Abstract
We explore the phylogenetic relationships among HIV sequences sampled from young adult black men who have sex with men (YAB-MSM), who are connected through peer referral/social ties and who attend common venues. Using 196 viral sequences sampled from the peripheral blood mononuclear cells of 10 individuals, our preliminary phylogenetic results indicate that these socially connected YAB-MSM are infected with distantly related viruses and provide no evidence for viral transmission between network members. Our results suggest that HIV-prevention strategies that target young adult MSM should extend beyond their network members and local community.
Keywords: : phylogenetic analysis, HIV sequence, young black MSM, social networks, peer referral
Introduction
Black men who have sex with men (MSM), as a subpopulation based on ethnicity, sex, and transmission category, now represent the largest number of new HIV-1 infections in the United States (11,201; 25% of all diagnosed cases in 2014).1 This alarming development echoes an increased black MSM infection rate that rose nearly 22% over the decade of 2005–2014.2 More pronounced is the disproportionate infection rate in young adult black MSM (YAB-MSM) (20–29 years old), which is approximately ninefold higher than similar white MSM on a per-capita basis (123.7 vs. 13.2 per 100,000).1
MSM sexual networks frequently overlap with social, peer, and venue-based networks.3,4 Previous literature indicates that YAB-MSM tend to be involved in an unique context of several extensive sexual networks with a high prevalence of HIV.5,6 Such extensive risk networks could be formed in social contexts by attending “risk spaces” or venues7,8 where MSM meet their social and sex partners. In other words, social places where YAB-MSM interact with their peers may serve as a proxy for membership in a densely connected high-risk sexual network among MSM.9,10 Additional work has demonstrated that there is overlap between MSM on social and sexual networks; these networks are dynamic, and sex and social ties often rapidly cycle, particularly in these younger age groups.11 Given such contextual factors, it is expected that social and sexual networks tend to overlap each other, especially among YAB-MSM.
Identifying those particular social interactions that drive YAB-MSM infection dynamics is important from an epidemiological perspective. Characterizing HIV-1 transmission patterns in this population can be challenging, however, as few studies are designed to integrate the epidemiological, viral genetic, and social network data that are required to establish such meaningful connections. Successful analysis of these factors could have a significant impact on the prevention, diagnosis, and management of the disease in this underserved population.
We addressed this complex issue using data obtained from a subset of YAB-MSM individuals embedded within a larger MSM network constructed with respondent-driven sampling (RDS). The 10 participants were all HIV-1 positive and were related through referrals and social connections. We sought to explore whether HIV-1 sequences derived from this relatively small RDS-subpopulation of infected YAB-MSM might be closely related and, if so, reveal evidence of recent transmission. We specifically explored this subset because these 10 individuals have a high rate of HIV infection, as well as close and overlapping social ties. If any recent transmission has occurred that can be predicted by the social network, it is most probable in this group. If recent transmission is not detected in this group, it is unlikely that social connections are related to infection dynamics across the network as a whole. To shed light on this issue, our study leveraged a clone-based phylogenetic analysis of integrated viral sequences present within infected peripheral blood mononuclear cells (PBMCs).12,13
Data and Methods
Study site and sampling
Samples were obtained from a subset of individuals (iMAN; see Acknowledgements section) embedded within a parent study (YMAP; see Acknowledgments section). iMAN examines the association between phylogenetic and social clustering via common venue affiliations of MSM in Houston, Texas. iMAN participants were recruited using an RDS method,14 whereby respondents were issued four vouchers to distribute among other young, local MSM. Study inclusion criteria were as follows: (i) identification as a male, (ii) between the ages of 17 and 29, (iii) reported having sex (oral or anal) with a man in the past 6 months, (iv) frequent visitors at one or more risk or preventive venues in the past 12 months, and (v) willing to provide informed consent and biological samples.
