Abstract
γ-Crystallins, abundant proteins of vertebrate lenses, were thought to be absent from birds. However, bird genomes contain well-conserved genes for γS and γN-crystallins. RTPCR detected spliced transcripts for both genes in chicken lens, with lower levels in cornea and retina/retinal pigment epithelium. The level of mRNA for γS in chicken lens was relatively very low even though the chicken crygs gene promoter had lens-preferred activity similar to that of mouse. Chicken γS was detected by a peptide antibody in lens, but not in other ocular tissues. Low levels of γS and γN proteins were detected in chicken lens by shotgun mass spectroscopy. Water soluble and insoluble lens fractions were analyzed and 1934 proteins (<1% false discovery rate) detected, increasing the known chicken lens proteome 30 -fold. Although chicken γS is well-conserved in protein sequence, it has one notable difference in leucine16, replacing a conserved surface glutamine in other γ-crystallins, possibly affecting solubility. However, L16 and engineered Q16 versions were both highly soluble and had indistinguishable circular dichroism, tryptophan fluorescence and heat stability(Tm values ~65C) profiles. L16 has been present in birds for over 100Myrand may have been adopted for a specific protein interaction in the bird lens. However, expression of all γ-crystallins in bird lenses has clearly been either eliminated or reduced during evolution. In view of this, the conservation of genes for γS and γN-crystallins in birds is surprising, suggesting they may have been preserved for some other role unrelated to the bulk properties of the lens.
Keywords: crystallin, eye, protein folding, promoter, proteomics, evolution
INTRODUCTION
The optical properties of the vertebrate eye lens are largely determined by high concentrations of soluble proteins, collectively known as crystallins, which accumulate without turnover throughout life [1–4]. Many different proteins have been recruited to serve as crystallins in different evolutionary lineages; however a core group, consisting of the α-crystallins, the β-crystallins and the γ-crystallins, is expressed in species from zebrafish to human. The α-crystallins belong to the superfamily of small heat shock proteins [5] while the β-crystallins and the γ-crystallins are related and belong to the βγ-crystallin superfamily, which has non -lens members in both prokaryotes and eukaryotes [6, 7]. The β-and γcrystallins arose in the earliest ancestor of the vertebrate lens [8] and through waves of gene duplication diversified into multigene families coding for proteins with repeats of a common structural motif [9]. However, while the multigene family of β-crystallins has orthologs throughout the vertebrates, the γcrystallins have been subject to much more evolutionary change, so that the major classes of fish γcrystallins, the γM-crystallins, have no orthologs in mammals [10]. Even within the mammals, some of the γ-crystallin genes have been lost in certain lineages, for example in primates and in guinea pig (Cavia porcellus) [10–12]. Studies on bird lens (mainly the chicken, Gallus gallus) found abundant α-crystallins and β-crystallins but did not detect γ-crystallins which were long thought to be completely absent from birds [13–15]. Instead, birds apparently replaced γ-crystallins with another protein, δ-crystallin, an example of the recruitment of a pre-existing protein, in this case the enzyme argininosuccinate lyase, as a crystallin [16].
Why replace γ-crystallins? It seems that these proteins are very highly specialized for the high protein content environment of the lens, particularly that in fish. They are a key part of the repertoire of proteins that allow a lens to achieve the very high refractive index needed for focusing under water. This function may be unneeded or even deleterious for species that need softer, accommodating lenses and, as a consequence, γ-crystallins have been changed and lost during vertebrate evolution in a way not seen in α-crystallins and β-crystallins. Furthermore, γ-crystallins have often been implicated in cataract, both congenital, due to mutations, and age-related. Some γ-crystallin-related cataracts are associated with formation of amyloid-like fibrillar deposits [17, 18].
Two members of the γ-crystallin family, γS and γN, are preserved from fish to mammals (10), however, although genes for both proteins are expressed in the mouse lens, mouse γN is only found at low levels while in humans CRYGN seems to be a pseudogene [10, 19]. In rodents γS protein is abundant in lens and has been found in photoreceptors and stressed retinal pigment epithelium [10, 20–22]. γS is also abundant in human lens [23] and is found in drusen (deposits basal to the RPE) in human age-related macular degeneration [24]. Previously we noticed that there are apparently intact gene sequences for crygs and crygn in the chicken genome (10). The possibility of a conserved, non-lens role for these two proteins in other species raises the question of a functional role for these γ-crystallins in the chicken. However, our investigation of the bird γ-crystallins suggests that although they are surprisingly well-conserved, they make no more than a low level contribution to the avian lens.
RESULTS
cDNA libraries
cDNA libraries derived from whole eye of E15 and P15 chicks were used for expressed sequence tag sequencing as part of the NEIBank project [25, 26]. The data for these libraries are available at http://neibank.nei.nih.gov/cgi-bin/libList.cgi?org=chicken. Among the transcripts sequenced were many for crystallins, most notably for δ-crystallin in the P15 library. The data gave a more complete cDNA sequence for chicken αA-crystallin (GenBank: EF408909.1), particularly the 3′UTR. Other sequences were consistent with those known for α -and β-crystallin gene transcripts; however no clones were identified for any γ-crystallins.
