Abstract
The common marmoset monkey (Callithrix jacchus; Cj) is an advantageous nonhuman primate species for modeling age-related disorders, including Parkinson's disease, due to their shorter life span compared to macaques. Cj-derived induced pluripotent stem cells (Cj-iPSCs) from somatic cells are needed for in vitro disease modeling and testing regenerative medicine approaches. Here we report the development of a novel Cj-iPSC line derived from adult marmoset fibroblasts. The Cj-iPSCs showed potent pluripotency properties, including the development of mesodermal lineages in tumors after injection to immunocompromised mice, as well as ectoderm and endoderm lineages after in vitro differentiation regimens, demonstrating differentiated derivatives of all three embryonic layers. In addition, expression of key pluripotency genes (ZFP42, PODXL, DNMT3B, C-MYC, LIN28, KLF4, NANOG, SOX2, and OCT4) was observed. We then tested the neural differentiation capacity and gene expression profiles of Cj-iPSCs and a marmoset embryonic stem cell line (Cj-ESC) after dual-SMAD inhibition. Exposure to CHIR99021 and sonic hedgehog (SHH) for 12 and 16 days, respectively, patterned the cells toward a ventralized midbrain dopaminergic phenotype, confirmed by expression of FOXA2, OTX2, EN-1, and tyrosine hydroxylase. These results demonstrate that common marmoset stem cells will be able to serve as a platform for investigating regenerative medicine approaches targeting the dopaminergic system.
Keywords: : induced pluripotent stem cells, Parkinson's disease, neural differentiation, nonhuman primate model
Introduction
Induced pluripotent stem cells (iPSCs) have become a powerful tool for regenerative medicine research since their initial derivation from mouse [1] and subsequently human fibroblasts [2,3]. Circumventing the limitations of human embryo-derived embryonic stem cells (ESCs), iPSC lines can be derived from adult tissues and can be used to study pluripotency, development, differentiation to mature cells and tissues, and disease modeling. In particular, a significant advantage of iPSCs over ESCs is their potential as a source for autologous cell-replacement therapies. iPSCs are hypothesized to be recognized as “self tissue” facilitating successful therapeutic delivery of host-derived iPSCs without immunological rejection.
Nonhuman primates are an invaluable resource for preclinical investigation of the safety and efficacy of iPSC-based therapies. In that regard, the common marmoset monkey (Callithrix jacchus; Cj) has been identified as an advantageous species for modeling age-related disorders, such as Parkinson's disease, due to their shorter life span compared to larger nonhuman primates [4]. To realize this opportunity, Cj-stem cell lines are needed as platform tools for in vitro phenotype characterization and regenerative medicine strategies. For example, neurons derived from Cj-stem cells can be genetically modified to model in vitro genetic neurological diseases and, by generating immortal cell lines, can be utilized for indefinite study and manipulation of these diseases.
The potential for neural differentiation has been reported from Cj-ESCs [5] and Cj-iPSCs derived from fetal fibroblasts [6–8], fetal liver cells [9], neonatal skin [10], and adult bone marrow [11]. There is also one report demonstrating direct reprogramming of marmoset embryonic skin to neuronal cells [12]. However, the derivation of Cj-iPSCs from adult skin fibroblasts and further patterning and differentiation to dopaminergic neurons have not been reported.
Fetal and newborn tissues are a useful experimental source of cells that are more easily manipulated and reprogrammed compared to adult fibroblasts, but they are not a realistic iPSC source for most human-directed applications. Furthermore, studies that have succeeded in deriving iPSCs from the marmoset did so via either retrovirus systems [6,9] or the reversible piggyback system [10,13] that also integrates into the host genome. However, using nonintegrating episomal vectors circumvents the concern of continued expression of exogenous reprogramming genes [14]. In addition, while differentiation of dopaminergic (DAergic) neurons has been successful in human and rhesus cells [15], this has not yet been achieved with marmoset stem cells, including patterning to become midbrain floor plate-derived DAergic neurons, which are the neurons that degenerate in PD.
The aim of this study was to fill these gaps by producing a Cj-iPSC line from adult marmoset skin fibroblasts using nonintegrating expression plasmids, generating a protocol for mature neuronal differentiation of Cj-iPSCs, and characterizing the expression of pluripotent and neural differentiation-related genes throughout the differentiation process of both Cj-ESCs and Cj-iPSCs.
Materials and Methods
iPSC derivation
All procedures involving animals were performed in accordance with the recommendations in the National Research Council Guide for the Care and Use of Laboratory Animals (2011) in an AAALAC accredited facility (Wisconsin National Primate Research Center, University of Wisconsin-Madison). Experimental procedures were approved by the Graduate School Institutional Animal Care and Use Committee of the University of Wisconsin-Madison.
A small strip of skin and subcutaneous tissue from an adult common marmoset (4 years old) was obtained during an unrelated procedure under anesthesia. The tissue was immediately plated down to individual wells of a six-well plate coated with gelatin. Once the emerging fibroblasts expanded sufficiently, expression plasmids (pEP4 E02S EN2K, pEP4 E02S ET2K, pCEP4-M2 L, and miRNA302 [16]) were electroporated into the fibroblasts with a Gene Pulser II (Biorad) at settings of 250 V, 950 μF in Opti-MEM I Reduced-Serum Medium (Life Technologies 31985-070).
For the first 3 days, the cells were fed with fibroblast medium consisting of DMEM/F12 (SH30023.01; Thermo Scientific), 10% fetal bovine serum (12476-024; Gibco), NEAA (nonessential amino acids) (11140-050; Gibco), and sodium pyruvate (13-115E; Lonza). On day 3, the medium was adjusted to a small-molecule medium consisting of Essential 6 (A1516401; Life Technologies), bFGF (100 μg/mL; WiCell), N2 (17502-048; Gibco), B27 (17504-044; Gibco), PD0325901 (10 mM, 40006; Stemgent), A 83-01 (50 mM, 2939; Tocris), CHIR99021 (20 mM, 4423; Tocris), LIF (10 ng/mL, 5283; Sigma), and Y-27632 (10 mM, 1254; Tocris). The cells were then fed with stem cell medium (Stem Cell Culture section) on day 15. When pluripotent stem cell colonies arose, as microscopically observed, they were picked and transferred to separate wells of a 24-well plate for expansion and cryopreservation.