In 2014, we conducted a survey of iMAN participants that focused on their sociodemographic characteristics, risk behaviors, and venue affiliation (both social and health venues). Participants could further nominate up to five social partners (i.e., people with whom participants share personal information) and up to five sexual partners (i.e., people with whom participants had sex in the prior 6 months). Using a fuzzy matching algorithm that uses partners' sociodemographic information,15 we constructed a network of participating identifiers and participating social and sexual referrals. Using this information, we selected a subpopulation of 10 HIV-1-infected YAB-MSM (designated as iMAN1–iMAN10) who were connected through the peer referral survey, which is illustrated in Figure 1.
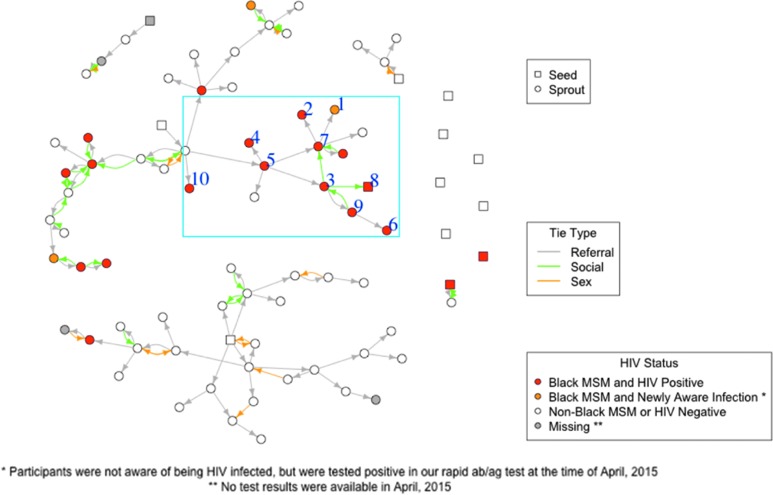
FIG. 1.
This figure illustrates the entire peer referral chain combined with social and sex connections that include all HIV-positive and HIV-negative young black and non-black MSM, whose data were retrieved in April 2015. The 10 individuals used for the current pilot study were embedded within this entire network. The numbered nodes (1–10) within the blue box were selected for our sample of 10 HIV-positive black MSM. A square node indicates seed (first-wave focal participants) based on RDS, and a circular node indicates sprout (second or more wave-chained contacts). A red node indicates HIV-positive black MSM; an orange node indicates newly infected black MSM, defined as being unaware of their HIV infection at the time of interview and being tested positive by the antibody/antigen quick test; and a white node indicates non-black MSM (including white, Hispanic, and Asian MSM) or HIV-negative black or non-black MSM. Gray indicates a peer referral connection, green indicates a self-reported social connection, and orange indicates a self-reported sex connection in the last 6 months. Social and sex connections were linked by a matching algorithm based on their partners' sociodemographic information, using a fuzzy matching algorithm. Arrows indicate the direction of nomination. No self-reported sex connections in the last 6 months were identified among the 10 individuals. No cell samples from the red circle node that is connected to node no. 7 through both peer referral and social ties were available. MSM, men who have sex with men; RDS, respondent-driven sampling.
Social network data
A peer-referral network based on social and sexual connections was constructed in the form of an adjacency matrix, with 1 as indicating a connection and 0, otherwise, which was then maximally symmetrized and visualized using igraph network visualization.16 Although sexual connections were considered, the 10 sampled YAB-MSM did not self-report any nonanonymous sexual behavior in the prior 6 months (Fig. 1). We also constructed two types of venue-based networks among the 10 YAB-MSM samples, with 1 as indicating the existence of at least 1 venue attended in common out of 20 social venues and 17 health venues, and 0, otherwise.
