Abstract
MicroRNAs are small RNAs that negatively regulate gene expression posttranscriptionally. Because changes in microRNA expression can promote or maintain disease states, microRNA-based therapeutics are being evaluated extensively. Unfortunately, the therapeutic potential of microRNA replacement is limited by deficient delivery vehicles. In this work, microRNAs are delivered in the absence of a protective vehicle. The method relies on direct attachment of microRNAs to folate (FolamiR), which mediates delivery of the conjugated microRNA into cells that overexpress the folate receptor. We show that the tumor-suppressive FolamiR, FolamiR-34a, is quickly taken up both by triple-negative breast cancer cells in vitro and in vivo and by tumors in an autochthonous model of lung cancer and slows their progression. This method delivers microRNAs directly to tumors in vivo without the use of toxic vehicles, representing an advance in the development of nontoxic, cancer-targeted therapeutics.
INTRODUCTION
The lack of efficient delivery vehicles is recognized as a major bottle-neck in microRNA (miRNA)–based therapeutics. Problems such as delivery-associated toxicity, poor transfection efficiency, systemic clearance, nonspecific biodistribution, and degradation in circulation are hindering the therapeutic potential of miRNAs (1). These vehicle-related defects may be overcome by eliminating the need for a vehicle, using a targeted vehicle-free delivery approach. Previously, this strategy has been developed for the delivery of antagomirs (antisense miRNA inhibitors) (2–6), which are used to sequester overexpressed miRNAs, and other antisense oligonucleotides that inhibit gene expression by hybridization via sequence complementarity (7, 8). However, a similar approach has not been successfully used to reintroduce clinically relevant miRNAs (mimics).
Vehicle-free delivery of antagomirs has advanced more rapidly than delivery of miRNA mimics because of modifications that can be made to the backbone of the antagomirs, which protect the molecule from degradation by serum ribonucleases. Although the backbone of antisense oligonucleotides and antagomirs can be modified to protect RNAs from degradation in circulation (4, 9–13), miRNA mimics are not amendable to major stabilizing modifications because the alterations prevent recognition and loading into Argonaute, a component of the RNA-induced silencing complex (RISC) (14, 15) that is essential for miRNA function. Therefore, generating a functional miRNA mimic that is completely resistant to endonucleases is not realistic. In light of this, miRNA mimics have been packaged into various protective delivery vehicles including nanoparticles (16–19), liposomes (20), micelles (21), and hydrogels (22).
Our previous work demonstrated that lentiviral- and liposomal-mediated delivery of the tumor-suppressive miRNA miRNA-34a (miR-34a) reduces tumor burden in non–small cell lung cancer (NSCLC) mouse models (23, 24). In addition to vehicle- and viral-mediated miRNA delivery, systemic injection of vehicle-free oligonucleotides has also been tested. However, this approach has proven problematic because of the pharmacokinetic and stability limitations associated with intravenous delivery, and thus either relies on local delivery or necessitates achieving a high oligonucleotide concentration that is often only seen in kidneys and liver (7). Although local delivery is an option, achieving delivery beyond sites that are accessible to local delivery, such as to micrometastatic lesions, is not achievable.
To overcome the challenge of nontargeted delivery, cell surface receptors specifically overexpressed on tumor cells can be exploited to further enhance miRNA mimic delivery beyond sites accessible by local delivery. Engineering the ligands that bind to those receptors to carry a functionally active miRNA could generate a molecule that could potentially be used to target miRNAs specifically to tumor cells (25). To be useful, the target receptor must meet two criteria: The receptor must be overexpressed on the cancer cell relative to normal cells, and the expression of the receptor must be sufficient to enable delivery of therapeutic quantities of a miRNA to the cancer cell. An attractive candidate that fulfills both of these criteria is the folate receptor (FR), which is overexpressed on many epithelial cancers, including cancers of the breast (26), lung (27), ovary (28), kidney (29), and colon (30), and various hematological malignancies such as acute myeloid leukemia (31). The presence of the FR on normal tissues appears to be limited in quantity, inconsequential for targeted drug applications, or inaccessible to blood-borne folates (25). The FR has a suitable ligand, vitamin B9 (folic acid), that is selective for the FR, binds to the FR with high affinity, and contains a derivatizable functional group for facile conjugation to imaging or therapeutic agents that does not interfere with binding to the receptor (32). Thus, FR/folate conjugate therapy has great potential for delivery of small RNAs such as miRNA or short interfering RNA (siRNA) (33, 34).
Successful folate-targeted delivery, with payloads as diverse as small radiopharmaceutical agents to large DNA-containing formulations, has been exemplified at both the preclinical and clinical levels (35). However, folate-mediated delivery of small RNAs lags behind because of the hypothesis that RNAs in circulation need to be protected from degradation. Various strategies have been pursued in the field of small RNA delivery to achieve protection, incorporating folate onto dendrimer, copolymer, or liposomal carrier vehicles (36, 37). These complexes have a very large size, which often leads to hampered penetration of target tissues because of the dense extracellular matrix found in most solid tumors (38).
Here, we provide evidence for a unique strategy that directly links miRNA mimics to the folate ligand, which we have termed FolamiRs. We reasoned that these smaller FolamiRs would perfuse solid tumors more easily than larger miRNA-encapsulating vehicles. One major concern with this approach is that the native form of small RNAs is relatively unstable in blood (8). In an effort to overcome this potential issue, yet retain activity of the mature strand, the passenger strand of the miRNA mimic was minimally modified with 2′-O-methyl RNA bases, which stabilize the RNA and increase nuclease resistance without impairing Argonaute loading (39). Furthermore, in our previous studies, it was shown that folate linked to rhodamine saturates a solid tumor after intravenous injection in less than 5 min (38, 40, 41). The speed by which the folate-conjugated molecules enter the tumor demonstrates that FolamiRs need only to survive in circulation for a very short period of time.
We present evidence for amethod that directly conjugates miR-34a to folate (FolamiR-34a) to deliver functional and minimally modified miRNAs specifically and rapidly to tumor tissue. We show that miR-34a is selectively targeted to the tumor, enters the tumorigenic cells, down-regulates target genes, and suppresses growth of tumors in vivo in mouse models of human lung and breast cancer.
RESULTS
Folate–miR-34a (FolamiR-34a) conjugation is achieved using click chemistry
When conjugating a drug to a ligand, a cleavable bridge is desired for delivery of therapeutic cargo because release of the unmodified cytotoxic agent is often required for maximal drug efficacy (42). Because no data were available to indicate if the conjugated folate would impair miRNA loading into Argonaute, both releasable and unreleasable chemistries were tested. The 5′ end of a modified passenger strand of miR-34a (miR-34a-3p) was conjugated to folate-dibenzocyclooctyne (Fol-DBCO) (see fig. S1, A to C, for Fol-DBCO synthesis and verification) using an unreleasable conformation (folate–miR-34a-3p: Fol-34a-3p) or a releasable conformation (folate–SS-miR-34a-3p: Fol-SS-34a-3p), which contains a reducible disulfide linkage (SS) between the miRNA and the folate molecule (fig. S2A). The mature strand of miR-34a (miR-34a-5p) was then annealed to the Fol-34a-3p or Fol-SS-34a-3p conjugate, generating FolamiR-34a duplexes (FolamiR-34a, unreleasable; FolamiR-SS-34a, releasable) (fig. S2A). Conjugates were identified using polyacrylamide gel electrophoresis (PAGE) (fig. S2B) and matrix-assisted laser desorption/ionization (MALDI) spectral analysis (fig. S3, A and B). To visualize FolamiR-34a in vivo, a near-infrared dye was conjugated to folate (NIR-Fol) (see fig. S4, A to C, for NIR-folate synthesis and verification). The miR-34a duplex was then conjugated to NIR-Fol or NIR-Fol-SS (fig. S5A), and the resulting conjugates were validated using PAGE (fig. S5, B and C) and MALDI spectral analysis (fig. S6, A and B). Our strategy for directly linking miR-34a to the folate ligand to generate FolamiRs that can deliver miRNAs specifically to cancer cells is shown in Fig. 1A.
