Abstract
In order to enable low-cost, high-throughput generation of cistrome and epicistrome maps for any organism, we developed DNA affinity purification sequencing (DAP-seq), a transcription factor (TF) binding site discovery assay that couples affinity-purified TFs with next-generation sequencing of a genomic DNA library. The method is fast, inexpensive, and more easily scaled than ChIP-seq. DNA libraries are constructed using native genomic DNA from any source of interest, preserving cell- and tissue-specific chemical modifications that are known to impact TF binding (such as DNA methylation) and providing increased specificity compared to in silico motif prediction methods such as protein binding microarrays (PBM) and systematic evolution of ligands by exponential enrichment (SELEX). The resulting DNA library is incubated with an affinity-tagged in vitro expressed TF, and TF-DNA complexes are purified using magnetic separation of the affinity tag. Bound genomic DNA is eluted from the TF and sequenced using next-generation sequencing. Sequence reads are mapped to a reference genome, identifying genome-wide binding locations for each TF assayed, from which sequence motifs can then be derived. A researcher with molecular biology experience should be able to follow this protocol, processing up to 400 samples per week.
Keywords: Transcription factor, TF, DAP-seq, ChIP-seq, regulatory DNA, cistrome, epicistrome, DNA methylation, transcriptional regulation
INTRODUCTION
The binding of TFs to specific locations in a genome results in dynamic transcriptional changes that drive a vast array of cellular processes, including development and environmental response. Disruption of TF binding sites (TFBS) has been associated with phenotypic diversity, including agriculturally important adaptive traits1 and various disease states such as cancer2. Characterizing the genome-wide binding profiles of individual TFs is essential for identifying the mechanisms underlying these changes. Furthermore, coordination between different TFs via local and/or proximal binding sites likely influences gene expression2. Therefore, characterizing TFBS for all TFs within an organism is crucial for expanding our knowledge of complex phenotypic traits and gene expression networks. Based on the genomes of well-characterized model systems, multicellular organisms dedicate a significant portion of their protein coding genes (6–8%) to the expression of between 1000–2500 DNA-binding transcription factors3. With the exception of data from ENCODE and modENCODE projects4–7, in vivo genome-wide TF location data is available for relatively few TFs. To expand this analysis for a wider range of organisms, scalable methods are needed for the low-cost and high-throughput examination of thousands of TFs.
Chromatin immunoprecipitation sequencing (ChIP-seq) is the current leading method for determining in vivo TFBS, capturing genomic sites bound by a given TF in a tissue-specific chromatin context. However, the method is limited in its throughput by the need to create gene-specific antibodies or tagged transgenic lines, which can be technically challenging and expensive8. Alternative methods are needed for capturing genome-wide binding data for many organisms. In vitro TFBS identification methods such as Protein Binding Microarrays (PBM) and High Throughput Systematic Evolution of Ligands by Exponential Enrichment (HT-SELEX) have achieved the highest throughput for deducing TF binding specificities in vitro9–11, but these methods use short synthetic oligonucleotides lacking secondary DNA modifications and genomic context, both important determinants of selective TF binding in vivo12–16. The DAP-seq technique15 described here employs an in vitro-expressed affinity-tagged TF in combination with high-throughput sequencing of a genomic DNA library, allowing for the generation of genome-wide binding site maps reflective of both local sequence context and DNA methylation status.
Overview of DAP-Seq
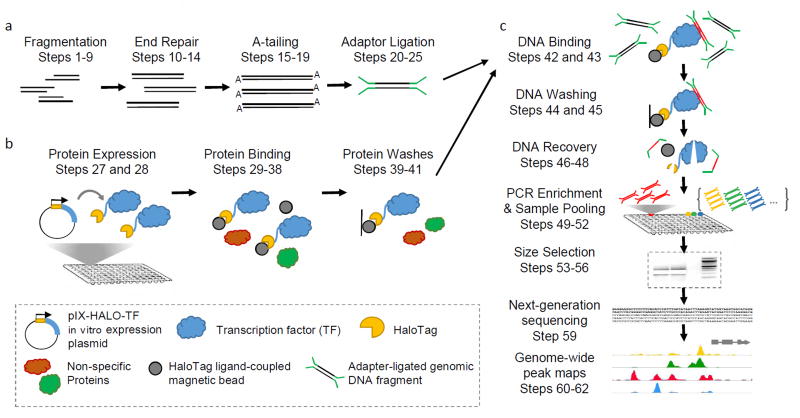
We developed a high-throughput TF-DNA binding assay called DAP-seq (DNA Affinity Purification and sequencing) that combines next-generation sequencing of a genomic DNA library with in vitro expression of affinity-purified TFs to generate cistrome and epicistrome maps for a wide range of species. A genomic DNA library is first prepared according to general library construction methods that include fragmentation of genomic DNA followed by ligation with Illumina-based sequencing adapters (Fig 1A). One genomic DNA library can be used for many individual DAP-seq experiments, increasing assay throughput and limiting library-to-library bias. To allow for high-throughput expression of affinity-tagged TFs, individual TF ORFs are transferred into the Gateway-compatible pIX-HALO expression vector, which contains an N-terminal HaloTag17 affinity tag sequence. The pIX-HALO-TF construct is then expressed using either a mammalian (reticulocyte) or plant (wheat germ) based in vitro transcription/translation coupled system (Fig 1B). Small-volume in vitro expression reactions produce an average of 50–1000 ng of HaloTag-fused protein in two hours, facilitating a rapid, high-throughput means of protein production. Once HaloTag-fused proteins are expressed, they are bound to magnetic HaloTag ligand (chloroalkane) beads and isolated from the expression system components (Fig 1B). Purified proteins are then combined with the adapter-ligated genomic DNA library and can bind to genomic DNA fragments in a sequence-specific manner (Fig 1C). After washing away the unbound fragments, the beads are boiled to denature the protein and release the DNA into solution. This DNA is then PCR amplified to attach multiplexing index sequences and enrich for TF-bound fragments. Samples are pooled and the resulting final library is sequenced using an Illumina sequencing platform (Fig 1C).
Figure 1. DAP-seq protocol overview.
(a) An adapter-ligated DNA library is prepared by shearing native genomic DNA into ~200bp fragments and ligating Illumina-based sequencing adapters onto the repaired ends. (b) Transcription factor (TF) ORF clones fused to the Halo affinity tag are expressed in vitro and bound to ligand-coupled beads, while non-specific proteins are washed away. (c) HaloTag-TF fusion proteins are incubated with adapter-ligated genomic DNA library, and unbound DNA fragments are washed away. Samples are heated to release TF-bound DNA and the recovered DNA is PCR amplified to attach indexed sequencing primers. Indexed DNA samples are subsequently combined and size selected to remove residual adapter dimers. Purified DNA libraries are sequenced using next-generation sequencing and the resulting genome-wide binding events are analyzed. Peaks shown are maize DAP-seq peaks viewed in the Integrative Genomics Viewer.
