The activity, subunit composition and mechanism of recruitment of TFIIIB to upstream DNA by TFIIIC has been highly conserved. As will be detailed in a later section, in the human system this activity has been referred to simply as TFIIIB or, more recently, TFIIIB-β (90–92).
TATA-binding protein
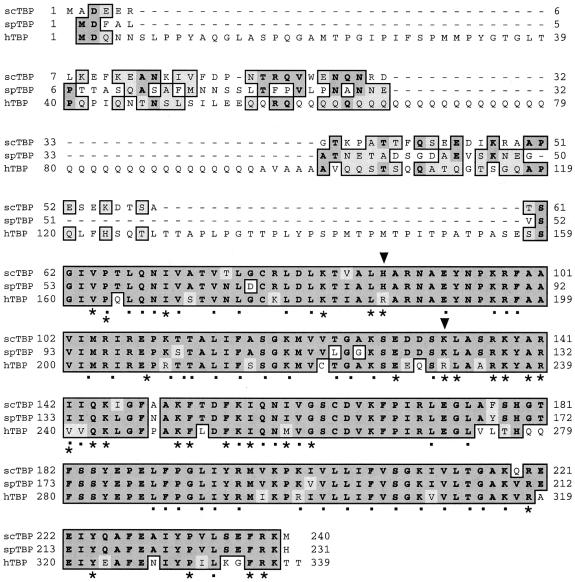
While TBP is required for transcription by all three nuclear RNA polymerases, B″ and Brf are pol III specific, although the latter is a homolog of the functionally analogous pol II initiation factor, TFIIB, and the archaeal initiation factor, TFB. Since human TBP could not substitute for S.cerevisiae TBP in genetic rescue experiments but S.pombe TBP could (below), the latter provided an important intermediate in obtaining recombinant hTBP (93,94). As alluded to above, TBP and Brf are physically associated in the absence of DNA and this presumably accounts at least in part for the ability of TBP to bind to upstream DNA in the absence of a TATA element (13,95,96; also see 97). TBPs are very highly conserved in their C-terminal regions, while the N-terminal regions are more species specific (Fig. 2). Studies in the S.cerevisiae and human systems revealed that the TBP residues that are important for interaction with Brf are located in the conserved region of TBP (87,98–100; for details see the TFIIIB–DNA model in 101). While all of the residues known to be important for Brf interaction are conserved in the two yeast TBPs, two of these residues are different in human TBP (Fig. 2). The amino acid corresponding to Lys133 and Lys124 in scTBP and spTBP, respectively, is an arginine, Arg231, in hTBP (Fig. 2). Although Lys→Arg would appear to be a conservative substitution, this difference is critical since substitution of just this single residue, Arg231→Lys, converts hTBP from inactive to active in supporting growth of TBP-deficient S.cerevisiae and in supporting pol III-dependent tRNA transcription in the S.cerevisiae in vitro system (102,103). These incisive studies indicate that the species-specific TBP function that is required for yeast viability which cannot be supplied by hTBP (but can be supplied by hTBP-R231K) is transcription of TATA-less (but not TATA-containing) genes, including tRNA genes (102,103). Moreover, this also suggests that TBP interacts with TATA elements in a different manner than with TATA-less promoters.
Figure 2.
Alignment of TBPs from S.cerevisiae, S.pombe and human. The alignment was performed by Clustal W (177). Asterisks designate residues involved in Brf interaction and dots designate residues involved in DNA binding. Inverted triangles designate the residues, which have been determined to be important for TBP interaction with Brf, but are not conserved, as described in the text.
The initiation factor IIIB subunit Brf
A candidate subunit of spTFIIIB, spBrf, was readily identified by BLAST (Table 2). Comparable to scBrf and hBRF, the N-terminal region of spBrf is related to TFIIB, while the C-terminal region of spBrf is homologous to the corresponding regions of scBrf and hBRF (74,96,104,105). The three Brf proteins are most homologous in their N-terminal regions, i.e. in their TFIIB-related domains, while their C-terminal domains, including the three conserved regions noted by others (104), are moderately conserved (Fig. 3).
Table 2. Comparison of TFIIIB subunits of S.pombe, S.cerevisiae and humana.
| S.pombe TFIIIB | S.cerevisiae homolog | Human homolog | . | S.pombe cosmid no. | |||||||||
| Subunit |
Mass (kDa) |
Length (amino acids) |
Subunit |
Length |
Identity (%) |
Similarity (%) |
Alignmentb |
Subunit |
Length |
Identity (%) |
Similarity (%) |
Alignmentb |
EMBL locusc |
| spTBP | 25 | 231 | scTBP | 240 | 93 | 95 | 179 | hTBP | 339 | 80 | 90 | 179 | SPAC30.12 |
| spBrf | 56 | 500 | scBrf | 596 | 37 | 52 | 601 | hBRF | 675 | 47 | 64 | 294 | SPBC13E7 |
| spB″ | 58 | 507 | scB″ | 594 | 34 | 57 | 138 | hB″ | 1388 | 35 | 55 | 148 | SPCC1919.14c |
aComparisons were conducted using the BLAST 2 Sequences program at NCBI with default parameters. The percent identity and percent similarity over the region of greatest match was recorded.
bThe number of positions in the alignment, including gaps.
cCosmid no./EMBL locus is accessible at http://www.sanger.ac.uk/Projects/S_pombe/FUNCAT/trna_syn.shtml.
Figure 3.
Alignment of Brf from S.cerevisiae, S.pombe and human, performed by Clustal W (177).
As a TF of central importance, scBrf interacts with several initiation-specific subunits of the pol III machinery, TBP, B″, TFC4p and the pol III subunits C34 and C17 (4,5,104,106,107). While the Brf–TFC4p interaction is thought to constitute a principal mechanism of scTFIIIB recruitment, the Brf–C34 interaction is similarly important for pol III recruitment (74,75,108), and these functions are conserved in the corresponding human orthologs (14). Conserved region II in the C-terminal domain of scBrf is important for interaction with C34 (100,101,104,106). It appears that Brf regions II and III (indicated by boxed areas labeled Brf 2 and Brf 3 in Fig. 3) function to bridge TBP and C34 (106). Two residues in the Brf 2 region of scBrf, L462 and D464, which appear critical for interaction with TBP, are conserved in spBrf (106), although, as will be detailed below, adjacent regions vary significantly in spBrf (Fig. 3).
The Brf protein in S.pombe is significantly different from the human and S.cerevisiae proteins in sequences flanking each of the three conserved regions (Fig. 3). While this is least apparent for the Brf 1 region, it is more significant for conserved regions Brf 2 and Brf 3. Sequences upstream of the Brf 1 and Brf 2 regions are conserved in S.cerevisiae and human but absent in the S.pombe Brf protein (Fig. 3). The significance of these differences remains to be discerned.
The initiation factor IIIB subunit B″
Sequence homology of spB″, scB″ and hB″ is limited to their SANT domains and surrounding regions (90). The SANT domain was initially identified in several proteins, including SWI3, ADA2, N-Cor and TFIIIB″, and had been compared to the myb DNA-binding domain (109). In particular, conserved Trp residues in the myb and SANT domains correspond to residues that contribute to the hydrophobic core structure of the myb domain (109). spB″ exhibits sequence homology to scB″ and hB″ in a region substantially more extensive than and including the SANT domain (90). spB″ and scB″ are more similar to each other than either is to hB″ in overall structure and other features; SpB″ and scB″ exhibit similar lengths with homology at their termini, lacking the very long extension at the C-terminus of hB″, and share both of the myb-related Trp residues (90). B″ contacts DNA both upstream and downstream of the TATA box, involving the highly conserved SANT domain region (110). Despite differences in structure outside the SANT domain, scB″ can functionally substitute for hTFIIIB-α or hB″ in a human cell-derived reconstituted in vitro transcription system (91,103).
scB″ has been well characterized biochemically. It functions in assembly of the pre-initiation complex and is necessary for formation of the heparin-resistant DNA–TFIIIB complex that reflects the stability of the pol III transcription complex (107,111). Indeed, multiple lines of evidence suggest that B″ serves as a scaffold that also provides one aspect of a clamp around the TBP–DNA complex, with another aspect provided by Brf; the resulting clamp may provide the physical basis for the renowned stability of the TFIIIB–DNA complex (54,100,107,110). Regions of scB″ required for TATA-dependent, TFIIIC-independent transcription of the scU6 gene are less extensive than the regions required for TFIIIC-dependent transcription of a TATA-less tRNA gene (107), consistent with contacts between TFC4p and B″ (66).
B″ does more than simply establish conditions to recruit pol III and stabilize the complex. scB″ also functions in upstream promoter melting, in a pol III-dependent manner, a fundamental step in transcriptional activation that is shared by other polymerases, including bacterial RNA polymerase (86,88). Evidence that B″ interacts directly with pol III has been noted (cited as unpublished observations in 107).
A TFIIIB-RELATED ACTIVITY, TFIIIB-α, IS USED FOR TYPE 3 GENE TRANSCRIPTION IN HIGHER EUKARYOTES BUT DOES NOT APPEAR IN YEAST
Unlike the situation in S.cerevisiae, in which a single TFIIIB complex directs transcription of tRNA, 5S rRNA and U6 snRNA genes, two different TBP-dependent TFIIIB activities exist in humans, hTFIIIB-α and hTFIIIB-β (92). As alluded to above, hTFIIIB-β is required for type 2 gene transcription and is composed of hTBP in association with hBRF and (loosely associated) hB″ (103) and appears to completely correspond in its subunit composition to S.cerevisiae TFIIIB. As reviewed above, U6 and other pol III-dependent genes use TFIIIC to recruit TFIIIB to the upstream DNA in S.cerevisiae. Since the S.cerevisiae U6 gene uses a B box to recruit TFIIIC, it appears similar to type 2 genes. In contrast, the type 3 genes use entirely upstream promoters, which have only been found in metazoa.