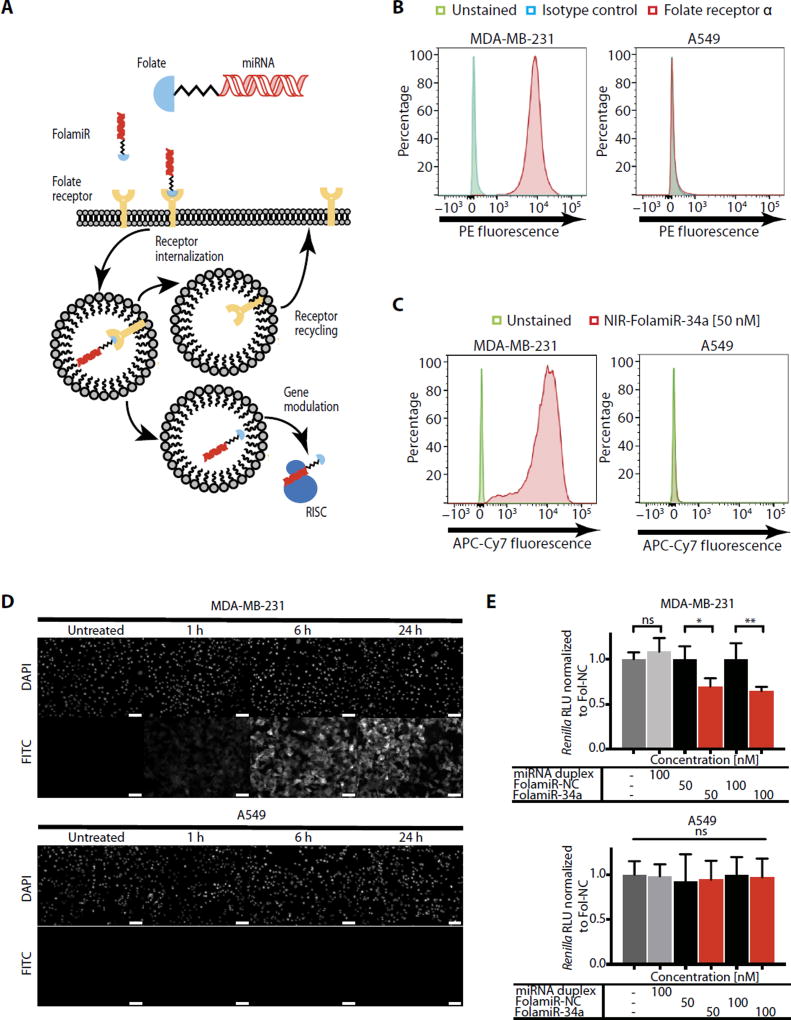
Fig. 1. Specificity of FolamiR uptake in cancer cells in culture.
(A) Proposed mechanism of action of FolamiRs. (B) Identification of FRα in FR+ MB-231 breast cancer cells and in FR− A549 lung cancer cells. Histograms represent overlaid flow cytometry data as a percentage of unstained, FRα+, and isotype control–stained cells. (C) NIR-FolamiR-34a uptake in FR+ MB-231 cells compared to FR− A549 cells. Histograms represent overlaid flow cytometry data as a percentage of unstained and NIR-FolamiR-34a (50 nM)–stained cells. (D) Fol-FITC uptake in FR+ MB-231 cells compared to FR− A549 cells. Scale bars, 50 µm. (E) Targeted silencing of miR-34a Renilla sensor using FolamiR in MB-231 and A549 cells in vitro. Data points were normalized to FolamiR-NC (negative control: scrambled miRNA) for each time point. Error bars: means ± SD. Each experiment corresponds to n = 3 with at least four technical replicates per treatment. **P < 0.01, *P < 0.05, one-way analysis of variance (ANOVA) and Bonferroni correction. ns, not significant.
FolamiR uptake is specific to cells overexpressing the FR
MDA-MB-231 (MB-231) breast cancer cells express detectable amounts of FR on the plasma membrane (43), making this cell line a plausible model for evaluating FolamiR activity. To verify expression of the FR, flow cytometric analyses were performed comparing MB-231 cells and A549 NSCLC cells (FR-negative control). MB-231 cells were confirmed to express FRα (Fig. 1B). These results were corroborated by analyzing the cellular uptake of NIR-FolamiR-34a using flow cytometry (Fig. 1C) and folate–fluorescein isothiocyanate (Fol-FITC) conjugate uptake using fluorescence microscopy (Fig. 1D). Both folate conjugates were taken up by the FR-positive (FR+) cell line MB-231 but not by the FR-negative (FR−) A549 cell line. This observation was functionally confirmed after treatment of MB-231 and A549 cell lines transiently expressing a miR-34a Renilla luciferase (Renilla) sensor with FolamiRs (Fig. 1E). The sensor includes a single miR-34a complementary binding site directly after the Renilla gene, allowing for monitoring of the posttranscriptional regulation of Renilla by miR-34a replacement. In both cell lines, the sensor was responsive to transfected miR-34a mimics (fig. S7); however, after FolamiR-34a exposure, the sensor was only down-regulated in MB-231 cells (Fig. 1E), suggesting that FolamiR targeting is dependent on FR-expressing cells. Together, these results suggest that MB-231 is an FR+ cell line that can take up FolamiR conjugates via FR interaction.
Release of folate ligand from FolamiR is not necessary for miRNA activity
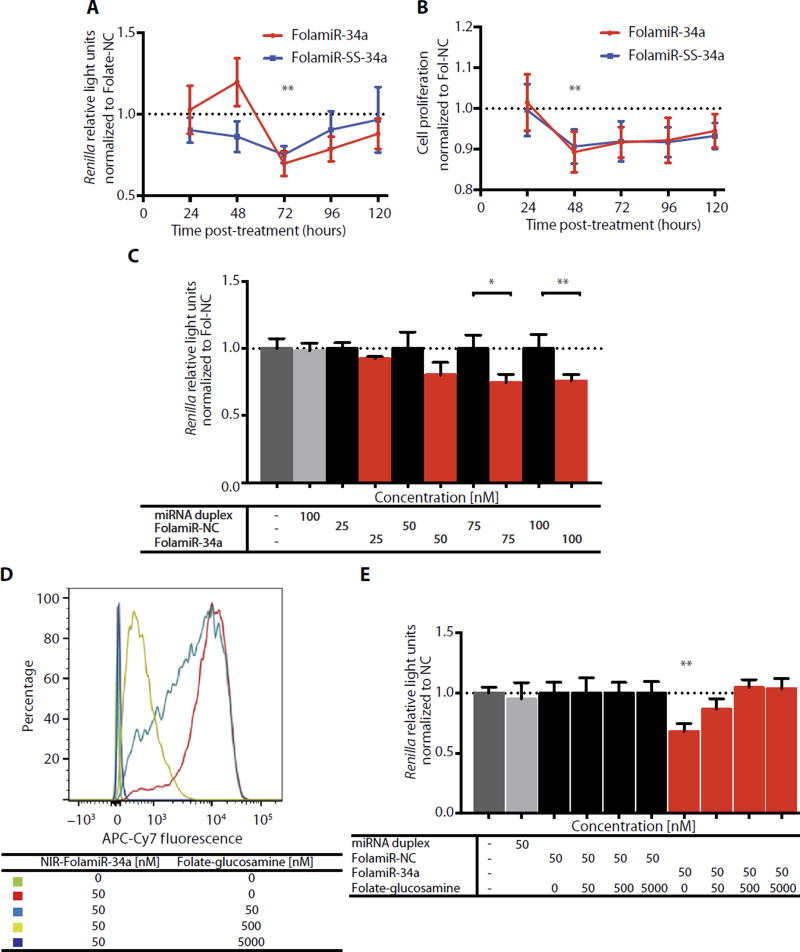
To monitor FolamiR-34a activity, MB-231 cells were generated to stably express the miR-34a Renilla sensor (MB-231 sensor) or a mutated version of the sensor that is unresponsive to miR-34a. A single clone with the highest expression of Renilla (fig. S8) was used to assess FolamiR-34a activity. When FolamiR-34a or FolamiR-SS-34a was added to the MB-231 sensor cells in the absence of transfection reagent, there was a decrease in Renilla activity 72 hours after exposure (Fig. 2A). Renilla activity rebounded 120 hours after exposure, likely because of replication-induced dilution of FolamiR-34a in the cells or degradation of FolamiR-34a. Proliferation of MB-231 sensor cells was reduced after a single FolamiR-34a treatment (Fig. 2B), which correlated with the reduction in Renilla activity. Surprisingly, both the releasable and unreleasable FolamiRs efficiently entered the cell and retained activity, suggesting that the conjugated folate does not interfere with loading of miR-34a-5p into Argonaute. To determine whether the sensor in the stable cell line responded to FolamiR-34a in a dose-dependent manner, MB-231 sensor cells were exposed to increasing concentrations of FolamiR-34a or control. A dose-dependent reduction in Renilla activity was only observed in cells treated with FolamiR-34a (Fig. 2C). To test whether cellular uptake of FolamiRs is dependent on FR expression on the cell membrane, in vitro competition assays were performed. Increasing the amount of folic acid glucosamine conjugate (folate-glucosamine) resulted in a dose-dependent reduction in cell-specific NIR-FolamiR-34a signal, indicating that folate-glucosamine competes with NIR-FolamiR-34a (Fig. 2D). Folate-glucosamine treatment abrogated the silencing effect of NIR-FolamiR-34a on the miR-34a Renilla sensor in a dose-dependent manner (Fig. 2E). To validate the concept as a delivery method of other small RNAs, besides miRNAs, an siRNA targeting firefly luciferase (siLuc2) was conjugated to folate. Firefly luciferase activity of MB-231 cells transiently expressing firefly luciferase was reduced after exposing the cells to Folate-siLuc2 (fig. S9), suggesting that folate-mediated delivery can be used to target other small RNAs to FR+ cells. Together, these results demonstrate that FolamiRs can deliver small RNAs that retain their function to cells overexpressing the FR and that delivery is dependent on FR expression.