Chicken Genome and Synteny
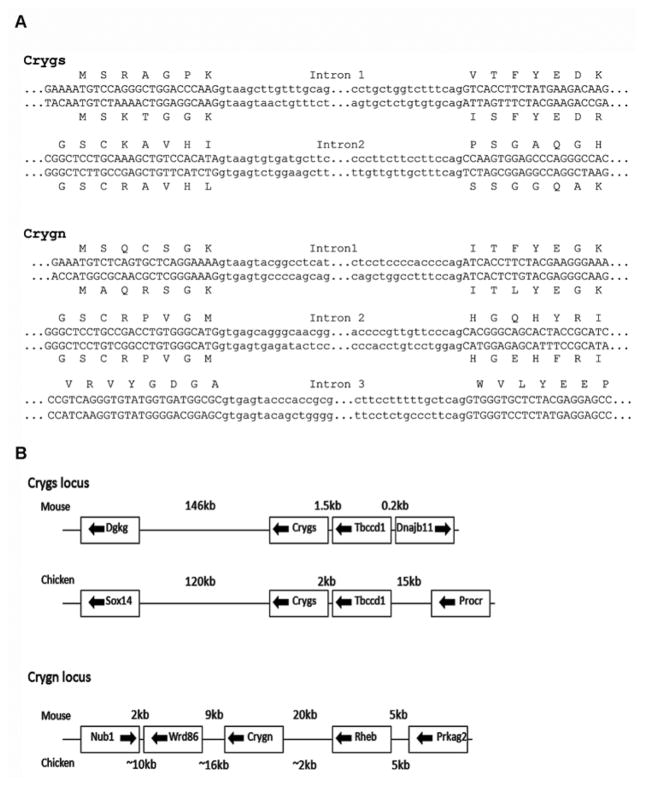
Homology searches of the chicken genome located genes for chicken orthologs of γS and γN on chromosomes 9 and 2 respectively. Both genes had complete open reading frames (ORFs), the typical exon structure for orthologous gene s in other species and normal splice junctions (Fig 1A).
FIGURE 1. Conservation of gene structure and linkage.
A: Exon/intron junctions of chicken (above) and mouse (below) genes for CRYGS and CRYGN. B: Comparison of the loci for CRYGS and CRYGN in mouse and chicken genomes. Approximate spacing of genes is indicated. Locations of putative orthologs of WDR86and RHEB in chicken were estimated from spliced EST positions in genome build galGal3.
Chicken CRYGS is closely linked to TBCCD1, a gene only ~2kb 5′ on the same strand (Fig 1B). In the human genome, TBCCD1 is only 1.5kb 5′ to CRYGS and orthologs in other mammals are similarly closely linked. Tbccd1(tubulin cofactor C domain containing 1) is a centrosomal protein that appears to be a key regulator of centrosome positioning and regulation [27]. Although the tight linkage to TBCCD1 is conserved, these two genes are not in a syntenic region and flanking genes are different from those in mammals.
Chicken CRYGN lies in a larger conserved syntenic region (Fig 1B). In mouse, crygn is 20kb downstream of Rheb (Ras homolog enriched in brain). In chicken, CRYGN is less than 2kb downstream of a transcribed region that in sequence closely matches RHEB, although the gene is not yet designated as such in the chicken genome build. Rheb protein directly binds and regulates TOR, a central regulator of cellular responses in metabolism, growth, stress and aging [28]. About 10kb 3′ to CRYGN in the chicken genome is a transcribed region with sequence similarity to mammalian WDR86(WD repeat domain 86) a gene of unknown function. Overall, it seems that the gene order NUB1/WDR86/CRYGN/RHEB/PRKAG2 is conserved in chicken and mammalian genomes.
Expression of chicken γ-crystallin transcripts in eye tissues
To detect spliced transcripts of chicken CRYGS and CRYGN, primers were designed and used in RT-PCR of RNA from P15 lens. Using primers for the ORFs of both chicken γS and γN, transcripts were amplified from lens, cornea and retina/rpe dissected from 5 week chicken eye (Fig 2A). As expected, transcripts of the appropriate size for both were detected in lens. Chicken γS also gave clear, but weaker, bands in cornea and retina/rpe. Chicken γN gave a weak band in cornea and a barely detectable band in retina/rpe.
FIGURE 2. Spliced transcripts and γS proteins in eye tissues;
A). RT-PCR of dissected eye tissues detected spliced transcripts retina/rpe (R), cornea (C), lens (L), no RT blank. B). Predicted amino acid sequences of chicken γS and γN aligned with mammalian orthologs. C). Western blot of chicken eye tissues with peptide antibody to chicken γS. M: markers; L: lens; R: retina; C: cornea; E: eyecup (rest of eye); recombinant chicken γS protein.
Products were cloned and sequenced and were found to correspond to spliced transcripts of both genes with complete open reading frames and predicted protein sequences similar to orthologs in mouse (Figure 2B). The experimentally verified versions of both γS and γN cDNAs were submitted to GenBank (Accessions: GQ980265 and GQ995487). There are single silent base differences between both cDNAs and the respective genome sequences that probably represent polymorphic variants in chickens. At this point, there appear to be no ESTs from any other chicken tissue libraries corresponding to the lens transcripts. However, a few transcripts from adult ovary or P16 whole embryo [29] are located in the long third exon of CRYGN. Unspliced or non-exonic transcript sequences have also been observed in the third intron of the human CRYGN gene, which may be related to a microRNA gene, MIR3907, located in that intron.
The protein sequences were examined for peptides that had potential to be antigenic and specific to both γS and γN and were used to raise polyclonal antibodies in rabbit. These were tested in western blots of P15 chicken eye tissues. The antibody to γS detected a very faint, sharp band at the expected size (~20kDa) only in lens (Fig 2D). No similar band was detected for γN in any tissue. Both antibodies were also used in attempts to localize proteins in sections of chicken eye by immunofluorescence; however no signals above background were detected.
To determine the relative level of expression of γS in the chicken lens, the expression levels of βB1, βA2 and γS in lens were measured for mouse and chicken using RTPCR (Tables 1 and 3). Using the βB1 levels of expression as the calibrator, there was approximately 10-fold less βA2 expression and 250-fold less γS expression in the chicken lens than in the mouse lens.
Table 1. RTPCR results for chicken and mouse crystallins.