Genomic DNA isolation, env amplification, cloning, and sequencing
To prevent contamination, sample mix-up, and bias, we blinded and processed samples one at a time. Genomic DNA was isolated from PBMCs using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer's protocol. To capture the viral sequence diversity present within a given individual, we performed four independent polymerase chain reactions that targeted the env region of HIV-1 from each sample, as previously described.17 Molecular clones were sequenced by the Sanger method, using forward and reverse M13 and internal EN2 and EN4 primers. Consensus sequences for 18–20 clones were obtained for each sample. The two most divergent sequences from each sample were blasted against GenBank to identify 20 control sequences, similar to that previously described.12,13
Phylogenetic analysis
Sequences were aligned with MAFFT Version 7.18 Models of sequence evolution were chosen, using Akaike's Information Criterion (AIC),19 as implemented in jModelTest version 2.20 The pool of possible models included 24 variants of the general-time-reversible21 class, with and without gamma-distributed rate variation across sites, and an estimated proportion of invariable sites.22 Best-fit models were selected, both assuming the entire alignment evolved according to a single process (unpartitioned) and allowing separate processes for each codon position (partitioned). Maximum likelihood (ML) and Bayesian phylogenetic analyses were performed, using AIC-chosen models both with and without partitioning by codon position.
Each ML analysis was performed using GARLI Version 2.0, with 32 independent search replicates and default search parameters.23 Topological support was estimated using 1,000 bootstrap replicates. Bootstrap proportions for each bipartition in the ML tree were summarized using SumTrees 4.0.0.24,25 Each Bayesian analysis was performed using MrBayes Version 3.2.5, with four independent runs and four Metropolis-coupled chains per run, and was run for 5 million generations.26,27 Additional ML and Bayesian analyses were conducted with a model that partitions parameters by codon position.28,29 All substitution model parameters, with the exceptions of topology and branch lengths, were unlinked across data subsets. All analyses were conducted both with and without missing data (gaps).30 We conducted further analyses, using two separate datasets that consisted of only first codon positions or second and third codon positions to look for evidence of positive selection's influencing the inferred topology.31 In all cases, recovered topologies and support values did not exhibit strong conflicts, so we report results only from the complete dataset and nonpartitioned model.
Results
Description of sample and network
Our cohort consisted of 10 YAB-MSM with an average age of 25 years [standard deviation (SD) = 2.2, min = 20, max = 28]. Of these, 30% had homeless history, 30% had experience with group sex, 30% used condoms inconsistently, 20% had experience in trading sex for money, 90% used online dating sites in the preceding 12 months, and 50% had been incarcerated at some point in their life. Participants had an average of seven sexual partners in the past 6 months (SD = 15, min = 1, max = 50) and visited an average of four health venues (SD = 2.5, min = 1, max = 9) and six risk/social venues (SD = 3.2, min = 1, max = 10). In addition, 70% were on HIV treatment, and the average viral load was 25,790 IU/ml (SD = 22,992, min = 358, max = 66,000). Figure 1 illustrates the broader RDS-derived network that consists of 69 young MSM with their respective connections. The 10 YAB-MSM samples included in this study are outlined in the figure and were connected through at least 1 common social and health venue.
Phylogenetic analysis
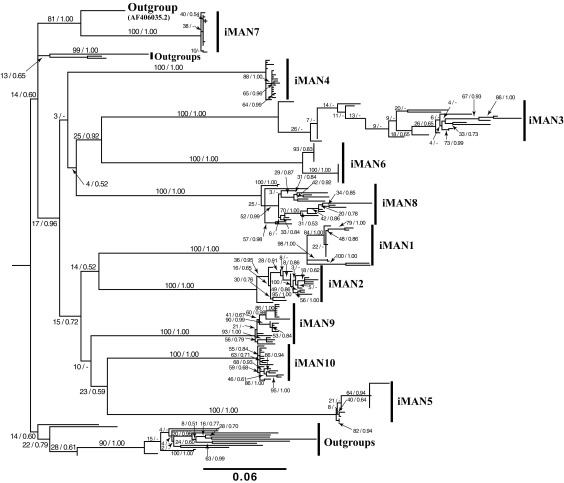
Figure 2 illustrates a ML phylogenetic tree that depicts the relationships among iMAN and control HIV-1 sequences, based on an unpartitioned model.