Fig. 2. Cellular response to FolamiRs in vitro.
(A) Targeted silencing of miR-34a Renilla sensor using FolamiRs in MB-231 breast cancer cells. Data points were normalized to FolamiR-NC (negative control: scrambled miRNA) for each time point. (B) Proliferation of MB-231 cancer cells as a function of FolamiR treatment (50 nM). Data points were normalized to FolamiR-NC for each time point. Error bars: means ± SD. Each experiment corresponds to n = 3 with at least three technical replicates per treatment. (C) Dose response of MB-231 cells to FolamiR-34a. Renilla values were measured 96 hours after treatment. Data points were normalized to FolamiR-NC. Duplex miRNA represents unconjugated miR-34a. Error bars: means ± SD. Each experiment corresponds to n = 3 with at least four technical replicates per treatment. (D) Displacement of NIR-FolamiR-34a binding from MB-231 cells (50 nM, 4°C) with increasing concentrations of folate-glucosamine conjugate. Histograms represent overlaid flow cytometry data as a percentage of unstained and NIR-FolamiR-34a–stained cells. (E) FolamiR-34a competition assay in MB-231 breast cancer cells in vitro. MiR-34a Renilla sensor response to FolamiR-34a (50 nM, 96 hours) in the presence of increasing concentrations of folate-glucosamine conjugate. Data points were normalized to FolamiR-NC (negative control: scrambled miRNA) for each experimental condition. Experiment corresponds to n = 3 with at least three technical replicates per treatment. *P < 0.05, **P < 0.01, one-way ANOVA with post hoc Bonferroni correction.
FolamiRs are active in vivo in an immunocompromised xenograft model of breast cancer
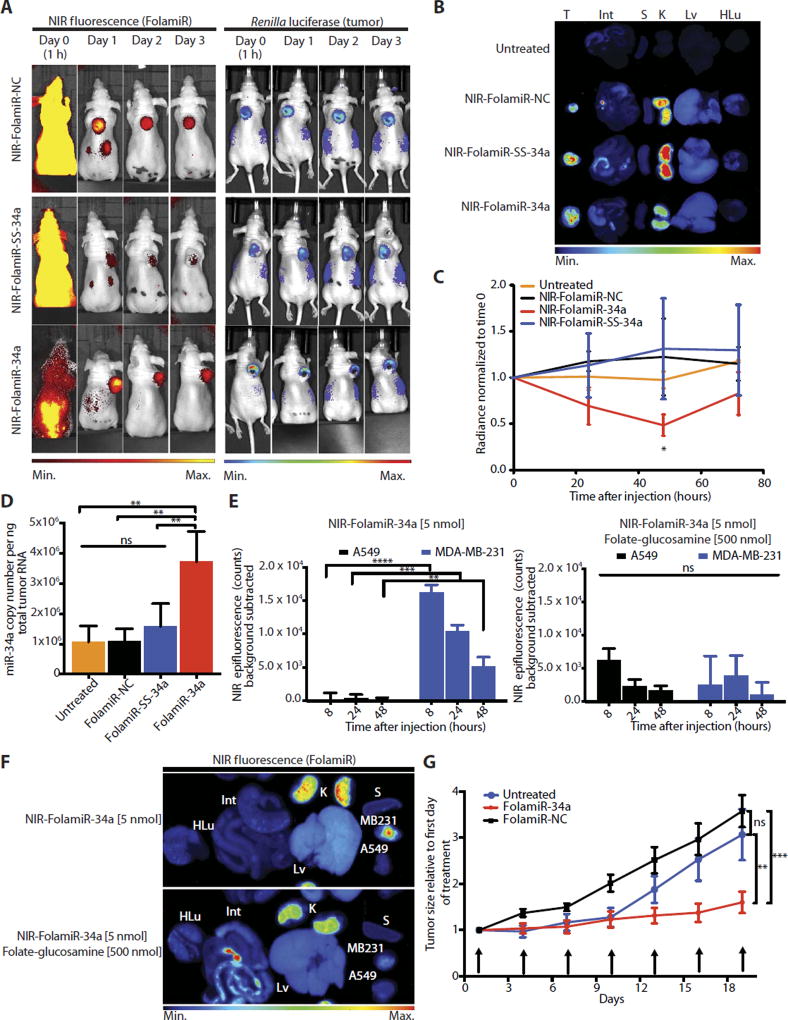
Transitioning miRNA delivery methods from cells in culture to in vivo models presents multiple challenges such as the potential for destabilization of the RNA in circulation due to serum ribonucleases, ineffective targeting to the tumor cells because of the tumor microenvironment, and poor uptake of the miRNA, all of which may reduce miRNA efficacy. To show that FolamiRs can overcome these challenges, a single dose (4 mg/kg, 5 nmol) of each NIR-Fol–tagged miRNA (NIR-FolamiR) was delivered via tail vein injection into animals with palpable MB-231 sensor cell xenografts. Fluorescence distribution and luciferase activity were measured to monitor targeting specificity, and as a surrogate for uptake and intercellular target repression, respectively. Twenty-four hours after injection, NIR-FolamiR was primarily retained in tumor tissues but cleared from the rest of the organism (Fig. 3A, left, NIR), including the liver (Fig. 3B, Lv). However, only the unreleasable NIR-FolamiR-34a induced Renilla knockdown in vivo (Fig. 3A, right, luciferase; quantified in Fig. 3C). After only a single injection of NIR-FolamiR-34a, Renilla expression was reduced by about 50%, which was greater than the reduction observed in the cell culture experiments (Fig. 2, A, C, and E). About 3.5 × 106 copies of miR-34a per nanogram of total RNA were present in the tumors harvested from mice treated with NIR-FolamiR-34a (Fig. 3D). In contrast, the copy number of miR-34a in the tumors harvested from mice treated with NIR-FolamiR-SS-34a was similar to the negative control animals, suggesting that the releasable folate conjugate may be degraded or prematurely reduced in circulation. To address this possibility, FolamiR conjugates were exposed to 50% serum. FolamiR-SS-34a was highly unstable in the presence of serum, whereas FolamiR-34a remained intact for more than 6 hours (fig. S10). FolamiR-34a was more stable than unconjugated miR-34a, suggesting that folate protects the miRNA from serum nucleases. To determine whether the FolamiRs bind specifically to FR in vivo, NIR-FolamiR-34a (4 mg/kg, 5 nmol) was injected intravenously in the presence or absence of 100-fold molar excess of folate-glucosamine in nude mice bearing FR+ MB-231 sensor cells engrafted on the right shoulder and FR− A549 cells engrafted on the left shoulder. The results indicate that only FR+ MB-231 tumors accumulate the FolamiR conjugate (Fig. 3, E and F) and that this FR-dependent accumulation can be blocked by an excess of folate-glucosamine (Fig. 3, E and F).
Fig. 3. FolamiR-34a inhibits the growth of MB-231 tumors in mice.
(A) Representative live imaging of female Nu/Nu congenic mice implanted with MB-231 sensor xenografts after intravenous injection of NIR-FolamiR-NC, NIR-FolamiR-SS-34a, or NIR-FolamiR-34a (4 mg/kg, 5 nmol). Left: NIR fluorescence distribution. Right: MiR-34a Renilla luciferase sensor signal. (B) Gross images of excised MB-231 tumors (T) and whole organs (Int, intestines; S, spleen; K, kidneys; Lv, liver; HLu, heart and lungs) visualized for fluorescence. (C) Effects of NIR-FolamiR-34a delivery on miR-34a Renilla sensor activity over time; data normalized to Renilla signal at day 0 (error bars: means ± SEM, n = 3). (D) miR-34a expression from excised MB-231 tumors measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR) 72 hours after injection with NIR-FolamiR conjugates (n = 3; error bars: means ± SD). (E) NIR epifluorescence quantification from live animals. Nude mice were implanted with A549 cells on the left shoulder and MB-231 cells on the right shoulder, and live imaging was conducted after intravenous injection of NIR-FolamiR-34a (4 mg/kg, 5 nmol) in the presence (right graph) or absence (left graph) of ≥100-fold molar excess of folate-glucosamine (n = 3, error bars: means ± SD). (F) Fluorescence distribution of excised organs and tumors from (E). A549: FR− tumor; MB-231: FR+ tumor. (G) Tumor size after FolamiR-34a treatment (n = 5; error bars: means ± SEM). Arrows represent treatment times (0.8 mg/kg, 1 nmol intravenous injection). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way (D and E) or two-way (B and G) ANOVA with Bonferroni correction.
