Abstract
The most recent guidelines have called for a significant shift towards viral load testing for HIV/AIDS management in developing countries; however point-of-care (POC) CD4 testing still remains an important component of disease staging in multiple developing countries. The advancements in micro/nanotechnologies and consumer electronics have paved the way for mobile healthcare technologies, and the development of point-of-care smartphone-based diagnostic assays for a variety of disease detection and treatment monitoring. Here, we report a simple, rapid (30 minutes) smartphone-based microfluidic chip for automated CD4 testing using a small volume (30 μL) of whole blood. The smartphone-based device includes an inexpensive (<$5) cellphone accessory and a functionalized disposable microfluidic device. We evaluated the performance of the device with spiked PBS samples and HIV-infected and uninfected whole blood, and compared the microfluidic chip results with the manual analysis and FACS results. Through t-tests, Bland-Altman analyses, and regression tests, we have shown a good agreement between the smartphone-based test and the manual and FACS analysis for CD4 count. The presented technology could have a significant impact in HIV management in developing countries through providing a reliable and inexpensive point-of-care CD4 testing.
TOC image
An inexpensive, rapid, and automated smartphone-based optical system for CD4+ T-cell count in whole blood.

Introduction
The number of people currently living with human immunodeficiency virus (HIV) is currently estimated to be 37 million, with 2.1 million new infections in 2015 alone. Widespread implementation of antiretroviral therapy (ART) has been highly effective in reducing mortality in HIV-infected individuals but the number of AIDS-related deaths is still more than 1.1 million every year1. CD4 testing and viral load measurements are critical parameters in identifying HIV-positive patients in need of ART and treatment monitoring. Although the recent ‘treat all’ policy of the World Health Organization (WHO) recommends earlier ART initiation regardless of CD4 count2, CD4 testing remains an important component of disease staging and plays a vital role in clinical disease management3.
Flow cytometry, the current gold standard method for counting CD4+ T-cells, uses a light beam focused on a stream of cells and several detectors, including a fluorescence detector, to identify cells of interest in a biological sample through capturing light scatter and fluorescent signals emitted from the fluorescence-labeled cells. Although this technology is accurate and sensitive,testiong currently available flow cytometers in the developed world settings are complex, time-consuming, bulky, expensive, labor-intensive, and cannot be easily implemented in resource-limited settings4. Point-of-care (POC) flow cytometry-based devices that have been developed recently are portable and less complex. These POC systems are robust, have achieved some of the critical criteria defined for POC testing, and have helped in improving patient care in resource-limited settings. However, they are still relatively expensive and their use outside of centralized laboratories in the developing world is further limited by electrical requirements and low throughput. Several other methods utilizing image-based, electrical sensing, ELISA, and centrifuge-based modalities have also been developed over the past few years for counting CD4+ T-cells at the POC4. These devices still have limitations for implementation in developing countries and to meet all the criteria required for a true POC CD4+ T-cell counter, i.e. ASSURED: affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end users. It has been estimated that only 13.7% of the available CD4 testing capacity was being utilized in 2013, with over 30% of the countries performing <1 test per patient and failing to meet the current recommendations. This underutilization is partially due to the need for reagents, maintenance requirements, and staff training. It has also been suggested that an unbalanced geographic distribution of CD4 count instruments might likely be responsible for poor utilization5.
Recent years have seen a rapid growth in mobile healthcare platforms due to advancements in mobile computing6. Their combination of mobile communication and computational power, packed into a small form factor device is a boon to the development of POC diagnostics7,8. Concurrently, microfluidics has shown a great promise in bringing POC testing to patients over the past decade9. Numerous approaches utilizing microfluidics and smartphone/cell phone capabilities for POC disease detection and management have been explored10–13. Here, we present an automated CD4 testing using small volume of whole blood by integrating advancements in consumer electronics and microfluidic chip fabrications (Fig. 1). We have evaluated the device with spiked PBS and whole blood samples drawn from healthy donors and HIV-infected subjects and compared the results with manual and FACS-based analyses.
Figure 1. Schematic comparing the process flow of CD4+T cell count using the smartphone-based system and FACS.
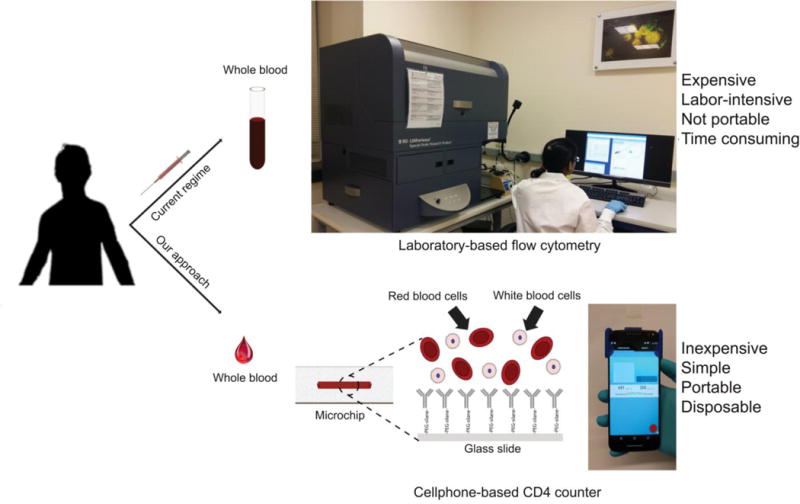
The smartphone-based system is portable, inexpensive, rapid (~30 mins), and easy-to-use compared to Fluorescence Activated Cell Sorting (FACS) which is expensive, lab-based, labor-intensive, and not portable.
