Abstract
Background
Chagas disease is a complex anthropozoonosis with distinct domestic and sylvatic mammal species acting as potential reservoirs. The diversity of vector species and their habitats are among the factors that hinder the control of the disease. Control programs periodically monitor the prevalence of T. cruzi infection in insect bugs through microscopical observation of diluted feces. However, microscopy presents limited sensitivity in samples with low parasite numbers, difficulties in examining all evolutionary stages of the insect and may in turn be limited to differentiate T. cruzi from other morphologically similar trypanosomatids. Here, we report two highly sensitive and accurate methodologies to infer T. cruzi infection rates and to quantify parasite load in the gut of field-collected triatomines.
Methods
Triatomines were manually collected in the period 2011–2012 and 2014–2015, in domestic, peridomestic or sylvatic habitats in rural areas of 26 municipalities, encompassing three distinct Brazilian biomes: Caatinga, Cerrado and Atlantic Rainforest. Following morphological and taxonomical identification, the search for flagellated protozoa was performed by optical microscopy. A conventional PCR targeting T. cruzi kDNA and a TaqMan qPCR directed to the parasite nuclear satellite DNA (SAT) were developed, both in multiplex, with the triatomine 12S subunit ribosomal RNA gene, used as internal amplification control. Both methods were used for detection (kDNA-PCR) and parasite load quantification (SAT-DNA-qPCR), to investigate T. cruzi infection in captured triatomines.
Results
The combined methods were assayed on a panel of 205 field-collected triatomine samples. Diagnostic analysis revealed 21% positivity for the kDNA-PCR, whereas microscopic examination enabled identification of T. cruzi in only 7.0% of the PCR-positive samples. Negative PCR results were confirmed by the absence of T. cruzi flagellates using microscopy. Caatinga biome yielded the highest T. cruzi infection rate (60%), followed by the Atlantic Rainforest and Cerrado with 7.1 and 6.1%, respectively. In addition, a wide range distribution of parasite load, varying from 8.05 × 10-2 to 6.31 × 1010 was observed with a median of 2.29 × 103 T. cruzi/intestine units. When parasite load was analyzed by triatomine species, a significantly higher median was found for Panstrongylus lutzi in comparison with Triatoma brasiliensis.
Conclusions
Our results demonstrate highly sensitive PCR-based methodologies to monitor T. cruzi infection in triatomines. In addition, the qPCR assay offers the possibility of further evaluation parasite load, as a promising biomarker of the vectorial capacity of triatomines in Chagas disease endemic areas.
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2343-x) contains supplementary material, which is available to authorized users.
Keywords: Trypanosoma cruzi, Multiplex PCR, Real-time qPCR, Triatomines, Natural infection, Parasite load
Background
Chagas disease (ChD) is an important neglected tropical illness caused by the flagellate protozoan Trypanosoma cruzi (Kinetoplastida: Trypanosomatidae). The protozoan is primarily transmitted by insects belonging to the subfamily Triatominae (Hemiptera: Reduviidae), including 18 genera and 151 species [1]. The disease is a complex anthropozoonosis with distinct domestic and sylvatic mammal species acting as potential reservoirs of the infection. Although considered neglected, ChD is endemic in South and Central Americas, and in some regions of the USA [2]. The disease has globally expanded, with more than 5 million people infected and approximately 70 million living at risk [3], and is now reported in the Americas, Europe, Australia and Asia due to the migration of infected individuals from endemic parts of the world [4].
The classical transmission form to humans, via triatomine feces, is correlated to the presence of T. cruzi infected bugs in domestic and/or peridomestic areas. The diversity of vector species and their habitats are among the factors that hinder the control of the disease. The importance of three genera, Triatoma, Rhodnius and Panstrongylus, lies in the fact that some of their members feed on humans and synanthropic mammals and may thus transmit T. cruzi [5]. In Brazil, four triatomine species play a direct role in the epidemiology of the disease: Triatoma brasiliensis Neiva, 1911; Panstrongylus megistus (Burmeister, 1835); Triatoma pseudomaculata Corrêa & Espínola, 1964 and Triatoma sordida (Stål, 1859). These species are found in sylvatic habitats despite being adapted to domestic and peridomestic areas. Other species identified in wild ecotopes, such as Triatoma costalimai Verano & Galvão, 1958; Triatoma wygodzinskyi Lent, 1951; Rhodnius neglectus Lent, 1954 and Triatoma rubrovaria (Blanchard, 1843), maintain an enzootic cycle involving wild mammals in a variety of terrestrial or arboreal biotopes [6]. In addition, Rhodnius brethesi Matta, 1919 has been reported as an active vector in the Amazon Region, considering the rates of T. cruzi transmission to people involved in plant extraction activities [7].
The risk of T. cruzi transmission in endemic areas mainly depends on the density of triatomine bugs and the prevalence of T. cruzi infection in insect vectors, humans and animal reservoirs [8]. The impact in reducing transmission by the vectors Triatoma infestans (Klug, 1834) and Rhodnius prolixus (Stål, 1859) has been observed particularly in the Southern Cone countries of South America and in Central America, respectively [9, 10]. Transmission prevention programs include insecticide-based campaigns, housing improvements, health education and blood donor screening [9]. Yet, the wide use of insecticides has created resistant triatomine populations [11–13].
The screening of T. cruzi infection in field-collected triatomines from different regions of Brazil is relevant for a better understanding of ChD epidemiology and for giving support to developing programs to control disease spreading. Furthermore, information regarding T. cruzi parasite load in triatomines could bring an important contribution to infer potential vectorial capacity of distinct species. Control programs periodically monitor the prevalence of T. cruzi infection in insect bugs through optical microscopy observation of diluted feces in the search for active trypanosomes. Although this method is inexpensive and widely used, it presents limited sensitivity in samples with low parasite numbers [14], difficulties in examining all evolutionary stages of the insect and may in turn be limited to differentiate T. cruzi from other morphologically similar trypanosomatids. In addition, it is a laborious and time-consuming procedure, where fresh insect examination is needed to detect live parasites [15].
Our group reported, for the first time, a standardized and accurate SYBR-Green qPCR assay to quantify parasite load levels in R. prolixus midgut and rectum, to investigate the role of cruzipain in the interaction of T. cruzi with the triatomine host [16]. In the present study, we propose a combination of two multiplex PCR-based assays (conventional and TaqMan real-time qPCR), as molecular diagnostic tools for the accurate detection and quantification of T. cruzi DNA in triatomines. The conventional PCR (qualitative) and qPCR (quantitative) were based on the parasite targets kinetoplast DNA or kDNA [17] and nuclear satellite DNA [18], respectively, together with the triatomine 12S subunit ribosomal RNA gene, as an internal control, in both assays. The generated data could bring more insight to investigate the association between PCR positivity and parasite load in field-collected triatomines and the risk for T. cruzi transmission to humans in endemic areas of Chagas disease.
Methods
Parasite culture and R. prolixus maintenance
Trypanosoma cruzi CL Brener (COLPROT 005), Dm28c (COLPROT 0010), INPA 4167 (COLPROT 0607) and Trypanosoma rangeli (Macias strain - COLPROT 0273) were obtained from the Coleção de Protozoários da Fundação Oswaldo Cruz, Rio de Janeiro, Brazil (Fiocruz, COLPROT, http://www.colprot.fiocruz.br). Epimastigote forms were grown in 3.7% BHI medium, containing 0.002% hemin and supplemented with 10% heat-inactivated FBS, at 28 °C for 4 days to reach late-log growth phase. Parasites were harvested by centrifugation (1500× g for 5 min at 25 °C), washed three times with 0.15 M NaCl, 0.01 M phosphate buffer pH 7.2 (PBS), prior to DNA extraction.
Rhodnius prolixus specimens were reared and maintained with controlled temperature and humidity conditions, as previously described [19]. The insects were obtained from the insectary of the Laboratório de Bioquímica e Fisiologia de Insetos, Instituto Oswaldo Cruz (Fiocruz), Rio de Janeiro, Brazil.
Field-collected triatomines
Insects were captured in the period 2011–2012 and 2014–2015, in 26 municipalities distributed in three Brazilian biomes (Caatinga, Cerrado and Atlantic Rainforest), where each one constitutes idiosyncratic landscape mosaics (Fig. 1). Caatinga, a biome of northeastern Brazil, is characterized by a semiarid climate with vegetation represented by a mosaic of xerophytic, deciduous, thorny scrub, with tree coverage lower than 60%. Cerrado biome is a heterogeneous, floristic savannah that covers more than 2 million km2 and extends from central Brazil to parts of Bolivia and Paraguay, representing the second largest South American biome. Atlantic Rainforest is formed by a set of forest formations and associated ecosystems such as salt marshes, mangroves and high fields, occupying an area c.13% of the country [20].
Fig. 1.