Next, a multiple-dosing study was performed to evaluate the efficacy of FolamiR-34a. MB-231 xenograft animals were treated with reduced doses of FolamiR-NC or FolamiR-34a (0.08, 0.4, and 0.8 mg/kg) every 3 days for a total of seven doses. Tumors in animals administered the control folate conjugate grew about 3.5-fold, whereas tumor size in animals treated with FolamiR-34a increased modestly (~1.5-fold) during the 20-day dosing period (Fig. 3G and fig. S11). Doses as low as 0.08 mg/kg (0.1 nmol) produced a striking reduction in tumor growth. Copy number of miR-34a in the excised tumor tissue was about 1.5-fold higher than that in the tumors extracted from mice administered the control (fig. S12A). There was no evidence of whole-organ toxicity or elevation in the serum cytokines interleukin-6 (IL-6) or tumor necrosis factor–α (TNF-α) in animals treated with FolamiR-34a (fig. S13A). These results are supported by a maximum tolerated dose (MTD) study performed in immunocompetent mice in which none of the mice dosed with FolamiR-34a presented with pathological signs of toxicity or significant changes in body weight up to the maximum dose tested of 26.64 mg/kg (33.3 nmol) (fig. S13B), indicating an MTD of >26.64 mg/kg.
FolamiR treatment has a therapeutic effect in an immunocompetent aggressive Kras;p53 NSCLC mouse model
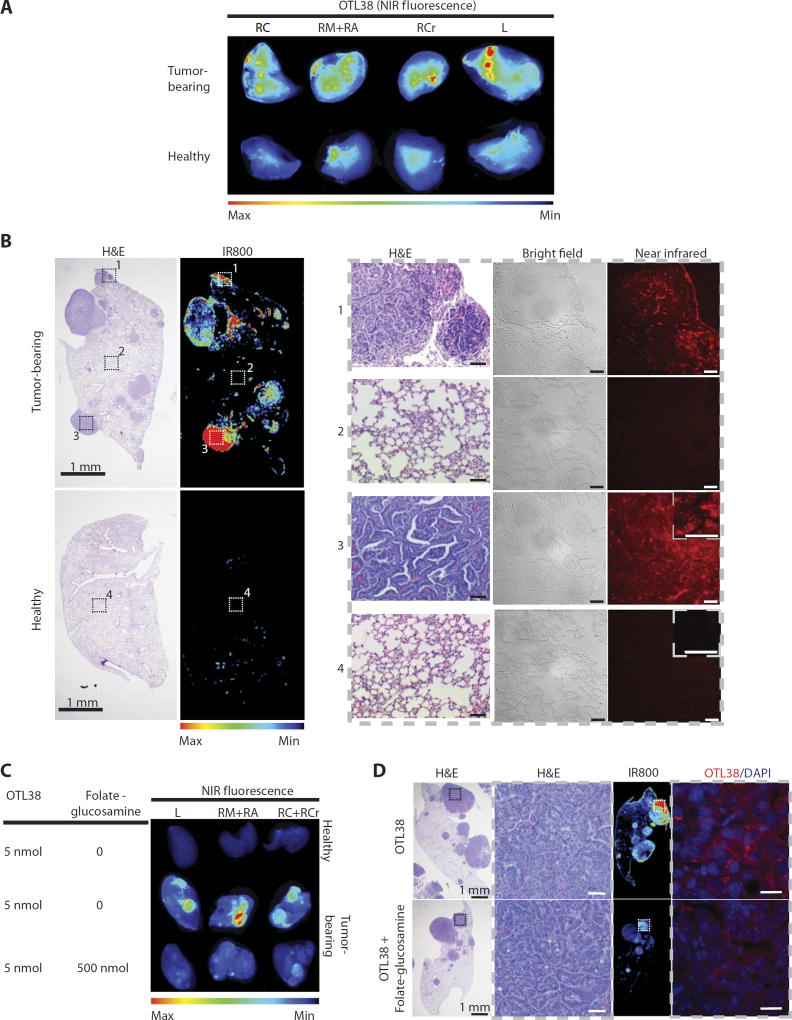
We speculated that the FolamiR strategy could be used to deliver miRNA mimics into other cancer models, with tumors growing in anatomically correct locations in the presence of an intact immune system. For that purpose, the aggressive KrasLSL-G12D/+; Trp53flx/flx NSCLC mouse model was tested for uptake and response to FolamiR-34a. The KrasLSL-G12D/+; Trp53flx/flx model accurately recapitulates not only NSCLC disease progression but also response and resistance to conventional therapies (44–46), and has previously been shown to be responsive to miR-34a replacement therapies (23, 24), supporting its use as a model for FolamiR efficacy. Because this model had not yet been validated for FR expression, pulmonary adenocarcinomas of this model were first evaluated for FR expression and tumor-specific uptake and retention of folate conjugates. Tumor-bearing animals were intravenously administered OTL38, a clinical version of folate conjugated to a NIR dye, which has recently been approved for a phase 3 clinical trial for image-guided surgical resection (NCT03180307) (32, 47, 48). The folate conjugate was preferentially retained in lung tumors and cleared from normal healthy tissues as observed at the gross organ level (Fig. 4A) and at the histological level (Fig. 4B). Even in tumor-bearing mice, nontumorigenic tissue was not targeted; however, OTL38 accumulated in small lesions and hyperplastic regions, highlighting the specificity of folate conjugates. Higher-magnification images indicate that the NIR signal is not an artifact of the cell density differences between healthy and malignant tissues; defined punctate signaling was observed in tumors after OTL38 administration, as has previously been observed because of receptor-mediated endocytosis of OTL38 (see insets in Fig. 4B). To determine whether OTL38 retention in pulmonary adenocarcinomas is mediated by its interaction with FR, an in vivo blockade assay was performed. OTL38 (5 nmol) was injected intravenously in the presence or absence of 100-fold molar excess of folate-glucosamine in mice bearing lung tumors. OTL38 retention in lung tumors was reduced by an excess of folate-glucosamine (Fig. 4, C and D), suggesting that OTL38 accumulation in tumors is dependent on the FR. These data confirm that the KrasLSL-G12D/+; Trp53flx/flx tumors specifically take up and retain OTL38, suggesting that FolamiR-34a should likewise accumulate in the tumor tissue.
Fig. 4. Murine KrasLSL-G12D/+;p53flx/flx lung adenocarcinomas express FR.
(A) NIR fluorescence imaging showing that ligand OTL38 (On Target Laboratories), an FRα-targeting ligand conjugated to a NIR dye, is preferentially retained in lung tumors and cleared from normal healthy lungs. KrasLSL-G12D/+;p53Flox/Flox mice were injected with 5 nmol of OTL38 8 weeks after tumor induction and sacrificed 24 hours after injection. Whole lungs were excised and imaged using LI-COR Odyssey CLx. A noninduced healthy mouse was used as a control. RC, right caudal lobe; RM, right medial lobe; RA, right accessory lobe; RCr, right cranial lobe; L, left lobe. (B) Histological and NIR images of right lung lobes from mice treated with OTL38. Left: Whole-organ low-magnification hematoxylin and eosin (H&E) and corresponding NIR images. Right: High-magnification H&E, bright-field, and NIR images of tumorous and healthy tissue corresponding to insets in (B). Scale bars, 50 µmand20 µm (inset). (C) Whole-organ NIR fluorescence images of excised lung lobes from mice bearing lung tumors treated with OTL38 (5 nmol) in the presence or absence of ≥100-foldmolar excess of folate-glucosamine (n = 3 per group). (D) Representative H&E and fluorescence images of tissues from (C). Insets on low-magnification H&E images correspond to adjacent high-magnification images of tumor tissue. Insets on whole-organ NIR fluorescence images correspond to adjacent high-magnification NIR images. H&E images represent the types of tissue shown in NIR images. Scale bars, 20 µm.