Standard DAP-seq libraries are generated using native genomic DNA that contains secondary modifications such as cytosine methylation. In order to observe binding in the absence of such features, we also developed a modified version of DAP-seq called ampDAP-seq, which uses a PCR-amplified genomic DNA library for protein binding. The amplification creates synthetic copies of the DNA strands, thereby removing all secondary DNA modifications. DAP-seq and ampDAP-seq binding profiles for a particular TF can then be compared to assess dataset-specific protein binding sites15. In addition, using the DAP-seq library as the source for the ampDAP-seq library ensures a low level of sample-to-sample bias.
Analysis of DAP-seq data
The binding site data generated by DAP-seq is robust and shares similarities to that produced by ChIP-seq. DAP-seq data can therefore be analyzed by standard peak-calling and motif-characterization software, facilitating downstream analysis such as target gene identification and integration with tissue-specific ChIP-seq datasets. For our DAP-seq data processing we typically only allow uniquely-mapped reads to be used in subsequent analyses, effectively masking repeat regions that may give false peaks. Additionally, the use of a negative control sample (pIX-HALO empty vector or input DNA) as background can substantially reduce or eliminate false peak signals. Direct comparison of multiple DAP-seq and ChIP-seq datasets for a handful of TFs has shown that DAP-seq peaks have a good rate of overlap with ChIP-seq peaks (36–81%) and are particularly pronounced in ChIP-seq peaks containing high motif scores (69–97%)15. As most DAP-seq peaks are predominantly associated with high-scoring motifs, these results suggest that many DAP-seq peaks likely correspond to direct in vivo TF binding targets. In contrast, most of the ChIP-seq peaks that do not overlap with DAP-seq peaks do not contain a detectable consensus motif and may thus result from indirect binding18. Comparison of DAP-seq and ChIP-seq datasets could therefore be used to find indirect binding sites, i.e., those identified in ChIP-seq datasets but not DAP-seq datasets.
Cell-line and tissue-specific DNA modifications are central features of multicellular development, and there are substantial differences in DNA methylation between different cell and tissue types in both humans and plants19,20. Understanding how these elements impact cellular activity is important for linking epigenetic changes with phenotypes. Integration of DAP-seq datasets with orthogonal datasets such as genome-wide DNA methylation maps can be used to assess the impact of DNA modifications on TF binding. Comparison of a leaf-specific genome-wide methylation map and hundreds of Arabidopsis DAP-seq TF datasets concluded that the binding activities of 76% of TFs were influenced by DNA methylation15. This analysis was further supported by comparison with ampDAP-seq datasets in which de novo TF binding events were observed when methylation was removed.
Advantages and limitations
A major advantage of DAP-seq compared to alternative approaches (e.g. ChIP-seq) is that it is easily scaled for high-throughput sample processing of entire TF ORF clone collections. No sample-specific reagents such as antibodies or gene-specific primers are needed, making a full TFome screen a straightforward, relatively low-cost endeavor. The sensitivity of next-generation sequencing means that the assay relies on relatively small amounts of both genomic DNA and affinity-tagged protein, which greatly accelerates the rate at which samples can be processed and information can be collected.
However, DAP-seq is subject to some technical limitations. Only ~30% of the 1,812 Arabidopsis TFs assayed in a recent large-scale cistrome mapping screen produced TFBS datasets that passed quality thresholds. Technical failures associated with high-throughput processing accounted for only about 10% of these assay failures, suggesting that additional factors influence the success rate of DAP-seq. The cause of these failures is unknown but seems to be dependent on TF-specific DNA binding properties related to the in vitro expressed protein or the absence of co-factors or other protein partners necessary for DNA binding. This is reflected in the finding that success rates tended to be consistently higher for some TF families (e.g. bZIP and NAC) than others (e.g. bHLH and MADS-box)15.
Other technical difficulties of DAP-seq arise most notably from issues related to inadequate amounts of protein. Low protein expression (often caused by an insufficient amount of expression plasmid DNA) can decrease DAP-seq success rates. It is therefore advisable to check the concentration of input plasmid prior to the expression reaction and, if large numbers of TFs are being processed, to normalize the amount of DNA added. The protein expression system used to generate affinity-tagged protein can also impact the success rate of DAP-seq. The pIX-HALO vector functions in both plant-based and mammalian in vitro expression systems, providing the option to vary the expression environment. In several cases where in vitro expressed Halo-Tag fusion proteins failed to produce successful DAP-seq datasets, GST-tagged fusion proteins expressed recombinantly in E.coli gave rise to high-quality DAP-seq data. Therefore, while this article focuses on performing high-throughput DAP-seq using in vitro expressed TFs, we have also included a brief supplemental protocol that describes the use of purified GST-fusion proteins expressed in E.coli on a smaller scale (Box 1).
BOX 1. Small-scale DAP-seq using E.coli expressed recombinant protein.
PROTEIN EXPRESSION AND PURIFICATION IN E. COLI – TIMING 1–2 days
-
1)
Perform standard purification of GST-tagged protein expressed in E.coli32,33. Typical culture volumes range from 2–500ml.
-
2)
Elute TF from glutathione-sepharose beads using excess reduced glutathione.
-
3)
Concentrate the protein if necessary and remove excess glutathione by dialysis or buffer exchange.
BINDING OF GST-FUSION PROTEIN – TIMING ~1.5hr
-
4)
Add 25µl MagneGST beads per sample to a 1.5ml microcentrifuge tube. Place on magnet and aspirate supernatant.
-
5)
Wash beads with 500 µl PBS. Place on magnet and aspirate supernatant. Repeat three times.
-
6)
Dilute 1–20 µl (~0.5–5µg) of purified GST-fusion protein in PBS for a final volume of 400 µl and add to washed MagneGST beads.
-
7)
Rotate for 1 hour at room temperature to bind TF to beads.
-
8)
Quick spin samples to collect liquid in tube bottom. Place on magnet and aspirate supernatant.
-
9)
Wash beads with 500 µl PBS + NP40 (0.005%). Place on magnet and aspirate supernatant. Repeat four times.
-
10)
Wash beads with 500 µl PBS. Place on magnet and aspirate supernatant. Repeat two times.
BINDING OF DNA TO PROTEINS – TIMING ~1.5 hr
-
11)
Resuspend beads in 40 µl PBS.
-
12)
Dilute 50–1000 ng of DNA library (steps 1–25 of PROCEDURE) in PBS for a final volume of 40µl. Add to resuspended beads from step 9.
-
13)
Rotate horizontally at room temperature for 1 hour
-
14)
Place tube on magnet and remove supernatant.
-
15)
Wash beads with 200 µl PBS + NP40 (0.005%). Place on magnet and aspirate supernatant. Repeat four times.
-
16)
Wash beads with 200 µl PBS. Place on magnet and aspirate supernatant. Repeat once more. On the final wash, transfer buffer and beads to a new tube.