A) Amplification efficiencies and crossing point values for chicken and mouse crybb1, cryba1 and crygs gene assays. Equal amounts of cDNA from either chicken or mouse lens were used. Additional assays for the cryba2 and crygs used 1:10 dilutions of the cDNA to check for consistency.
B) Relative expression values for mouse and chicken crystallin genes normalize to the crybb1 amount.
| A | |||
|---|---|---|---|
| Gene | Efficiency | Ct | |
| Chicken | Crybb1 | 1.917 | 13 |
| Chicken | Cryba2 | 1.985 | 14.38 |
| Chicken | Crygs | 1.946 | 18.23 |
| Chicken | Cryba2 1:10 | 1.985 | 16.85 |
| Chicken | Crygs 1:10 | 1.946 | 21.74 |
| Mouse | Crybb1 | 1.954 | 18.87 |
| Mouse | Cryba2 | 1.901 | 17.86 |
| Mouse | CrygS | 1.909 | 16.78 |
| Mouse | Cryba2 1:10 | 1.901 | 21.39 |
| Mouse | CrygS 1:10 | 1.909 | 19.82 |
| B | |||
|---|---|---|---|
| Crybb1 | Cryba2 | Crygs | |
| Mouse | 1 | 3.2 | 5.99 |
| Chicken | 1 | 0.24 | 0.025 |
Table 3.
Primers for RTPCR of Crystallins
Primer/probe sets for βB1, βA2, and γS crystallin transcripts of both mouse and chicken were designed using the Roche Universal ProbeLibrary Assay Design Center.
| Species | Gene | Left primer | Right primer | Universal Probe |
|---|---|---|---|---|
| Mouse | crybb1 | TGCCTCCTGGGAGCTACA | CTCCCCCGAGAATTCCAC | #15 |
| Mouse | cryba2 | AGTGGCCACCACAGCAAC | GAAGTTTTCCCCCTCAAACA | #56 |
| Mouse | crygs | GTCGCTGCAACTCCATTAGA | GGAGAAGTTGGGCCTTTCAT | #26 |
| Chicken | crybb1 | TGCACTATCCGTGTCTACCG | CCTCACAACGTCGGGTACA | #70 |
| Chicken | cryba2 | AGAACGGCGAGTTCAAGAAG | GGATTGGATCTGGTTGGTGT | #66 |
| Chicken | crygs | TTTGAAGCCACTGAAGACTGC | GTGGACCTCGCGAAAGTG | #25 |
Crygs gene promoter
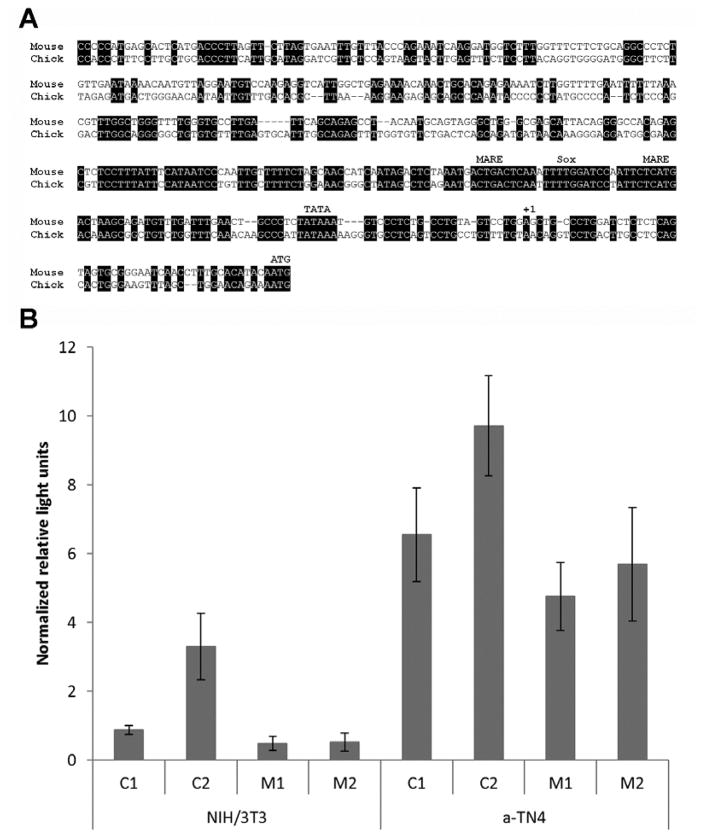
Alignment of 5′ flanking sequences for orthologous chicken, mouse and human genes for γS showed no evidence of major deletions or rearrangements (Fig 3A). Indeed, the overall level of conservation was comparable to that typically observed between functional genes in different species. The predicted core promoters included aligned TATA boxes and several well-conserved regions containing canonical binding sites for relevant factors such Maf and Sox family transcription factors. In these blocks of conservation, sequences that differ between chicken and mouse also generally differ between mouse and human. The equivalent regions of the chicken and mouse promoters, shown in Fig 3a, were cloned into luciferase reporter gene vectors. For both species, two constructs were made, with and without the 5′UTR of their respective transcripts, to test effects of UTR RNA structure on transcriptional or translation efficiency.
FIGURE 3. Conservation of crygs promoter sequence and function.
A) Alignment of promoters for chicken crygs and mouse crygs. +1 indicates the position chosen as the likely transcription start site and beginning of the 5′UTR. Conserved bases are highlighted and the positions of canonical maf-response elements (MARE), Sox family binding sites (Sox) and TATA boxes are indicated.
B) Promoter activity of chicken γS and mouse γS in NIH 3T3 and αTN4-1 cells. The activity was detected by Firefly signal and normalized by renilla signal in a dual luciferase assay. C1: chicken γS promoter without 5′-UTR, C2: chicken γS promoter with 5′-UTR, M1: mouse γS promoter without 5′ -UTR, M2: mouse γS promoter with 5′-UTR.