Materials and Methods
Microfluidic Device Design and Fabrication
The microfluidic device was fabricated using 3.175 mm thick clear cast Poly(methyl methacrylate) (PMMA) sheets (8560K239, McMaster-Carr), 100 μm thick optically clear double-sided adhesive (DSA) sheets (82610, 3M) and regular glass slides (48300-131, Corning). The PMMA was cut using a laser cutter (Universal Laser System, VLS 2.3) to form the inlet and the outlet of the microfluidic device. Similarly, the microchannels were cut onto the DSA using the laser cutter. The power, scan speed, and pulse per inch rate to cut PMMA sheets were set to 28.2 W, 2.5 mm/s, and 1000 pulses/inch, respectively. The power, scan speed, and pulse per inch rate to cut DSA sheets were set to 6 W, 15 mm/s, and 500 pulses/inch, respectively. The microchannel, inlet and outlet were designed using CorelDRAW®. The length and width of the channel were 40mm and 5 mm, respectively. The diameter of the inlet and outlet of the microchannels were 0.8 mm. The dimensions and the positioning of the microfluidic channel were designed to suit the smartphone-based optical setup such that the user will be able to image and test samples with ease. PMMA and double-sided adhesive film were cleaned with 70% ethanol and distilled water prior assembling the chip manually. The patterned DSA was sandwiched between a cut PMMA and modified glass slide (Fig. 2).
Figure 2. Microfluidic chip, optical attachment and assay design.
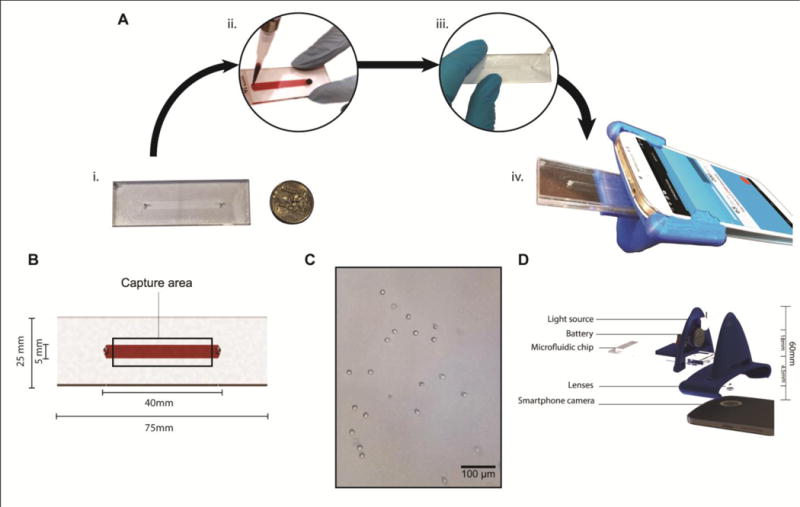
(A) The process flow showing the steps involved in performing the assay. (A.i) The disposable microfluidic chip is built and surface functionalized with anti-CD4 antibodies. (A.ii) ~30 μL of the sample is injected in the microfluidic chip. (A.iii) The chip is washed with PBS after an incubation of 30 mins. (A.iv) The microfluidic chip is inserted into the optical attachment and analysed using the smartphone application. (B) shows the dimensions of the microfluidic chip. (C) The actual image of captured CD4 cells on-chip using the smartphone system. (D) The exploded image of the smartphone attachment and the relevant dimensions. The total height of the attachment was 60 mm and the distance between the LED and the sample was 18 mm. The distance between the top surface of the microfluidic device and the first lens was.4.5 mm. The lenses were positioned adjacent to each other directly above the camera.
Surface modification of glass slides
Regular glass slides (25×75 mm) were cleaned with 70% ethanol and were dried using a nitrogen gun. To form the -OH functional groups, the slides were treated with oxygen plasma (100 mW, 15% oxygen) for 2:30 min in a plasma chamber (PE-50, Plasma Etch). Then, they were incubated with 10 mg/mL thiol PEG-silane (SH-PEG-Si) (PG2-SLTH-10k, NANOCS) in 5% ethanol for 30 minutes. Upon incubation, the glass slides were washed with 75% ethanol and were used for microfluidic chip fabrication.
Preparation of the microfluidic chip for CD4 capture
Followed by silanization of the glass slides, the microfluidic chip was fully assembled and activated with 1mg/mL 3-[2-Pyridyldithio] propionyl hydrazide (PI22301, Thermo-Fisher Scientific). Then, 30 μL of oxidized mouse monoclonal biotin anti-human CD4 antibody (0.5 mg/mL) (317406, BioLegend) was added to the microfluidic chip. The antibody was oxidized using the standard protocol in which antibody was mixed with 10 mM of sodium metaperiodate (71859, Sigma) and 0.1 M (adjusted to pH 5.5) sodium acetate (S2889, Sigma-Aldrich) and incubated at 4°C in dark for 20 min. The channel was washed three times with PBS to remove any excess reactants and kept at 4°C for further use.
CD4+ T-lymphocyte capture on the microfluidic platform
CD4+ T-cells were isolated from whole blood and suspended in PBS were used. To the prepared microfluidic device, 30 μL of isolated CD4 cells was added. It was incubated for 30 mins and was washed with PBS three times to remove any uncaptured cells. After capture, the microfluidic device was inserted into the optical setup and was analyzed using the smartphone application (Fig. 2). The dimension of the field-of-view using the smartphone system was ~634 × 475 μm. At the press of a button, our custom software application provides the equivalent concentration (cells/μL) of the sample. A total of 3 replicates were performed for each isolated CD4 sample tested and 2 replicates were performed for each whole blood test. Various concentrations of the suspension were prepared through dilutions and tested using the smartphone-based optical setup. In parallel, the number of cells for each sample was also assessed with an improved Neubauer hemocytometer (BR717805, Sigma-Aldrich) by a technician under a microscope.