The entirely upstream promoter elements of the human U6 snRNA gene, as well as the TFs involved in their recognition, and therefore the mechanisms of transcription complex assembly on the yeast and human U6 genes appear to be fundamentally different. In type 3 genes there is no B box. Instead, the upstream TF, SNAPc, binds to the proximal sequence element and recruits a distinct TFIIIB-like activity, referred to as hTFIIIB-α comprised of hTBP, hB″ and hBRFU/hTFIIIB50, and possibly other factors to the core promoter (90,91). A critical difference between conventional hTFIIIB and hTFIIIB-α is that the latter does not contain hBRF but instead contains a distinct homolog of hBRF referred to as BRFU or TFIIIB50 (90,91). For the purposes of this review it is noteworthy that while homologs of TBP, B″, Brf and TFIIB are readily apparent in the S.pombe and S.cerevisiae databases, a sequence corresponding to BRFU/TFIIIB50 could not be found in either yeast (Table 2 and unpublished observations).
TRANSCRIPTION FACTOR IIIA
The first eukaryotic TF purified to homogeneity and cloned was Xenopus TFIIIA, the prototypical C2H2 zinc finger protein (112–114; reviewed in 115). TFIIIA contains nine zinc fingers and binds extensively to >50 bp of 5S rDNA. This factor also binds 5S transcripts and this RNA-binding activity can be used as part of a feedback regulatory loop to control 5S rRNA synthesis in vivo (116–118). Saccharomyces cerevisiae TFIIIA (scTFIIIA) was cloned (119,120) and characterized biochemically (121–123). A comparison of the sequences of spTFIIIA, scTFIIIA and hTFIIIA (115,124) revealed that similarity is limited to their zinc finger domains (spTFIIIA is 30% identical and 45% similar to scTFIIIA and is 35% identical and 49% similar to hTFIIIA). Interestingly, spTFIIIA is characterized by unique features. Schizosaccharomyces pombe TFIIIA appears to contain 10 C2H2 zinc fingers as annotated in the database, compared to nine fingers identified in other TFIIIA molecules (115,124). While most zinc fingers in spTFIIIA exhibit similar spacing, the last two appear more distant, with little distance between the last finger and the C-terminus. It would be interesting to know how the additional finger in S.pombe affects DNA binding, transcription, RNA binding and/or feedback regulation (117).
RNA POLYMERASE III
The three eukaryotic nuclear RNA polymerases, historically referred to as polymerases I (A), II (B) and III (C), are principally involved in the transcription of large rRNA, mRNA and tRNA/5S rRNA, respectively (125). All three show homology: some subunits are polymerase specific but exhibit clear homology to subunits in one or both of the other polymerases; some subunits are actually shared, i.e. polypeptides produced from the same gene are distributed to two or three polymerases; some subunits are highly specific with no identifiable homolog in the other polymerases. The different compositions of the three polymerases, including their different but homologous two largest subunits, likely reflect their specialized functional requirements at the initiation, elongation and termination stages of the transcription cycle. Pol III appears the most complex of the nuclear RNA polymerases as it contains the largest number of subunits, 17 in S.cerevisiae as compared to 12 pol II subunits and 14 pol I subunits. Of the 17 scPol III subunits, five are shared among polymerases I, II and III (ABC27, ABC23, ABC14.5, ABC10α and ABC10β), another two are shared with pol I (AC19 and AC40), four are homologous to subunits found in pol I and/or pol II (C160, C128, C25 and C11) and six are unique to pol III (C82, C53, C37, C34, C31 and C17) with no apparent homologs in the other polymerases. Most of the latter group, the pol III-specific subunits, appear to function in recognizing the TFIIIC–TFIIIB–DNA initiation complex.
The crystal structure of a scPol II complex containing the two largest subunits together with eight additional subunits, several of which are shared with or are homologous to pol III subunits, has been solved (126). An interaction map of the scPol III subunits appears to exhibit good agreement with the crystal structure (66) and with crosslinking studies that located several subunits around the transcribed DNA (127). The core structure is very likely conserved in the pols III of many if not all eukaryotes.
Homologs of all 17 scPol III subunits (4,66,128) were identified in S.pombe by BLAST (Table 3). Among them, the two largest subunits (Rpc158 and Rpc130) are homologous to C160 and C128/RET1 in S.cerevisiae and also exhibit strong and extensive homology to the two largest subunits of RNA polymerases of bacteria, archaea, pol I and pol II. The largest subunit contains several highly conserved regions, including one that corresponds to the α-amanitin sensitivity domain (129). Since the pol II enzymes of many species are highly sensitive to α-amanitin, the action of this toxin has been investigated in the pol II system. Inhibition of transcriptional elongation by α-amanitin is associated with inhibition of factor-stimulated 3′→5′ hydrolytic cleavage of the elongating RNA (130), a cleavage activity that is intrinsic to all multisubunit polymerases including Escherichia coli RNA polymerase (131). As alluded to above, the pols III of S.cerevisiae, S.pombe and human exhibit distinct sensitivity profiles to α-amanitin; no sensitivity, intermediate sensitivity and greater sensitivity, respectively (6,8). Although the three pols III are highly conserved in the α-amanitin-sensitive domain, amino acid differences are noted upon alignment with each other and with pol II mutants that exhibit altered α-amanitin sensitivity (129,132–134), revealing candidate side chains in S.pombe pol III that may affect α-amanitin sensitivity. As will be reviewed in a later section, the pattern of differential α-amanitin sensitivity is correlated with the differential sensitivity of the three pols III to the minimal length termination signals used in these organisms (8).
Table 3. Comparison of pol III subunits of S.pombe, S.cerevisiae and humana.
| S.pombe pol III | S.cerevisiae homolog | Human homolog | Relationship to polsc | S.pombe cosmid no. | ||||||||
| Subunit |
Mass (kDa) |
Length (amino acids) |
Subunit |
Identity (%) |
Similarity (%) |
Alignmentb |
Subunit |
Identity (%) |
Similarity (%) |
Alignmentb |
|
EMBL locusc |
| Rpc158 | 158 | 1405 | C160 | 57 | 72 | 1455 | hRPC155 | 51 | 68 | 1407 | Conservedd I, II, III | SPBC651.08c |
| Rpc130 | 130 | 1165 | C128 | 71 | 83 | 1142 | nd | Conservedd I, II, III | SPAC4G9.08c | |||
| Rpc66 | 66 | 571 | C82 | 22 | 43 | 549 | hRPC62 | 32 | 53 | 188 | Specifice | pB1E7.00669 |
| Rpc39 | 39 | 348 | AC40 | 57 | 73 | 330 | hRPC40 | 50 | 68 | 307 | Sharedd I, III | SPBC1289.07c |
| Rpc53 | 37 | 330 | C53 | 23 | 47 | 163 | BN51 | 25 | 42 | 365 | Specific | SPCC18.07 |
| Rpc27 | 27 | 242 | C37 | 30 | 50 | 146 | nd | Specific | SPCC330.13 | |||
| Rpc34 | 34 | 301 | C34 | 42 | 62 | 263 | hRPC39 | 30 | 55 | 267 | Specifice | SPCC290.02 |
| Rpc24a | 24 | 210 | C31 | 28 | 44 | 164 | hRPC32 | 29 | 58 | 55 | Specifice | SPBC839.12 |
| Rpc16 | 16 | 142 | ABC23 | 85 | 93 | 81 | hRPC15 | 76 | 87 | 143 | Sharedd I, II, III | SPCC1020.04c |
| Rpc24b | 24 | 210 | ABC27 | 56 | 75 | 211 | hRPC25 | 43 | 63 | 210 | Sharedd I, II, III | SPAC23C4.15 |
| Rpc25 | 23 | 203 | C25 | 43 | 63 | 217 | nd | Conserved II, III | SPBC2G5.07c | |||
| Rpc19 | 14 | 125 | AC19 | 62 | 72 | 109 | hRPC16 | 58 | 78 | 79 | Sharedd I, III | SPAPYUL23.01 |
| Rpc14 | 14 | 125 | ABC14.5 | 37 | 54 | 144 | hRPC14 | 36 | 54 | 144 | Sharedd I, II, III | SPBC14C8.12 |
| Rpc11 | 13 | 109 | C11 | 48 | 63 | 111 | hRPC11 | 50 | 62 | 110 | Conserved I, II, III | SPAC22A12.05 |
| Rpc10 | 7.3 | 63 | ABC10α | 50 | 78 | 42 | hRPC10 | 56 | 78 | 50 | Sharedd I, II, III | SPBC19C2.03 |
| Rpc8 | 8.3 | 71 | ABC10β | 75 | 86 | 69 | hRPC8 | 71 | 85 | 67 | Sharedd I, II, III | SPAC1B3.12c |
| Rpc17 | 14.9 | 129 | C17 | 26 | 44 | 158 | hRPC17 | 31 | 52 | 129 | Specific | pB1E7.00669 |
nd, not determined.
aComparisons were conducted using the BLAST 2 Sequences program at NCBI with default parameters. The percent identity and percent similarity over the region of greatest match was recorded.
bThe number of positions in the alignment, including gaps.
cCosmid no./EMBL locus is accessible at http://www.sanger.ac.uk/Projects/S_pombe/FUNCAT/trna_syn.shtml.
dSubunits that may form a core enzyme complex comparable to the core pol II recently crystallized (126).
eSpecific subunits that form a subcomplex required for promoter-dependent initiation (see text).