DNA RECOVERY AND AMPLIFICATION – TIMING ~1.5hr
-
17)
Place tube on magnet and remove supernatant.
-
18)
Resuspend beads in 25 µl of EB.
-
19)
Place tube at 98°C for 10min.
-
20)
Place tube on magnet and aspirate supernatant containing eluted DNA.
-
21)
Proceed with PCR enrichment (steps 49–51 of PROCEDURE).
-
22)
Purify PCR products using the size selection gel extraction method described in steps 52–56 of PROCEDURE or perform a bead purification using AMPure XP beads or equivalent at a 1:1 ratio of beads to PCR product34 if individual purification of samples is preferred.
-
23)
Quantify library as described in step 52 of PROCEDURE.
Finally, although DAP-seq retains many of the tissue/cell line-specific secondary modifications and features present in genomic DNA, the effect of additional genomic elements (such as chromatin accessibility and histone modifications) are not reflected in DAP-seq datasets. This aspect allows visualization of global TF binding events in a chromatin-free context but fails to capture these important tissue-specific dynamics. One powerful way to overcome this limitation is to overlay tissue-specific chromatin accessibility information from methods such as DNase-seq, ATAC-seq, and MNase-seq on DAP-seq datasets21–23. Integration of DAP-seq and DNase hypersensitivity data from multiple Arabidopsis tissue types showed that DAP-seq captures in vivo binding sites that correspond to multiple tissue-specific binding events15. This type of analysis offers a cost-effective means to assess the TF binding landscape across many tissues and cell-types without having to perform thousands of individual ChIP-seq experiments.
Applications
We have successfully performed the DAP-seq method using Arabidopsis, maize15 and human (unpublished) TFs. As the method requires only TF clones and genomic DNA, it can be adapted for any organism with a sequenced genome. DAP-seq is particularly attractive for species where generation of transgenic lines for ChIP-seq is lengthy, costly, or technically challenging. DAP-seq can also be modified to study the effects of various perturbations such as nucleotide variation in DNA-binding domains (genetic variants) or TF binding variation due to DNA methylation (epigenomic variants)24. Furthermore, as the assay is in vitro, additional components or protein interaction partners can easily be added to assess their impact on DNA binding without interference from unknown factors. Finally, the comparative binding profiles of a particular TF can be studied using genomic DNA from different cell lines or tissue types containing source-specific DNA chemical modifications. We focus on cytosine methylation in this paper, but DAP-seq could also be used to study rarer modifications that also may affect TF binding25. The use of endogenous genomic DNA should preserve these modifications and allow them to be profiled in a similar fashion.
Experimental Design
Quality of Genomic DNA
The success of DAP-seq strongly depends on the quality of the genomic DNA library. We prepare genomic DNA from frozen tissue using a phenol:chloroform-based extraction protocol and quantify the amount of purified DNA using a fluorometric method such as Qubit (ThermoFisher). Using a Nanodrop to quantify genomic DNA for library preparation is not recommended as minor impurities in the purified DNA sample, particularly from plant material, can make quantification inaccurate. The library preparation protocol described below is based on a starting amount of 5 µg of genomic DNA. To scale up, multiply all reaction amounts proportionately. Before the TF affinity purification steps are performed, efforts should be made to ensure that the adapter-ligated library is of high quality. Failure to test the library could result in reduced amplification and motif/binding site recovery. We typically test 5 ng of the library by quantitative PCR (qPCR) to verify that the adapters have been ligated effectively and that the library will amplify. A protocol describing the qPCR steps is presented in Box 2.
BOX 2. Quality Control Check on gDNA Library.
CRITICAL: This experiment can be done using the KAPA Library Quantification Kit for Illumina Platforms (KK4824 for Universal qPCR master mix or kits optimized for specific thermocyclers), substituting the primer mix in the kit with Primer A and Primer B. Alternatively, we use the following qPCR setup that is consistent with subsequent steps in the preparation of ampDAP-seq libraries and final sequencing libraries. Only one qPCR verification (with replicates) is needed for each batch of library.
Verification of Adapter Ligation by Quantitative PCR (qPCR) and BioAnalyzer/agarose gel - TIMING ~60 min
Dilute the sample from Step 24 of PROCEDURE to 0.5 ng/uL.
- Use 10uL of diluted DNA library in the following PCR reaction. As negative controls, include one PCR reaction using DNA library prior to ligation and one with no template.
Component Amount per
reaction (µL)Final
amount/concentrationLigated DNA library (0.5 ng/uL) 10 5ng Water 2.6 - 5× Phusion HF Buffer 4 1× 10 mM dNTPs 1 500 µM Primer A (25 µM) 0.4 0.5 µM Primer B (25 µM) 0.4 0.5 µM Phusion DNA Polymerase (2000 U/mL) 0.4 2 U 10× SYBR Green I 1.2 0.6× - Load the sample and run the following program on a real-time PCR instrument. Select the ‘Absolute Quantification’ option with SYBR Green dye in the instrument software.
Cycle
NumberDenature Anneal Extend 1 95°C for 2 min --- --- 2 98°C for 30 s --- --- 3–32 98°C for 15 s 60°C for 30 s 72°C for 2 min 33 --- --- 72°C for 10 min Compare the amplification curves of the ‘adapter ligated library’ and ‘library without adapter’ samples. The ‘adapter ligated library’ should amplify to saturation (usually less than 12 cycles) while the ‘library without adapter’ should not amplify.
Run the qPCR products on an agarose gel or BioAnalyzer. We typically run a 1.5% agarose gel at 100 V for 45 min. The ‘adapter ligated library’ should be ~120 bp longer than the ‘library without adapter‘.
Quantity of DNA Library Input
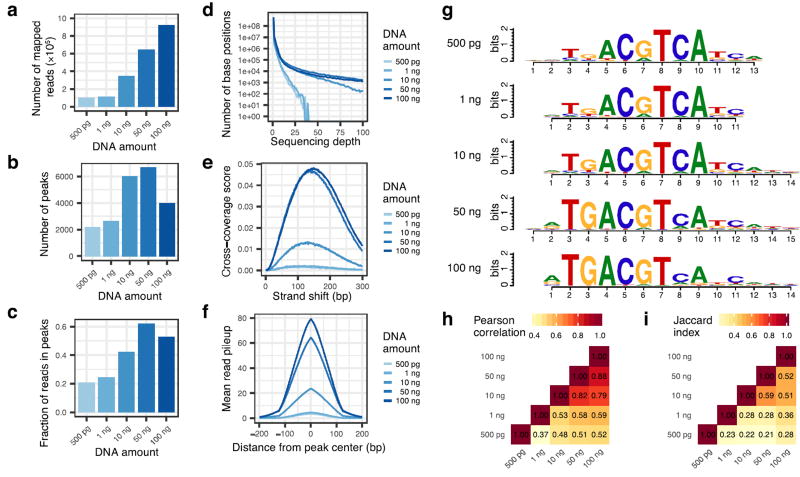
The amount of DNA library used in a DAP-seq experiment is highly dependent on genome size. For a relatively small genome (i.e. Arabidopsis thaliana) 30 ng is sufficient, but with larger genomes (e.g. human or maize) we find 100 ng produces better results. In order to establish an optimal input amount, we typically run a DNA library titration experiment using positive control proteins. Based on these data, we select an input amount that maximizes multiple criteria including number of peaks, percent reads in peaks (5% minimum), consistency of motif calls (with those published in literature and with experiments containing higher input amounts), and a quantitative correlation of reads in the union peak set of replicate experiments (Fig 2).