Similarly, fresh whole blood from HIV-negative patients (Research Blood Components) was tested along with whole blood samples from HIV-infected patients. For whole blood samples, hemocytometer counts were not performed. A Conventional Flow-cytometry system was used to establish the concentration of the CD4+ T-cells in whole blood samples.
In the case of HIV-positive samples, whole blood was obtained by venipuncture from participants prospectively enrolled in the HIV-1 Eradication and Latency (HEAL) Cohort, a longitudinal study of HIV-infected, antiretroviral therapy-treated, virologically suppressed participants followed at Brigham and Women’s Hospital and Massachusetts General Hospital. This study was approved by the Partners Human Research Committee. Participants of the HEAL cohort represented a convenient sample of participants meeting the HEAL inclusion criteria. Samples obtained were based on participant flow and no other sample selection criteria was in place for the study. All patients (HIV-positive and negative) provided informed consent for blood samples to be collected.
Whole blood lysis and FACS staining
The BD Pharm LyseTM (555899, BD Biosciences) lysing solution (10×) was first diluted 1:10 with distilled water such that the pH of the 1× solution ranged between 7.1–7.4. Prior to use, the temperature of the solution was brought to room temperature. 2 mL of 1× lysing solution was added to the tube containing 200 μL of a whole blood sample with 10 μL (50 μg/mL) of rat monoclonal FITC anti-human CD4 antibody (357406, BioLegend) and 10 μL (50 μg/mL) of mouse monoclonal Alexa Fluor 647 anti-human CD3 antibody (557 557706, BD Biosciences). The mixture was vortexed immediately after adding the lysing solution and was incubated at room temperature in dark for 15 minutes. After the incubation, the tubes were centrifuged at 200 g of relative centrifugal force (RCF) for 5 minutes. The supernatant was discarded and the pellet was re-suspended in 2 mL of PBS and centrifuged once more for 5 minutes at 200 g RCF. If there was significant amount of red blood cells present, the lysis steps were repeated. The supernatant was removed and discarded without disturbing the pellet. The pellet was re-suspended in 100 μL of 4% (v/v) paraformaldehyde solution in PBS and incubated for 30 minutes at 4°C. After the incubation, the tube was centrifuged at 300 g RCF for 10 minutes. The supernatant was discarded and the pellet was re-suspended in 200 μL of PBS. The sample was tested with Accuri C6 Flow cytometer.
CD4 isolation
Peripheral blood mononuclear cells (PBMCs) were isolated using the Ficoll-Histopaque density centrifugation method from a healthy donor in local blood banks. CD4+ T-cells were isolated from PBMCs using the EasySep Human CD4 T-cell enrichment kit (Stem Cell Technologies, 19052). CD4+ T-cells were cryopreserved for future experiments. Cells were later thawed and washed twice in complete Roswell Park Memorial Institute 1640 (RPMI) medium. They were washed and were suspended in PBS prior to the cellphone-based testing procedure.
Smartphone-based optical setup
The optical attachment for the smartphone (MotoX- XT1575, Motorola) was designed with a CAD software (Solidworks 2016, Dassault Systèmes). The optical element of the attachment composed of a pair of aspheric lenses harvested from pick-up heads of an external DVD (SDRW-08D2S, Asus) drive and an internal CD drive (GCC-4320B, LG). The sample was trans-illuminated by a broadband LED (IL041, Microtivity), which was powered by a 3V cell battery (CR1620, Panasonic). The device was designed to operate with the rear camera of the smartphone (Fig. 2). It attached to the smartphone through a slide on mechanism. Stoppers were designed on the attachment such that the optical axis of the camera lens would align itself with the smartphone’s rear camera. The effective focal point was established practically through trial and error. The system was designed to avoid manual focusing and errors that arise from it. The image magnification in our optical attachment was calculated through imaging a micrometer scale. The smartphone system can potentially detect 1 cell per field-of-view (~633.6 × 475 μm), which effectively translates to 33 cells/μL. The attachment was 3D printed from polylactic acid (PLA) using a desktop fused deposition modeling (FDM) 3D printer (Ultimaker 2 Extended, Ultimaker). The final optical attachment measured 6.1 × 8.3 × 3.1 cm and weighed ~22 g.
Software Application
The smartphone application was designed to take images of the sample, perform image processing and analyze the image to identify the number of cells in the field-of-view (FOV), in a single click. The phone, which was running on Android 6.0, performed the necessary image processing, detection and analysis. The application can be easily ported to the iOS as well. The application was developed using Android Studio using an open-source, external computer vision library (OpenCV- ver. 2.4.8). Using an adaptive thresholding algorithm in combination with other image processing algorithms, the software can assess the concentration of CD4+ T-cells. Adaptive thresholding was used to separate the foreground (CD4 cells) from the background. Additional Gaussian filters were implemented to reduce noise in the thresholded image. A size gate was used to remove any artifacts outside of the size range. The remaining detected objects were enumerated and the concentration was calculated by the smartphone application automatically. The image processing time on-phone to calculate CD4 concentration of a sample using the smartphone system can be <10 seconds.
Statistical Analysis
Passing-Bablok regression analysis14, Bland-Altman Analysis15, Mountain plot16, sensitivity and specificity measurements were assessed using Medcalc 14.8.1. Microsoft Excel was used in the classification accuracy calculations while Stata 13 was used as well.
Multiple t-tests and the least square linear regression analysis were performed using Graphpad Prism ver. 6.0. Statistical significance was determined using t tests for multiple comparisons with Holm-Sidak corrections and 5% alpha assuming consistent SD.