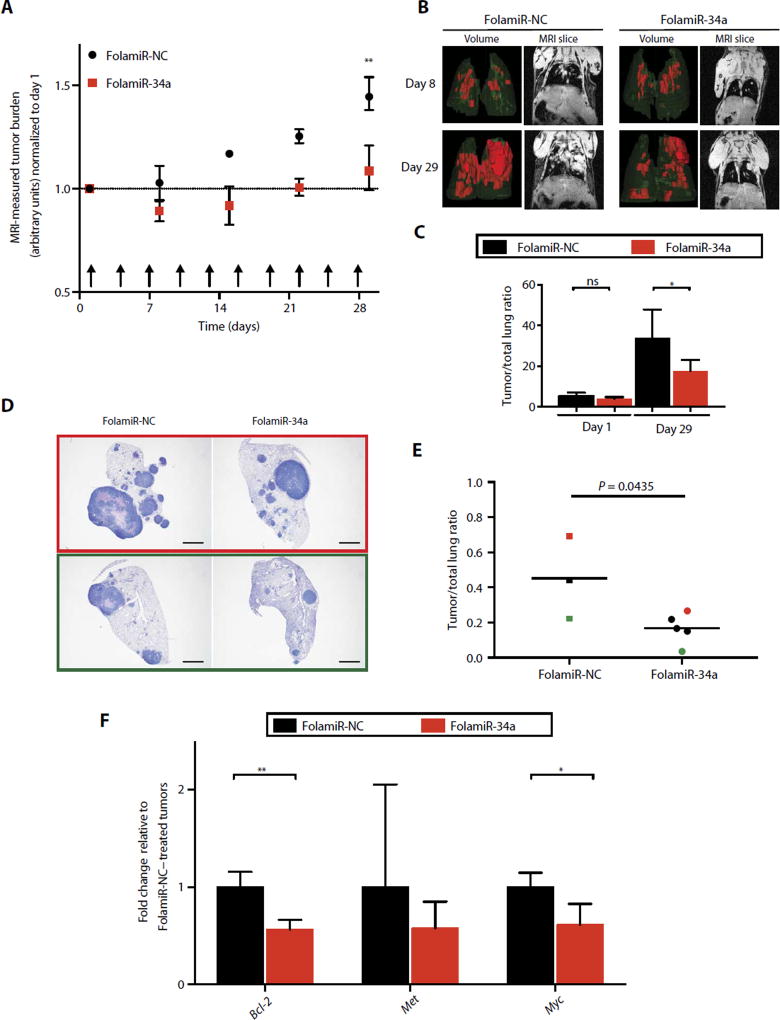
To determine whether the KrasLSL-G12D/+; Trp53flx/flx tumors are responsive to FolamiR-34a, tumor-bearing animals were intravenously administered FolamiR-34a (0.8 mg/kg, 1 nmol) every 3 days for a total of 10 doses. Tumor growth was monitored by magnetic resonance imaging (MRI) during the course of the study, and the resulting volume measurements generated from the MRI data indicated that FolamiR-34a reduced tumor growth compared to animals administered the control folate conjugate (Fig. 5, A to C). Tumor size was statistically unchanged in animals administered FolamiR-34a, whereas tumor size in FolamiR-NC–treated mice increased 1.5-fold relative to the first day of dosing. A similar response was observed at the histological level upon termination of the study (Fig. 5, D and E). Quantification of histological sections indicated a statistically significant, 1.8-fold reduction in tumor burden in lungs harvested from FolamiR-34a–treated mice relative to FolamiR-NC–treated mice. Similar to the miR-34a copy number increase observed in MB-231 xenografts, copy number of miR-34a in the excised lung tumor tissue was about threefold higher than in the tumors extracted from mice administered the control (fig. S12B). To validate thatmiR-34a was acting to repress endogenous target genes, transcripts of the miR-34a targets BCL-2, MET, and MYC were quantified. Both BCL-2 and MYC were statistically down-regulated in tumors harvested from mice admintered FolamiR-34a, confirming miR-34a activity on endogenous target genes (Fig. 5F).
Fig. 5. Targeted replacement of miR-34a via FolamiR reduces tumor size in a murine model of lung adenocarcinoma.
(A) MRI-measured tumor burden after FolamiR-34a treatment (FolamiR-NC, n = 3; FolamiR-34a, n = 5). Error bars represent means ± SEM. **P < 0.01, two-way ANOVA and Bonferroni post hoc tests. Arrows represent treatment times (0.8 mg/kg, 1 nmol intravenous injection; total 10). (B) Representative three-dimensional volume renderings of the lungs and MRI images of the thoracic region of mice treated with FolamiRs during (day 8) and at the end of treatment (day 29). (C) Tumor/whole lung volume ratios from (B) at the indicated times showing the percentage of lung volume occupied by tumors. Error bars represent means ± SD. *P < 0.05, one-way ANOVA. (D) Representative H&E-stained tissue of the left lobe of the lung from animals from each treatment group (FolamiR-NC, FolamiR-34a). Scale bars, 1 mm. (E) Overall tumor burden calculated from total tumor area averaged from three histological sections obtained from each treated animal relative to the total area of the lung. Bars indicate the median. FolamiR-NC: n = 3, FolamiR-34a: n = 5; unpaired t test. (F) miR-34a target genes (Met, Myc, and Bcl-2) evaluated by qRT-PCR of tumor tissue normalized to Actin and graphed relative to FolamiR-NC–treated tumors (FolamiR-NC: n = 3, FolamiR-34a: n = 5). Error bars represent means ± SD. *P < 0.05, unpaired t test.
DISCUSSION
miRNAs are exceptional candidates for combating many diseases such as cancer; however, their therapeutic power is dwarfed by the lack of safe, robust, specific, and efficient delivery methods. In theory, miRNAs are ideal therapeutic candidates: First, miRNAs are small and, because the kinetics of tumor uptake are inversely proportional to the size of the particle, miRNAs should easily penetrate the dense architecture of the tumor and the microenvironment (38). Second, because of basal expression of some therapeutically relevant miRNAs in noncancerous cells, slight changes in cellular miRNA concentrations imposed after miRNA delivery are unlikely to produce toxic side effects. However, these same doses may prove lethal for cancerous cells, especially in cells that are addicted to loss of the miRNA, which is reconstituted upon delivery (49). Finally, the pleiotropic and promiscuous nature of miRNAs could make a single miRNA therapy analogous to a multidrug chemotherapeutic cocktail by targeting various complementary pathways simultaneously (50). It is therefore unlikely that acquired resistance, which is an unforeseen hallmark of most targeted cancer therapeutics (51), would occur with miRNA therapy.
Although these strengths suggest that miRNA therapeutics may be powerful against diseases such as cancer, the power of miRNA therapeutic use is stifled because of lack of delivery methods that allow the miRNAs to reach and penetrate the targeted tissues without unwanted toxicity. Because of the low stability of RNA in serum, miRNAs have been packed inside various protective vehicles (16–22, 52, 53). Unfortunately, the added benefit of protecting the miRNA cargo comes with harmful side effects. One way to avoid vehicle-associated toxicity is to remove the vehicle entirely. This high-risk approach requires that the miRNA bases are heavily modified to protect the miRNA from nucleases or that the miRNA is targeted and internalized rapidly.
The data presented here support conjugating a therapeutically relevant miRNA directly to a targeting ligand whose receptor is overexpressed on cancerous cells. We show that miR-34a, when conjugated to folate, enters tumorigenic cells specifically, represses both exogenous and endogenous targets efficiently, and inhibits tumor growth in both immunocompromised and immunocompetent models with no signs of toxicity at the tested therapeutic dose. The data suggest that folate conjugated to the passenger miRNA strand protects the miRNA mimic from degradation without impairing loading into Argonaute. Although these findings are encouraging, some current limitations of ligand-mediated delivery will need to be addressed, such as endosomal sequestration of the therapeutic agent. Folate conjugates are taken up via the natural process of endocytosis. Thus, endosomal entrapment of FolamiRs could be a limiting step in achieving efficient targeting. We anticipate that inclusion of an intermolecular endosomal escape mechanism incorporated into second-generation FolamiRs could ensure robust cytosolic delivery of the therapeutic miRNAs. In addition, because of the heterogeneous nature of the tumor, delivery to cancer cells not expressing the FR is an additional concern. Although other folate conjugates have shown success in their ability to deliver the conjugated agent to tumor cells not expressing the receptor, whether or not a similar bystander effect occurs in association with FolamiR delivery will need to be evaluated.
Because miRNA therapeutics continue to advance, it is imperative that relevant delivery vehicles with limited toxicity are identified. The evidence presented here supports a new method for vehicle-free miRNA delivery. This approach has completely removed the toxic delivery vehicle without impairing miRNA delivery or activity. It is, thus, highly plausible that miRNAs could be delivered at high quantities and in the absence of unwanted toxicity to treat various diseases such as cancer. Although we focused specifically on miRNA and siRNA delivery, as RNA-based therapeutics are explored further, we anticipate that it will be plausible to conjugate any relevant small RNA, such as Piwi-interacting RNAs (piRNAs) and transfer RNA (tRNA) fragments, onto folate or other appropriate ligands to achieve fast and tumor-specific uptake.
MATERIALS AND METHODS
Study design
This study was designed to test whether conjugating a therapeutically relevant miRNA directly to a targeting ligand whose receptor is overexpressed on cancerous cells could be used to deliver functional miRNAs specifically to cancer cells. To investigate whether this strategy would work therapeutically, miR-34a was conjugated to a folate ligand to generate FolamiRs. The resulting FolamiRs were tested for specific targeting and antiproliferative activity in cell-based experiments, in an immunocompromised mouse model bearing human breast FR+ tumors, and in an immunocompetent mouse model of lung adenocarcinoma. A priori power analysis was used to estimate sample size requiring a statistical significance of 0.05, α µ 0.5, and 80% power. On the basis of the power calculation, the suggested number of animals to include in each treatment group was six. The expected effect size was then determined from small pilot studies. Because of a strong and significant reduction in Renilla reporter activity observed after treating three animals with FolamiR-34a (Fig. 3, A and B), the remaining three animals were not treated. In this case, the power of 0.5 accurately predicted significance using three animals with an α of <0.5. For all in vivo work, tumor burden was calculated either by caliper measurement for xenograft animals or by MRI for genetically engineered mouse model (GEMM) studies. Animals were then randomized before treatment such that the tumor burden and error in each treatment group were equivalent to all other treatment groups.