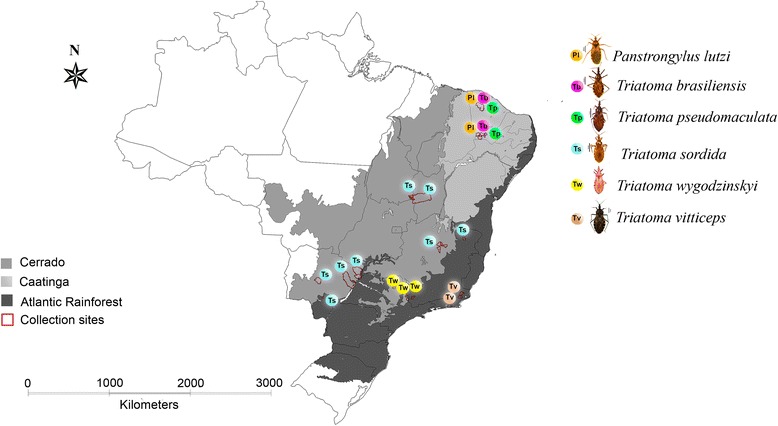
Representative map of Brazil indicating the distribution of triatomine collection sites and the studied biomes
Householders were informed of the aim of the research project, anticipated benefits and potential participation. Each environment was georeferenced and potential ecotopes, including dwellings and rock-piles, under tree bark and nests of rodents or birds, were investigated for the presence of triatomines. Manual collections with the use of forceps were carried out in both wild and dwellings sites. All captured bugs were placed in labelled plastic bags with folded filter paper inside and transported to the Laboratório Interdisciplinar de Vigilância Entomológica de Diptera e Hemiptera, Instituto Oswaldo Cruz (Fiocruz), Rio de Janeiro, Brazil, where they were identified to species [21, 22] and stage and collection site, followed by the search of flagellated protozoa by optical microscopy. Nymph instars were classified according to the identification of the adult stages found in the colony.
Optical microscopy
All live third-, fourth- and fifth-instar nymphs and adult triatomines were examined for T. cruzi infection by microscopic examination (ME) of fecal samples. One fecal drop from each live bug, obtained by abdominal compression, was diluted in one drop of sterile saline solution, NaCl 0.85% (c.50 μl) on a glass slide and examined fresh by microscopy, for active trypanosomes, at magnifications of 200–400×. For dead insects, midgut and rectum were dissected and soaked in a sterile Petri dish containing 2 ml saline solution. The homogenate (50 μl) was observed between slide and coverslip for T. cruzi search by optical microscopy. Forceps were rinsed in 10% bleach and 70% ethanol between extracting successive samples.
DNA extraction for molecular assays
Following ME, live insects used for molecular studies were placed in absolute ethanol until being dissected under a stereomicroscope to collect midgut and rectum. For dead insects, the intestinal content homogenates previously collected for ME were stored in sterile microtubes at 4 °C for DNA extraction. Non-infected R. prolixus specimens maintained in insectary were used as negative controls during DNA extraction of field-collected triatomines. Samples were pretreated for 2 h at 56 °C with 50 μl lysis buffer containing 10 mM Tris-HCl (pH 9.2), 1 mM EDTA and 150 μg/ml proteinase K (Sigma-Aldrich, St. Louis, MO, USA). After treatment, samples were centrifuged (9800× g, 15 min, 4 °C) and the supernatant was collected and diluted in TE buffer (1 mM Tris-HCl pH 9.2, 1 mM EDTA) to reach 200 μl total volume. DNA was purified from the lysate using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) with slight modifications [23]. Briefly, at the elution step, 100 μl AE buffer were added to the silica-membrane column and maintained at room temperature for 10 min, before DNA elution. DNAs were stored at -20 °C until use, and their purity and concentration were determined using a Nanodrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA) at 260/280 and 260/230 nm.
Multiplex conventional PCR (cPCR)
Conventional PCR assays were carried out in a final volume of 50 μl, containing: 5 μl DNA (20–25 ng), 5 μl 10× Taq Platinum buffer, 0.2 mM dNTPs, 4.5 mM MgCl2, 1.25 U Taq Platinum DNA polymerase (Life Technologies, Carlsbad, CA, USA), 200 nM 121/122 primers (T. cruzi kDNA) [17, 24] and 100 nM P2B/P6R primers (triatomine 12S rRNA gene). Trypanosoma cruzi and triatomine primers are described in Table 1. Amplifications were performed in the GeneAmp PCR System 9700 (Life Technologies), as follows: 94 °C for 12 min; 36 cycles at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 10 min. PCR products were loaded onto 2% (w/v) agarose gels and submitted to electrophoresis at 80 V for 40 min. Gels were stained with a Nancy-520 fluorescent dye (Sigma-Aldrich). This procedure allows for discrimination between T. cruzi and T. rangeli, and each PCR reaction was run with a positive control for T. cruzi and T. rangeli epimastigotes, and a reagent negative control (without DNA). In addition, a PCR multiplex positive control formed by mixing 1:1 DNA from one dissected midgut of non-infected adult R. prolixus with DNA of T. cruzi CL Brener-epimastigotes (102 cells) was used. The sensitivity and specificity of microscopy for detecting T. cruzi infection in triatomines were calculated using PCR as the gold standard.
Table 1.
Primer and probe sequences for cPCR and qPCR assays
| Target | Primer/Probes | Amplicon size (bp) | Sequence (5′–3′) | Reference |
|---|---|---|---|---|
| T. cruzi kinetoplast DNA (kDNA) | 121 (Forward) | 330 | AAATAATGTACGGG(T/G)GAGATGCATGA | [17, 24] |
| 122 (Reverse) | GGTTCGATTGGGGTTGGTGA ATATA | |||
| T. cruzi satellite DNA (SAT-DNA) | Cruzi 1 (Forward) | 165 | ASTCGGCTGATCGTTTTCGA | [18] |
| Cruzi 2 (Reverse) | AATTCCTCCAAGCAGCGGATA | |||
| Cruzi 3 (Probe) | FAM-TTGGTGTCCAGTGTGTG-NFQ-MGB | |||
| Triatomine 12S rRNA | P2B (Forward) | 163 | AAAGAATTTGGCGGTAATTTAGTCT | Present study |
| P6R (Reverse) | GCTGCACCTTGACCTGACATT | |||
| Triat (Probe) | VIC-TCAGAGGAATCTGCCCTGTA-NFQ-MGB |
Quantitative multiplex real-time PCR (qPCR)
The qPCR reactions were carried out in a final volume of 20 μl, containing 2 μl DNA (8–10 ng), 2× TaqMan® Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA), 600 nM cruzi1/cruzi2 primers and 250 nM Cruzi3 probe (FAM/NFQ-MGB) targeting T. cruzi nuclear satellite DNA (SAT-DNA), 300 nM P2B primer, 500 nM P6R primer and 150 nM Triat Probe (VIC/NFQ-MGB) (Applied Biosystems) directed to the 12S ribosomal subunit gene of triatomines. Sequences of both sets of primers and probes are presented in Table 1. The cycling conditions were as follows: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 58 °C for 1 min. Amplifications were performed in the ABI Prism 7500 Fast (Applied Biosystems). Standard calibration curves for T. cruzi and triatomine targets were constructed by serially diluting total DNA obtained from non-infected triatomine intestine samples (adults R. prolixus from insectary) spiked with 105 T. cruzi epimastigotes (Dm28c clone, TcI). The resulting DNA was serially diluted to a range of 105 to 0.5 T. cruzi equivalents and 5 to 0.002 triatomine intestine unit equivalents.
To determine the limit of detection (LOD) for T. cruzi DNA in triatomines, intestine samples were spiked with 20 to 0.00128 parasite equivalents and assayed in 12 replicates for three consecutive days. The LOD was calculated as the lowest parasitic load corresponding to ≥ 95% of PCR positive results, according to clinical and laboratory standard guidelines [25].
Statistical analysis
All qPCR assays were performed in experimental duplicate. Results were expressed as the mean of T. cruzi/intestine units. Statistical analysis of parasite load between triatomine species (Student’s t-test or Mann-Whitney rank sum test, and Analysis of Variance, ANOVA) were performed with SigmaPlot v12.0 software (Systat Software, Inc). The LOD of the real-time qPCR was determined by Probit regression analysis, using Minitab 15 Statistical Software (Minitab Inc., State College, PA).
Results
The main goal of this study is the development of PCR approaches to infer T. cruzi infection rates and parasite load in field-collected triatomines from Chagas disease endemic areas. A conventional multiplex PCR (cPCR) was developed targeting T. cruzi kinetoplast DNA minicircles (kDNA) and triatomine 12S rRNA gene, used as an internal amplification control, for the screening of T. cruzi DNA in triatomine samples and enabling its differentiation from T. rangeli. Further, we propose a quantitative TaqMan-based multiplex real-time PCR (qPCR), targeting T. cruzi nuclear satellite DNA (SAT-DNA - FAM/NFQ-MGB Probe) and triatomine 12S rRNA gene (VIC/NFQ-MGB Probe), to estimate the normalized parasite load, according to the DNA amount recovered from field-collected triatomine samples.
Taking into account the diversity of T. cruzi lineages and triatomine species circulating in Brazilian endemic areas, we first tested the multiplex cPCR with reconstituted samples consisted of DNA obtained from dissected midgut of R. prolixus specimens, maintained in insectary, mixed 1:1 with DNA extracted from 102 T. cruzi epimastigotes of the strains/clones Dm28c (TcI), INPA 4167 (TcIV) and CL Brener (TcVI) (Fig. 2a). The amplification of the variable regions of minicircles kDNA was observed in all tested T. cruzi strains/clones (330 bp fragment) and T. rangeli (330, 360 and 760 bp fragments), allowing the differentiation between both species. In the duplex PCR, we amplified successfully the 12S rRNA gene of triatomines (163 bp) in the same samples. In parallel, the performance of the new primers designed for the subfamily Triatominae, used as an internal amplification control, was assayed using distinct triatomine species not infected with T. cruzi: R. neglectus, T. costalimai, T. pseudomaculata, T. wygodizinskyi, T. sordida and R. prolixus (Fig. 2b). The characteristic 163 bp product was revealed in all triatomine samples.