RNA isolation
To characterize the gene expression profiles of undifferentiated Cj-ESCs and Cj-iPSCs, as well as at day 16, 30, and 42 of the neural differentiation protocol, RNA was extracted using RNA STAT-60 (CS-110; Amsbio). Nucleic acid concentration was determined using a NanoDrop, and RNA integrity was tested using a 2100 Bioanalyzer (Agilent).
cDNA synthesis
cDNA was synthesized using 0.5 μg of RNA. An RT2 First-Strand Synthesis Kit (330401; Qiagen) was used for the Qiagen RT2 Profiler arrays, while a SuperScript III First-Strand Synthesis System Kit (18080051; Invitrogen) was used for the custom qPCR characterization experiments. A genomic elimination step was completed before the reverse transcription reaction, which was followed by the RNA elimination step. Reactions without reverse transcriptase (NoRT) were used as negative controls for the PCRs.
Reverse transcription-PCR
To determine pluripotency gene expression, reverse transcription-PCR (RT-PCR) was performed on the RNA samples of both Cj-ESCs and Cj-iPSCs using primers for ZFP42, PODXL, DNMT3B, OCT4, SOX2, KLF4, c-MYC, NANOG, and LIN28 (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd). Reactions were set up using GoTaq Polymerase (m5138; Promega) using equivalent amounts of the reverse transcription reaction from 0.5 μg of RNA. The PCRs were run in a programmable thermocycler (PTC-100; MJ Research, Inc.) with an initial denaturing step of 95°C for 2 min, followed by 36 cycles programmed thus: denaturing, 95°C, 30 s; annealing, 55°C; extension, 72°C, 30 s; followed by a final extension of 72°C for 15 min. Samples were then loaded on 2% agarose gels and amplification of a single band of the predicted size was verified for each primer pair.
Quantitative-RT-PCR
For qRT-PCR, RNA samples were collected as described above (RNA Isolation section). RNA quality was tested on a bioanalyzer (UW-Biotech Center). cDNA was synthesized using a cDNA First-Strand Synthesis Kit (330404; Qiagen). qRT-PCR analysis was done using SYBR green (330523; Qiagen) and an RT2 Profiler Human Stem Cell Lineage Array Kit (PAHS-508ZF-12; Qiagen). Further analysis of genes of interest was accomplished using 0.5 μg of RNA from three separate biological replicates collected from several different differentiation regimens and primers designed to be 18–22 nucleotides, 40–50% G/C content, within one degree (Tm) of each other and between 58°C–62°C, and spanning an exon–exon junction when possible. A list of qRT-PCR primer sequences can be found in Supplementary Table S1. Controls for the qRT-PCR experiment included technical triplicates, no reverse transcriptase controls for each primer set, amplification of reference genes (GAPDH and RPLP0; GAPDH was chosen for dCt calculations), and an interassay control of GAPDH. All primer sets were screened for gene amplification specificity using the standardized PCR temperature parameters (Reverse Transcription-PCR section).
Teratoma formation
Teratoma formation was carried out by Applied StemCell, Inc. (Menlo Park, CA). Passage 23 Cj-ESCs, and passage 17 Cj-iPSCs were injected into the left kidney capsule and right testis of three 6-week-old, male, SCID-beige mice (Charles River, Wilmington, MA) per cell line. Each injection consisted of 1–2 million cells/site, in 30% Matrigel. Tumors were collected 56 and 55 days postinjection for the Cj-ESCs and Cj-iPSCs, respectively. For histological analysis, tumor tissues were fixed with 10% formalin overnight, embedded in paraffin, cut into 5 μm sections, and stained with hematoxylin and eosin.
Endoderm differentiation
Cj-iPSCs were screened for endodermal lineage potential through two separate methods. First, cells were differentiated to definitive endoderm using a StemDiff Definitive Endoderm Kit (TeSR-E8 Optimized; 05115; STEMCELL Technologies) and analyzed on day 4 and 5 postinduction. RNA was collected from control undifferentiated cells as well as 4 and 5 days after endoderm induction as described above. Expression of the definitive endoderm genes CXCR4, SOX17, and FOXA2 was assessed using qRT-PCR (n = 3).
Immunocytochemistry (ICC) was performed using anti-SOX17 to verify protein expression and efficiency of differentiation. In addition, Cj-iPSCs were allowed to spontaneously differentiate in DMEM supplemented with GlutaMAX (35050-061; Life Technologies) and chemically defined lipid concentrate (11905-031; Life Technologies) for 23 days. RNA was collected in undifferentiated cells, after day 7 of differentiation, and subsequently every 2 days. Expression of AFP was evaluated at each time point using RT-PCR.
Stem cell culture
Pluripotent stem cells were grown in growth factor-reduced Matrigel (83 ng/well, 354230; BD)-coated 35-mm culture plates and medium was replenished daily consisting of E8 medium (A14666SA; Life Technologies), E8 supplement (A15171-01; Life Technologies), nodal (100 ng/mL, 3218-ND; R&D Systems), GlutaMAX (35050-061; Life Technologies), chemically defined lipid concentrate (11905-031; Life Technologies), and reduced glutathione (1.94 μg/mL, G4251; Sigma). To passage the cells, the medium was aspirated and Accutase (SCR005; Thermo Fisher Scientific) was added. Cells were maintained at 5% CO2/95% room air and 37°C in a Series II Water Jacket incubator (Thermo Scientific).