Sample size for the whole blood-based study was estimated through a power analysis using the correlation coefficient-based sample size calculator available on Medcalc. The calculator yielded a minimum sample size of 8 when the expected co-efficient was set at 0.9 based on preliminary tests. The alpha and beta values were at 0.05 and 0.10, respectively.
Results
Optical attachment characterization
To evaluate the optical cellphone attachment and the software in accurately identifying and enumerating CD4+ T-cells on-chip, we used serially diluted spiked PBS samples (n=57) with cell concentrations ranging from 0 to 1640 cells/mL and compared the smartphone-based cell counting results with manual analysis using a hemocytometer. To perform these tests, we used non-functionalized microfluidic chips in order to evaluate the sensitivity of the device in identifying pre-isolated CD4+ T-cells in samples with a broad range of CD4+ T-cell concentrations. A Passing-Bablok analysis (n=57) on the results obtained by the cellphone-based device and manual analysis using a hemocytometer revealed an A intercept value of −25.11 and a B slope value of 0.97 (Fig. 3A). The confidence interval (CI) ranged from −73.57 to 12.66 for the A value and 0.88 to 1.07 for the B value. No systematic or proportional biases were observed. Similarly, the Bland-Altman analysis showed a mean bias of 41.06 cell/μL with a standard deviation (SD) of 154.4 cells/μL (Fig. 3B). The limits of agreement (LoA) ranged from −261.6 and 343.7. As an additional verification of the detection algorithm in accurately identifying and counting CD4+ T-cells on-chip, we also manually counted the number of cells in recorded images by the smartphone. A mountain plot analysis revealed that the smartphone-based and manual analyses for CD4+ T-cell counting were comparable, which showed a high accuracy of the smartphone-based device in counting pre-isolated CD4+ T-cells in non-functionalized microfluidic devices (Fig. 3C).
Figure 3. Performance of the smartphone-based device for CD4+ T cell counting using non-functionalized microfluidic chips.
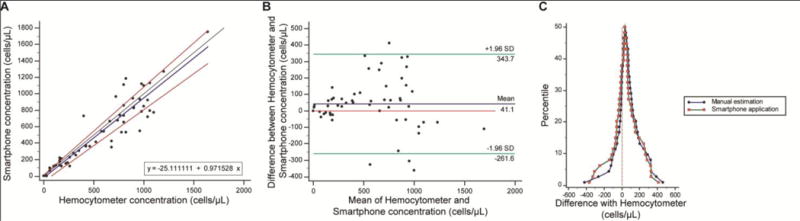
(A) Passing-Bablok and (B) Bland-Altman analyses to compare the concentration of CD4+ T-cells obtained from the smartphone-based CD4+ T cell counter using spiked PBS samples with CD4+ T cell concentrations between 0 cells/μL and 1640 cells/μL (n=57). The solid blue line represents the regression line, the black line represents the identity line, and the two red lines represent the confidence band of the regression line in the Passing-Bablok figure. In the Bland-Altman figure, the blue line shows the mean difference of the methods, and the green lines represent the 95% limits of agreements. (C) Mountain plots comparing the performance of the smartphone-based device and manual analysis using a hemocytometer.
Surface chemistry validation
To selectively capture CD4+ T-cells on-chip, the microfluidic chips were functionalized with anti-CD4 antibody. The glass substrate of the disposable microfluidic chip was functionalized with silane-polyethylene glycol (PEG)-thiol. The oxidized antibody was attached to the functionalized glass with 3-(2-pyridyldithio)propionyl hydrazide (PDPH) cross-linker (Fig. 4A). We used Fourier transform infrared spectroscopy (FTIR) to verify the surface functionalization and antibody immobilization on-chip. FTIR spectrum shows the formation of peaks at 1600–1700 cm−1 and 1550–1450 cm−1 wavelengths that correspond to amide I (C=O stretching and NH-bending), and amide II (N-H bending) vibrations, respectively, confirming the conjugation of the antibody to the surface of the microfluidic chip (Fig. 4B)17, 18. The O–H bending vibration at 1401 cm−1 and 1265 cm−1 wavelength, the C=O stretching vibration at 1742 cm−1 wavelength, and the C–O stretching vibration at 1107 cm−1 wavelength can be attributed to the presence of PEG19. In addition, the peaks at 823 cm−1 and 1052 cm−1 wavelengths, which are characteristic to Si-O-Si stretching and bending vibrations, indicate the successful salinization reaction in the developed protocol20.
Figure 4. Surface chemistry protocol and characterization.
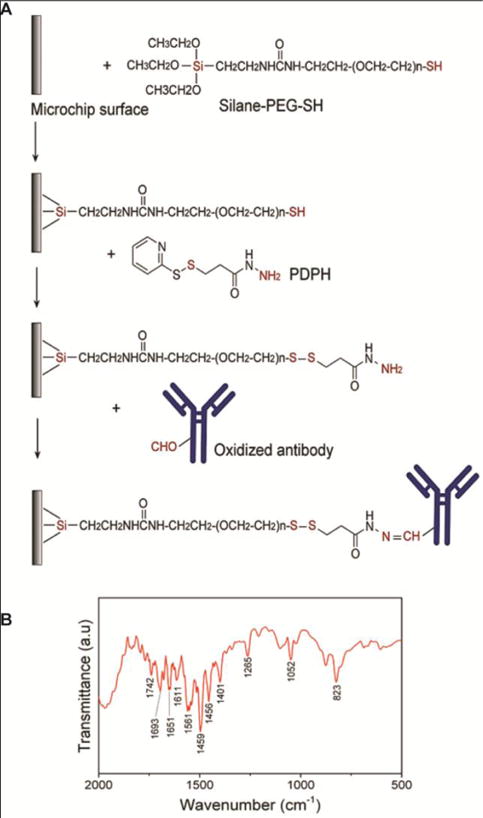
(A) Schematic illustration of the developed protocol for antibody conjugation to the surface of a microfluidic chip. The functional groups that are directly involved in the reactions are shown in red color in the figure, (B) FTIR spectrum of microfluidic chip surface confirming the conjugation of the antibody to the surface using the developed protocol
We established the optimal incubation time through capturing CD4+ T-cells with different incubation times (Fig. S1). A saturation of cell capture was achieved at 30 minutes, which was used as the incubation time for the smartphone-based microfluidic chip test validation. Further, we evaluated the CD4+ T-cell capture efficiency and specificity on functionalized microfluidic chips. These results showed cell capture efficiency and specificity of 84% and 95%, respectively (Fig. S2).