Fig. 2.
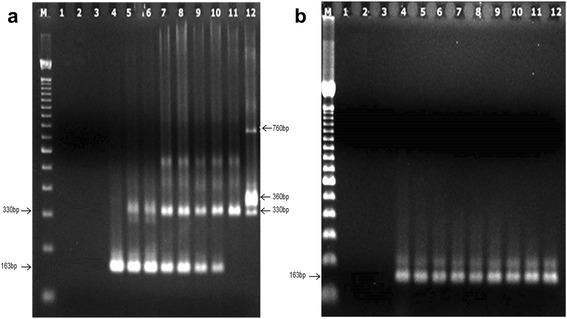
Assaying distinct T. cruzi strains/clones and triatomine species in the conventional multiplex PCR. a Reconstituted DNA samples (R. prolixus + T. cruzi DNAs) were used for the detection of distinct T. cruzi strains/clones with 121/122 primers (kDNA) in the multiplex format. Lane M: 100 bp DNA molecular marker; Lanes 1, 2: negative PCR controls (PCR reagents without DNA and ultra-pure water, respectively); Lane 3: DNA extraction negative control (extraction reagents + ultra-pure water); Lane 4: non-infected triatomine control (R. prolixus from insectary); Lanes 5, 6: DNA from R. prolixus midgut mixed with T. cruzi CL Brener (TcVI) DNA; Lanes 7, 8: DNA from R. prolixus midgut mixed with T. cruzi Dm28c (TcI) DNA; Lanes 9, 10: DNA from R. prolixus midgut mixed with T. cruzi INPA 4167 (TcIV) DNA; Lane 11: T. cruzi positive control (DNA obtained from 102 CL Brener epimastigotes); Lane 12: T. rangeli positive control (DNA obtained from 102 Macias strain epimastigotes). b Detection of distinct triatomine species using P2B/P6R primers (12S rRNA gene). Five individual midguts of each species were used as DNA source. Lane M: 100 bp DNA molecular marker; Lanes 1, 2: negative PCR controls (PCR reagents without DNA and ultra-pure water, respectively); Lane 3: DNA extraction negative control (extraction reagents in ultrapure water); Lanes 4, 5: R. neglectus; Lanes 6, 7: T. costalimai; Lane 8: T. pseudomaculata; Lane 9: T. wygodizinskyi; Lane 10: T. sordida; Lanes 11, 12: R. prolixus
In the present investigation, 205 field-collected triatomines were used to assess the performance of two PCR-based strategies for detection and quantification of T. cruzi parasites directly from intestinal content. The insects were collected in domestic, peridomestic or sylvatic habitats in rural areas encompassing three distinct Brazilian biomes. Following species identification, the triatomines were sorted according to the respective biome (Fig. 1): Caatinga (n = 55): T. pseudomaculata (26/55; 47.4%), T. brasiliensis (22/55; 40.0%) and P. lutzi (7/55; 12.6%); Cerrado (n = 66): T. sordida (41/66; 62.1%) and T. wygodizinskyi (25/66; 37.9%); Atlantic Rainforest (n = 84): T. sordida (54/84; 64.3%), T. wygodizinskyi (23/84; 27.4%) and Triatoma vitticeps (7/84; 8.3%). Using manual collections, most of the specimens (74.7%) were captured in peridomestic ecotopes in the three biomes. Only four T. brasiliensis adults (1.9% of all captured insects) were found in the intradomicile environment, and all T. wygodizinskyi specimens (23.4% of the captured insects) were from sylvatic habitats, collected with the use of forceps on the cracks of rocks. A detailed description of the captured triatomines is presented (Additional file 1: Table S1).
Parasitological and molecular diagnosis of field-collected triatomines
The diagnostic analysis to detect T. cruzi in the 205 field-collected triatomines revealed a kDNA-PCR positivity of 21% (43/205), and the flagellate was detected by optical microscopy in three out of the 43 PCR-positive samples (7.0%). All PCR-negative samples were also negative for the parasite presence by microscopy (Table 2).
Table 2.
Diagnostic results of T. cruzi infection generated by conventional multiplex PCR (cPCR) and optical microscopy in field-collected triatomines. Conventional PCR (cPCR) was considered as gold standard
| Field triatomine samples (n = 205) | ||
|---|---|---|
| cPCR + | cPCR - | |
| Microscopy + | 3 (7%) | – |
| Microscopy - | 40 (93%) | 162 (100%) |
| Total | 43 | 162 |
Figure 3 shows a representative gel of the results generated by the multiplex conventional PCR for the screening of T. cruzi natural infection in field-collected triatomines. The presence of the 163 bp fragment corresponding to the triatomine 12S rRNA gene, indicated no PCR inhibition in the analyzed insect samples. The absence of the 330 bp kDNA fragment (Lane 9) confirmed the sample as a “true negative” result for T. cruzi DNA considering the amplification of the internal control triatomine gene. In addition, T. rangeli DNA was not detected in field-collected triatomines, since no amplification of both kDNA fragments (360 and 760 bp) was observed in all analyzed samples.
Fig. 3.
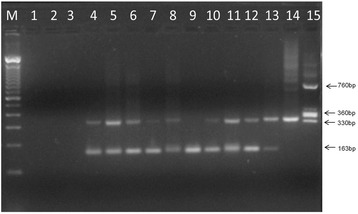
Representative gel for molecular detection of T. cruzi DNA in field triatomine samples by multiplex conventional PCR. Lane M: 100 bp molecular marker; Lanes 1, 2: PCR negative controls (PCR reagents without DNA and ultra-pure water, respectively); Lane 3: DNA extraction negative control (extraction reagents in ultrapure water); Lanes 4, 12: field triatomine samples; Lane 13: multiplex PCR positive control (R. prolixus midgut spiked with ~102 CL Brener epimastigotes); Lane 14: T. cruzi positive control (DNA extracted from 102 CL Brener epimastigotes); Lane 15: T. rangeli positive control (DNA extracted from 102 Macias strain epimastigotes)
When PCR-positive samples were analyzed by triatomine species (Fig. 4a), the highest infection rate was observed for P. lutzi (6/7, 85.7%), followed by T. pseudomaculata (15/26, 57.7%), T. brasiliensis (12/22, 54.5%), T. vitticeps (2/7, 28.6%), T. wygodzinskyi (3/48, 6.3%) and T. sordida (5/95, 5.3%). Caatinga biome yields the highest infection rate (60%) with 33 positive insects out of the 55 captured specimens. The positive kDNA-PCR samples corresponded to T. pseudomaculata (n = 15), T. brasiliensis (n = 12) and P. lutzi (n = 6). Concerning the Atlantic Rainforest and Cerrado biomes, the infection rates revealed by kDNA-PCR were lower, 7.1% (6 positive out of 84 collected insects) and 6.1% (4 positive out of 66 collected insects), respectively (Fig. 4b). The six positive specimens from the Atlantic Rainforest were T. wygodzinskyi (n = 3), T. vitticeps (n = 2) and T. sordida (n = 1), and four T. sordida samples were found positive in the Cerrado region.
Fig. 4.
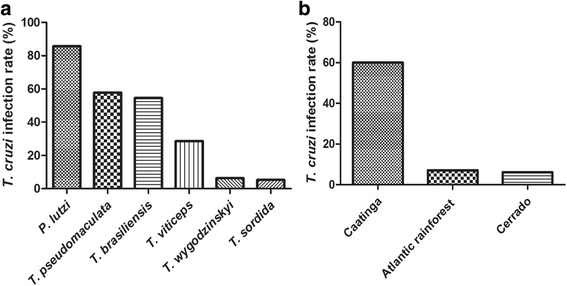
Trypanosoma cruzi detection among distinct triatomine species and biomes from Brazil. T. cruzi DNA detection was performed by multiplex cPCR, and the results were compared between distinct field-collected triatomine species (a) and the geographical regions or biomes where insects were captured (b)
Regarding microscopy observation, the three positive samples were related to T. pseudomaculata, T. brasiliensis and P. lutzi live adults, collected in Caatinga.
Trypanosoma cruzi DNA quantification in triatomines
To quantify the parasite load in positive kDNA-PCR triatomine samples, a TaqMan-based multiplex qPCR was developed, targeting T. cruzi nuclear satellite DNA (SAT-DNA-qPCR) and the triatomine 12S rRNA gene. Following our methodology, detection of T. cruzi DNA was linear from 105 to 0.5 parasite equivalents, using triatomine intestine samples (not infected) spiked with T. cruzi epimastigotes (Fig. 5a). Moreover, the detection of triatomine DNA was linear, ranging from 5 to 0.002 intestine unit equivalents (Fig. 5b). For the T. cruzi and triatomine targets in multiplex, PCR amplification efficiencies were 83.4 and 112.6% respectively, and the linearity coefficient (R2) was 0.99 for both targets, confirming the improved performance of the assay.
Fig. 5.