Figure 2. DAP-seq DNA library titration experiment with Arabidopsis transcription factor TGA5 (AT5G06960).
(a) Number of mapped reads increases with input library amount. (b) Number of peaks called is highest at 50 ng of input then decreases, likely due to increase in background noise, consistent with the reduction in fraction of reads in peaks (c). (d) Coverage histogram of the number of positions in the genome (y-axis) covered at each sequencing depth (x-axis) shows that input of 10, 50, 100 ng leads to enrichment (i.e., high sequencing depth) at a subset of base positions. Strand shift cross-coverage score (e) and mean read pileup at peaks (f), both indicators of the extent of enrichment at specific genomic locations, are much higher for 50 and 100 ng input. (g) The sequence motifs identified from the top 600 peaks of all experiments are very similar. Similarities between pairs of experiments, both quantitative (h – Pearson correlation of reads in peaks) and qualitative (i – Jaccard index of peak regions) show that experiments of 10, 50, and 100 ng identify similar genome-wide binding profiles. DNA library was prepared from A. thaliana ecotype Col-0. Following the DAP-seq protocol with the TF TGA5, the resulting libraries were sequenced on an Illumina HiSeq 4000 instrument with 100-bp paired-end reads. Reads were mapped using bowtie228 version 2.2.9 against the TAIR10 reference with default parameters. Reads aligned to nuclear chromosomes with MAPQ score ≥30 were used to call peaks by the GEM peak caller30 using only first read in the pair with parameters “--k_min 6 --k_max 20 --k_seqs 600 --outNP --outMEME --outJASPAR --k_neg_dinu_shuffle --t 11”. The top 600 peaks, ranked first by enrichment q-value then by fold enrichment, were used for de novo motif discovery by MEME-ChIP36 version 4.11.2. Fractions of reads in peaks, coverage histograms, strand shift cross-coverage scores, mean read pileup at peaks, and Pairwise Pearson Correlations were computed by the R package ChIPQC37 version 1.10.2. Motif logos were drawn by the R package motifStack38 version 1.16.2. Pairwise Jaccard Index were computed by BEDTools39 version 2.24.0.
AmpDAP-seq
An extra step can be added to the library preparation protocol to abolish the native methylation present in a genomic DNA library. We call this modified procedure ampDAP-seq as it involves amplifying a small amount of the adapter-ligated genomic DNA library to create synthetic copies of the DNA that are no longer methylated. This modification-free DNA library is then used in the standard DAP-seq procedure. Comparison of ampDAP-seq and DAP-seq binding events will reveal regions where the TF binds only when cytosine DNA methylation and other DNA modifications are absent. Such analysis reveals the impact of secondary modifications on DNA binding for a particular TF.
Controls
Both positive and negative controls are typically included on each 96-well plate of DAP-seq samples. For Arabidopsis samples, we typically use TGA5 (bZIP) and ANAC096 (NAC) as positive controls to judge the overall success rate for each plate, as these TFs give consistent results in repeated DAP-seq experiments. For a negative control, we use an empty vector sample expressing only the HaloTag or a HaloTag-GST fusion plasmid. This control will reveal non-specific peaks resulting from the DAP-seq procedure or from other proteins in the expression system that may be inadvertently carried through the bead-binding steps.
Protein expression
DAP-seq should work with any affinity-tagged TF. Both protein expression and subsequent binding to the beads (verified by Western blot; Box 3) are crucial for a successful experiment. For high-throughput processing of samples, we use the Gateway-compatible pIX-HALO in vitro expression vector,26,27 which has both T7 and SP6 promoters. This vector functions equally well in Promega’s wheat germ and rabbit reticulocyte TNT expression systems, and both have been used with success in DAP-seq. The HaloTag17 binds rapidly and irreversibly (covalently) to a synthetic chloroalkane ligand that is coupled to magnetic beads, giving rise to very low background. The pIX-HALO vector contains a C-terminal 6×His tag immediately following the Gateway recombination site. All TF clones tested in our screens contained endogenous stop codons, so we cannot assess whether expression of the 6×His tag would impact DNA binding or background levels. The pIX-HALO gateway destination vector is available from ABRC via the following link: http://www.arabidopsis.org/servlet/TairObject?id=1001200298&type=vector.
BOX 3. Quality Control Check on Fusion Protein Expression and Bead Binding.
To verify expression of full-length fusion protein and subsequent binding to the HaloTag beads, a Western blot can be performed.
In the first well, load a standard protein ladder.
In the second well, load 10% of the expression reaction sample from Step 36 of PROCEDURE (typically 5µL) + 1× LDS with 4% BME.
In the third well, load 10% of the supernatant after protein binding from Step 38 of PROCEDURE (typically 8µL) + 1× LDS with 4% BME.
Repeat steps 2 and 3 for all additional samples across successive wells.
Run gel, transfer to a nitrocellulose or PVDF membrane, and block non-specific sites as for a standard Western blot35.
Incubate membrane with a 1:1500 anti-HaloTag monoclonal antibody (1 mg/mL) for 1 hour at room temperature. After washing, incubate with a 1:100,000 dilution of anti-mouse secondary antibody (0.8 mg/mL) for 1 hour at room temperature.
Detect proteins with ECL Western Blotting Substrate according to the manufacturer’s specifications.
Verify the size of the protein in the expression reaction sample. It should be the size of the protein of interest plus the HaloTag (~34kDa).
Verify that there is a moderate decrease (preferably about 70% or more) in amount of protein between the expression reaction and the supernatant, indicating that at least some of the protein was bound to the beads.