CD4 enumeration on functionalized microfluidic chips using spiked PBS samples
We evaluated the cellphone-based device using functionalized microfluidic chips and PBS samples spiked with pre-isolated CD4+ T-cells (n=30) with cell concentrations ranging from 60 cells/μL to 1200 cells/μL. The results obtained from the cellphone-based system were compared with the results obtained through a manual analysis using a hemocytometer. A Passing-Bablok analysis revealed an A intercept value of −33.21 with a confidence interval of −87.11 to 17.37 and a B slope value of 0.93 with a confidence interval of 0.83 to 1.09 (Fig. 5A) (n=30). The cusum test showed no deviation from linearity with P=0.34. No systematic or proportional bias was observed. The Bland-Altman analysis revealed the mean bias to be −77.32 with an SD of 132.8, and the LoA ranged from −337.6 to 183.0 (Fig. 5B) (n=30).
Figure 5. Device evaluation for sensitivity and specificity using surface-modified microfluidic chip.
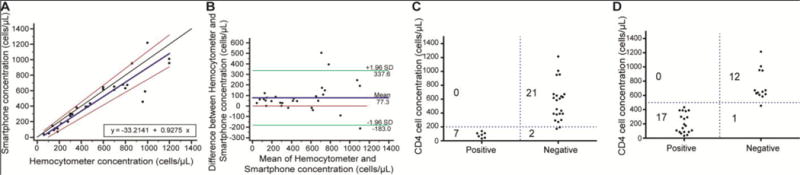
(A) Passing-Bablok and (B) Bland-Altman analyses to compare the concentration of CD4+ T-cells obtained from the smartphone-based CD4+ T cell counter when PBS samples with CD4+ T cell concentrations between 60 cells/μL and 1200 cells/μL (n=30) were used. The solid blue line represents the regression line, the black line represents the identity line, and the two red lines represent the confidence band of the regression line in the Passing-Bablok figure. In the Bland-Altman figure, the blue line shows the mean difference between the methods, and the green lines represent the 95% limits of agreement. The accuracy of the smartphone-based device for measuring CD4+ T cell concentration (C,D) was 93.3%, and 96.6% for 200 cells/μL and 500 cells/μL thresholds, respectively (n=30). The blue lines represent the threshold values for each criterion above which the sample is considered to be of normal quality. The threshold values were established based on the WHO guidelines for advanced disease treatment and ART initiation thresholds respectively.
We also evaluated the performance of the smartphone-based device in identifying samples with cell concentrations below the threshold of 200 cells/μL, and the sensitivity and specificity of the device were 100% and 91.30%, respectively (Fig. 5C). The accuracy of the smartphone-based system was 93.3%. Similarly, at the threshold of 500 cells/μL, the sensitivity and specificity were 100% and 92.31%, respectively with an accuracy of 96.66% (Fig. 5D). The results on diluted CD4-spiked samples suggested that the lowest detectable concentration as 60 cells/μL (Table S2, Fig. 5A).
Device evaluation using whole blood
The performance of the smartphone-based device was also evaluated using whole blood samples drawn from healthy donors and HIV-infected patients (n=11) by comparing the microfluidic chip results with the results obtained by FACS. We evaluated the linearity of the measurements by the smartphone-based device and FACS using a least squares regression (Fig. 6A). The straight-line equation yielded a slope value of 1.078 ± 0.093 and a y-intercept value of −50.78 ± 76.73. The R-squared value was 0.94 and the pearson correlation coefficient (r) was 0.97. Bland-Altman analysis (n=11) between the results obtained by the smartphone-based device and FACS showed a mean bias of −7.72 with an SD of 101.3 (Fig. 6B). The LoA ranged between −206.2 and 190.7. No systematic of proportional bias was observed.
Figure 6. Device evaluation using whole blood samples drawn from healthy donors and HIV-infected patients.
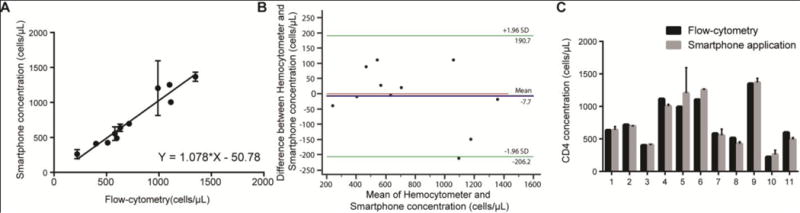
(A) Linear correlation between the results obtained by the smartphone-based system and FACS using whole blood (n=11). The pearson correlation coefficient was 0.97. (B) Bland-Altman analysis between the results obtained by FACs and the smartphone-based system. The blue line in this figure shows the mean difference of the methods, and the green lines represent the 95% limits of agreements (n=11). (C) Head-tohead comparison of concentration measurements made by both methods. Each value of x-axis represents a patient sample and t-tests revealed no statistical significance (P>0.05) between the two methods for all samples tested. Error bars in A and C represent the standard error of mean.