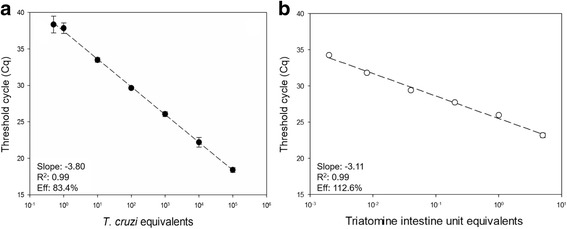
Reportable range for T. cruzi and triatomine intestine unit quantification by real-time qPCR. Multiplex TaqMan qPCR assays were carried out with serially diluted DNA extracted from reconstituted triatomine intestine samples containing T. cruzi epimastigotes, ranging from 105 to 0.5 T. cruzi equivalents (a) and 5 to 0.002 triatomine intestine unit equivalents (b). The slope, R2 and amplification efficiency (Eff) are indicated in the chart
The amplification signal for different triatomine species based on the 12S rDNA target, as well as the possible interference of blood-meal source in the multiplex real-time PCR assay, were investigated (Fig. 6, Table 3). There was no significant difference concerning the amplification of DNA extracted from one intestine of an adult insect (8–10 ng) from the species R. prolixus, R. neglectus, T. vitticeps and T. sordida, with Cq values varying from 24.54 ± 0.39 to 26.86 ± 1.37 (Kruskal-Wallis: H = 4.59, df = 3, P = 0.205). On the other hand, the interference of possible triatomine blood-meal sources was also investigated, and no amplification using DNA (20 ng) extracted from the blood of human, bat, cat, dog, opossum or rat was observed (Table 3).
Fig. 6.
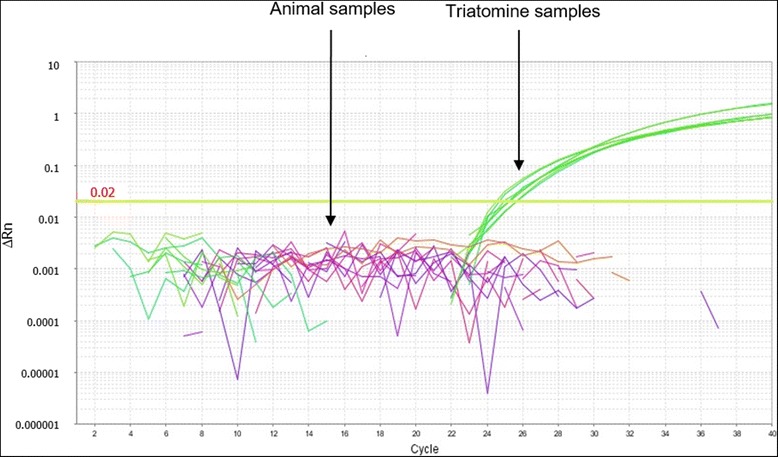
Detection of different triatomine species and interference of possible blood meal sources in the real time qPCR by analyzing the 12S rDNA gene. Representative amplification plots of DNA samples extracted from one intestine unit of different triatomine species (adults maintained in insectary) and absence of amplification for DNAs obtained from blood samples of possible meal sources for field-collected triatomines
Table 3.
Representation of Cq mean values for detection of the 12S rDNA gene by real-time PCR in distinct triatomine species and absence of amplification signal in animal blood samples
| Triatomine detection (1 intestine, 8–10 ng DNA) | Possible blood-meal source detection (blood, 20 ng DNA) | ||
|---|---|---|---|
| Triatomine species | Cq (Mean ± SD) | Origin | Cq (Mean ± SD) |
| Rhodnius prolixus | 25.54 ± 0.30 | Human | nd |
| Rhodnius neglectus | 26.55 ± 0.54 | Bat | nd |
| Triatoma vitticeps | 26.86 ± 1.37 | Cat | nd |
| Triatoma sordida | 24.54 ± 0.39 | Dog | nd |
| Opossum | nd | ||
| Rat | nd | ||
Abbreviations: C q threshold cycle, SD standard deviation, nd not detected
Table 4 presents the results of SAT-DNA qPCR positivity to estimate LOD for T. cruzi DNA in triatomine reconstituted samples. A LOD of 0.41 parasite equivalents was found and the threshold cycle (Cq) corresponding to the detection of one solely triatomine intestine, used as an internal reference, varied from 21.19 ± 0.39 to 23.06 ± 1.51 with the threshold set at 0.02.
Table 4.
SAT-DNA qPCR positivity to estimate the limit of detection (LOD) for T. cruzi DNA in triatomine samples
| Parasite load (T. cruzi equivalents) | Assay # | Positive samples (%) | Triatomine internal reference (Cq ± SD) |
|---|---|---|---|
| 20 | 1 | 12 (100) | 21.19 ± 0.39 |
| 2 | 12 (100) | ||
| 3 | 12 (100) | ||
| 4 | 1 | 12 (100) | 22.09 ± 1.85 |
| 2 | 12 (100) | ||
| 3 | 12 (100) | ||
| 0.8 | 1 | 12 (100) | 21.48 ± 0.86 |
| 2 | 12 (100) | ||
| 3 | 12 (100) | ||
| 0.16 | 1 | 10 (83.3) | 22.56 ± 1.40 |
| 2 | 8 (66.7) | ||
| 3 | 10 (83.3) | ||
| 0.032 | 1 | 11 (91.7) | 22.65 ± 1.12 |
| 2 | 10 (83.3) | ||
| 3 | 5 (41.7) | ||
| 0.0064 | 1 | 7 (58.3) | 23.06 ± 1.51 |
| 2 | 6 (50.0) | ||
| 3 | 6 (50.0) | ||
| 0.00128 | 1 | 1 (8.3) | 22.39 ± 0.76 |
| 2 | 1 (8.3) | ||
| 3 | – |
Abbreviations: C q threshold cycle, SD standard deviation
The multiplex qPCR assay was used to estimate T. cruzi load in 38 out of the 43 field-collected triatomines that gave positive results by the kDNA-PCR test. A wide range parasite load distribution varying from 8.05 × 10-2 to 6.31 × 1010 was observed, with a median of 2.29 × 103 (2.43 × 101–2.10 × 104) T. cruzi/intestine units (Fig. 7a). When parasite load was analyzed by triatomine species (Fig. 7b), a significantly greater median value was found for P. lutzi in comparison with T. brasiliensis (Shapiro-Wilk: P < 0.050, Mann-Whitney Rank Sum Test: U = 14, T (6,12) = 79, P = 0.044). It was possible to infer the parasite load for only one specimen of T. wygodzinskyi and T. vitticeps each and for 3 out of 5 positive T. sordida.
Fig. 7.
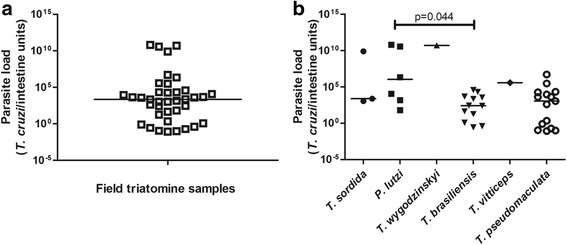
Distribution of parasite load in field triatomine samples. a Trypanosoma cruzi load estimated by multiplex real-time qPCR targeting the parasite nuclear satellite sequences (SAT-DNA-qPCR) and normalized by triatomine intestine units. The straight horizontal line indicates the median value. b Parasite loads and median values (horizontal lines) are indicated per triatomine species
Discussion
Molecular diagnostic tests have been improved in the last years, with the development of conventional and quantitative real-time PCR-based assays for detection and quantification of T. cruzi DNA in different biological specimens. These include human blood and serum [26–28], animal tissues [29] and feces or digestive tract of triatomine vectors [30, 31]. The establishment of precise methodologies able to detect and quantify T. cruzi load in field-collected triatomine samples, would be important for monitoring natural infection rates in Chagas disease vectors and its further application for molecular epidemiological studies. Here, we report the development and application of PCR and TaqMan real-time qPCR assays, both in multiplex, for the detection and quantification of T. cruzi load in field-collected triatomines.
PCR-based bug infection rates may be underestimated due to PCR inhibitors. For this, DNA extraction methods from feces or digestive tract need to be improved in order to increase PCR sensitivity. In this study, triatomine guts were pretreated in a lysis solution containing proteinase K for 2 h at 56 °C, before DNA extraction using a modified protocol based on silica columns. The absence of PCR inhibition was demonstrated by amplification of the internal control reference (triatomine 12S rRNA gene) in both conventional and real-time PCR assays. From the latter, it was possible to observe slight variation on Cq values, from 21.19 ± 0.39 to 23.06 ± 1.51, for samples corresponding to one adult triatomine intestine unit containing 20 or 0.0064 T. cruzi equivalents, respectively (Table 4).
The kDNA-based PCR screening of field-collected triatomines was shown to be more sensitive than microscopic examination (ME) with a positivity of 21%, whereas only 7% of positive PCR results were confirmed by ME of fecal drops of live insects. In addition, all negative PCR samples were microscopically negative for T. cruzi infection. It should be noticed that the three triatomine specimens for which ME was able to identify T. cruzi flagellates in the feces (3/205, 1.5%) represented live adult insects, whereas the molecular analysis was performed with DNA obtained from midgut and rectum of live and dead insects. These technical features could in part explain the differences in positivity between microscopy and PCR, considering the lower number of parasites in triatomine-diluted feces when compared with intestinal content homogenates used for DNA extraction, and thus yielding false negative results by the traditional ME diagnosis.