MATERIALS
Reagents
5 µg genomic DNA
96 TF ORFs in pIX-HALO vector
Elution Buffer (Qiagen, cat. no. 19086)
3 M NaOAc, pH 5.2 (Thermo Scientific, cat. no. PI17888)
100% Ethanol (200 proof; Sigma-Aldrich, cat. no. 792780)
End-It DNA End-Repair Kit (Epicentre, cat. no. ER81050)
100 mM dNTPs (Biopioneer, cat. no. MDM-4)
Klenow (3’–5’ exo-, NEB, cat. no. M0212)
T4 DNA Ligase (Promega, cat. no. M1804)
-
Y-adapter (Truncated Illumina TruSeq Adapter):
Strand A 5’ ACACTCTTTCCCTACACGACGCTCTTCCGATCT 3’;
Strand B 5’ P-GATCGGAAGAGCACACGTCTGAACTCCAGTCAC 3’ (where ‘P’ indicates a 5’ phosphate group)
-
25 µM PCR primers:
Primer A (Illumina TruSeq Universal Primer): 5’AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT 3’;
Primer B (Illumina TruSeq Index Primer): 5’ CAAGCAGAAGACGGCATACGAGAT-NNNNNN-GTGACTGGAGTTCAGACGTGTGCTCTTCCG 3’ (where ‘NNNNNN’ represents the 6mer index sequence for multiplexing)
Phusion High-Fidelity DNA Polymerase (NEB, cat. no. M0530)
TNT SP6 Coupled Wheat Germ Extract System (Promega, cat. no. L4130) or TNT SP6 Coupled Reticulocyte Lysate System (Promega, cat. no. L4600). Promega T7 systems can be used as well.
Recombinant RNasin® Ribonuclease Inhibitor (Promega, cat. no. N2515)
Magne HaloTag Beads (Promega, cat. no. G7282)
MagneGST Glutathione Particles (if using E.coli expressed recombinant protein, Promega, cat. no. V8611)
PBS + NP40, pH 7.4
Agarose (Biopioneer, cat. no. C0009)
Gel Extraction Kit (Qiagen, cat. no. 28704)
Qubit dsDNA High-Sensitivity (HS) Assay Kit (Life Technologies, cat. no. Q32851)
Anti-HaloTag® Monoclonal Antibody (optional, Promega, cat. no. G9211)
Equipment
S-series focused ultrasonicator (Covaris)
MicroTUBE holder (Covaris, cat. no. 500114)
Snap-Cap microTUBEs (Covaris, cat. no. 520045)
Microcentrifuge Tubes, 1.5 mL
Microcentrifuge
Thermocycler
Low-binding 96-well PCR plates (RPI, cat. no. 141314)
DynaMag-2 Magnet (Life Technologies, cat. no. 12321D)
Tube/Plate Rotator
Adhesive Foil (E&K Scientific, cat. no. T592100)
DynaMag-96 Side Magnet (Life Technologies, cat. no. 12331D)
Gel Electrophoresis Box
Qubit Fluorometer (Life Tech, cat. no. Q33216)
Reagent Setup
Ethanol, 70% (vol/vol) To 35 mL of 200 proof ethanol, add 15 mL of sterile water. Keep sealed when not in use. Prepare a fresh solution before each library preparation.
dATP, 1 mM Add 1µL of 100 mM dATP to 99 µL of sterile water. Store at −20°C for up to 1 yr.
dNTPs, 10 mM Add 1 mL each of 100 mM dATP, dTTP, dCTP, dGTP to 6 mL of sterile water. Aliquot into 1.5 mL tubes. Store at −20°C for up to 1 yr.
Annealed Y-adapter: Dilute each adapter strand to 100 µM with sterile water. Combine 150 µL of each oligo into 200 µL of sterile water (total of 500 µL). Incubate the mixture at 96°C for 2 minutes. Remove the tube from the heat source and cool to room temperature (22°C) on the bench top. Store at −20°C for up to 6 months.
PBS+NP40: Dissolve 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4 and 0.24 g KH2PO4 in 800 mL of sterile water. Adjust pH to 7.4 with HCl. Add water to 1 liter and sterilize by autoclaving. Add 200 µL of 25% NP40 and mix well. Store at room temperature for up to 1 month.
Equipment Setup
‘200-bp target peak size’ protocol: Set up a program on the Covaris focused-ultrasonicator with the following conditions: Duty Cycle, 10%; Intensity, 5; Cycles/Burst, 200; Time, 60s; Number of cycles, 3.
PROCEDURE
DNA Library Prep for DAP-seq – TIMING 8.5 hr
-
1)
FRAGMENTATION – TIMING ~5 min Dilute 5 µg of genomic DNA in Elution Buffer (EB) for a total of 125 µL.
-
2)
Transfer the entire sample into a Covaris microTUBE and sonicate using the ‘200-bp target peak size’ protocol with a Covaris S2 (see Equipment Setup).
-
3)
SAMPLE CLEANUP – TIMING ~60 min. Transfer the sonicated sample into a clean 1.5mL tube. Add 12.5µL 3M NaOAc (0.1× volume) and 250 µL cold 100% ethanol (2× volume). Vortex to mix.
-
4)
Incubate on ice or at −20°C for at least 15 min, but no longer than 24 hours.
-
5)
Centrifuge at maximum speed (20,000 g) for 20 minutes at 4°C. Discard supernatant by decanting.
-
6)
Wash pellet with 1 mL 70% ethanol. Centrifuge at maximum speed (20,000 g) for 10 minutes at 4°C. Discard supernatant by decanting.
-
7)
Quick spin for 5 sec at 3000 g and pipette off any remaining ethanol, being careful not to disturb the DNA pellet.
-
8)
Allow pellet to dry 10–15 min at room temperature (22°C) or 5–10min at 37°C. Be sure pellet is completely dry before resuspending. Allowing the pellet to dry too long may make it more difficult to resuspend, but should not harm the DNA.
-
9)Resuspend DNA pellet in 34 µL EB. Place at 37°C for 5 minutes to help dissolve DNA.
-
▪PAUSE POINT – Samples can be stored at 4°C overnight or at −20°C for 1 week.
-
▪
-
10)END REPAIR – TIMING ~45 min. Add the following reagents to set up the End Repair reaction:
Component Amount per reaction
(µL)Final
amount/concentration10× End-It buffer 5 1× 10mM dNTP mix 5 1mM 10 mM ATP 5 1mM End-It enzyme mix 1 - -
11)
Mix gently and quick spin for 5 sec at 3000 g. Incubate at room temperature for 45 min.
-
12)
SAMPLE CLEANUP – TIMING ~60 min. Add 5 µL 3 M NaOAc and 100 µL cold 100% ethanol to sample. Vortex to mix.
-
13)
Repeat steps 4–8 to perform ethanol precipitation.
-
14)Resuspend DNA in 32 µL EB. Place at 37°C for 5 minutes to aid resuspension.
-
▪PAUSE POINT – Samples can be stored at 4°C overnight or at −20°C for 1 week.
-
▪
-
15)A-TAIL REACTION– TIMING ~30 min. Add the following reagents to set up the A-tailing reaction:
Component Amount per
reaction (µL)Final
amount/concentration10× NEBuffer2 5 1× 1 mM dATP 10 200µM Klenow Fragment (3’–5’ exo- ; 5U/µL) 3 15 U -
16)
Mix gently and quick spin for 5 sec at 3000 g. Incubate at 37°C for 30 min.
-
17)
SAMPLE CLEANUP – TIMING ~60 min. Add 5 µL 3 M NaOAc and 100µL cold 100% ethanol to sample. Vortex to mix.