We also performed multiple t-tests with the Holm-Sidak correction assuming similar scatter to compare the results obtained by the smartphone-based device and FACS (Fig. 6C). These results showed no statistical significance (P>0.05) between all values measured by the smartphone-based device and FACS. The P values were 0.98, 0.93, 0.96, 0.62, 0.35, 0.50, 0.90, 0.69, 0.93, 0.86 and 0.62 for samples 1 through 11, respectively.
Discussion
ART has been improved significantly, however, early diagnosis of infection and retaining diagnosed HIV-infected patients in care using optimal drug regimens are still some of the key challenges in HIV management specifically in developing countries. In healthy individuals, CD4 count usually ranges from 500 to 1600 cells/μL. The CD4 count below 200 cells/μL is an indication of an advanced stage of the HIV infection. According to the WHO 2013 Consolidated ART guidelines, ART is recommended to be conditionally initiated for patients with CD4 count of 500 cells/μL or less and immediately initiated for HIV-infected people in serodiscordant partnerships, children up to 5 years of age, and people with active tuberculosis or with hepatitis B co-infection and chronic liver disease21. Only a few countries offer ART to all HIV-infected patients regardless of CD4 count22.19 countries allowed for immediate ART in HIV serodiscordant couples and 12 countries allowed for immediate ART for all breastfeeding and pregnant women23. In addition, the risk of death and mortality rate significantly increases in HIV-infected patients presenting with CD4 count less than 500 cells/μL and CD4 count has a critical role in decision-making for assessment and prophylaxis for major opportunistic infections3. Low CD4 count is prognostic for several diseases with high mortality rate including pneumocystis, jirovecii pneumonia, bacterial pneumonia, toxoplasmosis, cryptococcal meningitis, disseminated cytomegalovirus disease, and Mycobacterium avium complex. For example, 20% of AIDS-related deaths in low- and middle-income countries is due to Cryptococcal meningitis24. Therefore, ART for all HIV-infected patients irrespective of CD4 count is a recommendation rather than a rule and CD4 count remains a key diagnostic and ART monitoring tool. Standard flow cytometry-based methods used for CD4 count in the developed world cannot be easily implemented in resource-limited settings, as they are expensive, laboratory-based, complex, time-consuming, and labor-intensive. Therefore, there is an urgent need to develop novel POC technologies for rapid, inexpensive, and sensitive CD4 count testing appropriate for resource-limited settings.
POC CD4 testing has been shown to improve patient retention and enrollment. A study conducted in Mozambique showed that patient follow-up loss post CD4 testing and before ART initiation was nearly halved due to POC CD4 testing, and subsequently the ART initiation of enrolled patients was nearly doubled25. POC CD4 testing is especially useful in monitoring patients’ immune status and progression into advanced disease. A recent study showed that even by pricing the POC CD4 testings at 140% higher than the conventional methods for CD4 count, POC CD4 testing is more cost-effective than the conventional methods due to its ability in improving the linkage to care and life expectancy26. Furthermore, POC CD4 testing can drastically reduce the turnaround time currently required for patient baseline evaluations after HIV diagnosis and before ART initiation from on average 10.5 days to 0.1 days27. Similarly, it was shown that the time for ART initiation can be reduced to an average of 9 days from 31.5 days when POC CD4 testing was used as compared to the conventional CD4 testing. A need for semi-quantitative POC systems capable of distinguishing patients at the thresholds of 200 cells/μL has been emphasized recently as well28.
Here, we reported the development and evaluation of a smartphone-based microfluidic chip sensor for the accurate and sensitive quantification of CD4+ T-cells on disposable microfluidic chips functionalized with anti-CD4 antibody using small volume of whole blood samples. The cellphone-based device performance was evaluated using PBS samples spiked with pre-isolated CD4+ T-cells and whole blood drawn from healthy donors and HIV-infected patient subjects. The device results were verified by comparing with the results obtained using manual analysis and FACS. We also confirmed the accuracy of the microfluidic chip technology using whole blood samples from healthy donors and HIV-infected individuals. We have used a directional antibody conjugation technique for efficient cell capture on inexpensive disposable microfluidic chips. We obtained a sensitivity of 100% and specificity of >90% in accurately classifying samples based on 200 and 500 cells/μL thresholds. To put the performance of our device into context, the commercially available Alere PIMA™ has shown 94% and 96% sensitivities at 200 and 500 cells/μL thresholds, respectively29, 30. Similarly, the sensitivity of CyFlow® miniPOC was >95% at 200, 350 and 500 cells/μL thresholds31. It is important to keep in mind that the performance of these systems were observed in the field, and further field-validation is required with our presented system before direct comparisons can be made.
The currently available POC technologies have drastically improved care in resource-limited settings. However, most of these technologies are still limited to testing centers, thereby partially inducing under-utilization of the equipment. Few systems like Alere PIMA™, have improved coverage by supporting mobile testing sites, yet are still relatively expensive with the initial equipment ranging around $7500 with an additional $1200 for maintenance32. Our solution has the potential to reduce the costs significantly for CD4 testing. The smartphone-based system presented here requires a mass-producible and inexpensive microfluidic device and an optical smartphone attachment (<$5). The total material cost to fabricate the microfluidic chip was <$2, which includes $0.17 for Poly (methyl methacrylate), <1 cent for the double sided adhesive (DSA), $0.50 for glass slides, $0.18 for the antibody, and ~$1.06 for all other reagents. The material cost to fabricate the smartphone attachment was <$2.75, with ~$0.32 for the 3D printed smartphone accessory, $0.09 for an LED, $1 for the lenses, $0.6 for a battery, and $0.7 for the switches and wires.