Neural differentiation
Neural induction medium composed of DMEM/F12 (SH30023.01; Thermo Scientific), MEM-NEAA (11140-050; Gibco), N2 supplement (17502-048; Gibco), SB431542 (10 μM, 04-0010; Stemgent), and DMH1 (200 nM, 4126; Tocris) was added 1 day after passaging ∼20,000 cells/35-mm well. By day 8, colonies were of proper size, were lifted using dispase (2 mg/mL DMEM/F12), and grown in suspension as neurospheres in T75 low cell adhesion flasks for 20 days. The neurospheres were then dissociated and plated back down to Matrigel-coated coverslips and maintained in neural differentiation medium consisting of neural basal medium (21103-049; Gibco), MEM-NEAA (11140-050; Gibco), N2 supplement (17502-048; Gibco), B27 supplement (17504-044; Gibco), brain-derived neurotrophic factor (BDNF; 10 ng/mL, 450-02; Pepro tech), glial-derived neurotrophic factor (GDNF; 10 ng/mL, 450-10; Pepro tech), TGF-β3 (1 ng/mL, 100-36E; Pepro tech), ascorbic acid (200 μM, A0278; Sigma), and cyclic adenosine 3′,5′-monophosphate (cAMP; 1 μM, A9501; Sigma).
To further pattern cells to floor plate-derived midbrain dopaminergic neurons, the cells were exposed to neural induction medium with the addition of sonic hedgehog (SHH; 500 ng/mL, 464-SH; R&D Systems) and CHIR99021 (0.4 μM, 04-0004; Stemgent) and 8 days later, lifted using dispase (2 mg/mL DMEM/F12) and grown in suspension. After 12 or 16 days, CHIR99021 was removed and SHH was reduced to 20 ng/mL. On day 16 in all conditions, FGF8b (100 ng/mL, 100-25; Pepro tech) was added until day 28.
As an alternative method to pattern cells, SHH was removed 4 days after induction and purmorphamine (5 μM, 04-0009; Stemgent) was added. On day 16, CHIR99021 was removed, purmorphamine reduced to 0.2 μM, and FGF8b (100 ng/mL, 100-25; Pepro tech) was added.
Immunofluorescence
Immunofluorescent staining was performed to characterize pluripotent, patterning, neuronal and DAergic differentiation expression profiles. Coverslips with adherent cells were fixed at room temperature with 4% PFA for 20 min. After 3 × 10-min washes in PBS, nonspecific binding was blocked with PBS, 10% normal serum, and 0.2% Triton X-100 for 1 h at room temperature. Coverslips were then incubated with the primary antibody for 2.5 h at room temperature (Supplementary Table S2).
Cells were then rinsed 5 × 5 min with PBS and incubated with a secondary antibody solution containing PBS, 10% normal serum, Alexa Fluor (AF) 594 donkey anti-rabbit (A21207; Invitrogen, Grand Island, NY), Cy3 donkey anti-mouse (715-165-150; Jackson), AF 647 goat anti-rabbit (111-605-045; Jackson), AF 488 donkey anti-goat (705-545-003; Jackson), AF 488 donkey anti-rabbit (711-545-152; Jackson), or AF 488 donkey anti-mouse (A21202; Invitrogen) for 2 h at room temperature.
All secondary antibodies were used at a 1:1,000 dilution. Sections were then counterstained with DAPI to visualize nuclei. Negative controls were obtained by omitting the primary antibodies and using primary isotype controls in the immunostaining procedures (Supplementary Fig. S1). Undifferentiated and nonpatterned cells also served as internal positive and negative controls.
Data analysis
All graphs present mean ± SEM. All gene expression analyses were done using one-way ANOVA and Tukey's multiple comparison test with Prism 5 (GraphPad software, Inc.). Statistical significance was set at α < 0.05. ANOVA results are reported in Supplementary Table S3.
Results
Derivation of common marmoset iPSCs
Adult marmoset fibroblasts were electroporated with episomal plasmids transiently expressing the human pluripotent genes C-MYC, LIN28, KLF4, NANOG, SOX2, and OCT4, to reprogram to an induced pluripotent state (Cj-iPSCs). For direct comparison, we obtained a previously derived marmoset ESC line (Cj-ESC) [17]. Morphologically, both cell lines showed classic tightly packed colonies with defined borders, monolayer growth, and a high nucleus/cytoplasmic ratio.