A study using kDNA-PCR tests for screening of T. cruzi infection from fecal samples of field-collected triatomines in Argentina allowed the identification of 7.5% positive samples among those microscopically negative [32]. For few samples in which the presence of flagellates was evidenced by ME, both kDNA and nuclear satellite sequence of T. cruzi were not amplified. However, positive amplification of a 24Sα rDNA conserved fragment revealed infection with another trypanosomatid, suggesting that caution should be taken when diagnosing T. cruzi infection based only on ME of unstained fresh preparations of triatomine feces [32]. Similar evidence was described in a recent study in Brazil, in which, 46% of sylvatic T. brasiliensis collected in Rio Grande do Norte State were infected with T. cruzi-like parasites by ME of fecal drops, while using mini-exon PCR amplification, a lower infection prevalence was detected in the triatomine digestive tracts [33]. The authors suggested this was a result of misidentification of T. cruzi-like parasites as T. cruzi during direct ME thus generating false positive results for local Chagas disease control surveillance campaigns. Additionally, results obtained from first- and second-stage nymphs using ME are underestimated due to the difficulty in obtaining feces during these stages. This limitation is not observed when PCR is performed [15].
Differences in T. cruzi infection prevalence are observed between triatomine species [33, 34]. Regarding T. brasiliensis, the influence of feeding sources on the rate of T. cruzi infection in specimens collected in distinct sites and ecotopes in Rio Grande do Norte State has been demonstrated [33]. Triatoma brasiliensis is the most important vector species in northeastern Brazil, due to its wide geographical distribution, its rates of natural infection and capacity to inhabit both natural and human environments [35]. Triatoma pseudomaculata is the second epidemiologically more important species after T. brasiliensis in the northeast of Brazil [36]. Both are synanthropic species, endemic in the State of Ceará (Caatinga biome) and are easily found in municipalities where Chagas disease is reported [34, 37]. Triatoma pseudomaculata is more frequently found in peridomiciliary habitats and generally feeding on birds. Nevertheless, the domiciliary foci of this species have been observed in Ceará and Minas Gerais [38, 39]. Caatinga biome occupies the third position in triatomines diversity [40]. Subsistence agriculture is the main activity of the inhabitants, beyond cattle raising, mainly goats and the presence of chickens living free in peridomicile attracting blood-sucking insect vectors around human habitations [41]. In the present study, T. pseudomaculata and T. brasiliensis captured in the Caatinga biome, the State of Ceará, were the most frequent species captured in the region, with prevalence of 47.4 and 40%, respectively, and T. cruzi infection rates of 57.7 and 54.5%, respectively. The previous report in the municipality of Russas (Ceará) showed high rates of natural infection by T. cruzi in T. brasiliensis (90%) and T. pseudomaculata (80%) using PCR directed to the mini-exon region; although the authors considered these much higher than expected [42]. Triatoma brasiliensis is predominantly found in domestic environments of the semiarid region of Brazil [36]. Here, it is important to note that four out of 12 T. brasiliensis-positive specimens were captured in the intradomicile environment (one of these was also positive by microscopy), and the others were from the peridomicile environment. Triatoma brasiliensis was also found positive for T. cruzi in the domestic, peridomestic and wild environments in Tauá (Ceará State), with the highest infection rate of 14% in the peridomicile [43].
In our study, of the few specimens (n = 7) of P. lutzi captured in Caatinga, in the peridomicile environment, only one was negative for T. cruzi kDNA thus yielding the highest infection prevalence for this species when compared with all others (85.7%) (Fig. 4). Of the six positive adult specimens, five were found at the same site of collection (Additional file 1: Table S1). Panstrongylus lutzi is one of the native species of northeastern Brazil, and its geographical distribution coincides with the extension of the Caatinga biome. Mainly in Ceará and Pernambuco states, it is increasingly common the presence of adults in the intradomicile and generally presenting high rates of T. cruzi infection, elevating the epidemiological importance of this species [44]. Studies evaluating natural infections by T. cruzi in P. lutzi have been reported using parasitological search by microscopy; in one of these, T. cruzi flagellates were identified in 29.1% of the specimens found in the intradomicile, in 20 municipalities of Ceará [44]. As mentioned, the high positivity found in the present study for kDNA-PCR may have been overestimated due to the collection of a small number of P. lutzi specimens and the vast majority being infected.
Although the number of collected bugs in the Brazilian Atlantic Rainforest and Cerrado biomes was higher than in Caatinga, T. cruzi infection rates were lower (7.1 and 6.1%, respectively). Nowadays, in Brazil, T. sordida merits special attention due to its wide distribution (Atlantic Rainforest, Caatinga, Cerrado and Amazon biomes), tendency to invade domestic environments and vectorial competence [45]. This species is endemic in Cerrado, the main biome of Central Brazil, more frequently found in the peridomestic environment, especially in chicken coops, occupying areas where T. infestans has been eliminated. Triatoma sordida is usually considered as a secondary vector of T. cruzi and usually exhibits low rates of natural infection [46, 47]; this species is associated with the reinfestation of dwellings treated with insecticides [48], and is the most common synanthropic species captured in the Central-West region of Brazil [49]. These features are in agreement with our findings, where T. sordida was the main species captured in both, Cerrado and Atlantic Forest biomes, with frequencies of 62.1 and 64.3%, respectively. In the Cerrado, four specimens out of 41 yielded positive for T. cruzi kDNA (9.8% infection rate). In another investigation, 10.7% of T. sordida specimens captured in Mato Grosso do Sul were positive for flagellated protozoa, as determined by microscopic search, and 18.1% were positive for T. cruzi using kDNA-PCR [50]. The latter value was almost two-fold higher than that observed herein (9.8%), for specimens collected in different areas of the Cerrado biome, including the State of Mato Grosso do Sul, and none were found positive by direct microscopy. In the Atlantic Rainforest, despite the high prevalence of T. sordida captured in the peridomicile (64.3%), only one specimen out of 54 was positive for T. cruzi kDNA (1.9%). Overall, comparing the triatomine species evaluated herein, T. sordida revealed the lowest prevalence of T. cruzi infection (5.3%, Fig. 4a).
Triatoma wygodzinskyi is a rupicolous species, restricted to the sylvatic environment and has received scarce attention due to its limited epidemiological importance as a vector of T. cruzi [51]. This triatomine was initially described among a small number of specimens collected in southern Minas Gerais and São Paulo states [21]. Environmental information provided by remote sensors predicted a more extensive geographic distribution for T. wygodzinskyi, including the states of Rio de Janeiro, Paraná, Santa Catarina and Rio Grande do Sul [52]. In our study, this species was captured in sylvatic environments of Cerrado and Atlantic Rainforest biomes, with a prevalence of 37.9 and 27.4%, respectively, and presented three positive specimens for T. cruzi kDNA that were only found in the Atlantic forest (6.3% infection rate).
Triatoma vitticeps has a geographical distribution more restricted to the Atlantic Rainforest [40], with adult specimens being captured in rural areas in Rio de Janeiro, Bahia, Minas Gerais and Espírito Santo [53, 54]. The species colonizes peridomiciliary areas, increasing the risk of T. cruzi transmission to humans [54]. In a recent survey conducted in the northeastern region of Minas Gerais, T. vitticeps was present throughout almost all the municipalities, predominantly in its adult form, in the intradomicile [55]. In the present work, T. vitticeps corresponded to 8.3% of the specimens collected in the Atlantic Rainforest biome, in the peridomicile environments of two municipalities of Rio de Janeiro, and revealed an infection rate of 28.6% (two positives out of seven collected). In 1998, an investigation was conducted in the municipal district of Santa Maria Madalena, Rio de Janeiro State, where T. cruzi prevalence in T. vitticeps adult triatomines in the intradomicile was estimated as 79% [56]. The higher infection rate of 86.2% by flagellates similar to T. cruzi was found in the Espírito Santo State for sylvatic T. vitticeps; 47.4% of 116 analyzed insects were captured in the bedroom of residences of which 85% were infected [53]. In spite of the occasional invasion of houses by T. vitticeps in Rio de Janeiro State [56–59] and its recognized role in the vectorial transmission of Chagas disease in Espírito Santo State [60–62], the extended interval between feeding and defecation reduces the success for the classical vectorial route transmission and could in part contribute to the low incidence of Chagas disease in both states [62, 63].
To the best of our knowledge, to date there is no study comparing T. cruzi parasite load among field-collected triatomine species or per geographical area. Here, 38 of the 43 triatomines with positive results for T. cruzi kDNA were quantified by the developed TaqMan multiplex qPCR. Not all kDNA-PCR positive samples could be confirmed by SAT-DNA-qPCR, which is expected from the fewer copies of satellite DNA sequences in the T. cruzi DTU - TcI [64]. Although molecular characterization of T. cruzi populations was not performed in this study, it is known that TcI displays the first largest distribution in both, wild or domestic transmission cycles [65], beyond being very frequently found in concomitant infection with other DTUs in triatomine bugs and seemed to be the predominant isolate, even in mixed infections [66]. A higher sensitivity of kDNA compared to SAT-DNA-qPCR to evaluate human parasitaemia in Chagas disease patients has also been confirmed [27].