Promoter-reporter constructs were co-transfected into lens-derived mouse αTN4-1 and non-lens mouse NIH 3T3 cell lines with the control vector phRL-TK for normalization. Crystallin promoters of widely different species are dependent on the same suite of transcription factors [30] and function in similar ways in lens-cells of other species both in culture and in transgenic models (for example: [31–33]). This allows comparison of promoters from both chicken and mouse in mouse cells.
Both chicken and mouse CRYGScore promoters were active in lens -derived cells and, with lower expression in fibroblasts, showing tissue preference (Fig 3B). For both chicken and mouse constructs, inclusion of the 5′UTR elevated expression in lens. The mouse promoter constructs were essentially inactive in fibroblasts, as was the chicken core promoter lacking the 5′ UTR. Interestingly, inclusion of the chicken 5′ UTR also elevated non-lens expression, but this was still at a lower level than in lens-derived cells.
Relative abundance of crystallins in chicken lens proteome
Water soluble and water insoluble lens fractions were digested by trypsin, peptides separated using high pH reverse phase chromatography followed by conventional low pH reverse phase, and peptides identified using electrospray high-resolution mass spectrometry. Target/decoy protein database methods were used to control peptide identification errors to less than 1% and protein identifications were inferred from peptides using basic parsimony principles. A total of 1934 proteins (excluding common contaminants) were detected from both lens fractions at a 1% false discovery rate. More proteins (1746) were detected in the water-insoluble fraction than in the water-soluble fraction (1095). The high abundance of crystallins in the water-soluble fraction reduces sensitivity for detecting minor lens proteins. This increases the known chick lens proteome by about 30 -fold compared to previous studies [15, 34] and is the first characterization of water-insoluble lens proteins in avian lenses.
Analysis of shotgun mass spectroscopy of tryptic digests of whole chick lens was performed using the complete revised sequences for γS and γN. Eleventryptic peptides for γS and 10 tryptic peptides for γN were detected, confirming that the lens transcripts are translated (Table 2).
Table 2.
Peptides for γS and γN-crystallins
Fully tryptic γS and γN-crystallin peptides identified during shotgun analysis of lens protein digests of water-soluble and water insoluble fractions from 10-week-old chicken lenses.
| γS-crystallin peptides (residue #s) | γN-crystallin peptides (residue #s) |
|---|---|
| VTFYEDKNFLGR (8 – 19) | ITFYEGKCFSGR (8 – 19) |
| VTFYEDKNFLGRR (8 – 20) | KLEVCGTCSSFQQR (20 – 33) |
| RYECDADCPDFHTYLNR (20 – 36) | VQSGAWVCFDHPELHGQQFVLEHGEYPHWQR (44 – 74) |
| VEGGTWVAYERPNYSGNMYVLR (42 – 63) | M* GSCRPVGMHGQHYR (82 – 96) |
| VEGGTWVAYERPNYSGNM* YVLR (42 – 63) | MGSCRPVGM* HGQHYR (82 – 96) |
| RGEYPDYHHWMGLNDR (64 – 79) | MGSCRPVGMHGQHYR (82 – 96) |
| RGEYPDYHHWM* GLNDR (64 – 79) | IQLFEGSCFGGHSMELTEDCSCLR (97 – 120) |
| GEYPDYHHWMGLNDR (65 – 79) | IQLFEGSCFGGHSM* ELTEDCSCLR (97 – 120) |
| AVHIPSGAQGHIQVFEK (85 – 101) | GRGWEQPHVNAVR (121 – 133) |
| GDFGGQMFEATEDCPSILEECHFR (102 – 125) | GWEQPHVNAVR (123 – 133) |
| GDFGGQM* FEATEDCPSILEECHFR (102 – 125) | VYGDGAWVLYEEPNYGGR (134 – 151) |
| VLEGIWVFYEHPNYR (132 – 146) | AWQGFSANVQSVR (165 – 177) |
| GEYRQPVEWGAVTPAVQSFR (155 – 174) | AWQGFSANVQSVRR (165 – 178) |
| QPVEWGAVTPAVQSFR (159 – 174) |
denotes an oxidized amino acid form
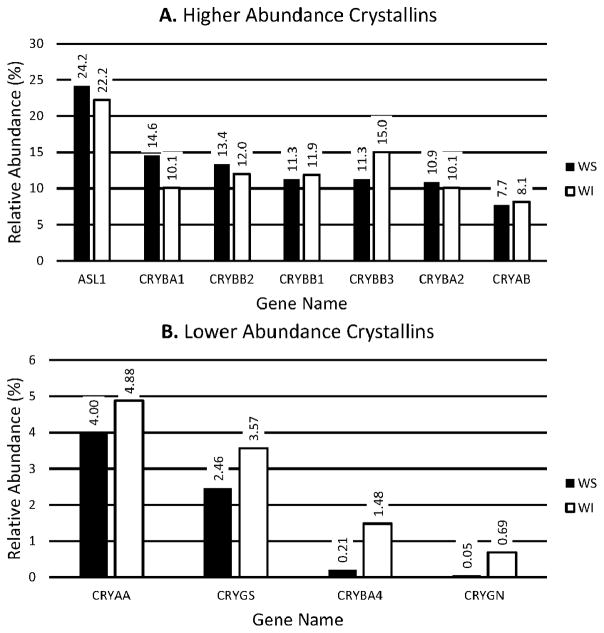
Since summed intensities of fragment ions in MS/MS data collected during shotgun proteomics analysis give estimates of relative protein abundance [35], the method was used to compare the relative weight percentages of γS and γN to other crystallins in the soluble fraction of 7–8 week old chicken lenses (Fig. 4). As expected, δ-crystallin comprised approximately 24% of the soluble crystallin abundance, while the next most abundant crystallin βB2 comprised 15%. The 3 least abundant crystallins were γS (2.5%), βA4 (0.21%) and γN (0.05%). The pelleted and washed fraction of the young chicken lens was also examined to determine if the low soluble concentration of these crystallins could be due to insolublization. However, the weight percentages of these crystallins in the insoluble fraction were similar to those in the soluble fractions: 3.6, 1.5, and 0.69% respectively.