Although α-amanitin inhibits factor-stimulated 3′→5′ RNA cleavage activity and mutations that confer resistance to α-amanitin have been observed only in the largest polymerase subunits, alterations of either of the two largest pol III subunits affects 3′→5′ RNA hydrolytic cleavage (129,135). This, and a wealth of other data, are consistent with the idea that the two largest subunits together define the catalytic center for RNA polymerization and mediate effects on initiation, elongation and termination (126,136). The second largest subunit, also known as Ret1p, was identified genetically by its effects on pol III termination (137,138). Ret1p plays a complex role in elongation and termination, apparently interacting with the nascent RNA as well as the template DNA (135,139,140).
Five S.pombe subunits (Rpc24, Rpc16, Rpc14, Rpc10 and Rpc8) are homologous to the S.cerevisiae common subunits, ABC27, ABC23, ABC14.5, ABC10α and ABC10β, which are shared by all three polymerases. In the recently solved pol II crystal structure ABC27 (rpb5) appears to form part of a jaw on the leading edge of the polymerase that is proposed to open and close on the advancing DNA as the polymerase moves along the template (126). A role for ABC27 in responding to activators has also been noted (141). ABC23 (rpb6) contributes to a clamp near the active site of the polymerase (126). The S.pombe homolog of ABC14.5 can substitute for the S.cerevisiae homolog in pol III if RPC160 is overexpressed (142). Consistent with this, the ABC14.5 (rpb8) subunit interacts with the largest pol III subunit, although no specific function has yet been ascribed to it (66,126). ABC10α and ABC10β would appear to associate with the AC19–AC40 heterodimer (below) as part of a conserved component of the polymerase (126). It should also be noted that although the above subunits are shared by the three nuclear polymerases, the pol III interaction map shows that most of these make contact with the two largest pol III-specific subunits (66). In addition, an interaction between the TFIIIB-recruiting subunit of TFIIIC (τ131) and ABC10α in vivo and in vitro has been noted (143). Thus, even though the shared subunits may provide similar functions in the three polymerases, their function could be modulated in a polymerase-specific manner.
Two S.pombe subunits, Rpc40 and Rpc19, are the apparent orthologs of the S.cerevisiae pol III subunits AC40 and AC19, which are homologous to the two pol II subunits Rpb3 (144) and Rpb11 (145), respectively. The S.pombe genes encoding both of these subunits can functionally replace the corresponding S.cerevisiae genes (146–148). AC19 and AC40 contain an α-motif, a short region homologous to a functionally important domain of the α subunit of E.coli RNA polymerase (149). In E.coli RNA polymerase the α subunit is a target for activators (150). This α-motif is also conserved in spRpc40 and spRpc19 (unpublished observation). The proposal that the AC40–AC19 heterodimer in S.cerevisiae represents a functional counterpart of the α2 homodimer of E.coli RNA polymerase may have been borne out by the pol II crystal structure (126). In this case the pol II homologs Rpb3 and Rpb11 were compared to the α2-like structure visualized on the face of the polymerase that is oriented towards upstream DNA.
Another small subunit, spRpc11, which is a homolog of scRPC11, was identified as part of a study that led to a wonderful insight into transcription termination by pol III (151). A S.cerevisiae pol III chimera in which the essential C11 subunit was replaced in vivo with S.pombe C11 exhibited a temperature-sensitive phenotype (151). Upon biochemical purification of the resulting pol III from S.cerevisiae the heterologous subunit, spC11, dissociates from the enzyme complex. This feature was exploited as part of an elegant marriage of genetics and biochemistry (151). C11 is conserved from yeast to humans and is homologous to the pol I subunit A12.2 and the pol II subunit B12.6. Most significantly, C11 was also noted to exhibit homology to TFIIS, a pol II transcription elongation factor that functions by stimulating the polymerase-intrinsic 5′→3′ RNA cleavage activity mentioned above. In this case the TFIIS-stimulated 3′→5′ cleavage activity allows pol II to overcome pausing and other blocks in elongation (reviewed in 131). In S.cerevisiae C11 stimulates the intrinsic pol III 3′→5′ RNA cleavage activity and in this case this is important for termination (151). It was proposed that C11 may function in the transition between RNA elongation and termination and that the C11-stimulated RNA cleavage activity of pol III removes kinetic barriers to termination (151). It is intriguing that C11 and C17, factors that function in termination and initiation, were found to interact in the yeast two hybrid assay (128).
The remaining six small subunits are pol III specific, homologs of which are not found in pol I or pol II. Three of these, Rpc66, Rpc34 and Rpc24, appear to be the orthologs of the S.cerevisiae pol III-specific subunits C82, C34 and C31 and the human pol III-specific orthologs hRPC62, hRPC39 and hRPC32, respectively, which are recruited by TFIIIB and function in initiation (136,152,153). These three subunits associate with each other and, under certain conditions, can be detected as a sub-complex dissociated from the core pol III enzyme (136,153,154). Human pol III lacking this sub-complex and yeast enzymes with mutations in C34 or C31 can polymerize RNA from non-specific template DNA and terminate appropriately, but are specifically deficient in initiation (108,136,154). Based on their homology it seems likely that Rpc66, Rpc34 and Rpc24 would form an orthologous initiation-related sub-complex in S.pombe.
As noted above in the section on TFIIIB, specific recruitment of pol III to the transcription start site is mediated in large part by the C34–scBrf interaction in S.cerevisiae and by the orthologous hRPC39–hBrf interaction in humans. This is consistent with a wealth of data, including photocrosslinking studies of a pre-initiation complex which revealed that a region of RPC34 is the most upstream of all of the detectable pol III subunits, in a position compatible with interaction with TFIIIB (127). Sequence comparisons revealed that the C34 residue, Lys135, which is important for the C34–scBrf interaction, is conserved in the S.pombe and human counterparts, Rpc34 and hRPC39, respectively (108; unpublished observations). Also noteworthy is that an acidic segment in C31, which has been implicated as functioning in transcription initiation, is also conserved in the S.pombe and human orthologs, spRpc31 and hRPC32 (154). Additional contacts that contribute to the network of communications that specify initiation include interactions of C17 with C31 and Brf (128). scRPC17 has recently been shown to interact with scBrf in vivo, suggesting that C17 may function in initiation (128).
One subunit, spRpc53, appears to be the ortholog of S.cerevisiae C53 and human hRPC53/BN51. Mapping of interaction domains revealed that scRPC53 interacts with TFC4p, the TFIIIC subunit that lies over the transcription start site (66), suggesting a role in initiation. In S.cerevisiae inactivation of C53 leads to G1 arrest (155). This cell cycle arrest can be reversed by supplementation with BN51 cDNA, which encodes the human RPC53 subunit (155,156). Indeed, mammalian BN51 cDNA was initially isolated on the basis of its effect on the cell cycle, as its inactivation leads to G1 arrest (156,157). Consistent with this, it was recently found that expression of BN51 (hRPC53) is regulated by the c-Myc oncoprotein (158). Although the scRPC37 and scRPC53 subunits have been found to interact, the functional significance of this remains to be determined (66).
A human pol III holoenzyme capable of transcribing the VA1 and tRNA genes has been affinity purified from a HeLa cell line using epitope-tagged BN51, the human homolog of RPC53 (159). This holoenzyme contains pol III, hTFIIIB, hTFIIIC and La antigen, but not hTFIIIA or pol II-related proteins (159). We used a similar approach to affinity purify S.pombe pol III, independently tagging Rpc53 and Ret1p. The affinity purified S.pombe pol III is able to conduct promoter-dependent initiation and recycling on a pre-assembled S.pombe transcription complex, but lacks holoenzyme activity (unpublished results). Efforts to isolate a pol III holoenzyme from S.cerevisiae have also been unsuccessful (4).
A PRESENT LIST OF S.POMBE POL III MACHINERY FOR WHICH THERE IS EVIDENCE OF EXPRESSION IN FISSION YEAST OR FUNCTIONAL COMPLEMENTATION IN S.CEREVISIAE
Table 4 lists the set of 26 pol III and associated TF subunits that can be identified in the S.pombe sequence database. Two S.cerevisiae-specific TFIIIC subunits were not found, while a human TFIIIC subunit not present in S.cerevisiae was found in the S.pombe database.
Table 4. Status of expression or function.
|
S.pombe |
Status |
S.cerevisiae gene |
Synonym(s) |
Reference |
| TBP | X,C | TBP1 | SPT15, TFIID | (90,91) |
| Brf | X | BRF1 | PCF4, TDS1 | Unpublished |
| B″ | X | TFC5 | TFIIIB″ | Unpublished |
| TFIIIA | N | TFC2 | PZF1 | |
| Sfc1 | X | TFC1 | τ95 | (7) |
| Sfc3 | X | TFC3 | τ138 | (7) |
| Sfc4 | X | TFC4 | PCF1, τ131 | (7) |
| Sfc6 | X | TFC6 | τ91 | (7) |
| Sfc9 | N | |||
| Rpc158 | N | RPC160 | C160 | |
| Rpc130 | X | RET1 | C128 | Unpublished |
| Rpc66 | N | RPC82 | C82 | |
| Rpc39 | C | RPC40 | AC40 | (146) |
| Rpc53 | X | RPC53 | C53 | Unpublished |
| Rpc27 | N | RPC37 | C37 | |
| Rpc34 | X | RPC34 | C34 | Unpublished |
| Rpc24a | N | RPC31 | C31 | |
| Rpc16 | C | RPO26 | ABC23 | (178) |
| Rpc24b | C | RPB5 | ABC27 | (179) |
| Rpc25 | N | RPC25 | C25 | |
| Rpc19 | C | RPC19 | AC19 | (146) |
| Rpc14 | C | RPB8 | ABC14.5 | (142) |
| Rpc11 | C | RPC11 | C11 | (151) |
| Rpc10 | C | RPC10 | ABC10α | (180) |
| Rpc8 | C | RPB10 | ABC10β | (181) |
| Rpc17 | N | RPC17 | C17 |
X, immunodetection as described in the text; C, complementation of S.cerevisiae; N, none reported.