To estimate parasite load in field-collected triatomines, a highly sensitive TaqMan multiplex qPCR was developed with a LOD of 0.41 T. cruzi/intestine units. This is in agreement with the one reported for other tissues and organisms such as human blood, also using T. cruzi nuclear satellite DNA as a target [67]. In addition, the assay enabled amplification of DNA from intestine samples of different triatomine species with similar Cq values, indicating a low variance in the 12S rDNA copy number in triatomine genomes, which contributes to the reproducibility of the assay. Also, possible blood-meal sources of field-collected triatomines, such as human, bat, cat, dog, opossum or rat did not interfere in the multiplex real-time PCR assay, showing the improved specificity of this methodology.
The observed dynamic extension provided linear quantification in at least 6-log range, for both T. cruzi and triatomines, allowing an accurate normalization of parasite load. This normalization is essential to compensate variations between samples, such as DNA amount, that depends on the developmental stage of the insect and the time post-feeding, beyond the quality of sample preservation and differences in size between species. Following our methodology, it is possible to perform a direct comparison between field samples from different species and geographical localities.
In spite of the wide range of parasite load values observed in the present study among field-collected triatomines (c.10-2–1010 T. cruzi/intestine units), a significant difference in the median values was only observed between P. lutzi and T. brasiliensis (P = 0.044, Fig. 7b). In two P. lutzi specimens, the number of T. cruzi/intestine units exceeded 1010, as also observed for one specimen of T. sordida and for the single T. wygodzinskyi sample; whereas for T. brasiliensis, the highest parasite load did not exceed 105 T. cruzi/intestine units (Fig. 7b). Studies on trypanosome-triatomine interactions may allow the identification of humoral and cellular mechanisms in the vectors that either facilitate or prevent the transmission of parasites in different vector species, acting as limiting factors for the development of the parasite [68–70]. The susceptibility of a vector depends on various factors, including digestive enzymes, hemolysins, agglutinins, microbiota and especially antimicrobial factors, which are potentially involved in regulating the development of T. cruzi in the gut [70]. Moreover, differential regulation of parasite populations between different strains of T. cruzi has also been demonstrated [70]. In spite of not presenting data of T. cruzi genotyping, our findings reproduce the magnitude of parasitism found in field-collected triatomines. This novel quantitative real-time PCR raises the possibility for further evaluating parasite load, as a promising biomarker of vectorial capacity, through investigating the correlation between the amount of T. cruzi found in different portions of the insect midgut and the triatomine ability to transfer the parasite during a blood-meal.
Conclusions
Our results present a novel multiplex PCR-based approach, as a combination of conventional and real-time PCR methodologies, to monitor T. cruzi infection in triatomines. Both methods can be successfully applied to epidemiological studies of Chagas disease surveillance, and for evaluating T. cruzi life-cycle and its colonization in the digestive tract of triatomine experimental models. Parasite load quantification in triatomines emerges as a potential biomarker to evaluate vectorial capacity.
Acknowledgments
The authors thank Ana Laura Carbajal for providing the captured insects of T. wygodzinskyi, Maria Angelica Cardoso for technical support, the COLPROT/Fiocruz for supplying the protozoa used, and the Laboratório de Bioquímica e Fisiologia de Insetos from the Oswaldo Cruz Institute/Fiocruz for providing the R. prolixus specimens and other triatomine species maintained in insectary. We also thank the Program for Technological Development in Inputs for Health (PDTIS-FIOCRUZ) for the facilities on the real-time PCR.
Funding
This work was supported by grants PROEP-Fiocruz n° 400,106/2011–6, Universal/CNPq n° 471,239/2011–9, DECIT/CNPq n° 467,201/2014–5, and FAPERJ n° E_03/2012. C Britto and CM d’Avila-Levy are researcher fellows of CNPq and FAPERJ (CNE, JCNE), and OC Moreira is a research fellow of FAPERJ (JCNE).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional file. Nucleotide sequences reported in this paper are available in the GenBank database under accession numbers: T. cruzi kDNA: U07846.1; T. cruzi satellite DNA: EU728662–EU728667; Triatomine 12S subunit ribosomal RNA: AF394519.1.
Abbreviations
- BHI
Brain heart infusion
- ChD
Chagas disease
- COLPROT
Coleção de Protozoários da Fundação Oswaldo Cruz, Rio de Janeiro, Brazil
- cPCR
Conventional PCR
- Cq
Threshold cycle
- DTU
Discrete typing unit
- Fiocruz
Oswaldo Cruz Foundation
- kDNA
Kinetoplast deoxyribonucleic acid
- LOD
Limit of detection
- NFQ-MGB
Non fluorescent quencher-minor groove binding
- qPCR
Quantitative real-time PCR
- R2
Coefficient of linearity
- SAT-DNA
Satellite deoxyribonucleic acid
- TcI, TcIV and TcVI
Trypanosoma cruzi discrete typing units I, IV and VI
- UFF
Fluminense Federal University
Additional file
Detailed description of field-collected triatomine specimens from 26 municipalities of three Brazilian biomes. (DOC 350 kb)
Authors’ contributions
Conceived and designed the experiments: OCM, SAOG, CMDAL and CB. Performed the experiments: TV, PFA, CML, DMS, LRA, FFM and JRSM. Analyzed the data: OCM, TV, PF, LRA, JRSM and CB. Wrote the paper: OCM, TV, SAOG, CML and CB. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2343-x) contains supplementary material, which is available to authorized users.
Contributor Information
Otacilio C. Moreira, Email: otacilio@ioc.fiocruz.br
Thaiane Verly, Email: thaiane.verly@ioc.fiocruz.br.
Paula Finamore-Araujo, Email: paulafinamore@gmail.com.
Suzete A. O. Gomes, Email: suzetearaujo@id.uff.br
Catarina M. Lopes, Email: aniratac@ioc.fiocruz.br
Danielle M. de Sousa, Email: misael@ioc.fiocruz.br
Lívia R. Azevedo, Email: livia_az@hotmail.com
Fabio F. da Mota, Email: fabio@ioc.fiocruz.br
Claudia M. d’Avila-Levy, Email: davila.levy@ioc.fiocruz.br
Jacenir R. Santos-Mallet, Email: jacenir@ioc.fiocruz.br
Constança Britto, Email: cbritto@ioc.fiocruz.br.
References
- 1.Justi SA, Galvão C. The evolutionary origin of diversity in Chagas disease vectors. Trends Parasitol. 2017;33(1):42–52. doi: 10.1016/j.pt.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roellig DM, Brown EL, Barnabé C, Tibayrenc M, Steurer FJ, Yabsley MJ. Molecular typing of Trypanosoma cruzi isolates, United States. Emerg Infect Dis. 2008;7:1123–1125. doi: 10.3201/eid1407.080175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO - World Health Organization. Chagas disease (American trypanosomiasis). 2017. http://www.who.int/mediacentre/factsheets/fs340/en/. Accessed 24 Aug 2017.
- 4.Tanowitz HB, Weiss LM, Montgomery SP. Chagas disease has now gone global. PLoS Negl Trop Dis. 2011;5:e1136. doi: 10.1371/journal.pntd.0001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO - World Health Organization. Control of Chagas disease. 2002 WHO Expert Committee Techn Rep Ser No. 905, Geneva, Switzerland.
- 6.Schofield CJ, Galvao C. Classification, evolution, and species groups within the Triatominae. Acta Trop. 2009;110:88–100. doi: 10.1016/j.actatropica.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Coura JR, Junqueira AV, Fernandes O, Valente S, Miles M. Emerging Chagas disease in the Amazonian Brazil. Trends Parasitol. 2002;18:171–176. doi: 10.1016/S1471-4922(01)02200-0. [DOI] [PubMed] [Google Scholar]
- 8.Cohen JE, Gürtler RE. Modeling household transmission of American trypanosomiasis. Science. 2001;293:694–8. [DOI] [PubMed]
- 9.Dias JC, Silveira AC, Schofield CJ. The impact of Chagas disease control in Latin America: a review. Mem Inst Oswaldo Cruz. 2002;97:603–612. doi: 10.1590/S0074-02762002000500002. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto K, Schofield CJ. Elimination of Rhodnius prolixus in central America. Parasit Vectors. 2012;5:45. doi: 10.1186/1756-3305-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picollo MI, Vassena C, Santo Orihuela P, Barrios S, Zaidemberg M, Zerba E. High resistance to pyrethroid insecticides associated with ineffective field treatments in Triatoma infestans (Hemiptera: Reduviidae) from northern Argentina. J Med Entomol. 2005;42:637–642. doi: 10.1093/jmedent/42.4.637. [DOI] [PubMed] [Google Scholar]
- 12.Lardeux F, Depickère S, Duchon S, Chavez T. Insecticide resistance of Triatoma infestans (Hemiptera, Reduviidae) vector of Chagas disease in Bolivia. Tropical Med Int Health. 2010;15:1037–1048. doi: 10.1111/j.1365-3156.2010.02573.x. [DOI] [PubMed] [Google Scholar]
- 13.Depickère S, Buitrago R, Siñani E, Baune M, Monje M, et al. Susceptibility and resistance to deltamethrin of wild and domestic populations of Triatoma infestans (Reduviidae: Triatominae) in Bolivia: new discoveries. Mem Inst Oswaldo Cruz. 2012;107(8):1042–1047. doi: 10.1590/S0074-02762012000800013. [DOI] [PubMed] [Google Scholar]
- 14.Lardeux F, Aliaga C, Depickère S. Bias due to methods of parasite detection when estimating prevalence of infection of Triatoma infestans by Trypanosoma cruzi. J Vector Ecol. 2016;41(2):285–291. doi: 10.1111/jvec.12224. [DOI] [PubMed] [Google Scholar]
- 15.Pizarro JC, Lucero DE, Stevens L. PCR reveals significantly higher rates of Trypanosoma cruzi infection than microscopy in the Chagas vector, Triatoma infestans: high rates found in Chuquisaca, Bolivia. BMC Infect Dis. 2007;27:66. doi: 10.1186/1471-2334-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uehara LA, Moreira OC, Oliveira AC, Azambuja P, Lima APCA, Britto C, et al. Cruzipain promotes Trypanosoma cruzi adhesion to Rhodnius prolixus midgut. PLoS Negl Trop Dis. 2012;6(12):e1958. doi: 10.1371/journal.pntd.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturm NR, Degrave W, Morel C, Simpson L. Sensitive detection and schizodeme classification of Trypanosoma cruzi cells by amplification of kinetoplast minicircle DNA sequences: use in diagnosis of Chagas disease. Mol Biochem Parasitol. 1989;33(3):205–214. doi: 10.1016/0166-6851(89)90082-0. [DOI] [PubMed] [Google Scholar]
- 18.Piron M, Fisa R, Casamitjana N, Lopez-Chejade P, Puig L, Vergés M, et al. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop. 2007;103:195–200. doi: 10.1016/j.actatropica.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Azambuja P, Garcia ES. Care and maintenance of triatomine colonies. In: Crampton JM, Beard CB, Louid C, editors. Molecular biology of insect disease vectors: a methods manual. London: Chapman and Hall; 1997. pp. 56–64. [Google Scholar]
- 20.MMA - Ministério do Meio Ambiente. Biomas do Brasil. 2016. http://www.mma.gov.br/biomas. Accessed 3 Dec 2016.