FIGURE 4. Relative weight abundances of crystallins in 10 week-old chicken lenses.
The summed fragment ion intensities from all MS/MS spectra assigned to each crystallin in large-scale shotgun proteomics for water soluble (white bars) and insoluble (black bars) fractions of chicken lens were used to estimate the relative percent weight abundances of each crystallin. High and low abundance crystallins are shown in separate panels with different scales.
Implications for protein sequence and structure in chicken γS
Based on the predicted sequence of chicken γS (Fig 2B) and the structure of mouse γS [36], a homology model for the protein was constructed (Fig 5A). Overall the protein sequence of chick γS fits the known template structure of mouse γS well. Most sequence differences between mouse and chicken γS are conservative and in many cases the variants are present in equivalent positions in other γcrystallins. However, the predicted pI for the protein is 6.3, reflecting a difference in the content of strongly basic residues, making it rather more acidic than other γ-crystallins, for example, the mouse ortholog has a pI of 7.2. This could potentially contribute to differences in solubility or protein interactions.
FIGURE 5. Sequence and evolution of avian γS-crystallins.
A). Homology model of chicken γS. Model was produced using Rosetta [49] and visualized using Chimera [50]. Main chain tracing of the homology model of chicken γS based on the coordinates of the mouse ortholog. Positions of some non-conserved residues that might have effects on solubility, folding or stability are shown. B). Alignment of γS sequences for birds and selected species predicted from genome sequences. Aligned using Clustal as implemented in MEGA [51]. B). C) Evolutionary tree for γS protein sequences. Q16 is present in reptiles and mammals. L16 is present in birds. Tree produced using neighbor joining in MEGA.
Chicken γS differs from most other species with an RR dipeptide at positions 64–65 (numbered not counting the initiator methionine which is removed in γ-crystallins, however this dipeptide is seen in other γ-crystallins (such as mouse γA–F), and the dipeptide is not strongly conserved in the γS of other species [10]. Cysteine 123 is an unusual residue at this position, but this is not a strongly conserved region of sequence among γ-crystallins. C123 is located in a partially helical loop region connecting βstrands c–d in the third motif of the protein in the C-terminal domain (Fig 5A). Depending on the precise conformation in this region, the cysteine could be vulnerable to oxidation and intermolecular disulfide formation. However, as modeled, the side chain of the residue is buried and “safe”.
One notable but enigmatic difference in the bird sequence is leucine 16 (L16) in chicken γ S. Most other γ-crystallins, and all other known γ S sequences (including those in reptiles), have glutamine (Q16) at this position [10], which is in the middle of the tip of the β-hairpin of the first structural motif of the protein; part of a key structure that defines this superfamily. L16 is also found in all predicted γS ORFs derived from genome sequence for birds (Fig 5B, C) suggesting that this substitution occurred more than 100Myr ago [37]. The basis for the strong conservation of Q16 in non-avian species and its substitution in birds is not at all clear. However, single residue changes in surface residues of γ crystallins can have profound effects on solubility and can indeed lead to cataracts [19]. Consequently we decided to compare the properties of wild type L16 chicken γS with engineered Q16.
Recombinant proteins
The ORFs of both chicken γS and γN were cloned into a pET vector for expression in E. coli host, a system that has been used many times for different γ-crystallins [38, 39]. The identity of the recombinant proteins was confirmed by mass spectroscopy of tryptic peptides and t he peptide antibody for chicken γS detected the recombinant proteins cleanly, confirming the antibody specificity. Both L16 and Q16 proteins could be concentrated to 40mg/ml, remaining soluble.
In contrast, we were unable to express any detectable recombinant γN, in spite of numerous attempts to optimize and to select for higher expressing clones. The peptide antibody for γN failed to detect any recombinant proteinin bacterial whole cell extract.
Structure and stability of chicken γS
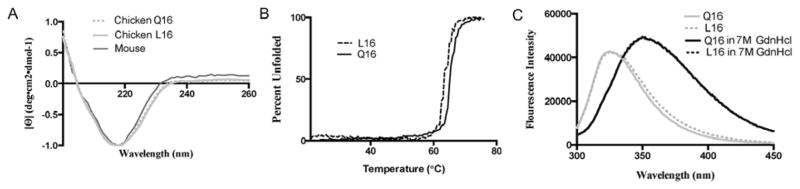
CD for both L16 and Q16 chicken γS gave profiles consistent with predominantly β-sheet secondary structure, similar to other γ-crystallins (Fig 6A). Thermal stability was monitored using the magnitude of the 220nm minimum in CD corresponding to organized β-sheet during heating (Fig 6B). Both L16 and Q16 had similar Tm values, close to 65C, similar that previously observed for mouse γS [40]. Most γ-crystallins contain paired tryptophans in each domain which exhibit significant quenching and blue shifting of fluorescence emission in the fully folded protein which is lost upon unfolding in a chemical denaturant [41]. These residues are conserved in chicken γS and fluorescence spectroscopy for both versions showed quenching of the Trp-emission, consistent with a canonical tertiary fold in both cases (Fig 6C).
FIGURE 6. Structure and stability of recombinant chicken γS proteins.