FIG. 2.
This figure illustrates a ML phylogenetic tree that depicts the relationships among iMAN and control HIV-1 sequences, based on an unpartitioned model. Bipartition support values give bootstrap proportions on the left and Bayesian posterior probabilities on the right. ML, maximum likelihood.
All viral clones from a given individual were recovered as monophyletic with very strong support. Short branches within clades from each individual suggest generally little intrahost viral diversity. Notable exceptions to this pattern are viruses sampled from iMAN2, iMAN3, and iMAN8. Variation in viral diversity across samples may be driven by a variety of factors, including treatment regime and time since infection; this information, however, was unavailable.
No evidence (i.e., paraphyly of viruses sampled from one person with respect to those sampled from another) was found to suggest a direction of the transmission of viruses between individuals.13 In addition, viruses sampled from different individuals were quite divergent (mean average divergence between individuals = 0.36, SD = 0.05, min = 0.27, max = 0.48), as evidenced by long branches' subtending viral clades from different individuals. Support for relationships among viruses sampled from different individuals was quite low across all branches, suggesting that these viruses do not exhibit the structure expected for a recent transmission cluster.
Viral samples from one individual (iMAN7) were not placed in the clade with the rest of the case samples and, instead, were inferred to be most closely related to GenBank outgroup sequence AF406035.2 with moderately strong support (Fig. 2). Major phylogenetic patterns, notably the monophyly of viruses from each individual, long branches subtending each intraindividual clade, and weak support for relationships among viruses from different individuals, were consistent across all analyses and data subsets.
Discussion
Our phylogenetic results provide preliminary evidence that the sampled viruses of 10 YAB-MSM who were connected through RDS-driven interactions did not constitute a recent transmission cluster. Combined with the fact that respondents did not indicate sexual relationships among network participants, these findings suggest several important considerations for understanding HIV transmission dynamics in this community. Social networks among YAB-MSM are dynamic in nature.32 Therefore, current network connections may only weakly correlate with those network connections that were relevant for viral transmission in the past. In addition, viral genome evolution over time could obscure older transmission events or more distant connections in transmission chains. Overall, our results suggest that the identification of transmission chains among network members who belong to a larger RDS network may be challenging. This finding is in agreement with several other studies that found only a small fraction of individuals with associations between RDS social networks and phylogenetic-based transmission networks.33,34
iMAN7 harbored viral sequences most closely related to a control sequence derived from a study of men in other city. This relationship is not unexpected, as control sequences were chosen to be closely related to sampled viral sequences in this study. In addition, iMAN7 had several risk behaviors that may have increased the probability of transmission from outside the network, including 50 sex partners, the highest attendance at risk venues, inconsistent condom usage, experience with group sex, and online dating site use. Because iMAN7 is directly connected to half of our sample of 10 through one or more social connections, further investigation into our larger social network of YAB-MSM is warranted.
We note several limitations to our study. Because we characterized only 10 YAB-MSM who were identified within a larger RDS network, our power to detect rare instances of transmission between individuals closely connected in a network may be low. Furthermore, because our study did not include all HIV-positive cases present within the larger network, we cannot eliminate the possibility that alternative transmission patterns might be found in a larger sample set. Moreover, participants may have omitted other chain referrals as sex partners, or we may have missed other individuals within the chain who were not part of the venue. Self-reports are prone to response bias and inherently exclude the existence of sexual relationships in anonymous settings.
While interpreting negative findings can be challenging, we believe that our results are informative in understanding the complex dynamics of MSM social networks. Previous studies have suggested that phylogenetically clustered YAB-MSM are not geographically clustered35,36 and possibly supported by a more general population.36 One factor that contributes to this geographic heterogeneity may be the increased use of Internet resources as a venue for finding MSM partners.36 In fact, the majority of our cohort had used “hook-up apps.” This study lends preliminary support to the hypothesis that, even within what would seem to be a very narrow segment of the larger community, sexual partnering does not occur primarily among those who share the same space. HIV-intervention strategies that focus on YAB-MSM may need to utilize more than race and geography to prove effective.