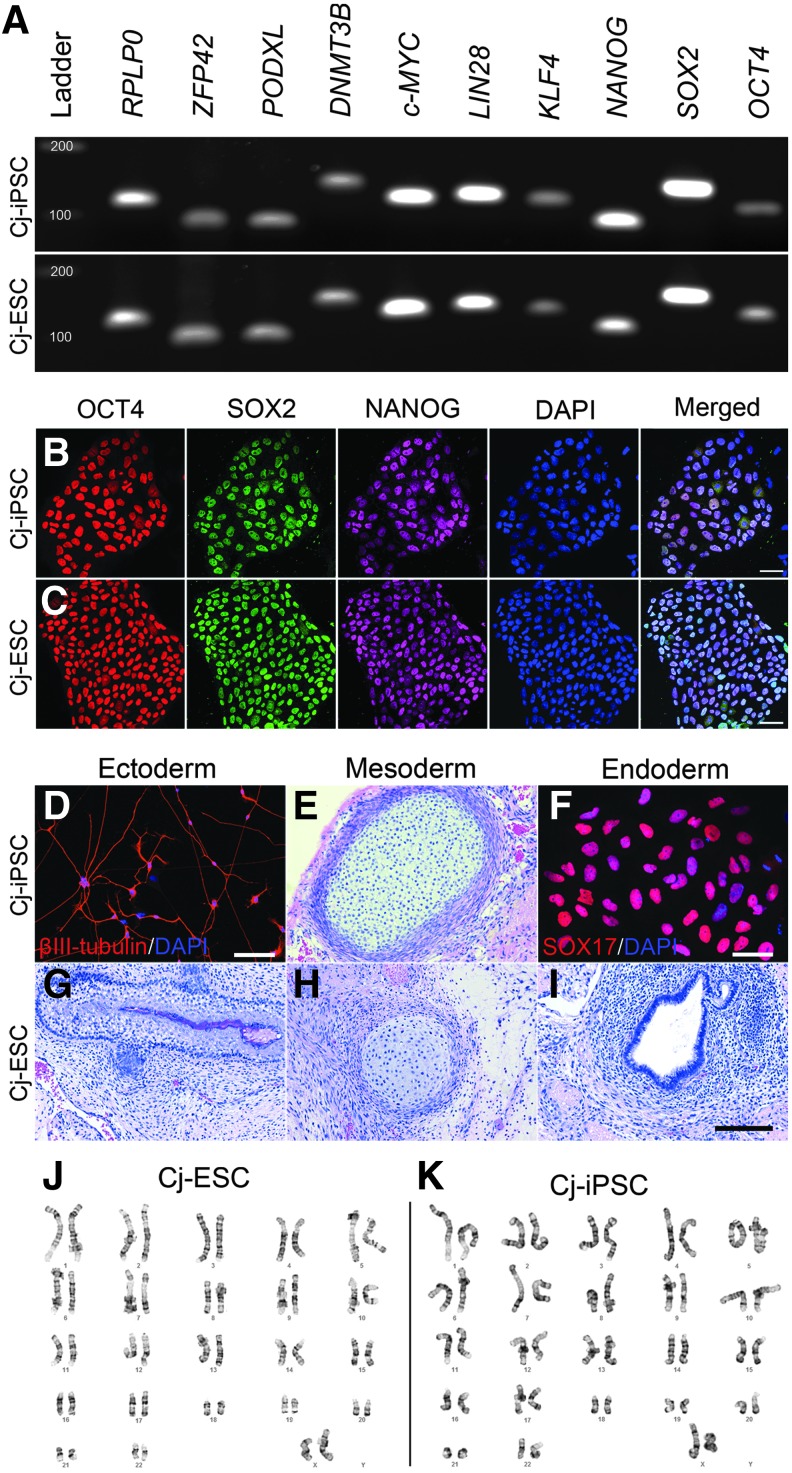
To determine whether the reprogrammed cells expressed endogenous pluripotent genes, exon-spanning primers for RT-PCR were designed. Subsequent Sanger sequencing of OCT4, SOX2, NANOG, KLF4, LIN28, and c-MYC amplicons verified the expression of marmoset pluripotency genes, and not the human episomal reprogramming plasmids. Both the Cj-ESCs and Cj-iPSCs expressed endogenous mRNAs for the pluripotency genes ZFP42, PODXL, DNMT3B, C-MYC, LIN28, KLF4, NANOG, SOX2, and OCT4 (Fig. 1A).
FIG. 1.
Cj-ESCs and Cj-iPSCs are pluripotent. (A) RT-PCR detection of endogenous pluripotent mRNAs; ZFP42, PODXL, DNMT3B, C-MYC, LIN28, KLF4, NANOG, SOX2, and OCT4. RPLP0 was used as a reference gene. (B, C) Both cell lines coexpress OCT4/SOX2/NANOG. After neural differentiation, Cj-iPSCs formed (D) βIII-tubulin-positive neurons. Tumors from the Cj-iPSCs formed (E) mesoderm (cartilage). After directed differentiation to definitive endoderm (F), SOX17 was expressed. Teratomas from Cj-ESCs formed (G) ectoderm (epithelium), (H) mesoderm (cartilage), and (I) endoderm (glands). Both Cj-ESC (J) and Cj-iPSC (K) lines had normal karyotype. Scale bars: (B, C, F), 50 μm; (D, E, G–I), 100 μm. Cj-ESC, Cj-embryonic stem cell; Cj-iPSC, Cj-derived induced pluripotent stem cell; RT-PCR, reverse transcription-PCR.
To verify protein localization, triple immunofluorescent staining for OCT4, SOX2, and NANOG was completed. The results demonstrate that individual cells within the pluripotent colonies coexpressed NANOG, OCT4, and SOX2 transcription factors in both Cj-iPSCs (Fig. 1B) and Cj-ESCs (Fig. 1C).
When injected into the kidney capsule and testis of immunocompromised mice, the Cj-iPSCs (passage 17; 55 days postinoculation) produced tumors containing structures arising from differentiated mesoderm (Fig. 1E); ectoderm and endoderm derivatives were not clearly identified. As a result, to further evaluate the potential to derive ectoderm and endoderm, the Cj-iPSCs were exposed to neuronal differentiation, directed endoderm differentiation, and in vitro spontaneous differentiation protocols (Supplementary Fig. S2). It was confirmed that the Cj-iPSCs could differentiate to both βIII-tubulin-positive neurons (Fig. 1D) and definitive endoderm expressing CXCR4, SOX17, FOXA2, and AFP (Fig. 1F and Supplementary Fig. S2A–F). The Cj-ESCs (passage 23; 56 days postinoculation) produced teratomas expressing typical differentiated derivatives of all three germ layers (Fig. 1G–I), confirming their pluripotent developmental potential. Karyotype analysis showed normal chromosomes for both cell lines (Fig. 1J, K).
Marmoset-derived stem cells differentiate to neurons
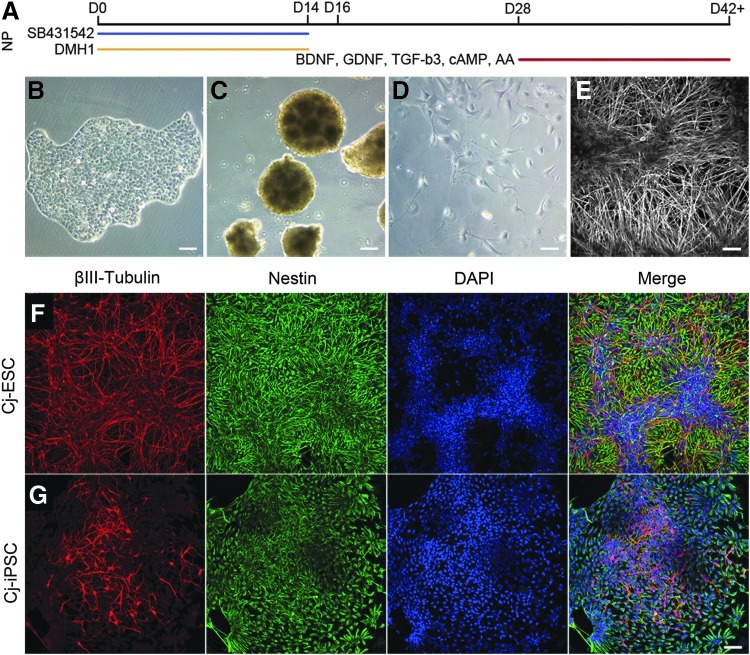
To evaluate the potential of adult marmoset fibroblast-derived pluripotent stem cells to differentiate to neurons, we implemented a basic dual-SMAD inhibition, nonpatterned neural induction protocol using SB431542 and DMH1, followed by growth factor-mediated terminal differentiation using BDNF, GDNF, TGF-β3, as well as cAMP and ascorbic acid (Fig. 2A). Figure 2 illustrates the progression from a representative pluripotent stem cell colony (Fig. 2B) to suspended neurospheres after lifting of colonies from the culture surface (Fig. 2C), dissociated cells plated for terminal differentiation (Fig. 2D), and extensive process extension after continued adherent culture (Fig. 2E). ICC revealed that nearly all plated cells expressed nestin, an intermediate filament protein expressed in neural progenitor cells, βIII-tubulin, a microtubule protein specific to neurons, or both (Cj-ESC, Fig. 2F; Cj-iPSCs, Fig. 2G).