The limited dependence on infrastructure availability can improve decentralization especially in low and middle-income countries. The setup does not require a power outlet and can function with a 3.3V cell battery. The microfluidic chips can be adapted to avoid the need for refrigeration. We have previously developed an on-chip surface chemistry protocol to preserve multi-layered surfaces of immuno-functionalized microfluidic devices with immobilized antibodies using trehalose33. Trehalose is a naturally occurring stress response factor and which helps in maintaining cellular integrity under harsh environmental conditions34–36. We were able to demonstrate that functionalized microfluidic chips treated with trehalose retained the functional efficiency at high temperatures (50 °C) and humidity (85%) with negligible difference compared to fresh microfluidic chips. Similar protocol can be used for the surface chemistry scheme used in this study to develop microchips for CD4+ T cells capture in whole blood with long shelf life. Another solution is freeze-drying the surface chemistry to prolong stability and shelf life of the microfluidic chip.
There is currently an accelerated growth in smartphone adoption in low and middle-income countries. The number of smartphone users nearly doubled in the last two years just in Africa37. Similarly, in India, cheaper android smartphones have pushed the country to become the second largest user base in the world above the United States, making it one of the largest growing smartphone market38. With a fast growing user base, smartphone-based diagnostic alternatives can potentially be used with little to no training39, 40. The work we presented here is an example of how smartphones can be used for HIV/AIDS management in resource-limited settings.
Supplementary Material
Acknowledgments
We would like to thank Dheerendranath Battalapalli, Vignesh Natarajan and Prudhvi Thirumalaraju for their valuable assistance and fruitful conversations. We would also like to thank Francoise Giguel for her help with logistics and Qianjing He for her help in acquiring blood samples from patients. Research reported in this publication was partially supported by the National Institute of Health under award numbers R01AI118502 and P30ES000002; Brigham and Women’s Hospital and Harvard Medical School through Bright Futures Prize and Innovation Evergreen Award; National Sciences and Engineering Research Council of Canada (NSERC) through NSERC Postdoctoral fellowship (M.S.); Harvard T.H. Chan School of Public Health, Harvard Center for Environmental Health through Harvard NIEHS Grant; Harvard CFAR (P30 AI060354) and American Board of Obstetrics and Gynecology, American College of Obstetricians and Gynecologists, American Society for Reproductive Medicine, Society for Reproductive Endocrinology and Infertility through ASRM Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Author contributions
H.S. conceived the method. M.K.K., H.J.P and H.S. planned the experiments. M.K.K., M.S and H.S. designed the 3D printed attachment and M.K.K. fabricated it. M.K.K. developed the android application. H.J.P. and M.K.C. designed and fabricated the microfluidic chips. M.S.D designed the surface chemistry. H.J.P., M.K.K., S.K, V.Y. and M.K.C. performed the experiments. B.S aided in cell-staining optimization. H.J.P., M.K.C., W.A, and M.K.K. analysed the data. B.E. performed the CD4 isolation and provided the cells for the experiments; A.M.T. provided the HIV-positive patient samples. A.M.T., J.Z.L. and D.R.K. provided supervision and logistical support. M.K.K. and H.S. wrote the manuscript. All authors have given approval to the final version of the manuscript.
Competing financial interests:
The authors declare no competing financial interests.
Notes and references
- 1.UNAIDS. Global AIDS Update. World Health Organization; Geneva, Switzerland: May, 2016. [Google Scholar]
- 2.WHO. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. 2nd. World Health Organization; 2016. [PubMed] [Google Scholar]
- 3.Ford N, Meintjes G, Pozniak A, Bygrave H, Hill A, Peter T, Davies MA, Grinsztejn B, Calmy A, Kumarasamy N, Phanuphak P, deBeaudrap P, Vitoria M, Doherty M, Stevens W, Siberry GK. Lancet Infect Dis. 2015;15:241–247. doi: 10.1016/S1473-3099(14)70896-5. [DOI] [PubMed] [Google Scholar]
- 4.Shafiee H, Wang S, Inci F, Toy M, Henrich TJ, Kuritzkes DR, Demirci U. Annual Review of Medicine. 2015;66:387–405. doi: 10.1146/annurev-med-092112-143017. [DOI] [PubMed] [Google Scholar]
- 5.Habiyambere V, Ford N, Low-Beer D, Nkengasong J, Sands A, Gonzalez MPerez, Fernandes P, Milgotina E. PLoS medicine. 2016;13:e1002088. doi: 10.1371/journal.pmed.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinhubl SR, Muse ED, Topol EJ. Science translational medicine. 2015;7:283rv283. doi: 10.1126/scitranslmed.aaa3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosa ASM, Yoo I, Sheets L. BMC Medical Informatics and Decision Making. 2012;12:67. doi: 10.1186/1472-6947-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis T, Synowiec C, Lagomarsino G, Schweitzer J. Bulletin of the World Health Organization. 2012;90:332–340. doi: 10.2471/BLT.11.099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez FA. Bioanalysis. 2013;5:1–3. doi: 10.4155/bio.12.307. [DOI] [PubMed] [Google Scholar]