Acknowledgments
All phylogenetic analyses were conducted using high-performance computing resources provided by Louisiana State University (HPC@LSU). J.M.B. was supported by NSF Grants D.E.B.-1355071 and D.B.I.-1262571, in addition to startup funds from the Louisiana State University College of Science and Department of Biological Sciences. L.M.C. was supported by NSF Grant D.E.B.-1355071 to J.M.B. Data were collected and network analyses were conducted at the University of Texas Health Science Center at Houston (UTHealth), and K.F., L.Y.H., and J.A.S were supported by the National Institutes of Health/NIMH 1R01MH100021 (YMAP: Young Men's Affiliation Project of HIV risk and prevention venue) NIH/NIGMS 1R21GM113694 (iMAN: integrated Molecular & Affiliation Network analysis of HIV transmission), the National Institutes of Health/NIDA 1R01DA039934, and startup Texas State funds from School of Public Health, UTHealth.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Centers for Disease Control and Prevention: HIV surveillance report, 2014. Available at: www.cdc.gov/hiv/library/reports/surveillance (2015), accessed July27, 2016
- 2.Centers for Disease Control and Prevention: Trends in U.S. HIV diagnoses, 2005–2014. Available at: www.cdc.gov/nchhstp/newsroom/docs/factsheets/hiv-data-trends-fact-sheet-508.pdf (2016), accessed July27, 2016
- 3.Tieu HV, Liu TY, Hussen S, et al. : Sexual networks and HIV risk among black men who have sex with men in 6 US cities. PLoS One 2015;10:e0134085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan PA, Hogan JW, Huang A, et al. : Phylogenetic investigation of a statewide HIV-1 epidemic reveals ongoing and active transmission networks among men who have sex with men. J Acquir Immune Defic Syndr 2015;70:428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurt CB, Beagle S, Leone PA, et al. : Investigating a sexual network of black men who have sex with men: Implications for transmission and prevention of HIV infection in the United States. J Acquir Immune Defic Syndr 2012;61:515–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention: Increase in newly diagnosed HIV infections among young black men who have sex with men—Milwaukee County, Wisconsin, 1999–2008. Morb Mortal Wkly Rep 2011;60:99–102 [PubMed] [Google Scholar]
- 7.Frost SD: Using sexual affiliation networks to describe the sexual structure of a population. Sex Transm Infect 2007;83(Suppl 1):i37–i42 [DOI] [PubMed] [Google Scholar]
- 8.Fujimoto K, Williams ML, Ross MW: Venue-based affiliation network and HIV risk behavior among male sex workers. Sex Transm Dis 2013;40:453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens SC, Fann CK, Strona FV, et al. : Identifying syphilis risk networks through venue attendance in San Francisco. Sex Transm Dis 2014;41:333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amirkhanian YA: Social networks, sexual networks and HIV risk in men who have sex with men. Curr HIV/AIDS Rep 2014;11:81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider JA, Cornwell B, Jonas A, et al. : Network dynamics of HIV risk and prevention in a population-based cohort of young Black men who have sex with men. Netw Sci 2017;5:1–29 [Google Scholar]
- 12.Metzker ML, Mindell DP, Liu XM, et al. : Molecular evidence of HIV-1 transmission in a criminal case. Proc Natl Acad Sci.U S A 2002;99:14292–14297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scaduto DI, Brown JM, Haaland WC, et al. : Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Nati Acad Sci 2010;107:21242–21247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heckathorn DD: Respondent-driven sampling: A new approach to the study of hidden populations. Soc Probl 1997;44:174–199 [Google Scholar]