-
18)
Repeat steps 4–8 to perform ethanol precipitation.
-
19)
Resuspend DNA in 30 µL EB. Place at 37°C for 5 minutes to aid resuspension.
-
20)ADAPTER LIGATION -TIMING ~180 min. Add the following reagents to set up the adapter ligation:
Component Amount per
reaction (µL)Final amount/concentration T4 DNA Ligase 10× Buffer 5 1× 30 µM Y-Adapter 10 6µM T4 DNA Ligase (3U/µL) 5 15 U -
21)Mix gently and quick spin for 5 sec at 3000g. Incubate at room temperature for 3hr.
-
▪PAUSE POINT – Samples can be stored at 4°C overnight or at −20°C for 1 week.
-
▪
-
22)
SAMPLE CLEANUP AND QUANTIFICATION – TIMING ~130 min. Add 5 µL 3 M NaOAc and 100 µL cold 100% ethanol to sample. Vortex to mix.
-
23)
Repeat steps 4–8 to perform ethanol precipitation.
-
24)
Resuspend DNA in 31 µL EB. Place at 37°C for 5 minutes to aid resuspension.
-
25)Measure the DNA concentration of 1 µL of sample using the Qubit dsDNA HS assay according to manufacturer’s specifications. Test 5 ng of the adapted library by qPCR to verify that the adapters have bound correctly and the library will amplify (Box 2).
-
▪CRITICAL STEP For 5 µg of starting material, expect to recover a final amount of 2.5–5 µg of DNA library. As long as you have enough DNA for your DAP-seq experiments (30–100ng per reaction) and have confirmed its quality, you may continue with the protocol.
-
▪PAUSE POINT – Samples can be stored at 4°C overnight or at −20°C for 1 week.
-
▪TROUBLESHOOTING ?
-
▪
-
26)
(OPTIONAL) AMPLIFY DNA TO REMOVE METHYLATION AND CREATE AMPDAP LIBRARY – TIMING ~120 min. Follow protocol in Box 4
BOX 4. AMPLIFY DNA TO REMOVE METHYLATION AND CREATE AMPDAP LIBRARY.
- Use 15ng of DNA library in the following PCR reaction (scale up as necessary):
Component Amount per
reaction (µL)Final amount/
concentrationLigated DNA x 15 ng Water 34.5 - x - 5× Phusion HF Buffer 10 1× 10 mM dNTPs 2.5 500 µM Primer A (25 µM) 1 0.5 µM Primer B (25 µM) 1 0.5 µM Phusion DNA Polymerase (2000U/mL) 1 2 U -
▪CRITICAL STEP Be sure to record which index on primer B you use to amplify each sample. This should be the same primer you use in the DAP-seq enrichment step (step 54).
-
▪
- Load the samples into a thermocycler and run the following program:
Cycle
NumberDenature Anneal Extend 1 95°C for 2 min --- --- 2 98°C for 30 s --- --- 3–13 98°C for 15 s 60°C for 30 s 72°C for 2 min 14 --- --- 72°C for 10 min -
▪CRITICAL STEP We have found that eleven cycles of PCR provides a good balance between yield and amplification bias. Performing fewer than eleven cycles does not produce adequate amounts of library for subsequent steps while a greater number of cycles can generate higher amplification bias that could interfere with peak analysis.
-
▪
Pool replicate wells together into a 1.5 mL tube. Add 10% volume 3M NaOAc and 200% volume cold 100% ethanol to the sample. Vortex to mix.
Follow steps 4–8 of PROCEDURE to perform ethanol precipitation.
Resuspend DNA in 31 µL EB. Place sample at 37°C for 5 minutes to aid resuspension.
- Use 1 µL of sample to measure the DNA concentration using the Qubit HS dsDNA assay according to manufacturer’s specifications.
-
▪PAUSE POINT – Samples can be stored at 4°C overnight or at −20°C for 1 week.
-
▪TROUBLESHOOTING ?
-
▪
DAP-seq Protocol – TIMING 7.5 hr
-
27)PROTEIN EXPRESSION – TIMING ~120 min. Assemble a 50 µL TNT expression reaction in a 96-well PCR plate according to the manufacturer’s specifications, using 1 µg of pIX-Halo-ORF plasmid DNA per reaction.
-
▪CRITICAL STEP While we generally find that adding between 400–2500 ng of plasmid DNA does not affect the success of a DAP-seq experiment, using less than 400ng of plasmid DNA will greatly decrease the chance of a successful DAP-seq experiment.
-
▪
-
28)
Mix gently by pipetting and incubate for 2 hr at 30°C.
-
29)
BINDING OF HALO-FUSION PROTEIN – TIMING ~70 min. Transfer 1 mL of Magne ® HaloTag ® Beads to a clean 1.5 mL tube.
-
30)
Place tube on magnetic rack and pipette off buffer once the solution has cleared and all the beads have been drawn to the magnet.
-
31)
Remove tube from magnetic rack and wash beads with 1 mL PBS+NP40. Pipette up and down to mix thoroughly.
-
32)
Repeat steps 30–31 twice more for a total of three washes.
-
33)
Place tube on magnetic rack and pipette off buffer. Remove tube from magnetic rack and add 950µ L PBS+NP40. This should yield a total volume of around 1mL with the beads.
-
34)Aliquot 10 µL of washed beads to each well of a 96-well PCR plate using a multi-channel pipette.
-
▪CRITICAL STEP Be sure to thoroughly mix beads by pipetting up and down before preparing aliquots. The beads settle very quickly and will be unevenly distributed if not constantly mixed.
-
▪
-
35)
Add 30 µL PBS+NP40 to each well for a total of 40 µL.
-
36)
Add 40 µL of expression reaction to each well of the plate. Save remaining 10µL for QC analysis (Box 3) and store at 4°C overnight or −20°C for up to 1 month.
-
37)Securely seal 96-well plate with adhesive foil and rotate at room temperature for 1 hr. It should be rotated end-over-end to keep the beads in solution.
-
▪CRITICAL STEP The beads must stay in solution to increase binding surface area. If the beads sit together at the bottom of the well, only the top beads will be accessible for binding.
-
▪TROUBLESHOOTING ?
-
▪
-
38)
Quick spin the plate for 5 sec at 3000 g to ensure samples are at the bottom of the wells. Place plate on 96-well magnetic rack. Transfer supernatant to a clean PCR plate for later QC analysis (Box 3). Saved supernatant can be stored at 4°C overnight or −20°C for 1 week.
-
39)
PROTEIN WASHES – TIMING ~20 min Remove plate from magnetic rack and add 85 µL PBS+NP40. Allow the beads to fall naturally through the buffer and settle in the well.
-
40)
Place plate on the magnetic rack and remove the supernatant.
-
41)
Repeat steps 39–40 twice for a total of three washes.