- 21.Lent H, Wygodzinsky P. Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas disease. Bull Amer Mus Nat Hist. 1979;163:127–520. [Google Scholar]
- 22.Galvão C, Dale C. Chaves de identificação para adultos. In: Galvão C, organizador. Série Zoologia. Guias e Manuais de Identificação: Vetores da doença de Chagas no Brasil. Curitiba: Sociedade Brasileira de Zoologia; 2015. p. 171–208.
- 23.Moreira OC, Ramírez JD, Velázquez E, Melo MF, Lima-Ferreira C, Guhl F, et al. Towards the establishment of a consensus real-time qPCR to monitor Trypanosoma cruzi parasitemia in patients with chronic Chagas disease cardiomyopathy: a substudy from the BENEFIT trial. Acta Trop. 2013;125(1):23–31. doi: 10.1016/j.actatropica.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Wincker P, Britto C, Pereira JB, Cardoso MA, Oelemann W, Morel CM. Use of a simplified polymerase chain reaction procedure to detect Trypanosoma cruzi in blood samples from chronic chagasic patients in a rural endemic area. Am J Trop Med Hyg. 1994;51:771–777. doi: 10.4269/ajtmh.1994.51.771. [DOI] [PubMed] [Google Scholar]
- 25.Burd EM. Validation of laboratory-developed molecular assays for infectious disease. Clin Microbiol Rev. 2010;23(3):550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schijman AG, Bisio M, Orellana L, Sued M, Duffy T, Mejia Jaramillo AM, et al. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis. 2011;5(1):e931. doi: 10.1371/journal.pntd.0000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramírez JC, Cura CI, Moreira OC, Lages-Silva E, Juiz N, Velázquez E, et al. Analytical validation of quantitative real-time PCR methods for quantification of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. J Mol Diagn. 2015;17:605–615. doi: 10.1016/j.jmoldx.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melo MF, Moreira OC, Tenório P, Lorena V, Lorena-Rezende I, Oliveira Júnior W, et al. Usefulness of real-time PCR to quantify parasite load in serum samples from chronic Chagas disease patients. Parasit Vectors. 2015;8:154. doi: 10.1186/s13071-015-0770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minuzzi-Souza TTC, Nitz N, Knox MB, Reis F, Hagström L, Cuba Cuba CA, et al. Vector-borne transmission of Trypanosoma cruzi among captive Neotropical primates in a Brazilian zoo. Parasit Vectors. 2016;9:39. doi: 10.1186/s13071-016-1334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dias FA, Guerra B, Vieira LR, Perdomo HD, Gandara ACP, Do Amaral RJV, et al. Monitoring the parasite load in the digestive tract of Rhodnius prolixus by combined qPCR analysis and imaging techniques provides new insights into trypanosome life cycle. PLoS Negl Trop Dis. 2015;9(10):e0004186. doi: 10.1371/journal.pntd.0004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saavedra M, Zulantay I, Apt W, Castillo J, Araya E, Martínez G, et al. Quantification by real-time PCR of Trypanosoma cruzi DNA in samples of Triatoma infestans used in xenodiagnosis of chronic Chagas disease patients. Parasit Vectors. 2016;9:382. doi: 10.1186/s13071-016-1664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcet PL, Duffy T, Cardinal MV, Burgos JM, Lauricella MA, Levin MJ, et al. PCR-based screening and lineage identification of Trypanosoma cruzi directly from faecal samples of triatomine bugs from northwestern Argentina. Parasitology. 2006;132(1):57–65. doi: 10.1017/S0031182005008772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almeida CE, Faucher L, Lavina M, Costa J, Harry M. Molecular individual-based approach on Triatoma brasiliensis: inferences on triatomine foci, Trypanosoma cruzi natural infection prevalence, parasite diversity and feeding sources. PLoS Negl Trop Dis. 2016;10(2):e0004447. doi: 10.1371/journal.pntd.0004447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarquis O, Borges-Pereira J, Mac Cord JR, Gomes TF, Cabello PH, Lima MM. Epidemiology of Chagas disease in Jaguaruana, Ceará, Brazil. I. Presence of triatomines and index of Trypanosoma cruzi infection in four localities of a rural area. Mem Inst Oswaldo Cruz. 2004;99:263–270. doi: 10.1590/S0074-02762004000300004. [DOI] [PubMed] [Google Scholar]
- 35.Costa J, Almeida CE, Dotson EM, Lins A, Vinhaes M, Silveira AC, et al. The epidemiologic importance of Triatoma brasiliensis as a Chagas disease vector in Brazil: a revision of domiciliary captures during 1993–1999. Mem Inst Oswaldo Cruz. 2003;98(4):443–449. doi: 10.1590/S0074-02762003000400002. [DOI] [PubMed] [Google Scholar]
- 36.Silveira AC, Feitosa VR, Borges R. Distribuição de triatomíneos domiciliados no período 1975/1983 no Brasil. Rev Bras Malariol Trop. 1984;36:15–312. [PubMed] [Google Scholar]
- 37.Coutinho CF, Souza-Santos R, Teixeira NF, Georq I, Gomes TTF, Boia MN, et al. An entomoepidemiological investigation of Chagas disease in the state of Ceará, northeast region of Brazil. Cad Saúde Publ. 2014;30:785–793. doi: 10.1590/0102-311X00176512. [DOI] [PubMed] [Google Scholar]
- 38.Freitas SPC, Lorosa ES, Rodrigues DCS, Freitas ALC, Gonçalves TCM. Feeding patterns of Triatoma pseudomaculata in the state of Ceará, Brazil. Rev Saúde Publ. 2005;39(1):27–32. doi: 10.1590/S0034-89102005000100004. [DOI] [PubMed] [Google Scholar]
- 39.Machado de Assis GF, Azeredo BV, de la Fuente AL C, Diotaiuti L, de Lana M. Domiciliation of Triatoma pseudomaculata (Corrêa e Espínola 1964) in the Jequitinhonha valley, state of Minas Gerais. Rev Soc Bras Med Trop. 2007;40:391–396. doi: 10.1590/S0037-86822007000400003. [DOI] [PubMed] [Google Scholar]
- 40.Gurgel-Gonçalves R, Galvão C, Costa J, Peterson A. Geographic distribution of Chagas disease vectors in Brazil based on ecological niche modeling. J Trop Med. 2012;2012:1–15. doi: 10.1155/2012/705326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarquis O, Carvalho-Costa FA, Toma HK, Georg I, Burgoa MR, Lima MM. Eco-epidemiology of Chagas disease in northeastern Brazil: Triatoma brasiliensis, T. pseudomaculata and Rhodnius nasutus in the sylvatic, peridomestic and domestic environments. Parasitol Res. 2012;110:1481–1485. doi: 10.1007/s00436-011-2651-6. [DOI] [PubMed] [Google Scholar]
- 42.Gumiel M, Mota FF, Rizzo VS, Sarquis O, Castro DP, Lima MM, et al. Characterization of the microbiota in the guts of Triatoma brasiliensis and Triatoma pseudomaculata infected by Trypanosoma cruzi in natural conditions using culture independent methods. Parasit Vectors. 2015;8:245. doi: 10.1186/s13071-015-0836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bezerra CM, Cavalcanti LPG, Souza RCM, Barbosa SE, Xavier SCC, Jansen AM, et al. Domestic, peridomestic and wild hosts in the transmission of Trypanosoma cruzi in the Caatinga area colonised by Triatoma brasiliensis. Mem Inst Oswaldo Cruz. 2014;109(7):887–898. doi: 10.1590/0074-0276140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caranha L, Lorosa ES, Rocha DS, Jurberg J, Galvão C. Feeding sources evaluation of Panstrongylus lutzi (Neiva & Pinto, 1923) (Hemiptera: Reduviidae: Triatominae) in the state of Ceará. Rev Soc Bras Med Trop. 2006;39(4):347–351. doi: 10.1590/S0037-86822006000400006. [DOI] [PubMed] [Google Scholar]
- 45.Diotaiuti L. Triatomine-vectors of Trypanosoma cruzi infection. In: Teixeira A, Vinaud M, Maria Castro A. (Eds.) Emerging Chagas Disease. Bentham Science Publishers; 2009. p. 24-39.