We have used two approaches to identify proteins corresponding to predicted S.pombe proteins: (i) detection by immunoblotting using antibodies raised against synthetic peptides corresponding to the N- and/or C-termini of the predicted subunit; (ii) isolation of the corresponding gene or cDNA and epitope tag cloning followed by expression in S.pombe and detection with anti-epitope or anti-peptide antibody (7). In some cases we detected one predicted protein after co-immunoprecipitation with antibody directed to another pol III-associated factor (7; unpublished observations).
Table 4 indicates expression in S.pombe (designated X) or functional complementation of a S.cerevisiae strain that lacks the corresponding subunit (designated C) for 18 of the S.pombe subunits. The former category, evidence of expression in S.pombe, includes four TFIIIC subunits, all three of the scTFIIIB subunits and three subunits of pol III, while the latter category, complementation, includes eight subunits of pol III plus TBP. To our knowledge, neither Brf, B″, TFIIIA nor any TFIIIC subunits derived from S.pombe has been reported to complement a S.cerevisiae strain.
POL III-RELATED TFs NOT IDENTIFIED IN THE S.POMBE DATABASE
Human SNAPc, a complex of five subunits, and Oct-1 are specifically required for type 3 gene transcription (34). No apparent homologs of the SNAPc subunits, Oct1 or BRFU/TFIIIB50 could be identified in S.pombe, consistent with the A box–B box promoter (i.e. type 2-like) structure of Schizosaccharomyces U6 snRNA genes (160). This is also consistent with the idea that there are no entirely upstream type 3 genes in S.pombe, as is the case in S.cerevisiae. In addition, no homologs of the hTFIIIC-associated terminator-binding protein NF1 or the S.cerevisiae TFIIIC subunits TFC7 or TFC8 could be found.
CHROMATIN-RELATED FACTORS
Three subunits of human TFIIIC2 (hTFIIIC220, hTFIIIC110 and hTFIIIC90) each harbor intrinsic histone acetylation activity (HAT) that enables TFIIIC to combat the repressive effects of chromatin (71,161). These hTFIIIC subunits exhibit the least homology with the scTFIIIC and spTFIIIC subunits. Human TFIIIC220 is not detectable in either yeast, hTFIIIC90 exhibits low level homology with the putative Sfc9p but not with any S.cerevisiae protein and, as noted previously, hTFIIIC110 and TFC6p appear related only when Sfc6p is included in the comparison (7). There is no evidence of HAT activity in recombinant Sfc6p or in affinity purified S.pombe TFIIIC (7), apparently similar to the lack of HAT activity in scTFIIIC (4). Yet, in S.cerevisiae the B box–scTFIIIC relationship is comparable to an enhancer–activator system that is able to relieve chromatin-mediated repression of U6 snRNA transcription in vitro and in vivo (53,162,163). It is important to recall here that the A box–B box distance in the U6 gene is unusually long (>200 bp) as compared to tRNA genes (typically <70 bp). Indeed, a nucleoprotein complex appears to organize this gene in a manner that facilitates TFIIIC-dependent binding of TFIIIB (164). Recent work has identified the HMG1 protein Nhp6 as essential for U6 transcription in yeast (51,165). This HMG1 protein appears to structure the scU6 gene (SNR6) in vivo in a manner that supports TFIIIC-dependent recruitment of TFIIIB, presumably overcoming the otherwise occlusive effect of the lengthy A box–B box distance (51). In S.cerevisiae U6 may be the only pol III-dependent gene that requires Nhp6 for essential transcription, again consistent with its unusually long A box–B box spacing (51), although effects of Nhp6Bp on decreased levels of certain tRNAs have also been noted (165).
Interestingly, although the S.pombe U6 snRNA gene also contains A box and B box promoter elements, the B box is within a relatively short distance of the A box, in an intron (160), such that the functional promoter is similar to that of S.pombe tRNA genes. Therefore, although a Nhp6 homolog is present in S.pombe, it is not clear that it would be required for or involved in U6 RNA expression in fission yeast. Likewise, while a nucleosome upstream of the core promoter of the human U6 (type 3) gene is positioned between the PSE and the DSE and facilitates activator function (47,166), this would not necessarily be expected for the S.pombe U6 snRNA gene since the latter uses an internal B box element for transcriptional activation (M.Hamada and R.J.Maraia, unpublished observations).
TERMINATION BY POL III
As alluded to in the Introduction, since multiple lines of evidence suggest that termination and re-initiation appear to be stimulated by TFIIIC-associated and other components in the human but not the S.cerevisiae system, this aspect of RNA production may represent a significant divergence in the pol III systems in these organisms. Examination of termination signal recognition by the human, S.cerevisiae and S.pombe pols III revealed that S.pombe is intermediate between human and S.cerevisiae (8; Fig. 1). In addition, hints of qualitative differences in the termination mechanisms are also apparent (8). Evidence from in vivo and in vitro experiments indicated that fewer dT residues are required to signal pol III termination in S.pombe as compared to S.cerevisiae (8). This is consistent with earlier studies in S.cerevisiae (167) and the fact that several 5S rRNA genes in S.pombe end in four consecutive T residues, significantly fewer T residues than in S.cerevisiae (168; R.J.Maraia, unpublished observations). Saccharomyces cerevisiae pol III appears to require a minimum of six or seven dT residues for efficient termination, while vertebrate pol III can terminate very efficiently at four dT residues (2,167,169). For vertebrate pol III the identity of the nucleotides surrounding a T4 tract can have substantial effects on termination and termination-associated activities that include RNA 3′ processing and re-initiation, while such effects have not been reported for yeast pol III (26,169–172). Thus, in addition to the difference in the minimal dT length that can serve as a terminator in these species, there may be additional differences in the mechanisms used for termination and termination-associated activities in the three species (8), although this remains to be explored further.
While it is intriguing to consider that differential sensitivity of S.pombe, S.cerevisiae and human pols III to α-amanitin correlates with the sensitivity of these polymerases to a minimal length termination signal, the mechanistic significance of this observation remains to be explored (8). Although the three pols III are highly conserved in the α-amanitin-sensitive domain, as are the pols II from various organisms, alignment with pol II mutants with altered α-amanitin sensitivity (129,132–134) reveals candidate amino acids in the S.pombe pol III that may affect α-amanitin sensitivity. Therefore, site-directed mutagenesis can be used to examine the potential relationship between α-amanitin sensitivity and pol III termination. Since termination can be an important point of regulation and differences in this phase of transcription may afford opportunities for differential regulation (173), this aspect of pol III transcription deserves further attention.
As mentioned above, TFIIIC1, topoisomerase I, PC4, NF1 and La protein have all been reported to stimulate termination and re-initiation by human pol III (17,19,23–26). In addition, the effects of termination signal flanking sequences on termination, post-transcriptional 3′ cleavage of the nascent RNA and re-initiation by pol III have been documented and in some cases demonstrated to be dependent on the activity of La (23,25,26,172). Although evidence of termination-dependent facilitated recycling of scPol III has been reported, this activity has not been linked to TFIIIC (16).
In summary, evidence for three classes of mechanistic differences in the pol III termination systems of yeasts and humans has been emerging: (i) the minimal length of the dT tract length that signals termination; (ii) the effects of flanking sequence on dT tract function; (iii) the effects of TFIIIC-associated and other factors on termination and/or re-initiation. The striking evolutionary divergence in the downstream TFIIIC subunits noted above and in Figure 1, in conjunction with the apparent species-specific termination-related activities of pol III noted above (23,25,26,172,174), suggests that these downstream TFIIIC subunits may have co-evolved with differences in pol III termination and in humans, and perhaps to some extent in yeasts, can accommodate versatile and regulatory transcriptional systems (175). The possibility that TFIIIC subunits may be involved in pol III termination can be explored in the genetically tractable system represented by S.pombe.
CONCLUDING REMARKS
We have surveyed the genomic and protein sequence databases of S.pombe for pol III, TFIIIA, TFIIIB and TFIIIC components. All 17 subunits of pol III, plus TFIIIA, all three of the scTFIIIB (equivalent to hTFIIIB-β) subunits and five TFIIIC subunits (three homologs of S.cerevisiae subunits and two homologs of subunits present in humans but not S.cerevisiae) were identified. There is a striking degree of similarity in the pol III systems in all three organisms, most readily apparent in the pol III, TFIIIA, TFIIIB and those TFIIIC subunits that interact near the transcription initiation site, while the subunits that interact with downstream regions of the tRNA gene, including the transcription termination element, are more divergent (Fig. 1). Although the polymerases from these organisms exhibit a high degree of homology, they also exhibit mechanistic variances in termination and termination-associated activities. This raises the possibility that these differences may be mediated in part by the divergent downstream TFIIIC subunits.
The human pol III system also includes an additional class of factors not found in either yeast that function in the transcription of metazoan-specific type 3 genes, which include a distinct Brf-homologous protein known as BRFU/TFIIIB50, a component of hTFIIIB-α that has no apparent homolog in either S.pombe or S.cerevisiae. Therefore, in overall complexity the S.pombe TFIIIB/pol III gene system appears most similar to that of S.cerevisiae since it lacks type 3 genes and the TFIIIB-α components that are used for type 3 gene-specific transcription in higher eukaryotes.
A total of 26 predicted polypeptides in the S.pombe database that correspond to pol III and associated TFs were identified. In comparison to the S.cerevisiae and human systems it would appear that this may represent the complete set of core pol III factors in fission yeast. Experimental data derived in part from expression in S.pombe and in part from complementation of a defect in the corresponding S.cerevisiae is available for 18 of the 26 predicted S.pombe proteins. Recent findings regarding some apparently unique aspects of the S.pombe pol III system, including the widespread use of upstream TATA elements as mentioned in the Introduction, leave open the possibility that additional pol III-associated factors and mechanisms that are not presently apparent may also emerge from the study of fission yeast.