- 10.Kanakasabapathy MK, Sadasivam M, Singh A, Preston C, Thirumalaraju P, Venkataraman M, Bormann CL, Draz MS, Petrozza JC, Shafiee H. Science translational medicine. 2017:9. doi: 10.1126/scitranslmed.aai7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng D, Mudanyali O, Oztoprak C, Isikman SO, Sencan I, Yaglidere O, Ozcan A. Lab Chip. 2010;10:1787–1792. doi: 10.1039/c003477k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, Yaglidere O, Su TW, Tseng D, Ozcan A. Lab Chip. 2011;11:315–322. doi: 10.1039/c0lc00358a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg B, Cortazar B, Tseng D, Ozkan H, Feng S, Wei Q, Chan RY, Burbano J, Farooqui Q, Lewinski M, Di Carlo D, Garner OB, Ozcan A. ACS Nano. 2015;9:7857–7866. doi: 10.1021/acsnano.5b03203. [DOI] [PubMed] [Google Scholar]
- 14.Passing H, Bablok Journal of clinical chemistry and clinical biochemistry. Zeitschrift fur klinische Chemie und klinische Biochemie. 1983;21:709–720. doi: 10.1515/cclm.1983.21.11.709. [DOI] [PubMed] [Google Scholar]
- 15.Bland JM, Altman DG. Lancet (London, England) 1986;1:307–310. [PubMed] [Google Scholar]
- 16.Krouwer JS, Monti KL. European journal of clinical chemistry and clinical biochemistry: journal of the Forum of European Clinical Chemistry Societies. 1995;33:525–527. [PubMed] [Google Scholar]
- 17.Costantini F, Nascetti A, Scipinotti R, Domenici F, Sennato S, Gazza L, Bordi F, Pogna N, Manetti C, Caputo D, Cesare GD. RSC Advances. 2013 doi: 10.1039/C3RA46058D. [DOI] [Google Scholar]
- 18.Wu Y, Zuo F, Zheng Z, Ding X, Peng Y. Nanoscale research letters. 2009;4:738–747. doi: 10.1007/s11671-009-9314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Feng L, Yang H, Xin G, Li W, Zheng J, Tian W, Li X. Physical chemistry chemical physics: PCCP. 2012;14:13233–13238. doi: 10.1039/c2cp41988b. [DOI] [PubMed] [Google Scholar]
- 20.Zhao S, Zhang J, Zhu M, Zhang Y, Liu Z, Ma Y, Zhu Y, Zhang C. Journal of Materials Chemistry B. 2014 doi: 10.1039/C4TB01287A. [DOI] [Google Scholar]
- 21.WHO. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection Recommendations for a public health approach. Geneva: 2013. [PubMed] [Google Scholar]
- 22.NIH. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. D o H a H Services; 2013. [Google Scholar]
- 23.Nelson LJ, Beusenberg M, Habiyambere V, Shaffer N, Vitoria MA, Montero RG, Easterbrook PJ, Doherty MC. AIDS. 2014;28(Suppl 2):S217–224. doi: 10.1097/QAD.0000000000000239. [DOI] [PubMed] [Google Scholar]
- 24.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 25.Jani IV, Sitoe NE, Alfai ER, Chongo PL, Quevedo JI, Rocha BM, Lehe JD, Peter TF. The Lancet. 2011;378:1572–1579. doi: 10.1016/S0140-6736(11)61052-0. [DOI] [PubMed] [Google Scholar]
- 26.Hyle EP, Jani IV, Lehe J, Su AE, Wood R, Quevedo J, Losina E, Bassett IV, Pei PP, Paltiel AD, Resch S, Freedberg KA, Peter T, Walensky RP. PLoS medicine. 2014;11:e1001725. doi: 10.1371/journal.pmed.1001725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vojnov L, Markby J, Boeke C, Harris L, Ford N, Peter T. PLoS One. 2016:11. doi: 10.1371/journal.pone.0155256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford N, Meintjes G, Vitoria M, Greene G, Chiller T. Current Opinion in HIV and AIDS. 2017;12:123–128. doi: 10.1097/COH.0000000000000348. [DOI] [PubMed] [Google Scholar]
- 29.Rathunde L, Kussen GM, Beltrame MP, Dalla Costa LM, Raboni SM. Int J STD AIDS. 2014;25:956–959. doi: 10.1177/0956462414526862. [DOI] [PubMed] [Google Scholar]
- 30.Scott LE, Campbell J, Westerman L, Kestens L, Vojnov L, Kohastsu L, Nkengasong J, Peter T, Stevens W. BMC Med. 2015;13:168. doi: 10.1186/s12916-015-0396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wade D, Diaw PA, Daneau G, Diallo AA, Mboup S, Dieye TN, Kestens L. PLoS One. 2015;10:e0116663. doi: 10.1371/journal.pone.0116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larson B, Schnippel K, Ndibongo B, Long L, Fox MP, Rosen S. PLoS One. 2012:7. doi: 10.1371/journal.pone.0035444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asghar W, Yuksekkaya M, Shafiee H, Zhang M, Ozen MO, Inci F, Kocakulak M, Demirci U. Sci Rep. 2016;6 doi: 10.1038/srep21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpinelli J, Krämer R, Agosin E. Applied and Environmental Microbiology. 2006;72:1949–1955. doi: 10.1128/AEM.72.3.1949-1955.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kempf B, Bremer E. Archives of Microbiology. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 36.Crowe LM. Comparative Biochemistry and Physiology-Part A: Molecular & Integrative Physiology. 2002;131:505–513. doi: 10.1016/s1095-6433(01)00503-7. [DOI] [PubMed] [Google Scholar]
- 37.Intelligence G. The Mobile Economy Africa 2016. London, United Kingdom; 2016. [Google Scholar]
- 38.Newzoo . Global Mobile Market Report 2017. Amsterdam, Netherlands: 2017. [Google Scholar]
- 39.Kanakasabapathy MK, Sadasivam M, Singh A, Preston C, Thirumalaraju P, Venkataraman M, Bormann CL, Draz MS, Petrozza JC, Shafiee H. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK. Sci Transl Med. 2015;7:273re271. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