Preparation of folate-miRNAs (FolamiRs)
MiRNA duplexes were constructed using two RNA oligonucleotides: denoted as miR-34a-5p guide strand and miR-34a-3p passenger strand (Integrated DNA Technologies). The miR-34a-3p passenger strand comprises a 20-nucleotide (nt) RNA oligo double-modified with an azide linker on the 5′ end and 2′-O-methyl RNA bases on the 3′ end. The miR-34a-5p guide strand comprises a 22-nt RNA oligo with minimal modifications on the 3′ end with 2′-O-methyl RNA bases. A scrambled miRNA (negative control) synthesized with the same modifications was used to form a control duplex. A bi-orthogonal click reaction was performed between Fol-DBCO and azide-modified antisense miR-34a (or scramble) (fig. S1A and fig. S2A). For experiments with siLuc2, the oligos with the following sequences were used: sense strand, 5′-GGACGAGGACGAGCACUUCUU-3′; antisense strand, 5′-GAAGUGCUCGUCCUCGUCCUU-3′. Click reaction (54) was performed at a 1:10molar ratio (azide oligo/Fol-DBCO) at room temperature in water for 8 hours and then cooled to 4°C for 4 hours. Unconjugated folate was removed from the reaction using Oligo Clean & Concentrator (Zymo Research) per the manufacturer’s instructions. Conjugation was verified using 15% tris base, acetic acid, EDTA (TAE) native PAGE and MALDI spectral analysis. For NIR-folate compound conjugation, an additional verification was done using LI-COR Odyssey CLx (LI-COR Biosciences).
After conjugation, the miR-34a-5p guide strand was annealed to the folate passenger strand and NIR-folate passenger strand conjugates. In brief, folate–miR-34a-3p and miR-34a-5p were mixed in an equal molar ratio (1:1; final concentration, 5 µM each) in annealing buffer [10 mM tris buffer (pH 7) (Sigma) supplemented with 50 mM NaCl (Sigma) and 1 mM EDTA (Sigma)], incubated at 95°C for 5 min, ramp-cooled to room temperature over a period of 1 hour, and then stored at −80°C.
Flow cytometry
FR+ MB-231 cells and FR− A549 cells grown as described previously were detached by trypsinization and washed twice in ice-cold phosphate-buffered saline (pH 7.4) and resuspended to a density of 1 × 107 cells/ml in serum-free medium. Cell viability was determined by trypan blue exclusión, and cells were only used if the viability of cells was >80%. Next, flow cytometric analyses were performed following standard protocols. In brief, 1 × 106 cells were incubated with phycoerythrin (PE) anti-FOLR1 antibody (catalog no. 908303, BioLegend) or matched isotype antibody (catalog no. 400213, BioLegend) as a control and analyzed by flow cytometric analysis using LSRFortessa flow cytometer (BD Biosciences). Data were analyzed using FlowJo software v10 (Tree Star Inc.). Functionality of the FR was confirmed, first, by incubating MB-231 and A549 cells with NIR-FolamiR-34a (50 nM) followed by flow cytometric analyses as described above and, second, by microscopy analysis of cells incubated with Fol-FITC (55) at a final concentration of 50 nM. Cells were evaluated at different time points using an Olympus IX73 microscope equipped with a 1.25× objective, Olympus DP80 camera, and CellSens 1.11.
In vitro FolamiR delivery
MB-231 cells (HTB-26) and A549 NSCLC cells (CCL-185), both mycoplasma-free as determined by testing for mycoplasma contamination via MycoAlert Mycoplasma Detection Kit (Lonza), were grown in RPMI 1640 medium, no folic acid (Life Technologies) supplemented with 10% fetal bovine serum (Sigma), penicillin (100 U/ml), and streptomycin (100 µg/ml) (HyClone, GE Healthcare Life Sciences), and maintained at 37°C in 5% CO2. Authentication of MB-231 cells was performed using short tandem repeat profiling [American Type Culture Collection (ATCC)]. A549 cells were purchased directly from ATCC, and thus, their authentication has been confirmed.
For luciferase reporter experiments, a miR-34a sensor plasmid was generated by inserting the antisense sequence to miR-34a into the 3′-untranslated region of Renilla luciferase in the vector (psiCHECK, Promega). MiR-34a–specific silencing was confirmed in MB-231 cells by transiently transfecting a miR-34a sensor or a mutated miR-34a sensor. MiR-34a sensor–expressing cells were transfected with a miR-34a mimic using Lipofectamine RNAiMax (Life Technologies) to confirm silencing mediated by exogenous miRNA. To generate stable clones, MB-231 cells were seeded in six-well plates at a density of 1 × 106 cells per well and were transfected with 2 µg of miR-34a sensor plasmid using Lipofectamine 2000 (Life Technologies). Stable clones were selected using hygromycin B (500 µg/ml; HyClone, GE Healthcare Life Sciences) as a selection marker. Single clones were evaluated for Renilla expression, and the clone with the highest Renilla expression was selected.
MB-231 sensor cells were seeded into 96-well plates containing FolamiR-34a, FolamiR-SS-34a, or FolamiR-NC (negative control) in folic acid and serum-free RPMI medium for a final concentration of 50 nM. Untreated and unconjugated duplex miRNA were included as controls. Renilla luciferase values were obtained 24, 48, 72, 96, and 120 hours after incubation using the Renilla Glo Luciferase kit (Promega) following the manufacturer’s instructions. Renilla levels were normalized to FolamiR-NC for each time point. Experiments were performed three times, with technical triplicates for each condition.
Flank tumor establishment
For single-dose studies, subcutaneous tumors were induced in female Nu/Nu (NU-Foxn1nu; Charles River) congenic mice (6 weeks; n = 5) by subcutaneous injection of 5 × 106 MB-231 sensor cells suspended in 200 µl of Matrigel (Corning). For longitudinal studies, parental MB-231 cells were used. Because of the observation that rodents present high plasma and tissue levels of 5-methyl-tetrahydrofolate, the naturally occurring form of folate (around 10-fold higher than in humans) (56), mice were maintained on folic acid–deficient diet (TD.95247, Envigo) 2 weeks before tumor implantation and during the experiment series. A folate-deficient diet has shown to reduce folate levels to physiological levels seen in humans (57). To determine tumor growth, individual tumors were measured using a vernier caliper, and tumor volume was calculated as follows: tumor volume (mm3) = width × (length2) × 2−1. Animals were excluded if tumors had not reached a volume of 150 cm3 by the time of treatment. For single-dose experiments, animals were injected intravenously with 5 nmol of FolamiRs after acquisition of luminescent and florescence signals (day 0, Fig. 1A). For multiple-dosing experiments, animals were randomized into experimental arms by minimizing the differences in their mean tumor size. When tumor volume reached ~200 mm3, animals were treated with intravenous injections of the indicated molar concentration of FolamiR every 3 days. All experimental protocols were approved by the Purdue Animal Care and Use Committee and were in compliance with National Institutes of Health (NIH) guidelines for animal use.
Induction of tumor formation in Kras;p53 mice
Induction of tumor formation in KrasLSL-G12D/+; Trp53flx/flx (FVB.129 background) double-mutant mice (6 to 10 weeks old) was performed on the basis of the method of DuPage et al. (45). In brief, lung-specific transgene activation was achieved via intratracheal delivery of adenoviral particles (106 plaque-forming units) encoding for Cre recombinase. Tumors were allowed to preform for 8 weeks before experiments.
In vivo blocking of FR
Subcutaneous tumors were induced in female Nu/Nu (NU-Foxn1nu; Charles River) congenic mice (6 weeks; n = 3) following injection of 5 × 106 FR+ MB-231 sensor cells (right side) and FR human A549 cells (left side) suspended in 200 µl of Matrigel (Corning). Tumors were allowed to form, and mice bearing A549 and MB-231 tumors of similar size were included in the experiment. For the Kras;p53 mouse model, tumors were allowed to form for 8 weeks after transgene activation (n = 3). Tumor formation was monitored using MRI. Competition studies were performed in mice (n = 3) by coadministration, via the tail vein, of 5 nmol of NIR-FolamiR-34a for the xenograft model or 5 nmol of OTL38 for the Kras;p53 mouse model in the presence or absence of 500 nmol (≥100-fold molar excess) of folic acid glucosamine. Folic acid glucosamine conjugate was used because of its increased solubility at low pH compared to folic acid and to prevent precipitation in the kidneys (58). In vivo whole-animal imaging and ex vivo tissue distribution studies were performed as described above.
Statistics
For two-group analysis, a two-tailed Student’s t test was used to examine group differences. Two-way or one-way ANOVA with post hoc Bonferroni correction was used for multigroup comparison using Prism statistical package (version 7, GraphPad Software). Error bars represent either means ± SD or means ± SEM as denoted in the figure legends. Statistically significant P values are indicated in figures and/or legends. Individual-level data are reported in table S1.
Supplementary Material
Materials and Methods
Fig. S1. Fol-DBCO ligand synthesis and LC-MS spectral analysis.
Fig. S2. Evaluation of folate-miRNA conjugation measured by 15% native TAE PAGE.
Fig. S3. MALDI spectra of FolamiRs.
Fig. S4. NIR-folate ligand synthesis and LC-MS spectral analysis.
Fig. S5. Evaluation of NIR-FolamiR conjugation measured by 15% native TAE PAGE.
Fig. S6. MALDI spectra of NIR-FolamiR conjugates.
Fig. S7. miR-34a Renilla luciferase sensor response to miR-34a transfection.