-
42)BINDING OF DNA TO PROTEINS – TIMING ~70 min. Resuspend beads in 40 µL PBS+NP40. Add 30–100 ng DNA library (sample from step 24 for DAP-seq or step 26 for ampDAP-seq) to each reaction and bring the volume up to 80 µL with EB.
-
▪TROUBLESHOOTING ?
-
▪
-
43)
Securely seal the 96-well plate with adhesive foil and rotate at room temperature for 1 hr.
-
44)
DNA WASHES – TIMING ~20 min Place plate on magnetic rack and remove supernatant.
-
45)Repeat steps 39–40 four times to wash the beads. On the last wash, transfer buffer and beads to a new plate.
-
▪CRITICAL STEP This step washes away excess DNA that is not bound to proteins. Doing fewer washes may result in higher background.
-
▪CRITICAL STEP Transferring the beads to a new plate eliminates possible non-specific DNA carry-over that could lead to higher background.
-
▪
-
46)DNA RECOVERY AND AMPLIFICATION – TIMING ~100 min. Place plate on magnet. Remove the supernatant and resuspend the beads in 30 µL EB.
-
▪PAUSE POINT – Samples may be stored at 4° overnight.
-
▪
-
47)
Seal well with adhesive foil and place plate in thermocycler at 98°C for 10 min.
-
48)
Immediately place on ice for 5 min.
-
49)Assemble the following PCR reaction in a clean PCR plate, aliquoting 25ul to each well:
Component Amount per
reaction (µL)Final
amount/concentrationWater 9.5 - 5× Phusion HF Buffer 10 1× 10 mM dNTPs 2.5 500 µM Primer A (25 µM) 1 0.5 µM Primer B (25 µM) 1 0.5 µM Phusion DNA Polymerase (2000 U/mL) 1 2 U -
▪CRITICAL STEP Using a unique index for each sample will allow you to easily pool samples for sequencing. Be sure to record the index used for each sample as this will be necessary for pooling and eventual demultiplexing.
-
▪
-
50)
Quick spin DNA-containing plate for 5 sec at 3000 g and place on magnetic rack. Transfer 25 µL of supernatant into each well of the PCR reaction plate.
-
51)Seal with adhesive foil, vortex, and quick spin for 5 sec at 3000 g. Place in thermocycler and run the following program:
Cycle
NumberDenature Anneal Extend 1 95°C for 2 min --- --- 2 98°C for 30 s --- --- 3–22 98°C for 15 s 60°C for 30 s 72°C for 2 min 23 --- --- 72°C for 10 min -
▪PAUSE POINT – Samples can be stored at 4°C overnight or at −20°C for at least 6 months.
-
▪
-
52)POOLING AND SIZE SELECTION – TIMING ~100 min Combine 5 µL of each sample into a 1.5 mL tube. Seal and store the rest of the PCR plate for later analysis of individual samples or new pools if desired. The plate can be kept at −20°C for up to 6 months.
-
▪CRITICAL STEP Do not pool samples with the same index. You will not be able to demultiplex them in your sequencing data.
-
▪CRITICAL STEP The number of samples that can be pooled together will depend on the species. For example, 96 Arabidopsis samples can be pooled together, but human sample pools should contain fewer samples each since they will require greater sequencing depth.
-
▪
-
53)Aliquot approximately 60 µL of the pool into a new 1.5 mL tube and add 12 µL of 6× gel loading dye. Load the sample across two large wells (~35 µL each) of a 1% (wt/vol) agarose gel containing 0.005% (vol/vol) ethidium bromide. CAUTION Ethidium bromide is mutagenic and should be handled with care.
-
▪CRITICAL STEP More of the pool can be loaded into the gel if desired, but we have found this to yield a sufficient amount of DNA for sequencing.
-
▪
-
54)
Run at 100 V for 20 min, or until smear has separated from primer dimer (~125bp).
-
55)Using a new scalpel blade, cut out a ~200–400 bp DNA smear from gel.
-
▪TROUBLESHOOTING ?
-
▪
-
56)
Extract DNA using the Qiagen Gel Extraction Kit, with a final elution in 31 µL EB.
-
57)Use 1 µL of sample to measure the DNA concentration in a Qubit dsDNA HS assay.
-
▪PAUSE POINT – Finished samples can be stored at −20°C for at least 6 months.
-
▪TROUBLESHOOTING ?
-
▪
-
58)
(OPTIONAL) PROTEIN EXPRESSION & BINDING ANALYSIS – TIMING ~1 day Perform a Western Blot using the aliquots from step 36 & 38 to verify proper expression and binding. Follow protocol in Box 3.
DAP-seq data analysis
-
59)NEXT-GEN SEQUENCING Run the sample on an Illumina sequencer according to the manufacturer’s specifications. We find both 75-bp and 100-bp single-end read runs to be sufficient.
-
▪CRITICAL STEP The number of reads required will depend on the characteristics of the genome (size, repeat content, etc.).
-
▪
-
60)
READ ALIGNMENT Align FASTQ files to a reference genome using a standard short read mapping software such as bowtie228. Read trimming and quality/repeat read filtering may also be necessary depending on data quality and the reference genome.
-
61)
PEAK CALLING Use the mapped read files (SAM or BAM format) to identify peaks using a peak calling software such as MACS229 or GEM30. If you ran a negative control sample, it can be used for background subtraction in the peak calling program.
-
62)
PEAK ANALYSIS Examine the mapped reads and peak files (BED or narrowPeak format) in a genome browser such as Integrative Genomics Viewer31 and compare peaks to the negative control sample. Successful DAP-seq experiments will typically give over 5% reads in peaks and produce peaks with a significant enrichment over background. Enriched motifs such as those produced by the MEME motif discovery software can be compared to comprehensive databases of known TF binding sites9.