A) Far-UV CD spectra of mouse, chicken L16 and chicken Q16 γS have very similar spectra and predicted contents of secondary structure.
B) Thermal stability of secondary structure in in chicken L16 and chicken Q16 γS measured by magnitude of 220nm minimum characteristic of β-sheet.
C) Trp fluorescence of chicken L16 and chicken Q16 γS in folded and chemically denatured (7M GdnHCl) states. Both show similar profiles, indicating quenching in the folded state and a shift in the maximum emission upon unfolding.
DISCUSSION
The evolution of the eye lens in terrestrial vertebrates has been accompanied by a decrease in both the number of genes for γ-crystallins and in the overall contributions of γ-crystallins to total lens protein. Fish have large numbers of genes for different types of γ-crystallins and, as a class, γ-crystallins are highly abundant in the dense, high refractive index fish lens [10]. Mammals with mainly diurnal habits, such as primates, need more flexible, accommodating lenses and have reduced or lost expression of some γ-crystallins, while nocturnal, burrowing rodents have comparatively high levels of γ-crystallins [11]. This is taken to the extreme in birds, which have the most flexible lenses, and in which the replacement of γ-crystallins is almost complete, indeed this class of crystallin was long thought to be absent from birds [13–15]. Indeed, the chicken genome shows no evidence of genes related to the γA-F cluster of mammals or the γM-crystallin families of fish; however, genome sequence shows that chickens and other birds retain genes for γS and γN[10].
γS and γN families are both represented by duplicated genes in zebrafish [10]. In mouse, the single gene for γN is expressed at low levels in lens and retina/rpe, but in humans γN appears to be nonfunctional [10] suggesting that its role in mammals is dispensable. In contrast, γS is expressed at very high levels in both mouse and human lens and has also been found in mouse retina and RPE[20, 21, 42, 43], and even in drusen (deposits basal to RPE) in humans [24]. A functional role for γS in stabilization of actin cytoskeleton has also been suggested from examination of a knockout mouse model and the ability of recombinant mouse γS to stabilize F-actin in vitro[44].
In the chicken genome, genes for γS and γN have well-conserved canonical gene structure and complete open reading frames. Even the core promoter of the chicken γS gene is well-conserved in sequence and in reporter assays in lens-derived and non-lens cells it has a level of tissue-preferred activity similar to that of the mouse ortholog. Indeed, both genes are expressed in chicken eye, with spliced transcripts detectable by RT-PCR in lens and at lower levels in cornea and retina/rpe. However, compared to mouse, the level of γS mRNA is significantly reduced. Since the promoter is capable of initiating transcription and the gene is capable of producing mature, spliced transcripts in lens, the relative silencing of the chicken γS gene may be due to one or more other mechanisms, such as epigenetic inhibition.
Shotgun proteomics analysis for tryptic peptides shows that both γS and γN proteins are present in the mature chicken lens. γN is at very low levels. At 2.4%, γS appears to be abundant compared with non-crystallin, non-structural proteins, but it is lower than its orthologues in mammals. It is possible that the peptide abundance may overestimate the true abundance of γS in chicken lens. Previous 2D gel analysis of chicken lens showed no spot corresponding to an unaccounted protein of this abundance [15]. Apparent abundance of proteins by shotgun analysis can be influenced by efficiency of tryptic digestion and the characteristics of tryptic peptides. Overall, it seems safe to conclude that γN makes very little contribution to chicken lens while γS is present but at lower levels than in mammals.
Although chicken γS is generally well conserved, it has one notably non-conservative change from other members of the family. Hydrophobic leucine occupies position 16 at the tip of the folded βhairpin of the first Greek key motif of the protein, a structural position occupied by the polar residue glutaminein other γ-crystallins. (This position is also conserved as polar in the final fourth motif of most β- and γ-crystallins although the other two motifs are more variable [10]). While there is no obvious rationale for the strict conservation of glutamine at this position in most γ-crystallins, the substitution of hydrophobic leucine might be expected to have an effect on solubility, a key functionality of crystallins. Indeed, several inherited cataracts are due to single residue substitutions of surface residues in γcrystallins [19]. We reasoned that lower expression of γS in birds could be associated with decreased solubility. However, replacement of L16 with Q16 did not have any major effect on solubility of recombinant proteins: both could be concentrated to 40mg/ml without precipitation, and weighted spectral counting by mass spectrometry suggested that the abundance of γS in chicken lens was similar in both water-soluble and insoluble fractions. The L16/Q16 difference also had no major effect on secondary structure or tertiary fold of freshly prepared proteins, as measured by CD and Trp fluorescence. Both versions of the protein also showed similar thermal stability, with Tm values in the same range as previously measured for mouse γS [40].
L16 is present in the predicted γS ORFs in other bird genomes, suggesting that it has been present for over 100Myr. It is possible that early in bird evolution a protein-protein interaction between γS and some other lens component found in birds led to selective pressure for the change to L16. A similar argument has been made to account for changes in otherwise conserved residues in the enzyme LDH-B in most species in which it also served as ε-crystallin [45, 46].
Given their generally low expression, and considering that genes for other γ-crystallins have been completely deleted from the avian genome, it is interesting that these two γ-crystallin genes have been so well conserved for so long. They may have other functional roles that do not require the high level expression of γ-crystallins in fish or mammals. It is also possible that both genes have been retained at least partly because they are under the selective protection of other overlapping transcriptionally active regions. The non-functional human CRYGN gene for γN contains a miRNA gene in one intron while the human CRYGS also contains an enigmatic, transcriptionally active region in an intron [47]. It is not known whether such transcriptionally active regions are also present in birds. However, it is clear that both crygs and crygn are in syntenic regions conserved from mammals to birds and maintain close linkage to their upstream neighbors (TBCCD1 and RHEB respectively) which encode proteins implicated in key cellular processes [27, 28]. Perhaps the crystallin genes have no essential roles in themselves but are protected from deletion by the selective pressures that preserve neighboring or embedded genes?