FIG. 2.
Neural differentiation of marmoset stem cells. (A) Schematic for NP dual-SMAD inhibition protocol using SB431542 and DMH1. Representative images throughout neural differentiation showing (B) pluripotent cell colonies, d0–8; (C) neurospheres, d8–28; (D) plated dissociated neurospheres, d30; (E) extensive process extension, d42+. The Cj-ESC (F) and Cj-iPSC (G) lines were efficiently differentiated to neurons shown by nestin (green) and βIII-tubulin (red) expression; DAPI (blue). Scale bars: (B–G), 100 μm. NP, nonpatterned.
Characterization of gene expression throughout neural differentiation
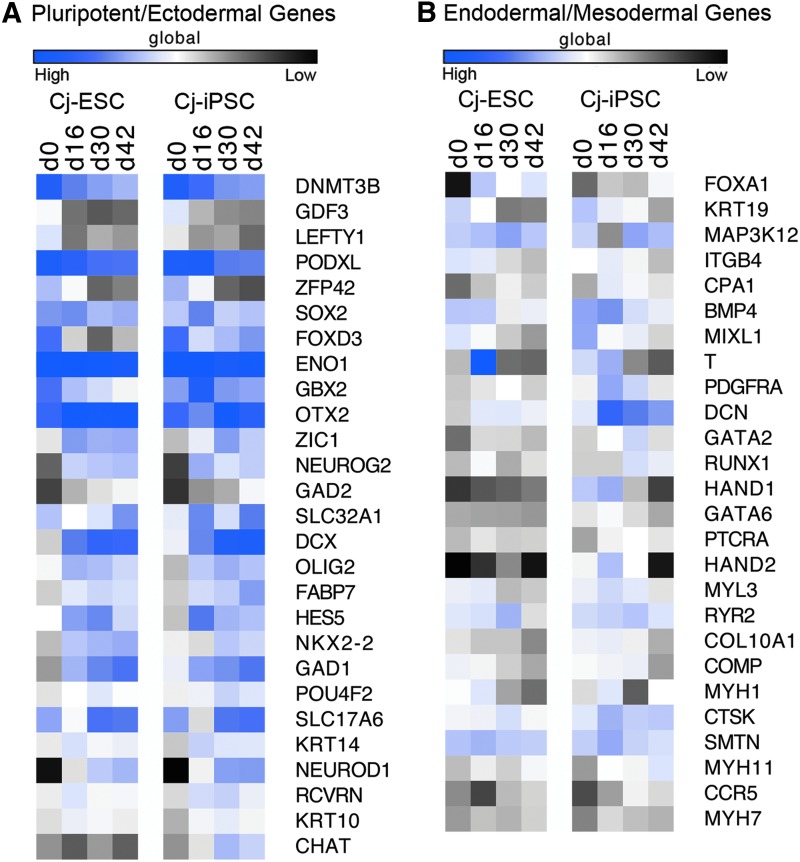
An RT2 human cell lineage qPCR profiler array was first used to generally characterize gene expression in the marmoset pluripotent cells. Data from mRNA samples collected on day 0 (d0), d16, d30, and d42 of neural differentiation in both Cj-ESC and Cj-iPSC lines are represented by respective Ct values (n = 1) integrated as a global heat map (Fig. 3). Data from the profiler array from d0 to d42 showed that the pluripotency genes DNMT3B, GDF3, LEFTY1, PODXL, and ZFP42 are downregulated during differentiation in both Cj-ESCs and Cj-iPSCs. The ectodermal germ layer genes OTX2, ZIC1, and NEUROG2 were upregulated, while FOXD3 and GBX2 were either downregulated or maintained (GBX2, Cj-iPSCs). Both lines showed maintenance of the ectodermal progenitor genes SOX2 and ENO1 and upregulation of GAD2, SLC32A1, DCX, OLIG2, FABP7, HES5, and NKX2.2. There was also upregulation of ectodermal terminal differentiation genes GAD1, SLC17A6, NEUROD1, and CHAT (Cj-iPSCs only), but little to no change of expression of POU4F2, KRT14, RCVRN, or KRT10.
FIG. 3.
RT2 Profiler Cell Lineage Array in Cj-ESCs and Cj-iPSCs throughout neural differentiation. (A) Global heat map of pluripotent and ectodermal lineage-related genes. (B) Global heat map of endoderm and mesoderm lineage-related genes. Blue indicates higher levels of expression compared to black.
For nonectodermal-related genes, both cell lines had a slight upregulation of the endodermal germ layer gene FOXA1 and downregulation of the endoderm progenitor gene KRT19. There was no change in the Cj-ESCs of germline mesoderm genes T, PDGFRA, DCN, GATA2, RUNX1, HAND1, and GATA6, mesoderm progenitor genes PTCRA and HAND2, or the mesoderm terminal differentiation genes MYL3, RYR2, COL10A1, COMP, MYH1, CTSK, SMTN, MYH11, CCR5, and MYH7. However, both cell lines showed downregulation of BMP4 and MIXL1. In addition, the Cj-iPSCs showed downregulation of T, HAND1, and HAND2 and upregulation of DCN. Overall, both cell lines appear relatively similar in gene expression profile, with the exception of some nonectodermal genes in the Cj-iPSCs.