- 15.Shah NS, Iveniuk J, Muth SQ, et al. : Structural bridging network position is associated with HIV status in a younger Black men who have sex with men epidemic. AIDS Behav 2014;18:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csardi G, Nepusz T: The igraph software package for complex network. Inter J Complex Syst 2006;54:1695 [Google Scholar]
- 17.Metzker ML, Ansari-Lari MA, Liu XM, et al. : Quantitation of mixed-base populations of HIV-1 variants by automated DNA sequencing with BODIPY dye-labeled primers. Biotechniques 1998;25:446–447, 450–442, 454, passim. [PubMed] [Google Scholar]
- 18.Katoh K, Standley DM: MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 2013;30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akaike H: A new look at the statistical model identification. IEEE Trans Autom Contr 1974;19:716–723 [Google Scholar]
- 20.Darriba D, Taboada GL, Doallo R, et al. : jModelTest 2: More models, new heuristics and parallel computing. Nat Methods 2012;9:772–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavaré S: Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci 1986;17:57–86 [Google Scholar]
- 22.Yang Z: Molecular Evolution: A Statistical Approach. Oxford University Press, Oxford, 2014 [Google Scholar]
- 23.Zwickl DJ: Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation, The University of Texas at Austin, 2006 [Google Scholar]
- 24.Sukumaran J, Holder MT: DendroPy: A Python library for phylogenetic computing. Bioinformatics 2010;26:1569–1571 [DOI] [PubMed] [Google Scholar]
- 25.Sukumaran J, Holder M: SumTrees: Summarization of split support on phylogenetic trees, version 1.0. 2. Part of the DendroPy phylogenetic computation library, version 2.6. 1. Available at http://packages.python.org/DendroPy (2009). Accessed on April7th, 2017
- 26.Ronquist F, Huelsenbeck JP: MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 27.Altekar G, Dwarkadas S, Huelsenbeck JP, et al. : Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 2004;20:407–415 [DOI] [PubMed] [Google Scholar]
- 28.Nylander JA, Ronquist F, Huelsenbeck JP, et al. : Bayesian phylogenetic analysis of combined data. Syst Biol 2004;53:47–67 [DOI] [PubMed] [Google Scholar]
- 29.Brown JM, Lemmon AR: The importance of data partitioning and the utility of Bayes factors in Bayesian phylogenetics. Syst Biol 2007;56:643–655 [DOI] [PubMed] [Google Scholar]
- 30.Lemmon AR, Brown JM, Stanger-Hall K, et al. : The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Syst Biol 2009;58:130–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doyle VP, Andersen JJ, Nelson BJ, et al. : Untangling the influences of unmodeled evolutionary processes on phylogenetic signal in a forensically important HIV-1 transmission cluster. Mol Phylogenet Evol 2014;75:126–137 [DOI] [PubMed] [Google Scholar]
- 32.McFadden R, Bouris A, Voisin D, et al. : Dynamic social support networks of younger black men who have sex with men with new HIV infection. AIDS care 2014;26:1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis AM, Murillo W, Hernandez A, et al. : Social network–based recruitment successfully reveals HIV-1 transmission networks among high-risk individuals in El Salvador. J AcquiImmune Defic Syndr 2013;63:135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SS, Tam DKP, Tan Y, et al. : An exploratory study on the social and genotypic clustering of HIV infection in men having sex with men. AIDS 2009;23:1755–1764 [DOI] [PubMed] [Google Scholar]
- 35.Oster AM, Wieganda RE, Sioneana C, et al. : Understanding disparities in HIV infection between black and white MSM in the United States. AIDS 2011;25:1103–1112 [DOI] [PubMed] [Google Scholar]
- 36.Lubelchek RJ, Hoehnen SC, Hotton AL, et al. : Transmission clustering among newly diagnosed HIV patients in Chicago, 2008 to 2011: Using phylogenetics to expand knowledge of regional HIV transmission patterns. J Acqui Immune Defic Syndr 2015;68:46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]