Finally, while the major purpose of this study was to investigate chicken γ-crystallins, the large chicken lens proteomics dataset appearing in the supplementary data should prove useful in future studies, especially as the lens proteomes of other species are more widely explored [30]. Comparative analyses of lens proteomes may help explain how evolution has produced lenses uniquely suited to each species, and provide important clues regarding the functions of specific lens proteins, which like the γ-crystallins, may differ greatly in abundance between species.
MATERIALS AND METHODS
Chicken eye cDNA Libraries and Sequencing
Eyes from E15 embryo and P15 hatched White Leghorn chickens were stored in RNALater (Ambion, TX) until RNA was extracted. The cDNA libraries were prepared by Bioserve (Beltsville MD) in the pCMVSPORT6 vector (Invitrogen, Carlsbad CA) as previously described [23]. Clones were sequenced and processed as NEIBank web pages [25] [http://neibank.nei.nih.gov/cgi-bin/libList.cgi?org=chicken].
Chickens for these procedures were euthanized according to protocols approved by the Beltsville Area Animal Care and Use Committee of USDA.
PCR methods
Primers were designed to amplify the complete transcript of chicken γS-crystallin (CGS5UTR:TGCCTCCAGCACTGGGAAGTTTAGC;CGSPOLYA:TCTGAGTGACTCAAGTGCTTT ATTG);
the ORF of chicken γS-crystallin (CGSATG: GAAAATGTCCAGGGCTGGACCCAAG; CGSSTOP: CCCACCATTTACTCAGCGATGCTG);
and the ORF of γN (CGN1: AATGTCTCAGTGCTCAGGAAAAATC; CGN2: GTCTCAGCTAGAAGTAGTTGATA
AC). These were used to amplify cDNAs from P15 lens RNA by RT-PCR. Primers for the ORFs were also used to amplify transcripts from dissected lens, retina/RPE and cornea from 5 week chicken eye.
Quantitative PCR
The relative amount of γS transcript in chicken lens was determined by comparing the expression levels of βB1, βA2, and γS crystallins in the mouse lens to the relative levels in the chicken lens. The amount of transcript was determined by qPCR using TaqMan-type assays in a Roche LightCycler using the Roche LightCycler TaqMan Master kit (Roche Applied Sciences, [www.roche-applied-science.com]). Primer/probe sets for βB1, βA2, and γS crystallins of both mouse and chicken were designed using the Roche Universal ProbeLibrary AssayDesign Center [https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp?id=UP030000} (Table 3).
cDNA from chicken and mouse lens RNA was generated and the efficiency of amplification for each assay was determined by fitting a curve to the crossing threshold values (Ct) for serial dilutions of target sample for each assay. The relative expression levels of the genes were calculated using the Ct values generated from identical starting quantities of cDNA; The relative ratio of two transcripts =Eq−ΔCt,q/Ecb−ΔCt,cb where Eq is the efficiency of amplification for the target (βa2 or γS), Ecb is the efficiency of amplification for the calibrator (βB1), Ct,q is the crossing threshold of the target (βA2 or γS), and Ct,cb is the crossing threshold of the calibrator (βB1).
Bioinformatics
Chicken crystallin sequences were identified from genome sequence using Blast searches at NCBI (http://ncbi.nlm.nih.gov) and BLAT [48] searches at UCSC (http://genome.ucsc.edu). 5′ coding exons not detectable by BLAT were located by inspection. Homology modeling of chicken γS was performed using Rosetta [49], based on the coordinates of mouse γS [36] PDB ID: 1ZWO. The structure was visualized using Chimera [50]. Phylogenetic analysis of the complete coding sequences of γS proteins was performed using Clustal alignment and the Neighbor Joining method in the MEGA package [51].
Antibody and Immunochemical Methods
Chicken γS peptide, gS-1: Acetyl-CKAVHIPSGAQGHIQVFEK-amide, and chicken γN peptide gN-1: CRPVGMHGQHYRIQLFEGS-amide were synthesized at Twentyfirst Century Biochemicals, Inc. (Marlboro, MA), conjugated to carrier and used to immunize rabbits. Western blots were performed on dissected tissues from 5 week chicken eye using 1:1000 dilutions of primary antibodies following standard methods [10].
Mass spectrometric identification and relative quantitation of chicken lens crystallins
Soluble proteins were isolated from 7–8 week-old freshly dissected chicken lenses after overnight shipping on ice (Pel-Freez, Rogers, AK). A total of 6 lenses ranging in wet weight between 51 and 68 mg were homogenized in 1.0 ml of 20 mM phosphate buffer (pH 7.0), and 1.0mM EGTA and centrifugated at 16 000 × g for 15min. The insoluble pellet was then resuspended in 1 ml of homogenizing buffer, centrifuged again, and the soluble fraction added to the initial supernatant. This was repeated a total of 3 times. The pellet was then suspended with a probe sonicator using 3 × 5 sec burst on ice, and the protein content determined using a BCA assay and bovine serum albumin as a standard (Thermo Scientific). 100 μg portions of soluble and insoluble fractions were then dried by vacuum centrifugation, resuspended in digestion buffer containing 8 M urea, proteins reduced/alkylated, and digested overnight with trypsinas previous [52]. Peptides were then injected onto a NanoEase XBridge BEH130 5 μm particle 0.3 × 50 mm C18 column (Waters) using a mobile phase containing 10 mM ammonium formate (pH 10) with a 3 μl/min flow rate. Peptides were than eluted by sequential injection of 20 μl volumes of the same mobile phase, but containing 14, 20, 22, 24, 26, 28, 30, 40, and 90% acetonitrile. Eluted peptides were then diluted at a tee with a mobile phase containing 0.1% formic acid at a flow rate of 24 μl/min to acidify samples and dilute acetonitrile concentrations. Peptides were then separated by second dimension reverse phase C18 chromatography at low pH by trapping peptides on an Acclaim PepMap 0.1 × 20 mm NanoViper C18 peptide trap (Thermo Scientific), then separation using a PepMap RSLC C18, 2 μm particle, 75 μm × 25 cm EasySpray column (Thermo Scientific) using a 7.5 –30% acetonitrile gradient over 90 min in a mobile phase containing 0.1% formic acid and a 300 nl/min flow rate using a Dionex NCS-3500RS UltiMate RSLCnano UPLC system. Tandem mass spectrometry data was collected using an Orbitrap Fusion Tribrid mass spectrometer configured with an EasySpray NanoSource (Thermo Scientific). Survey scans were performed in the Orbitrap mass analyzer at 120,000 resolution, data-dependent MS2 scans in the linear ion trap collected in top speed mode with dynamic exclusion enabled and using higher energy induced dissociation following isolation using the instrument’s quadrupole. Further details of instrument settings are found in Supplementary Materials.