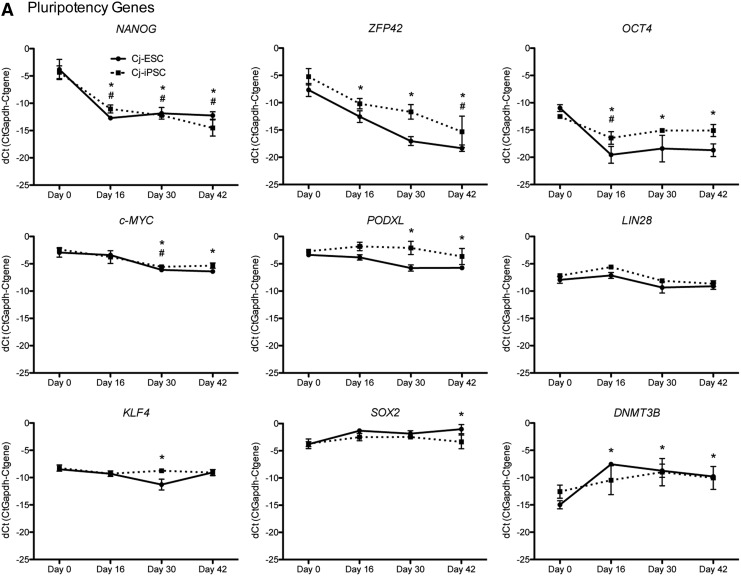
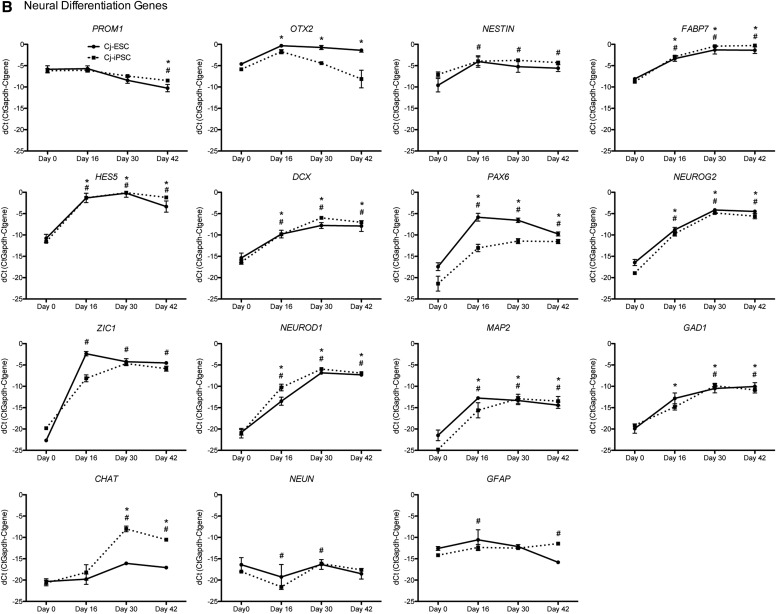
To verify the relative quantitative expression differences of genes across differentiation time points (n = 3 per time point), selected genes from the array (DNMT3B, PODXL, ZFP42, SOX2, OTX2, ZIC1, NEUROG2, DCX, FABP7, HES5, GAD1, NEUROD1, and CHAT) and additional pluripotency (OCT4, NANOG, KLF4, C-MYC, and LIN28), neural progenitor (PROM1, NESTIN, and PAX6), and neuronal/glial differentiation (MAP2, NEUN, and GFAP) genes were analyzed (Fig. 4). Relatively high expression of the pluripotency genes (Fig. 4A) NANOG, ZFP42, and OCT4 was observed at d0 followed by a decrease in expression beginning at d16. While C-MYC appeared to decrease slightly over time, PODXL, LIN28, and KLF4 remained relatively constant throughout differentiation and the neural stem cell gene SOX2 increased slightly along with DNMT3B.
FIG. 4.
qRT-PCR of pluripotent and neural differentiation genes. (A) Pluripotent genes: in both Cj-ESCs and Cj-iPSCs, pluripotent genes NANOG, ZFP42, and OCT4 decrease significantly throughout differentiation, while c-MYC, PODXL, LIN28, KLF4, SOX2, and DNMT3B present very slight deviations from d0 levels. (B) Neural differentiation genes: Cj-ESCs and Cj-iPSCs present similar gene profiles for PROM1, NESTIN, FABP7, HES5, DCX, NEUROG2, ZIC1, NEUROD1, MAP2, GAD1, NEUN, and GFAP with each gene either increasing or presenting no change. The Cj-iPSCs presented less PAX6 until d42 when the expression in the Cj-ESCs was reduced to a similar level. The Cj-iPSCs also had decreased OTX2 expression beginning at d30, which continued to d42. CHAT expression was increased in the Cj-iPSCs beginning at d30, which was not observed in Cj-ESCs. *, #P < 0.05 for Cj-ESCs and Cj-iPSCs, respectively, when compared to day 0. For full ANOVA results, see Supplementary Table S2.
The neural gene PROM1 decreased slightly, while OTX2, NESTIN, FABP7, HES5, DCX, PAX6, NEUROG2, ZIC1, NEUROD1, MAP2, and GAD1 increased in expression in both Cj-ESCs and Cj-iPSCs starting at d16 and were either maintained or further increased through d42 (Fig. 4B). However, OTX2 in the Cj-iPSCs decreased on d30 and d42 and PAX6 in the Cj-ESCs also decreased slightly between d16 and d42. CHAT expression increased in the CJ-iPSCs at d30, but not in the Cj-ESCs matching the observation from the profiler data. There was no increase in expression of the astrocytic marker GFAP.