Fig. S8. Evaluation of MB-231 miR-34a sensor cells.
Fig. S9. Folate-mediated delivery of functional siLuc2.
Fig. S10. Serum stability of FolamiRs and duplex RNA oligos.
Fig. S11. In vivo tumor growth response to dose titration of FolamiR-34a.
Fig. S12. miR-34a copy number in tumors treated with FolamiR-34a in mice.
Fig. S13. Serum cytokines and maximum tolerated FolamiR dose study in mice.
Table S1. Individual-level data.
Acknowledgments
We thank S. Humphrey, S. Myoung, and P. McCown for insightful discussion and for editing the manuscript. We would also like to acknowledge the following Purdue University Cores for their services and assistance: MRI Core, Bindley Imaging Core, Biological Evaluation Core, and Histology Research Laboratory, a core facility of the NIH-funded Indiana Clinical and Translational Science Institute. We also thank On Target Laboratories, LLC (West Lafayette, IN) for providing us with OTL38.
Funding: This research was supported by a Pathway to Independence Grant (R00CA178091 to A.L.K.), the Purdue Center for Cancer Research (P30CA023168), the Indiana Clinical and Translational Sciences Institute (funded in part by grant #UL1 TR001108 from the NIH), National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award (to A.L.K.), and Endocyte Grant (#200766 to P.S.L.).
A.L.K. and P.S.L. are inventors on provisional U.S. patent application 67538-01 submitted by Purdue University that covers methods and uses of targeted delivery of miRNAs. P.S.L. owns shares in Endocyte, which develops small-molecule drug conjugates using the folate-targeting technology discussed in the article.
Footnotes
Author contributions: E.A.O., P.S.L., and A.L.K. designed the study. E.A.O., S.T., L.R., P.S.L., and A.L.K. analyzed the data. E.A.O. and A.L.K. wrote the manuscript. E.A.O., L.R., and A.L.K. edited the manuscript. E.A.O. performed the cell-based and in vivo studies. E.A.O. performed the MRI studies and analysis. S.T. and L.R. synthesized folate compounds and performed MALDI and LC-MS experiments. E.A.O. performed serum stability assays and agarose gel analysis. S.T. assisted with in vivo validation. L.T.L. performed postnecropsy pathological analysis. A.L.K. and P.S.L. conceived and supervised the project.
Competing interests: The remaining authors declare that they have no competing interests.
References
- 1.Wang H, Jiang Y, Peng H, Chen Y, Zhu P, Huang Y. Recent progress in microRNA delivery for cancer therapy by non-viral synthetic vectors. Adv. Drug Deliv. Rev. 2015;81:142–160. doi: 10.1016/j.addr.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 2.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with “antagomirs”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 3.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 4.Gebert LFR, Rebhan MAE, Crivelli SEM, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014;42:609–621. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, Saltzman WM, Slack FJ. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2014;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sepp-Lorenzino L, Ruddy M. Challenges and opportunities for local and systemic delivery of siRNA and antisense oligonucleotides. Clin. Pharmacol. Ther. 2008;84:628–632. doi: 10.1038/clpt.2008.174. [DOI] [PubMed] [Google Scholar]
- 8.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 10.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raal FJ, Santos RD, Blom DJ, Marais AD, Charng M-J, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, Chasan-Taber S, Tribble DL, Flaim JD, Crooke ST. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: A randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 12.Miner P, Wedel M, Bane B, Bradley J. An enema formulation of alicaforsen, an antisense inhibitor of intercellular adhesion molecule-1, in the treatment of chronic, unremitting pouchitis. Aliment. Pharmacol. Ther. 2004;19:281–286. doi: 10.1111/j.1365-2036.2004.01863.x. [DOI] [PubMed] [Google Scholar]
- 13.Mulamba GB, Hu A, Azad RF, Anderson KP, Coen DM. Human cytomegalovirus mutant with sequence-dependent resistance to the phosphorothioate oligonucleotide fomivirsen (ISIS 2922) Antimicrob. Agents Chemother. 1998;42:971–973. doi: 10.1128/aac.42.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Højland T, Abramov M, Van Aerschot A, Odadzic D, Smicius R, Haas J, Andree C, Barman J, Wenska M, Srivastava P, Zhou C, Honcharenko D, Hess S, Müller E, Bobkov GV, Mikhailov SN, Fava E, Meyer TF, Chattopadhyaya J, Zerial M, Engels JW, Herdewijn P, Wengel J, Kjems J. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corey DR. Chemical modification: The key to clinical application of RNA interference? J. Clin. Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin PT, Shah BP, Lee K-B. Combined magnetic nanoparticle-based microRNA and hyperthermia therapy to enhance apoptosis in brain cancer cells. Small. 2014;10:4106–4112. doi: 10.1002/smll.201400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao L, Patel PC, Alhasan AH, Giljohann DA, Mirkin CA. Nucleic acid-gold nanoparticle conjugates as mimics of microRNA. Small. 2011;7:3158–3162. doi: 10.1002/smll.201101018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng CJ, Saltzman WM. Polymer nanoparticle-mediated delivery of microRNA inhibition and alternative splicing. Mol. Pharm. 2012;9:1481–1488. doi: 10.1021/mp300081s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endo-Takahashi Y, Negishi Y, Nakamura A, Ukai S, Ooaku K, Oda Y, Sugimoto K, Moriyasu F, Takagi N, Suzuki R, Maruyama K, Aramaki Y. Systemic delivery of miR-126 by miRNA-loaded Bubble liposomes for the treatment of hindlimb ischemia. Sci. Rep. 2014;4:3883. doi: 10.1038/srep03883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittal A, Chitkara D, Behrman SW, Mahato RI. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials. 2014;35:7077–7087. doi: 10.1016/j.biomaterials.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 22.Conde J, Oliva N, Atilano M, Song HS, Artzi N. Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment. Nat. Mater. 2015;15:353–363. doi: 10.1038/nmat4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasinski AL, Kelnar K, Stahlhut C, Orellana E, Zhao J, Shimer E, Dysart S, Chen X, Bader AG, Slack FJ. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015;34:3547–3555. doi: 10.1038/onc.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72:5576–5587. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srinivasarao M, Galliford CV, Low PS. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nat. Rev. Drug Discov. 2015;14:203–219. doi: 10.1038/nrd4519. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Wang J, Tacha DE, Li P, Bremer RE, Chen H, Wei B, Xiao X, Da J, Skinner K, Hicks DG, Bu H, Tang P. Folate receptor α associated with triple-negative breast cancer and poor prognosis. Arch. Pathol. Lab. Med. 2014;138:890–895. doi: 10.5858/arpa.2013-0309-OA. [DOI] [PubMed] [Google Scholar]
- 27.Cagle PT, Zhai Q, Murphy L, Low PS. Folate receptor in adenocarcinoma and squamous cell carcinoma of the lung: Potential target for folate-linked therapeutic agents. Arch. Pathol. Lab. Med. 2013;137:241–244. doi: 10.5858/arpa.2012-0176-OA. [DOI] [PubMed] [Google Scholar]
- 28.Kalli KR, Oberg AL, Keeney GL, Christianson TJH, Low PS, Knutson KL, Hartmann LC. Folate receptor alpha as a tumor target in epithelial ovarian cancer. Gynecol. Oncol. 2008;108:619–626. doi: 10.1016/j.ygyno.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal. Biochem. 2005;338:284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Vlashi E, Low P. Folate-linked drugs for the treatment of cancer and inflammatory diseases. Subcell. Biochem. 2012;56:163–179. doi: 10.1007/978-94-007-2199-9_9. [DOI] [PubMed] [Google Scholar]
- 31.Lynn RC, Poussin M, Kalota A, Feng Y, Low PS, Dimitrov DS, Powell DJ., Jr Targeting of folate receptor β on acute myeloid leukemia blasts with chimeric antigen receptor-expressing T cells. Blood. 2015;125:3466–3476. doi: 10.1182/blood-2014-11-612721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dam GM, Themelis G, Crane LMA, Harlaar NJ, Pleijhuis RG, Kelder W, Sarantopoulos A, de Jong JS, Arts HJG, van der Zee AGJ, Bart J, Low PS, Ntziachristos V. Intraoperative tumor-specific fluorescence imaging in ovarian cancer by folate receptor-α targeting: First in-human results. Nat. Med. 2011;17:1315–1319. doi: 10.1038/nm.2472. [DOI] [PubMed] [Google Scholar]