NOTE ADDED IN PROOF
Very limited sequence homology has recently been discerned between hTFIII220, TFC3p and Sf3p (176).
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Roberts and N. Martin for encouraging this review, members of the Sanger Center for analysis and annotation of S.pombe DNA, V. Wood for kindly answering our inquiries, E.P. Geiduschek, M. Riva, D. Schulman, D. Setzer and LMGR members for comments and discussion, R. Moir, M. Teichmann and P. Thuriaux for comments on the manuscript and M. Hamada for valuable contributions to the S.pombe pol III system.
References
- 1.Willis I.M. (1993) RNA polymerase III. Genes, factors and transcriptional specificity. Eur. J. Biochem., 212, 1–11. [DOI] [PubMed] [Google Scholar]
- 2.Geiduschek E.P. and Tocchini-Valentini,G.P. (1988) Transcription by RNA polymerase III. Annu. Rev. Biochem., 57, 873–914. [DOI] [PubMed] [Google Scholar]
- 3.Kassavetis G.A., Bardeleben,C., Bartholomew,B., Braun,B.R., Joazeiro,C.A.P., Pisano,M. and Geiduschek,E.P. (1994) In Conaway,R.C. and Conaway,J.W. (eds), Transcription: Mechanisms and Regulation. Raven Press, New York, NY, pp. 107–126.
- 4.Chedin S., Ferri,M.L., Andrau,J.C., Jourdain,S., Lefebvre,O., Wermer,M., Carles,C. and Sentenac,A. (1998) The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harbor Symp. Quant. Biol ., 63, 381–389. [DOI] [PubMed] [Google Scholar]
- 5.Paule M.R. and White,R.J. (2000) Transcription by RNA polymerases I and III. Nucleic Acids Res., 28, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodicker F., Ossenbuhl,F., Michels,D. and Benecke,B.J. (1999) Faithful in vitro transcription by fission yeast RNA polymerase III reveals unique α-amanitin sensitivity. Gene Expr., 8, 165–174. [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Hamada,M. and Maraia,R.J. (2000) Isolation and cloning of four subunits of a fission yeast TFIIIC complex that includes an ortholog of the human regulatory protein TFIIIC-β. J. Biol. Chem., 275, 31480–31487. [DOI] [PubMed] [Google Scholar]
- 8.Hamada M., Sakulich,A.L., Koduru,S.B. and Maraia,R.J. (2000) Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J. Biol. Chem., 275, 29076–29081. [DOI] [PubMed] [Google Scholar]
- 9.Dieci G., Percudani,R., Giuliodori,S., Bottarelli,L. and Ottonello,S. (2000) TFIIIC-independent in vitro transcription of yeast tRNA genes. J. Mol. Biol., 299, 601–613. [DOI] [PubMed] [Google Scholar]
- 10.Kunkel G.R., Maser,R.L., Calvert,J.P. and Pederson,T. (1986) U6 small nuclear RNA is transcribed by RNA polymerase III. Proc. Natl Acad. Sci. USA, 83, 8575–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiduschek E.P. and Kassavetis,G.A. (1992) RNA polymerase III transcription complexes. In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 1, pp. 247–280.
- 12.Joazeiro C.A., Kassavetis,G.A. and Geiduschek,E.P. (1996) Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev., 10, 725–739. [DOI] [PubMed] [Google Scholar]
- 13.Kassavetis G.A., Joazeiro,C.A., Pisano,M., Geiduschek,E.P., Colbert,T., Hahn,S. and Blanco,J.A. (1992) The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell, 71, 1055–1064. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh Y.J., Wang,Z., Kovelman,R. and Roeder,R.G. (1999) Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol. Cell. Biol., 19, 4944–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassavetis G.A., Braun,B.R., Nguyen,L.H. and Geiduschek,E.P. (1990) S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell, 60, 235–245. [DOI] [PubMed] [Google Scholar]
- 16.Dieci G. and Sentenac,A. (1996) Facilitated recycling pathway for RNA polymerase III. Cell, 84, 245–252. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z. and Roeder,R.G. (1998) DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol. Cell, 1, 749–757. [DOI] [PubMed] [Google Scholar]
- 18.Oettel S., Hartel,F., Kober,I., Iben,S. and Seifart,K.H. (1997) Human transcription factors IIIC2, IIIC1 and a novel component IIIC0 fulfil different aspects of DNA binding to various pol III genes. Nucleic Acids Res., 25, 2440–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Bai,L., Hsieh,Y.J. and Roeder,R.G. (2000) Nuclear factor 1 (NF1) affects accurate termination and multiple-round transcription by human RNA polymerase III. EMBO J., 19, 6823–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshinaga S.K., Boulanger,P.A. and Berk,A.J. (1987) Resolution of human transcription factor TFIIIC into two functional components. Proc. Natl Acad. Sci. USA, 84, 3585–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arrebola R., Manaud,N., Rozenfeld,S., Marsolier,M.C., Lefebvre,O., Carles,C., Thuriaux,P., Conesa,C. and Sentenac,A. (1998) Tau91, an essential subunit of yeast transcription factor IIIC, cooperates with tau138 in DNA binding. Mol. Cell. Biol., 18, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z. and Roeder,R.G. (1996) TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol. Cell. Biol., 16, 6841–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan H., Sakulich,A.L., Goodier,J.L., Zhang,X., Qin,J. and Maraia,R.J. (1997) Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell, 88, 707–715. [DOI] [PubMed] [Google Scholar]
- 24.Gottlieb E. and Steitz,J.A. (1989) Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J., 8, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maraia R.J., Kenan,D.J. and Keene,J.D. (1994) Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol. Cell. Biol., 14, 2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodier J.L. and Maraia,R.J. (1998) Terminator-specific recycling of a B1-Alu transcription complex by RNA polymerase III is mediated by the RNA terminus-binding protein La. J. Biol. Chem., 273, 26110–26116. [DOI] [PubMed] [Google Scholar]
- 27.Lin-Marq N. and Clarkson,S.G. (1998) Efficient synthesis, termination and release of RNA polymerase III transcripts in Xenopus extracts depleted of La protein. EMBO J., 17, 2033–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weser S., Bachmann,M., Seifart,K.H. and Meißner,W. (2000) Transcription efficiency of human polymerase III genes in vitro does not depend on the RNP-forming autoantigen La. Nucleic Acids Res., 28, 3935–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannon G.J., Chubb,A., Maroney,P.A., Hannon,G., Altman,S. and Nilsen,T.W. (1991) Multiple cis-acting elements are required for RNA polymerase III transcription of the gene encoding H1 RNA, the RNA component of human RNase P. J. Biol. Chem., 266, 22796–22799. [PubMed] [Google Scholar]
- 30.Hernandez N. (1992) Transcription of snRNA genes and related genes. In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 281–313.
- 31.Maraia R.J., Sakulich,A.L., Brinkmann,E. and Green,E.D. (1996) Gene encoding human Ro-associated autoantigen hY5 RNA. Nucleic Acids Res., 24, 3552–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maraia R.J., Sasaki-Tozawa,N., Driscoll,C.T., Green,E.D. and Darlington,G.J. (1994) The human Y4 small cytoplasmic RNA gene is controlled by upstream elements and resides on chromosome 7 with all other hY scRNA genes. Nucleic Acids Res., 22, 3045–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lobo S.M. and Hernandez,N.T. (1994) Transcription of snRNA genes by RNA polymerases II and III. In Conaway,R.C.C. and Conaway,J.W. (eds), Transcription: Mechanisms and Regulation. Raven Press, New York, NY, pp. 127–159.