Patterning floor plate-derived midbrain dopaminergic neurons
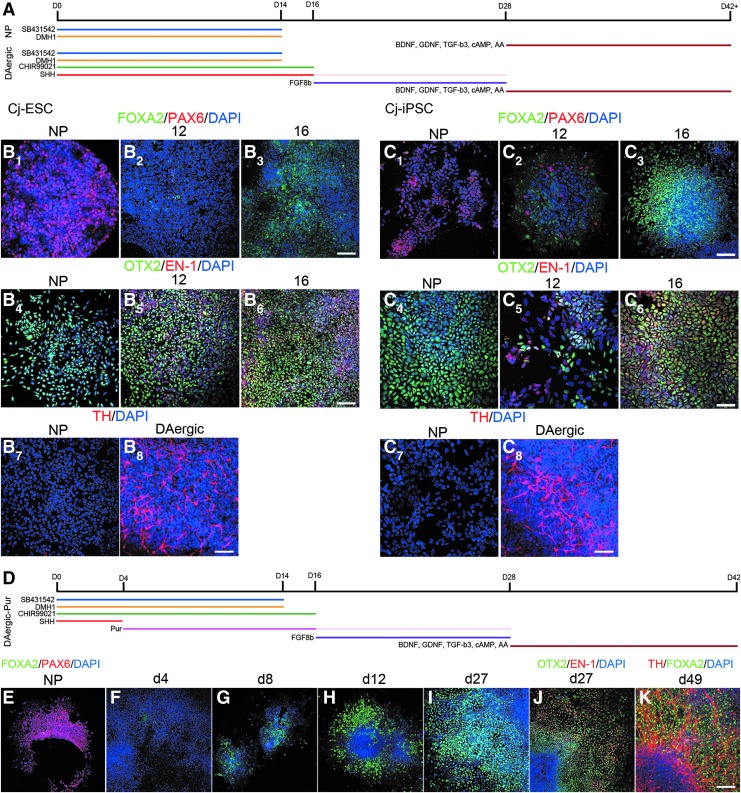
Cells exposed to a combination of the ventralizing morphogen SHH and the small-molecule CHIR99021, in addition to SB431542 and DMH1 (Fig. 5A), express regional markers representative of floor plate midbrain progenitor cells. Cj-ESC and Cj-iPSC cultures were patterned using an adapted protocol developed for human and rhesus cells [15].
FIG. 5.
Patterning floor plate-derived midbrain dopaminergic neurons. (A) Time line illustrating the protocol for basic NP dual-SMAD differentiation and DAergic patterning (DAergic). Expression of Cj-ESC (B) and Cj-iPSC (C) markers after 12 or 16 days of patterning. The nonpatterned controls default to a dorsal forebrain phenotype judged by PAX6+/FOXA2− (B1, C1) and OTX2+/EN1− (B4, C4). Sufficient ventralization was observed after 16 days of patterning compared to 12 days in both cell lines (B2,3, C2,3). Posteriorization to a midbrain fate was sufficient and maintained after 12 and 16 days of patterning, respectively, in both lines (B5,6, C5,6). The Cj-ESC and Cj-iPSC lines only produced TH+ neurons after the DAergic protocol (B7,8, C7,8). Replacing SHH with the small-molecule purmorphamine from day 4 to 28 (E) led to a visible increase in FOXA2 expression by day 8 and 12 with maintenance after day 27, with no change in posteriorization patterning (F–J). FOXA2 was maintained after 49 days and observed in TH+ neuronal nuclei (K). Scale bars: B1, C4–6, 50 μm; B2–8, C1–3,7,8 (E–K), 100 μm.
To derive floor plate progenitors, SHH was used at a high concentration (500 ng/mL) for 12 or 16 days to mimic ventralization observed in the mammalian neural tube development. This induced the floor plate marker FOXA2 expression, while repressing the more dorsal-expressing transcription factor PAX6 most efficiently in cells that received SHH for 16 days (Fig. 5: Cj-ESCs, B1–B3; Cj-iPSCs C1–C3).
Midbrain progenitors were simultaneously patterned using a posteriorizing small-molecule CHIR99021 for 12 or 16 days. Cells that received CHIR99021 coexpressed the midbrain/forebrain marker OTX2 with the midbrain/hindbrain marker EN1 indicative of midbrain derivation by 12 days (Fig. 5: Cj-ESCs, B4–B6; Cj-iPSCs C4–C6). The addition of FGF8b (100 ng/mL) from day 16 to 28 followed by 2 weeks of plated differentiation media resulted in tyrosine hydroxylase (TH) expressing neurons. DAergic neurons were present in both Cj-ESCs (Fig. 5B7, B8) and Cj-iPSCs (Fig. 5C7, C8) that had been patterned.
Ventralization with the small-molecule purmorphamine beginning at day 4 (Fig. 5D) led to similar results, but FOXA2 was observed to be expressed starting at day 8 with relatively higher efficiency by day 12 (Fig. 5E–H). At day 27, regional markers were analyzed and both ventralization (Fig. 5I) and posteriorization (Fig. 5J) were sufficient. By day 49, DAergic neurons were present with maintenance of FOXA2 expression (Fig. 5K).
Discussion
The data presented in this article demonstrate that adult common marmoset skin fibroblasts can be reprogrammed to cells displaying the properties of iPSCs. Furthermore, both Cj-ESCs and Cj-iPSCs can be efficiently differentiated to neurons as well as patterned to have a floor plate-derived midbrain dopaminergic phenotype. Thus, it is feasible to derive marmoset iPSCs for in vitro experiments on neural differentiation, as well as cell lines for new approaches using marmoset models for regenerative medicine research.
The reprogramming of adult marmoset fibroblasts presented visible colonies developing by 17 days after electroporation of the episomal expression plasmids. One difficulty encountered was the expansion of adult-derived fibroblasts before electroporation. Compared to fetal-derived cells, the growth rate of adult-derived fibroblasts is often slower. Interestingly, however, the reprogrammed Cj-iPSCs grow at a comparable rate to the Cj-ESCs, overcoming the slower expansion time of the parental fibroblasts. Although the Cj-ESCs produced teratomas, the Cj-iPSCs did not generate discernable tissues of all three germ layers. However, further in vitro differentiation of Cj-iPSCs into ectoderm and endoderm lineages validated the pluripotency of the cell line. The differentiation of the marmoset cells using the dual-SMAD inhibition protocol was very efficient and easily replicated.