- 46.Diotaiuti L, Paula OR, Falcão PL, Dias JCP. Avaliação do programa de controle vetorial da doença de Chagas em Minas Gerais, Brasil, com referência especial ao Triatoma sordida. Bol Oficina Sanit Panam. 1995;118:211–219. [Google Scholar]
- 47.Oliveira AWS, Silva IG. Distribuição geográfica e indicadores entomológicos de triatomíneos sinantrópicos capturados no estado de Goiás. Rev Soc Bras Med Trop. 2007;40:204–208. doi: 10.1590/S0037-86822007000200011. [DOI] [PubMed] [Google Scholar]
- 48.Pessoa GCD, Obara MT, Rezende JC, de Mello BV, Ferraz ML, Diotaiuti L. Deltamethrin toxicological profile of peridomestic Triatoma sordida in the north of Minas Gerais, Brazil. Parasit Vectors. 2015;8:263–269. doi: 10.1186/s13071-015-0873-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pereira JM, de Almeida PS, de Sousa AV, de Paula AM, Machado RB, Gurgel-Gonçalves R. Climatic factors influencing triatomine occurrence in central-West Brazil. Mem Inst Oswaldo Cruz. 2013;108:335–341. doi: 10.1590/S0074-02762013000300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cominetti MC, Almeida RF, Gonçalves GM, Andreotti R. Monitoring Trypanosoma cruzi infection in triatomines using PCR in Mato Grosso do Sul, Brazil. Rev Soc Bras Med Trop. 2013;3:277–280. doi: 10.1590/0037-8682-0024-2013. [DOI] [PubMed] [Google Scholar]
- 51.Barretto MP, Ribeiro RD. Estudo sobre reservatórios e vetores silvestres do Trypanosoma cruzi. Observações sobre a ecologia do Triatoma arthurneivai Lent & Martins, 1940 (Hemiptera, Reduviidae) Rev Bras Biol. 1981;41:317–320. [PubMed] [Google Scholar]
- 52.de la Fuente AL C, Porcasi X, Noireau F, Diotaiuti L, Gorla DE. The association between the geographic distribution of Triatoma pseudomaculata and Triatoma wygodzinskyi (Hemiptera: Reduviidae) with environmental variables recorded by remote sensors. Infect Genet Evol. 2009;9:54–61. doi: 10.1016/j.meegid.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 53.Santos GRL, Ferreira GEM, Ferreira AL. Natural infection of Triatoma vitticeps (Stal, 1859) with flagellates morphologically similar to Trypanosoma cruzi (Chagas, 1909) in Espírito Santo state. Rev Soc Bras Med Trop. 2006;39(1):89–91. doi: 10.1590/S0037-86822006000100019. [DOI] [PubMed] [Google Scholar]
- 54.Souza RDCM, Diotaiuti L, Lorenzo MG, Gorla DE. Analysis of the geographical distribution of Triatoma Vitticeps (Stal, 1859) based on data of species occurrence in Minas Gerais, Brazil. Infect Genet Evol. 2010;10(6):720–726. doi: 10.1016/j.meegid.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 55.Dias JVL, Fernandes EG, Pires HHR, Dias JCP. Occurrence and distribution of triatomines (Hemiptera: Reduviidae) in municipalities of the Northeastern region of Minas Gerais state, Brazil. Rev Soc Bras Med Trop. 2016;49(4):502–507. doi: 10.1590/0037-8682-0142-2016. [DOI] [PubMed] [Google Scholar]
- 56.Gonçalves TCM, Oliveira E, Dias LS, Almeida M, Nogueira WO, Pires FDA. An investigation on the ecology of Triatoma vitticeps (Stal, 1859) and its possible role in the transmission of Trypanosoma cruzi, in the locality of Triunfo, Santa Maria Madalena municipal district, state of Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 1998;93(6):711–717. doi: 10.1590/S0074-02761998000600002. [DOI] [PubMed] [Google Scholar]
- 57.Lorosa ES, Valente MVMP, Cunha V, Lent H, Jurberg J. Foco de doença de Chagas em Arcádia, Estado do Rio de Janeiro, Brasil. Mem Inst Oswaldo Cruz. 2003;98:885–887. doi: 10.1590/S0074-02762003000700004. [DOI] [PubMed] [Google Scholar]
- 58.Lorosa ES, Santos CM, Juberg J. Foco da doença de Chagas em São Fidélis, no Estado do Rio de Janeiro. Rev Soc Bras Med Trop. 2008;41(4):419–420. doi: 10.1590/S0037-86822008000400020. [DOI] [PubMed] [Google Scholar]
- 59.Sangenis LHC, Saraiva RM, Georg I, Castro L, Lima VS, Roque ALR, et al. Autochthonous transmission of Chagas disease in Rio de Janeiro state, Brazil: a clinical and eco-epidemiological study. BMC Infect Dis. 2015;15:4. doi: 10.1186/s12879-014-0732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dias JCP, Feitosa VR, Filho ANF, Rodrigues VLC, Alencar AS, Sessa PA. Fonte alimentar e potencial vetorial de Triatoma vitticeps (Stal, 1859) com relação à doença de Chagas humana no estado do Espírito Santo, Brasil. Mem Inst Oswaldo Cruz. 1989;84:165–173. doi: 10.1590/S0074-02761989000800032. [DOI] [Google Scholar]
- 61.dos Santos CB, Ferreira AL, Leite GR, Ferreira GEM, Rodrigues AAF, Falqueto A. Peridomiciliary colonies of Triatoma vitticeps (Stal, 1859) (Hemiptera, Reduviidae, Triatominae) infected with Trypanosoma cruzi in rural areas of the state of Espírito Santo, Brazil. Mem Inst Oswaldo Cruz. 2005;100:471–473. doi: 10.1590/S0074-02762005000500004. [DOI] [PubMed] [Google Scholar]
- 62.dos Santos CB, Leite GR, Sessa PA, Falqueto A. Dynamics of feeding and defecation in Triatoma vitticeps (Stål, 1859) (Hemiptera, Reduviidae, Triatominae) and its potential in the transmission of Trypanosoma cruzi. Mem Inst Oswaldo Cruz. 2006;101:543–546. doi: 10.1590/S0074-02762006000500010. [DOI] [PubMed] [Google Scholar]
- 63.Gonçalves TCM, Victório VMN, Jurberg J, Cunha V. Biologia do Triatoma vitticeps (Stål, 1859) em condições de laboratório (Hemiptera: Reduviidae: Triatominae). I Ciclo evolutivo. Mem Inst Oswaldo Cruz. 1988;83:519–523. doi: 10.1590/S0074-02761988000400020. [DOI] [PubMed] [Google Scholar]
- 64.Duffy T, Bisio M, Altcheh J, Burgos JM, Diez M, Levin MJ, et al. Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in chagas disease patients. PLoS Negl Trop Dis. 2009;3(4):e419. doi: 10.1371/journal.pntd.0000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jansen AM, Xavier SCC, Roque ALR. The multiple and complex and changeable scenarios of the Trypanosoma cruzi transmission cycle in the sylvatic environment. Acta Trop. 2015;151:1–15. doi: 10.1016/j.actatropica.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 66.Araujo CAC, Waniek PJ, Jansen AM. TcI/TcII co-infection can enhance Trypanosoma cruzi growth in Rhodnius prolixus. Parasit Vectors. 2014;7:94. doi: 10.1186/1756-3305-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duffy T, Cura CI, Ramirez JC, Abate T, Cayo NM, Parrado R, et al. Analytical performance of a multiplex real-time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl Trop Dis. 2013;7(1):e2000. doi: 10.1371/journal.pntd.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vallejo GA, Guhl F, Schaub GA. Triatominae-Trypanosoma cruzi/T. rangeli: vector-parasite interactions. Acta Trop. 2009;110:137–147. doi: 10.1016/j.actatropica.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Garcia ES, Ratcliffe NA, Whitten MM, Gonzalez MS, Azambuja P. Exploring the role of insect host factors in the dynamics of Trypanosoma cruzi - Rhodnius prolixus interactions. J Insect Physiol. 2007;53:11–21. [DOI] [PubMed]
- 70.Garcia ES, Genta FA, Azambuja P, Schaub GA. Interactions between intestinal compounds of triatomines and Trypanosoma cruzi. Trends Parasitol. 2010;26(10):499–505. doi: 10.1016/j.pt.2010.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional file. Nucleotide sequences reported in this paper are available in the GenBank database under accession numbers: T. cruzi kDNA: U07846.1; T. cruzi satellite DNA: EU728662–EU728667; Triatomine 12S subunit ribosomal RNA: AF394519.1.