- 34.Henry R.W., Ford,E., Mital,R., Mittal,V. and Hernandez,N. (1998) Crossing the line between RNA polymerases: transcription of human snRNA genes by RNA polymerases II and III. Cold Spring Harbor Symp. Quant. Biol., 63, 111–120. [DOI] [PubMed] [Google Scholar]
- 35.Yoon J.B., Murphy,S., Bai,L., Wang,Z. and Roeder,R.G. (1995) Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol., 15, 2019–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry R.W., Mittal,V., Ma,B., Kobayashi,R. and Hernandez,N. (1998) SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev., 12, 2664–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mittal V. and Hernandez,N. (1997) Role for the amino-terminal region of human TBP in U6 snRNA transcription. Science, 275, 1136–1140. [DOI] [PubMed] [Google Scholar]
- 38.Bai L., Wang,Z., Yoon,J.B. and Roeder,R.G. (1996) Cloning and characterization of the β subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerases II and III. Mol. Cell. Biol., 16, 5419–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhlman T.C., Cho,H., Reinberg,D. and Hernandez,N. (1999) The general transcription factors IIA, IIB, IIF and IIE are required for RNA polymerase II transcription from the human U1 small nuclear RNA promoter. Mol. Cell. Biol., 19, 2130–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobo S.M. and Hernandez,N. (1989) A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell, 58, 55–67. [DOI] [PubMed] [Google Scholar]
- 41.Mattaj I.W., Dathan,N.A., Parry,H.D., Carbon,P. and Krol,A. (1988) Changing the RNA polymerase specificity of U snRNA gene promoters. Cell, 55, 435–442. [DOI] [PubMed] [Google Scholar]
- 42.Lescure A., Tebb,G., Mattaj,I.W., Krol,A. and Carbon,P. (1992) A factor with Sp1 DNA-binding specificity stimulates Xenopus U6 snRNA in vivo transcription by RNA polymerase III. J. Mol. Biol., 228, 387–394. [DOI] [PubMed] [Google Scholar]
- 43.Murphy S., Yoon,J.B., Gerster,T. and Roeder,R.G. (1992) Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol., 12, 3247–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danzeiser D.A., Urso,O. and Kunkel,G.R. (1993) Functional characterization of elements in a human U6 small nuclear RNA gene distal control region. Mol. Cell. Biol., 13, 4670–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaub M., Myslinski,E., Schuster,C., Krol,A. and Carbon,P. (1997) Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J., 16, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittal V., Ma,B. and Hernandez,N. (1999) SNAP(c): a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev., 13, 1807–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao X., Pendergrast,S. and Hernandez,N. (2001) A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol. Cell, 7, 539–549. [DOI] [PubMed] [Google Scholar]
- 48.Brow D.A. and Guthrie,C. (1990) Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev., 4, 1345–1356. [DOI] [PubMed] [Google Scholar]
- 49.Eschenlauer J.B., Kaiser,M.W., Gerlach,V.L. and Brow,D.A. (1993) Architecture of a yeast U6 RNA gene promoter. Mol. Cell. Biol., 13, 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts S., Colbert,T. and Hahn,S. (1995) TFIIIC determines RNA polymerase III specificity at the TATA-containing yeast U6 promoter. Genes Dev., 9, 832–842. [DOI] [PubMed] [Google Scholar]
- 51.Kruppa M., Moir,R.D., Kolodrubetz,D. and Willis,I.M. (2001) Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol. Cell, 7, 309–318. [DOI] [PubMed] [Google Scholar]
- 52.Joazeiro C.A., Kassavetis,G.A. and Geiduschek,E.P. (1994) Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol. Cell. Biol., 14, 2798–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burnol A.F., Margottin,F., Huet,J., Almouzni,G., Prioleau,M.N., Mechali,M. and Sentenac,A. (1993) TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature, 362, 475–477. [DOI] [PubMed] [Google Scholar]
- 54.Persinger J., Sengupta,S.M. and Bartholomew,B. (1999) Spatial organization of the core region of yeast TFIIIB–DNA complexes. Mol. Cell. Biol., 19, 5218–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruet A., Camier,S., Smagowicz,W., Sentenac,A. and Fromageot,P. (1984) Isolation of a class C transcription factor which forms a stable complex with tRNA genes. EMBO J., 3, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manaud N., Arrebola,R., Buffin-Meyer,B., Lefebvre,O., Voss,H., Riva,M., Conesa,C. and Sentenac,A. (1998) A chimeric subunit of yeast transcription factor IIIC forms a subcomplex with tau95. Mol. Cell. Biol., 18, 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deprez E., Arrebola,R., Conesa,C. and Sentenac,A. (1999) A subunit of yeast TFIIIC participates in the recruitment of TATA-binding protein. Mol. Cell. Biol., 19, 8042–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanson R.N., Conesa,C., Lefebvre,O., Carles,C., Ruet,A., Quemeneur,E., Gagnon,J. and Sentenac,A. (1991) Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor tau (TFIIIC). Proc. Natl Acad. Sci. USA, 88, 4887–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marck C., Lefebvre,O., Carles,C., Riva,M., Chaussivert,N., Ruet,A. and Sentenac,A. (1993) The TFIIIB-assembling subunit of yeast transcription factor TFIIIC has both tetratricopeptide repeats and basic helix–loop–helix motifs. Proc. Natl Acad. Sci. USA, 90, 4027–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lefebvre O., Carles,C., Conesa,C., Swanson,R.N., Bouet,F., Riva,M. and Sentenac,A. (1992) TFC3: gene encoding the B-block binding subunit of the yeast transcription factor IIIC. Proc. Natl Acad. Sci. USA, 89, 10512–10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willis I., Schmidt,P. and Soll,D. (1989) A selection for mutants of the RNA polymerase III transcription apparatus: PCF1 stimulates transcription of tRNA and 5S RNA genes. EMBO J., 8, 4281–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parsons M.C. and Weil,P.A. (1992) Cloning of TFC1, the Saccharomyces cerevisiae gene encoding the 95-kDa subunit of transcription factor TFIIIC. J. Biol. Chem., 267, 2894–2901. [PubMed] [Google Scholar]
- 63.Braun B.R., Bartholomew,B., Kassavetis,G.A. and Geiduschek,E.P. (1992) Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J. Mol. Biol., 228, 1063–1077. [DOI] [PubMed] [Google Scholar]
- 64.Bartholomew B., Kassavetis,G.A. and Geiduschek,E.P. (1991) Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol., 11, 5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartholomew B., Kassavetis,G.A., Braun,B.R. and Geiduschek,E.P. (1990) The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J., 9, 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flores A., Briand,J.F., Gadal,O., Andrau,J.C., Rubbi,L., Van Mullem,V., Boschiero,C., Goussot,M., Marck,C., Carles,C., Thuriaux,P., Sentenac,A. and Werner,M. (1999) A protein–protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA, 96, 7815–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen Y., Igo,M., Yalamanchili,P., Berk,A.J. and Dasgupta,A. (1996) DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol. Cell. Biol., 16, 4163–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kovelman R. and Roeder,R.G. (1992) Purification and characterization of two forms of human transcription factor IIIC. J. Biol. Chem., 267, 24446–24456. [PubMed] [Google Scholar]
- 69.Lagna G., Kovelman,R., Sukegawa,J. and Roeder,R.G. (1994) Cloning and characterization of an evolutionarily divergent DNA-binding subunit of mammalian TFIIIC. Mol. Cell. Biol., 14, 3053–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinn E., Wang,Z., Kovelman,R. and Roeder,R.G. (1995) Cloning and characterization of a TFIIIC2 subunit (TFIIIC beta) whose presence correlates with activation of RNA polymerase III-mediated transcription by adenovirus E1A expression and serum factors. Genes Dev., 9, 675–685. [DOI] [PubMed] [Google Scholar]
- 71.Hsieh Y.J., Kundu,T.K., Wang,Z., Kovelman,R. and Roeder,R.G. (1999) The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol. Cell. Biol., 19, 7697–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.L’Etoile N.D., Fahnestock,M.L., Shen,Y., Aebersold,R. and Berk,A.J. (1994) Human transcription factor IIIC box B binding subunit. Proc. Natl Acad. Sci. USA, 91, 1652–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sethy I., Moir,R.D., Librizzi,M. and Willis,I.M. (1995) In vitro evidence for growth regulation of tRNA gene transcription in yeast. A role for transcription factor (TF) IIIB70 and TFIIIC. J. Biol. Chem., 270, 28463–28470. [DOI] [PubMed] [Google Scholar]
- 74.Khoo B., Brophy,B. and Jackson,S.P. (1994) Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev., 8, 2879–2890. [DOI] [PubMed] [Google Scholar]
- 75.Chaussivert N., Conesa,C., Shaaban,S. and Sentenac,A. (1995) Complex interactions between yeast TFIIIB and TFIIIC. J. Biol. Chem., 270, 15353–15358. [DOI] [PubMed] [Google Scholar]
- 76.Ruth J., Conesa,C., Dieci,G., Lefebvre,O., Dusterhoft,A., Ottonello,S. and Sentenac,A. (1996) A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J., 15, 1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 77.Rameau G., Puglia,K., Crowe,A., Sethy,I. and Willis,I. (1994) A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol. Cell. Biol., 14, 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sethy I. and Willis,I.M. (1995) Recessive mutations in the second largest subunit of TFIIIC suggest a new step in RNA polymerase III transcription. Gene Expr., 5, 35–47. [PMC free article] [PubMed] [Google Scholar]
- 79.Moir R.D., Sethy-Coraci,I., Puglia,K., Librizzi,M.D. and Willis,I.M. (1997) A tetratricopeptide repeat mutation in yeast transcription factor IIIC131 (TFIIIC131) facilitates recruitment of TFIIB-related factor TFIIIB70. Mol. Cell. Biol., 17, 7119–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marzouki N., Camier,S., Ruet,A., Moenne,A. and Sentenac,A. (1986) Selective proteolysis defines two DNA binding domains in yeast transcription factor tau. Nature, 323, 176–178. [DOI] [PubMed] [Google Scholar]
- 81.Schultz P., Marzouki,N., Marck,C., Ruet,A., Oudet,P. and Sentenac,A. (1989) The two DNA-binding domains of yeast transcription factor tau as observed by scanning transmission electron microscopy. EMBO J., 8, 3815–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lefebvre O., Ruth,J. and Sentenac,A. (1994) A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5 S RNA synthesis. Identification of two classes of suppressors. J. Biol. Chem., 269, 23374–23381. [PubMed] [Google Scholar]