- 33.Thomas M, Kularatne SA, Qi L, Kleindl P, Leamon CP, Hansen MJ, Low PS. Ligand-targeted delivery of small interfering RNAs to malignant cells and tissues. Ann. N. Y. Acad. Sci. 2009;1175:32–39. doi: 10.1111/j.1749-6632.2009.04977.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang K, Wang Q, Xie Y, Mor G, Sega E, Low PS, Huang Y. Receptor-mediated delivery of siRNAs by tethered nucleic acid base-paired interactions. RNA. 2008;14:577–583. doi: 10.1261/rna.739308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leamon CP, Parker MA, Vlahov IR, Xu L-C, Reddy JA, Vetzel M, Douglas N. Synthesis and biological evaluation of EC20: A new folate-derived, 99mTc-based radiopharmaceutical. Bioconjug. Chem. 13:1200–1210. doi: 10.1021/bc0200430. [DOI] [PubMed] [Google Scholar]
- 36.Jones SK, Lizzio V, Merkel OM. Folate receptor targeted delivery of siRNA and paclitaxel to ovarian cancer cells via folate conjugated triblock copolymer to overcome TLR4 driven chemotherapy resistance. Biomacromolecules. 2016;17:76–87. doi: 10.1021/acs.biomac.5b01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Dou B, Bao Y. Efficient targeted pDNA/siRNA delivery with folate–low-molecular-weight polyethyleneimine–modified pullulan as non-viral carrier. Mater. Sci. Eng. C Mater. Biol. Appl. 2014;34:98–109. doi: 10.1016/j.msec.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 38.Vlashi E, Kelderhouse LE, Sturgis JE, Low PS. Effect of folate-targeted nanoparticle size on their rates of penetration into solid tumors. ACS Nano. 2013;7:8573–8582. doi: 10.1021/nn402644g. [DOI] [PubMed] [Google Scholar]
- 39.Altmann K-H, Dean NM, Fabbro D, Freier SM, Geiger T, Hanera R, Hiisken DA, Martina P, Monia BPB, Miiller M, Natt F, Nicklind P, Phillipsd J, Pieles U, Sasmor H, Moser HE. Second generation of antisense oligonucleotides: From nuclease resistance to biological efficacy in animals. Chimia. 1996;50:168–176. [Google Scholar]
- 40.Yang J, Chen H, Vlahov IR, Cheng J-X, Low PS. Characterization of the pH of folate receptor-containing endosomes and the rate of hydrolysis of internalized acid-labile folate-drug conjugates. J. Pharmacol. Exp. Ther. 2007;321:462–468. doi: 10.1124/jpet.106.117648. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, Chen H, Vlahov IR, Cheng J-X, Low PS. Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13872–13877. doi: 10.1073/pnas.0601455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Zhao X, Chen J, Chen J, Kuznetsova L, Wong SS, Ojima I. Mechanism-based tumor-targeting drug delivery system. Validation of efficient vitamin receptor-mediated endocytosis and drug release. Bioconjug. Chem. 2010;21:979–987. doi: 10.1021/bc9005656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meier R, Henning TD, Boddington S, Tavri S, Arora S, Piontek G, Rudelius M, Corot C, Daldrup-Link HE. Breast cancers: MR imaging of folate-receptor expression with the folate-specific nanoparticle P1133. Radiology. 2010;255:527–535. doi: 10.1148/radiol.10090050. [DOI] [PubMed] [Google Scholar]
- 44.Singh M, Lima A, Molina R, Hamilton P, Clermont AC, Devasthali V, Thompson JD, Cheng JH, Bou Reslan H, Ho CCK, Cao TC, Lee CV, Nannini MA, Fuh G, Carano RAD, Koeppen H, Yu RX, Forrest WF, Plowman GD, Johnson L. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat. Biotechnol. 2010;28:585–593. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 45.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue W, Meylan E, Oliver TG, Feldser DM, Winslow MM, Bronson R, Jacks T. Response and resistance to NF-кB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov. 2011;1:236–247. doi: 10.1158/2159-8290.CD-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelderhouse LE, Chelvam V, Wayua C, Mahalingam S, Poh S, Kularatne SA, Low PS. Development of tumor-targeted near infrared probes for fluorescence guided surgery. Bioconjug. Chem. 2013;24:1075–1080. doi: 10.1021/bc400131a. [DOI] [PubMed] [Google Scholar]
- 48.Okusanya OT, DeJesus EM, Jiang JX, Judy RP, Venegas OG, Deshpande CG, Heitjan DF, Nie S, Low PS, Singhal S. Intraoperative molecular imaging can identify lung adenocarcinomas during pulmonary resection. J. Thorac. Cardiovasc. Surg. 2015;150:28–35.e1. doi: 10.1016/j.jtcvs.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du T, Zamore PD. Beginning to understand microRNA function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- 51.Lopez JS, Banerji U. Combine and conquer: Challenges for targeted therapy combinations in early phase trials. Nat. Rev. Clin. 2016;14:57–66. doi: 10.1038/nrclinonc.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, King PD, Weis SM, Cheresh DA. MicroRNA-132–mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol. Pharmaceutics. 2011;8:1381–1389. doi: 10.1021/mp2002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia W, Hilgenbrink AR, Matteson EL, Lockwood MB, Cheng J-X, Low PS. A functional folate receptor is induced during macrophage activation and can be used to target drugs to activated macrophages. Blood. 2009;113:438–446. doi: 10.1182/blood-2008-04-150789. [DOI] [PubMed] [Google Scholar]
- 55.van der Wilt CL, Backus HHJ, Smid K, Comijn L, Veerman G, Wouters D, Voorn DA, Priest DG, Bunni MA, Mitchell F, Jackman AL, Jansen G, Peters GJ. Modulation of both endogenous folates and thymidine enhance the therapeutic efficacy of thymidylate synthase inhibitors. Cancer Res. 2001;61:3675–3681. [PubMed] [Google Scholar]
- 56.Van Der Heijden JW, Oerlemans R, Dijkmans BAC, Qi H, Van Der Laken CJ, Lems WF, Jackman AL, Kraan MC, Tak PP, Ratnam M, Jansen G. Folate receptor β as a potential delivery route for novel folate antagonists to macrophages in the synovial tissue of rheumatoid arthritis patients. Arthritis Rheum. 2009;60:12–21. doi: 10.1002/art.24219. [DOI] [PubMed] [Google Scholar]
- 57.Lu Y, Stinnette TW, Westrick E, Klein PJ, Gehrke MA, Cross VA, Vlahov IR, Low PS, Leamon CP. Treatment of experimental adjuvant arthritis with a novel folate receptor-targeted folic acid-aminopterin conjugate. Arthritis Res. Ther. 2011;13:R56. doi: 10.1186/ar3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 59.Marks IS, Kang JS, Jones BT, Landmark KJ, Cleland AJ, Taton TA. Strain-promoted“click” chemistry for terminal labeling of DNA. Bioconjug. Chem. 2011;22:1259–1263. doi: 10.1021/bc1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gent YYJ, Weijers K, Molthoff CFM, Windhorst AD, Huisman MC, Smith DEC, Kularatne SA, Jansen G, Low PS, Lammertsma AA, van der Laken CJ. Evaluation of the novel folate receptor ligand [18F]fluoro-PEG-folate for macrophage targeting in a rat model of arthritis. Arthritis Res. Ther. 2013;15:R37. doi: 10.1186/ar4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu Y, Xu L-C, Parker N, Westrick E, Reddy JA, Vetzel M, Low PS, Leamon CP. Preclinical pharmacokinetics, tissue distribution, and antitumor activity of a folate-hapten conjugate–targeted immunotherapy in hapten-immunized mice. Mol. Cancer Ther. 2006;5:3258–3267. doi: 10.1158/1535-7163.MCT-06-0439. [DOI] [PubMed] [Google Scholar]
- 62.Orellana EA, Kasinski AL. Sulforhodamine B (SRB) assay in cell culture to investigate cell proliferation. Bio. Protoc. 2016;6:e1984. doi: 10.21769/BioProtoc.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krupnick AS, Tidwell VK, Engelbach JA, Alli VV, Nehorai A, You M, Vikis HG, Gelman AE, Kreisel D, Garbow JR. Quantitative monitoring of mouse lung tumors by magnetic resonance imaging. Nat. Protoc. 2012;7:128–142. doi: 10.1038/nprot.2011.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tidwell VK, Garbow JR, Krupnick AS, Engelbach JA, Nehorai A. Quantitative analysis of tumor burden in mouse lung via MRI. Magn. Reson. Med. 2012;67:572–579. doi: 10.1002/mrm.22951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Fig. S1. Fol-DBCO ligand synthesis and LC-MS spectral analysis.
Fig. S2. Evaluation of folate-miRNA conjugation measured by 15% native TAE PAGE.
Fig. S3. MALDI spectra of FolamiRs.
Fig. S4. NIR-folate ligand synthesis and LC-MS spectral analysis.
Fig. S5. Evaluation of NIR-FolamiR conjugation measured by 15% native TAE PAGE.
Fig. S6. MALDI spectra of NIR-FolamiR conjugates.
Fig. S7. miR-34a Renilla luciferase sensor response to miR-34a transfection.
Fig. S8. Evaluation of MB-231 miR-34a sensor cells.
Fig. S9. Folate-mediated delivery of functional siLuc2.
Fig. S10. Serum stability of FolamiRs and duplex RNA oligos.
Fig. S11. In vivo tumor growth response to dose titration of FolamiR-34a.
Fig. S12. miR-34a copy number in tumors treated with FolamiR-34a in mice.
Fig. S13. Serum cytokines and maximum tolerated FolamiR dose study in mice.
Table S1. Individual-level data.