TIMING
Genomic DNA LIBRARY PREPARATION: ~9.5 hr (11.5 hr for ampDAP library)
Steps 1 and 2, genomic DNA fragmentation: ~5 min per sample
Steps 3–9, fragmentation cleanup: ~60 min
Steps 10 and 11, end repair of fragmented DNA: ~45 min
Steps 12–14, end repair cleanup: ~60 min
Steps 15 and 16, A-tailing of 3’ ends: ~30 min
Steps 17–19, A-tail cleanup: ~60 min
Steps 20 and 21, adapter ligation: ~180 min
Steps 22–25, ligation cleanup and quantification: ~130 min
Step 26 (optional), amplification of library and cleanup: ~120 min
DNA AFFINITY PURIFICATION: ~8.5 hr
Steps 27–28, protein expression: ~130 min
Steps 29–38, binding of Halo-fusion protein: ~70 min
Step 39–41, protein washes: ~20 min
Steps 42–43, binding of DNA library to isolated protein: ~70 min
Steps 44–45, DNA washes: ~20 min
Steps 46–51, DNA recovery and amplification: ~100 min
Steps 52–57, pooling and size selection: ~100 min
ANTICIPATED RESULTS
Concentrations of DNA libraries after adapter ligation usually range from around 100–150 ng/µL, recovering about 60–90% of the original genomic DNA amount. Regardless of concentration, if a library passes qPCR quality testing, it can be used in the DAP-seq procedure. Final concentrations of DAP-seq samples can vary, but typical amounts range from 3–30 ng/µL. Any degree of amplification following the PCR enrichment step suggests sufficient amounts of DNA were present after the affinity purification step: however, it is not a guarantee that the DAP-seq experiment was successful. To assess whether a particular DAP-seq experiment worked well, the DNA must be sequenced and evaluated for the presence of binding peaks (Figure 1). The target number of sequencing reads will depend on the size of the genome. For Arabidopsis samples (125 Mb genome size), we typically pool 48 samples per lane on an Illumina HiSeq 2500 and 96 samples per lane on an Illumina HiSeq 4000, aiming for 2–4 million reads per DAP-seq sample. Extrapolating from these numbers, samples from human (3 Gb genome size) or other large genomes such as maize (2.5 Gb) should be pooled in groups of 16 samples per lane on an Illumina HiSeq 4000, with an aim of 20 million reads per individual sample. A Western Blot can be performed to verify protein expression and binding. This is not required, but may help in troubleshooting for particular TFs. Running equal amounts of the expression reaction and the supernatant from the bead binding step should show expression of the correct size fusion protein and a relative decrease in protein amount for the bead-bound supernatant sample, indicating bead retention.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting Table
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 25 | Recovery of adapter-ligated DNA is lower than expected (<50% original input) | DNA loss during cleanup steps may be responsible. | Try making a new library, taking extra care during precipitations, or using a different cleanup method (ex. Qiagen PCR purification columns or AMPure beads). |
| 26 (Box 4) | Little to no amplification of ampDAP library | Incomplete or failed adapter ligation will result in the inability of your DNA to amplify. | It is not recommended to continue with this library. For future preparations, try a new adapter mix. |
| 37 | Beads clump while rotating | Concentration of protein is likely high. | You can try adding more beads if you wish. Otherwise you may continue with the protocol without much negative impact. |
| 42 | No peaks observed with 30–100 ng DNA | Protein expression/binding is not consistent. DNA library cannot be properly amplified. | Try the titration experiment again with a different protein. Remake the DNA library. You may try the experiment with up to 1 µg DNA to see if it makes a difference. |
| 55 | No DNA smear is visible on the gel | Poor amplification or failed protein/DNA binding. | You may proceed with caution, cutting around 200–400 bp, but expect a low amount of final DNA recovery. |
| 57 | Little to no DNA recovery | Poor or failed protein expression or unamplifiable DNA library. | Run a Western Blot of your expression reaction to verify Halo-fusion protein expression. Include some of the sample from step 38 to verify binding to the beads. Run a qPCR of the ligated gDNA to assess quality. |
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB1024999) and the Gordon and Betty Moore Foundation (GBMF3034) to J.R.E, and from the National Science Foundation to A.G. (IOS1114484 and IOS1546873). J.R.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Proposed tweet: New protocol on using DAP-seq to map genome-wide transcription factor binding sites from #JoeEcker lab
Supporting papers
O’Malley and Huang et al., (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. 165, 1280–1292 Cell.
Kawakatsu, T. et al. Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell (2016). doi: 10.1016/j.cell.2016.06.044
AUTHOR CONTRIBUTIONS:
R.C.O and J.R.E. designed the original protocol. R.C.O., A.B., S.C.H., M.G., and A.G. modified and updated the protocol to its current state. J.R.N. performed all the sequencing. A.B., M.G., S.C.H., and J.R.E. wrote the manuscript with contributions from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Swinnen G, Goossens A, Pauwels L. Lessons from Domestication: Targeting Cis-Regulatory Elements for Crop Improvement. Trends in Plant Science. 2016 doi: 10.1016/j.tplants.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Deplancke B, Alpern D, Gardeux V. The Genetics of Transcription Factor DNA Binding Variation. Cell. 2016 doi: 10.1016/j.cell.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Babu MM, Luscombe NM, 3, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Niu W, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21:245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Negre N, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009 Jun;18:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weirauch MT, et al. Determination and Inference of Eukaryotic Transcription Factor Sequence Specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jolma A, et al. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Jolma A, et al. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527 doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- 12.Domcke S, et al. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 13.Hu S, et al. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013;2013 doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghav SK, et al. Integrative Genomics Identifies the Corepressor SMRT as a Gatekeeper of Adipogenesis through the Transcription Factors C/EBPB and KAISO. Mol. Cell. 2012;46:335–350. doi: 10.1016/j.molcel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 15.O’Malley RC, et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016 doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dror I, Golan T, Levy C, Rohs R, Mandel-Gutfreund Y. A widespread role of the motif environment in transcription factor binding across diverse protein families. Genome Res. 2015 doi: 10.1101/gr.184671.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Los GV, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008 doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 18.Worsley Hunt R, Wasserman WW. Non-targeted transcription factors motifs are a systemic component of ChIP-seq datasets. Genome Biol. 2014;15:412. doi: 10.1186/s13059-014-0412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakatsu T, et al. Unique cell-type-specific patterns of DNA methylation in the root meristem. 2016 doi: 10.1038/NPLANTS.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song L, Crawford GE. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010 doi: 10.1101/pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzo JM, Sinha S. In: Epidermal Cells: Methods and Protocols. Turksen K, editor. Springer; New York: 2014. pp. 49–59. [DOI] [Google Scholar]
- 24.Kawakatsu T, et al. Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell. 2016;166:492–506. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K, Zhao BS, He C. Nucleic Acid Modifications in Regulation of Gene Expression. Cell Chemical Biology. 2016;23:74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–7. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yazaki J, et al. Mapping transcription factor interactome networks using HaloTag protein arrays. doi: 10.1073/pnas.1603229113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Mahony S, Gifford DK. High Resolution Genome Wide Binding Event Finding and Motif Discovery Reveals Transcription Factor Spatial Binding Constraints. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper S, Speicher DW. Methods in molecular biology (Clifton, N.J.) 2011;681:259–280. doi: 10.1007/978-1-60761-913-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Structural Genomics Consortium et al. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urich Ma, Nery JR, Lister R, Schmitz RJ, Ecker JR. MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat. Protoc. 2015;10:475–83. doi: 10.1038/nprot.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallagher S, Chakavarti D. Immunoblot analysis. J. Vis. Exp. 2008;2:2008. doi: 10.3791/759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machanick P, Bailey TL. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll TS, Liang Z, Salama R, Stark R, de Santiago I. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front. Genet. 2014;5 doi: 10.3389/fgene.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jianhong Ou, Lihua Julie Zhu. MotifStack: Plot Stacked Logos for Single or Multiple DNA, RNA and Amino Acid sequence. 2015 [Google Scholar]
- 39.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]