- 83.Buratowski S. and Zhou,H. (1992) A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell, 71, 221–230. [DOI] [PubMed] [Google Scholar]
- 84.Colbert T. and Hahn,S. (1992) A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev., 6, 1940–1949. [DOI] [PubMed] [Google Scholar]
- 85.Lopez-De-Leon A., Librizzi,M., Puglia,K. and Willis,I.M. (1992) PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell, 71, 211–220. [DOI] [PubMed] [Google Scholar]
- 86.Kassavetis G.A., Kumar,A., Letts,G.A. and Geiduschek,E.P. (1998) A post-recruitment function for the RNA polymerase III transcription-initiation factor IIIB. Proc. Natl Acad. Sci. USA, 95, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shen Y., Kassavetis,G.A., Bryant,G.O. and Berk,A.J. (1998) Polymerase (Pol) III TATA box-binding protein (TBP)-associated factor Brf binds to a surface on TBP also required for activated Pol II transcription. Mol. Cell. Biol., 18, 1692–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kassavetis G.A., Letts,G.A. and Geiduschek,E.P. (1999) A minimal RNA polymerase III transcription system. EMBO J., 18, 5042–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kassavetis G.A., Nguyen,S.T., Kobayashi,R., Kumar,A., Geiduschek,E.P. and Pisano,M. (1995) Cloning, expression and function of TFC5, the gene encoding the B″ component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc. Natl Acad. Sci. USA, 92, 9786–9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schramm L., Pendergrast,P.S., Sun,Y. and Hernandez,N. (2000) Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev., 14, 2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teichmann M., Wang,Z. and Roeder,R.G. (2000) A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50 and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc. Natl Acad. Sci. USA, 97, 14200–14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Teichmann M. and Seifart,K.H. (1995) Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J., 14, 5974–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fikes J.D., Becker,D.M., Winston,F. and Guarente,L. (1990) Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature, 346, 291–294. [DOI] [PubMed] [Google Scholar]
- 94.Hoffmann A., Horikoshi,M., Wang,C.K., Schroeder,S., Weil,P.A. and Roeder,R.G. (1990) Cloning of the Schizosaccharomyces pombe TFIID gene reveals a strong conservation of functional domains present in Saccharomyces cerevisiae TFIID. Genes Dev., 4, 1141–1148. [DOI] [PubMed] [Google Scholar]
- 95.Huet J., Conesa,C., Manaud,N., Chaussivert,N. and Sentenac,A. (1994) Interactions between yeast TFIIIB components. Nucleic Acids Res., 22, 3433–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Z. and Roeder,R.G. (1995) Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc. Natl Acad. Sci. USA, 92, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.White R.J. and Jackson,S.P. (1992) Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell, 71, 1041–1053. [DOI] [PubMed] [Google Scholar]
- 98.Cormack B.P. and Struhl,K. (1993) Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science, 262, 244–248. [DOI] [PubMed] [Google Scholar]
- 99.Hernandez N. (1993) TBP, a universal eukaryotic transcription factor? Genes Dev., 7, 1291–1308. [DOI] [PubMed] [Google Scholar]
- 100.Colbert T., Lee,S., Schimmack,G. and Hahn,S. (1998) Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol. Cell. Biol., 18, 1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kassavetis G.A., Kumar,A., Ramirez,E. and Geiduschek,E.P. (1998) Functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol. Cell. Biol., 18, 5587–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cormack B.P., Strubin,M., Stargell,L.A. and Struhl,K. (1994) Conserved and nonconserved functions of the yeast and human TATA-binding proteins. Genes Dev., 8, 1335–1343. [DOI] [PubMed] [Google Scholar]
- 103.Teichmann M., Dieci,G., Huet,J., Ruth,J., Sentenac,A. and Seifart,K.H. (1997) Functional interchangeability of TFIIIB components from yeast and human cells in vitro. EMBO J., 16, 4708–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kassavetis G.A., Bardeleben,C., Kumar,A., Ramirez,E. and Geiduschek,E.P. (1997) Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol. Cell. Biol., 17, 5299–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hahn S. and Roberts,S. (2000) The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Genes Dev., 14, 719–730. [PMC free article] [PubMed] [Google Scholar]
- 106.Andrau J.C., Sentenac,A. and Werner,M. (1999) Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol., 288, 511–520. [DOI] [PubMed] [Google Scholar]
- 107.Kumar A., Kassavetis,G.A., Geiduschek,E.P., Hambalko,M. and Brent,C.J. (1997) Functional dissection of the B″ component of RNA polymerase III transcription factor IIIB: a scaffolding protein with multiple roles in assembly and initiation of transcription. Mol. Cell. Biol., 17, 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brun I., Sentenac,A. and Werner,M. (1997) Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J., 16, 5730–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aasland R., Stewart,A.F. and Gibson,T. (1996) The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci., 21, 87–88. [PubMed] [Google Scholar]
- 110.Shah S.M., Kumar,A., Geiduschek,E.P. and Kassavetis,G.A. (1999) Alignment of the B″ subunit of RNA polymerase III transcription factor IIIB in its promoter complex. J. Biol. Chem., 274, 28736–28744. [DOI] [PubMed] [Google Scholar]
- 111.Kumar A., Grove,A., Kassavetis,G.A. and Geiduschek,E.P. (1998) Transcription factor IIIB: the architecture of its DNA complex and its roles in initiation of transcription by RNA polymerase III. Cold Spring Harbor Symp. Quant. Biol., 63, 121–129. [DOI] [PubMed] [Google Scholar]
- 112.Engelke D.R., Ng,S.Y., Shastry,B.S. and Roeder,R.G. (1980) Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell, 19, 717–728. [DOI] [PubMed] [Google Scholar]
- 113.Ginsberg A.M., King,B.O. and Roeder,R.G. (1984) Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell, 39, 479–489. [DOI] [PubMed] [Google Scholar]
- 114.Miller J., McLachlan,A.D. and Klug,A. (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J., 4, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wolfe S.A., Nekludova,L. and Pabo,C.O. (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct., 29, 183–212. [DOI] [PubMed] [Google Scholar]
- 116.Andrews M.T. and Brown,D.D. (1987) Transient activation of oocyte 5S RNA genes in Xenopus embryos by raising the level of the trans-acting factor TFIIIA. Cell, 51, 445–453. [DOI] [PubMed] [Google Scholar]
- 117.Rollins M.B., Del Rio,S., Galey,A.L., Setzer,D.R. and Andrews,M.T. (1993) Role of TFIIIA zinc fingers in vivo: analysis of single-finger function in developing Xenopus embryos. Mol. Cell. Biol., 13, 4776–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brow D.A. and Geiduschek,E.P. (1987) Modulation of yeast 5 S rRNA synthesis in vitro by ribosomal protein YL3. A possible regulatory loop. J. Biol. Chem., 262, 13953–13958. [PubMed] [Google Scholar]
- 119.Woychik N.A. and Young,R.A. (1992) Genes encoding transcription factor IIIA and the RNA polymerase common subunit RPB6 are divergently transcribed in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 89, 3999–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Archambault J., Milne,C.A., Schappert,K.T., Baum,B., Friesen,J.D. and Segall,J. (1992) The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J. Biol. Chem., 267, 3282–3288. [PubMed] [Google Scholar]
- 121.Pizzi S., Dieci,G., Frigeri,P., Piccoli,G., Stocchi,V. and Ottonello,S. (1999) Domain organization and functional properties of yeast transcription factor IIIA species with different zinc stoichiometries. J. Biol. Chem., 274, 2539–2548. [DOI] [PubMed] [Google Scholar]
- 122.Rowland O. and Segall,J. (1996) Interaction of wild-type and truncated forms of transcription factor IIIA from Saccharomyces cerevisiae with the 5 S RNA gene. J. Biol. Chem., 271, 12103–12110. [DOI] [PubMed] [Google Scholar]
- 123.Rowland O. and Segall,J. (1998) A hydrophobic segment within the 81-amino-acid domain of TFIIIA from Saccharomyces cerevisiae is essential for its transcription factor activity. Mol. Cell. Biol., 18, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Drew P.D., Nagle,J.W., Canning,R.D., Ozato,K., Biddison,W.E. and Becker,K.G. (1995) Cloning and expression analysis of a human cDNA homologous to Xenopus TFIIIA. Gene, 159, 215–218. [DOI] [PubMed] [Google Scholar]
- 125.Roeder R.G. and Rutter,W.J. (1969) Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature, 224, 234–237. [DOI] [PubMed] [Google Scholar]
- 126.Cramer P., Bushnell,D.A., Fu,J., Gnatt,A.L., Maier-Davis,B., Thompson,N.E., Burgess,R.R., Edwards,A.M., David,P.R. and Kornberg,R.D. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science, 288, 640–649. [DOI] [PubMed] [Google Scholar]
- 127.Bartholomew B., Durkovich,D., Kassavetis,G.A. and Geiduschek,E.P. (1993) Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol., 13, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ferri M.L., Peyroche,G., Siaut,M., Lefebvre,O., Carles,C., Conesa,C. and Sentenac,A. (2000) A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol., 20, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Thuillier V., Brun,I., Sentenac,A. and Werner,M. (1996) Mutations in the α-amanitin conserved domain of the largest subunit of yeast RNA polymerase III affect pausing, RNA cleavage and transcriptional transitions. EMBO J., 15, 618–629. [PMC free article] [PubMed] [Google Scholar]
- 130.Chafin D.R., Guo,H. and Price,D.H. (1995) Action of α-amanitin during pyrophosphorolysis and elongation by RNA polymerase II. J. Biol. Chem., 270, 19114–19119. [DOI] [PubMed] [Google Scholar]
- 131.Wind M. and Reines,D. (2000) Transcription elongation factor SII. Bioessays, 22, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bartolomei M.S. and Corden,J.L. (1987) Localization of an α-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol. Cell. Biol., 7, 586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bartolomei M.S. and Corden,J.L. (1995) Clustered α-amanitin resistance mutations in mouse. Mol. Gen. Genet., 246, 778–782. [DOI] [PubMed] [Google Scholar]