Assessment of gene expression in the Cj-ESCs and Cj-iPSCs demonstrated very similar changes in pluripotent and neural differentiation profiles throughout the 42-day dual-SMAD inhibition protocol. The human cell lineage RT2-Profiler results suggested that both marmoset cell lines are similar with the exception of some nonectodermal gene expression in the Cj-iPSCs. After further qPCR analysis, similarity between lines was confirmed, although differences were seen with most notable examples in OTX2, PAX6, and CHAT (Fig. 4).
The patterning protocol for generation of DAergic neurons previously used for human and rhesus cells [15] needed to be adapted for the marmoset cells. Initial testing using 12 days of incubation with SHH showed insufficient ventralization judged by the loss of PAX6, but lack of FOXA2 expression. After increasing the length of exposure to 16 days, FOXA2 expression was elevated compared to the 12-day exposure protocol. The response after 16 days is similar to that found in the rhesus cell time line, while human cells were adequately ventralized by 12 days. Whereas posteriorizing with CHIR99021 for 12 days in the marmoset cells was sufficient, comparable to the human cell protocol.
Substituting SHH for the small-molecule purmorphamine, an SHH agonist, at 0.5 μM for only days 4–16 further optimized ventralization assessed by FOXA2 expression after just 12 days of exposure. FOXA2+/PAX6− colonies were evident by day 8. The first TH-positive cells were visualized on day 42. Maintaining the cells for an additional 7–14 days increased the abundance of TH-positive neurons.
One concern of using the suspended neurosphere method is nonhomogenous diffusion of morphogens and small molecules to cells within the spheres. Although patterning using the suspension protocol from day 8 to 28 was observed to lead to a rather homogeneous expression of intended regional markers (ie, FOXA2 and EN-1), it is likely that keeping the cells in a monolayer for as long as 12–16 days during initial patterning may minimize any reagent diffusion differences, yielding higher gene conversion efficiencies and potentially reducing the length of time required for the introduction of the ventralizing and posteriorizing factors. However, for long-term culture, the neurosphere method with intermittent trituration seems ideal for maintaining an expandable neuroprogenitor population.
Although this proof of principle study demonstrates that both Cj-ESCs and Cj-iPSCs can be patterned to DAergic neurons, follow-up studies are warranted to optimize the protocol and quantify efficiency using additional reprogrammed lines. In addition, verification of dopamine release and mature electrophysiological properties would need to be demonstrated before utilization in cell replacement strategies.
Our results on the pluripotency and differentiation potential of marmoset-derived stem cells further validate the common marmoset as a viable species for regenerative medicine and disease modeling purposes. The successful application of patterning floor plate-derived dopaminergic neurons from adult marmoset fibroblast-derived Cj-iPSCs is critical to support Parkinson's disease-related studies.
Supplementary Material
Acknowledgments
This research was supported by NIH grants R24OD019803 to TGG and MEE, P51OD011106 to the WNPRC, pilot funding from UL1TR000427 (ICTR UW-Madison, Clinical and Translational Science Award), the NINDS T32GM750738 neuroscience training program (S.C.V.), and bridge funding from the UW-Madison Office of the Vice Chancellor for Research and Graduate Education. The authors acknowledge Dr. Ruth Sullivan and Troy Thoong for their assistance with teratoma analysis.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Takahashi K, Okita K, Nakagawa M. and Yamanaka S. (2007). Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2:3081–3089 [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 4.Schultz-Darken N, Braun KM. and Emborg ME. (2016). Neurobehavioral development of common marmoset monkeys. Dev Psychobiol 58:141–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada H, Okada Y, Ibata K, Ebise H, Ota S, Tomioka I, Nomura T, Maeda T, Kohda K, et al. (2012). Efficient derivation of multipotent neural stem/progenitor cells from non-human primate embryonic stem cells. PLoS One 7:e49469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Zhang Y, Mishra A, Tardif SD. and Hornsby PJ. (2010). Generation of induced pluripotent stem cells from newborn marmoset skin fibroblasts. Stem Cell Res 4:180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farnsworth SL, Qiu Z, Mishra A. and Hornsby PJ. (2013). Directed neural differentiation of induced pluripotent stem cells from non-human primates. Exp Biol Med (Maywood) 238:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu Z, Mishra A, Li M, Farnsworth SL, Guerra B, Lanford RE. and Hornsby PJ. (2015). Marmoset induced pluripotent stem cells: Robust neural differentiation following pretreatment with dimethyl sulfoxide. Stem Cell Res 15:141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomioka I, Maeda T, Shimada H, Kawai K, Okada Y, Igarashi H, Oiwa R, Iwasaki T, Aoki M, et al. (2010). Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors, including Lin28. Genes Cells 15:959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debowski K, Warthemann R, Lentes J, Salinas-Riester G, Dressel R, Langenstroth D, Gromoll J, Sasaki E. and Behr R. (2015). Non-viral generation of marmoset monkey iPS cells by a six-factor-in-one-vector approach. PLoS One 10:e0118424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiedemann A, Hemmer K, Bernemann I, Gohring G, Pogozhykh O, Figueiredo C, Glage S, Schambach A, Schwamborn JC, Blasczyk R. and Muller T. (2012). Induced pluripotent stem cells generated from adult bone marrow-derived cells of the nonhuman primate (Callithrix jacchus) using a novel quad-cistronic and excisable lentiviral vector. Cell Reprogram 14:485–496 [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z, Kohda K, Ibata K, Kohyama J, Akamatsu W, Yuzaki M, Okano HJ, Sasaki E. and Okano H. (2014). Reprogramming non-human primate somatic cells into functional neuronal cells by defined factors. Mol Brain 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, et al. (2009). piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin and Thomson JA. (2009). Human induced pluripotent stem cells free of vector and transgene sequences. Science 324:797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xi J, Liu Y, Liu H, Chen H, Emborg ME. and Zhang SC. (2012). Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells 30:1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howden SE, Maufort JP, Duffin BM, Elefanty AG, Stanley EG. and Thomson JA. (2015). Simultaneous reprogramming and gene correction of patient fibroblasts. Stem Cell Rep 5:1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP. and Hearn JP. (1996). Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol Reprod 55:254–259 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.