- 134.Coulter D.E. and Greenleaf,A.L. (1985) A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J. Biol. Chem., 260, 13190–13198. [PubMed] [Google Scholar]
- 135.Bobkova E.V., Habib,N., Alexander,G. and Hall,B.D. (1999) Mutational analysis of the hydrolytic activity of yeast RNA polymerase III. J. Biol. Chem., 274, 21342–21348. [DOI] [PubMed] [Google Scholar]
- 136.Wang Z. and Roeder,R.G. (1997) Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev., 11, 1315–1326. [DOI] [PubMed] [Google Scholar]
- 137.James P., Whelen,S. and Hall,B.D. (1991) The RET1 gene of yeast encodes the second-largest subunit of RNA polymerase III. Structural analysis of the wild-type and ret1-1 mutant alleles. J. Biol. Chem., 266, 5616–5624. [PubMed] [Google Scholar]
- 138.Shaaban S.A., Krupp,B.M. and Hall,B.D. (1995) Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol. Cell. Biol., 15, 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bobkova E.V. and Hall,B.D. (1997) Substrate specificity of the RNase activity of yeast RNA polymerase III. J. Biol. Chem., 272, 22832–22839. [DOI] [PubMed] [Google Scholar]
- 140.Shaaban S.A., Bobkova,E.V., Chudzik,D.M. and Hall,B.D. (1996) In vitro analysis of elongation and termination by mutant RNA polymerases with altered termination behavior. Mol. Cell. Biol., 16, 6468–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miyao T. and Woychik,N.A. (1998) RNA polymerase subunit RPB5 plays a role in transcriptional activation. Proc. Natl Acad. Sci. USA, 95, 15281–15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Voutsina A., Riva,M., Carles,C. and Alexandraki,D. (1999) Sequence divergence of the RNA polymerase shared subunit ABC14.5 (Rpb8) selectively affects RNA polymerase III assembly in Saccharomyces cerevisiae. Nucleic Acids Res., 27, 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dumay H., Rubbi,L., Sentenac,A. and Marck,C. (1999) Interaction between yeast RNA polymerase III and transcription factor TFIIIC via ABC10alpha and tau131 subunits. J. Biol. Chem., 274, 33462–33468. [DOI] [PubMed] [Google Scholar]
- 144.Azuma Y., Yamagishi,M. and Ishihama,A. (1993) Subunits of the Schizosaccharomyces pombe RNA polymerase II: enzyme purification and structure of the subunit 3 gene. Nucleic Acids Res., 21, 3749–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kimura M., Ishiguro,A. and Ishihama,A. (1997) RNA polymerase II subunits 2, 3 and 11 form a core subassembly with DNA binding activity. J. Biol. Chem., 272, 25851–25855. [DOI] [PubMed] [Google Scholar]
- 146.Shpakovski G.V. and Shematorova,E.K. (1999) Rpc19 and Rpc40, two α-like subunits shared by nuclear RNA polymerases I and III, are interchangeable between the fission and budding yeasts. Curr. Genet., 36, 208–214. [DOI] [PubMed] [Google Scholar]
- 147.Imai K., Imazawa,Y., Yao,Y., Yamamoto,K., Hisatake,K., Muramatsu,M. and Nogi,Y. (1999) The fission yeast rpa17+ gene encodes a functional homolog of AC19, a subunit of RNA polymerases I and III of Saccharomyces cerevisiae. Mol. Gen. Genet., 261, 364–373. [DOI] [PubMed] [Google Scholar]
- 148.Imazawa Y., Imai,K., Fukushima,A., Hisatake,K., Muramatsu,M. and Nogi,Y. (1999) Isolation and characterization of the fission yeast gene rpa42+, which encodes a subunit shared by RNA polymerases I and III. Mol. Gen. Genet., 262, 749–757. [DOI] [PubMed] [Google Scholar]
- 149.Lalo D., Carles,C., Sentenac,A. and Thuriaux,P. (1993) Interactions between three common subunits of yeast RNA polymerases I and III. Proc. Natl Acad. Sci. USA, 90, 5524–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Busby S. and Ebright,R.H. (1999) Transcription activation by catabolite activator protein (CAP). J. Mol. Biol., 293, 199–213. [DOI] [PubMed] [Google Scholar]
- 151.Chedin S., Riva,M., Schultz,P., Sentenac,A. and Carles,C. (1998) The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev., 12, 3857–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Werner M., Hermann-Le Denmat,S., Treich,I., Sentenac,A. and Thuriaux,P. (1992) Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol. Cell. Biol., 12, 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Werner M., Chaussivert,N., Willis,I.M. and Sentenac,A. (1993) Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem., 268, 20721–20724. [PubMed] [Google Scholar]
- 154.Thuillier V., Stettler,S., Sentenac,A., Thuriaux,P. and Werner,M. (1995) A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J., 14, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mann C., Micouin,J.Y., Chiannilkulchai,N., Treich,I., Buhler,J.M. and Sentenac,A. (1992) RPC53 encodes a subunit of Saccharomyces cerevisiae RNA polymerase C (III) whose inactivation leads to a predominantly G1 arrest. Mol. Cell. Biol., 12, 4314–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ittmann M., Greco,A. and Basilico,C. (1987) Isolation of the human gene that complements a temperature-sensitive cell cycle mutation in BHK cells. Mol. Cell. Biol., 7, 3386–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ittmann M.M. (1994) Cell cycle control of the BN51 cell cycle gene which encodes a subunit of RNA polymerase III. Cell Growth Differ., 5, 783–788. [PubMed] [Google Scholar]
- 158.Greasley P.J., Bonnard,C. and Amati,B. (2000) Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res., 28, 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang Z., Luo,T. and Roeder,R.G. (1997) Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev., 11, 2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Frendewey D., Barta,I., Gillespie,M. and Potashkin,J. (1990) Schizosaccharomyces U6 genes have a sequence within their introns that matches the B box consensus of tRNA internal promoters. Nucleic Acids Res., 18, 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kundu T.K., Wang,Z. and Roeder,R.G. (1999) Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol. Cell. Biol., 19, 1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Marsolier M.C., Tanaka,S., Livingstone-Zatchej,M., Grunstein,M., Thoma,F. and Sentenac,A. (1995) Reciprocal interferences between nucleosomal organization and transcriptional activity of the yeast SNR6 gene. Genes Dev., 9, 410–422. [DOI] [PubMed] [Google Scholar]
- 163.Burnol A.F., Margottin,F., Schultz,P., Marsolier,M.C., Oudet,P. and Sentenac,A. (1993) Basal promoter and enhancer element of yeast U6 snRNA gene. J. Mol. Biol., 233, 644–658. [DOI] [PubMed] [Google Scholar]
- 164.Gerlach V.L., Whitehall,S.K., Geiduschek,E.P. and Brow,D.A. (1995) TFIIIB placement on a yeast U6 RNA gene in vivo is directed primarily by TFIIIC rather than by sequence-specific DNA contacts. Mol. Cell. Biol., 15, 1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lopez S., Livingstone-Zatchej,M., Jourdain,S., Thoma,F., Sentenac,A. and Marsolier,M.-C. (2001) High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol. Cell. Biol., 21, 3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Stunkel W., Kober,I. and Seifart,K.H. (1997) A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol. Cell. Biol., 17, 4397–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Allison D.S. and Hall,B.D. (1985) Effects of alterations in the 3′ flanking sequences on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J., 4, 2657–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mao J., Appel,B., Schaack,J., Sharp,S., Yanada,H. and Soll,D. (1982) The 5S RNA genes of Schizosaccharomyces pombe. Nucleic Acids Res., 10, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bogenhagen D.F. and Brown,D.D. (1981) Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell, 24, 261–270. [DOI] [PubMed] [Google Scholar]
- 170.Gunnery S., Ma,Y. and Mathews,M.B. (1999) Termination sequence requirements vary among genes transcribed by RNA polymerase III. J. Mol. Biol., 286, 745–757. [DOI] [PubMed] [Google Scholar]
- 171.Mazabraud A., Scherly,D., Muller,F., Rungger,D. and Clarkson,S.G. (1987) Structure and transcription termination of a lysine tRNA gene from Xenopus laevis. J. Mol. Biol., 195, 835–845. [DOI] [PubMed] [Google Scholar]
- 172.Maraia R.J., Chang,D.Y., Wolffe,A.P., Vorce,R.L. and Hsu,K. (1992) The RNA polymerase III terminator used by a B1-Alu element can modulate 3′ processing of the intermediate RNA product. Mol. Cell. Biol., 12, 1500–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Richardson J.P. (1993) Transcription termination. Crit. Rev. Biochem. Mol. Biol., 28, 1–30. [DOI] [PubMed] [Google Scholar]
- 174.Maraia R.J. (1996) Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc. Natl Acad. Sci. USA, 93, 3383–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Goodier J.L., Fan,H. and Maraia,R.J. (1997) A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol. Cell. Biol., 17, 5823–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rozenfeld S. and Thuriaux,P. (2001) Genetic interactions within TFIIIC, the promoter-binding factor of yeast RNA polymerase III. Mol. Gen. Genet., 265, 705–710. [DOI] [PubMed] [Google Scholar]
- 177.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.McKune K. and Woychik,N.A. (1994) Functional substitution of an essential yeast RNA polymerase subunit by a highly conserved mammalian counterpart. Mol. Cell. Biol., 14, 4155–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shpakovski G.V., Gadal,O., Labarre-Mariotte,S., Lebedenko,E.N., Miklos,I., Sakurai,H., Proshkin,S.A., Van Mullem,V., Ishihama,A. and Thuriaux,P. (2000) Functional conservation of RNA polymerase II in fission and budding yeasts. J. Mol. Biol., 295, 1119–1127. [DOI] [PubMed] [Google Scholar]
- 180.Shpakovskii G.V. and Lebedenko,E.N. (1997) Molecular cloning and characterization of cDNA of the rpc10+ gene encoding the smallest subunit of nuclear RNA polymerases of Schizosaccharomyces pombe (in Russian). Bioorg. Khim., 23, 441–448. [PubMed] [Google Scholar]
- 181.Shpakovskii G.V., Lebedenko,E.N. and Thuriaux,P. (1997) Cloning of cDNA for RNA polymerase subunit from the fission yeast Schizosaccharomyces pombe by heterospecific complementation in Saccharomyces cerevisiae (in Russian). Bioorg. Khim., 23, 110–117. [PubMed] [Google Scholar]