Peak lists in MS2 format [53] were created using MSConvert (ProteoWizard version 3.0.6705) [54] and in-house scripts. Peptides were identified using Comet [55] version 2014.2 rev. 2. Searches used either a Chicken Ensembl v77 protein database (16,366 sequences) or a UniProt Gallus gallus reference proteome (17,623 sequences downloaded 12/12/2014 from www.uniprot.org). Databases also included contaminants and reversed decoy sequences. Ensembl databases were reformatted to include protein descriptions by using information from BioMart and in-house scripts. Search parameters were: 2.5 Da average parent ion tolerance, 1.0005 Da monoisotopic fragment ion tolerance, variable oxidation of methionine (M+15.994), fixed alkylation of cysteine (C+57.02), and trypsin enzymatic cleavage with a maximum of 2 missed cleavages. Peptides were filtered to 1% false discovery rates (FDR) using the PAW pipeline[34]. Identified proteins were matched between the Ensembl results and the Uniprot results using a local installation of BLAST [56, 57] and in -house scripts. Some minor crystallins were not well annotated in Uniprot and lens fiber major intrinsic protein (MIP26) was missing from Ensembl.
Proteins were inferred from peptides using parsimony principles where identical peptide sets were reported as protein groups and peptide subsets were not reported. A minimum of two distinct peptides per protein was required for protein identification. An additional processing step used extended parsimony principles to group highly homologous proteins into families. For water-insoluble proteins, there were 1705 identified protein groups (0.6% FDR). Due to the relatively higher crystallin abundances, only 1039 protein groups (0.5% FDR) were identified for the water-soluble fraction. Relative crystallin abundances within each solubility fraction were estimated using MS2-intensity-weighted spectral counts, which have been shown to be more accurate than simple spectral counts [35]. Complete instrument settings, search parameters, and pipeline processing steps are included in the Supplemental data along with complete lists of identified peptides, identified proteins, and quantitative information.
Promoter analysis
5′ regions of mouse and chicken genes for γS were extracted from genome sequences and aligned using ClustalW implemented in BioEdit. Based on estimated transcription start sites (+1), promoter fragments for both genes were amplified from about −400bp 5′ with or without inclusion of the 5′ UTR of each transcript (i.e. to +0 or +80) using the following primers: Chicken:
Chpr-400: CTCGAGCCACCCTTTCCTTGCTGCACCCT;
Chpr+80: AAGCTTTCTGTTCCAGCTAAACTTCCCA;
Chrpr+1: AAGCTTAACAGGCAGGACTGAGGCACC;
Mouse:
Mspr-400: CTCGAGCCCCCATGAGCACTCATGACCCT;
Mspr+80: AAGCTTGTATGTGCAAAGGTTGATTCCCG;
Mspr+1: AAGCTTCTCCAGGACTACAGGCAGAGGGA.
Products were sequenced and cloned into the XhoI/HindIII sites of the pGL4.10 [luc2] luciferase vector (Promega, Madison, WI). Promoter constructs lacking (−400 to +1) or including (−400 to +80) 5′ UTR sequence were designated as C1 or C2 for chicken γS, and M1 or M2 for mouse γS.
The constructs were tested in αTN4-1 (mouse lens and NIH 3T3 (mouse fibroblast) cells. Cultured cells were plated 24 hours prior to transfection into 6-well culture dishes at 80–90% confluence and maintained at 5% CO2, 37°C. For transfections, 1 μg of DNA was mixed with 6 μl X-tremeGENE 9 transfection reagent (Roche, Mannheim, Germany) and serum-free media to a total volume of 100 μl. The complex was then added to the cells in the culture dishes in a drop-wise manner. The pGL4.10 construct was co-transfected with phRL-TK, an internal control vector containing the renilla luciferase gene (Promega, Madison, WI) to normalize transfection efficiency. The Dual-luciferase® Reporter Assay System (Promega, Madison, WI) was used to measure luciferase activity. Twenty four hours after transfection, the cells were gently rinsed with phosphate buffered saline and harvested with Passive Lysis Buffer. 100 μl of cell lysates were loaded into 96-well black microplate. 50 μl of LAR II reagent was added and the firefly luminescence was read using POLARstar OPTIMA microplate fluorimeter (BMG LABTECH, Cary, NC). After 20 minutes, 100 μl of Stop&Glo® reagent was added to the lysates and renilla luciferase luminescence was read. Duplicate luminescence values of firefly luciferase were normalized with renilla for each construct within an experiment and each experiment was repeated three times.
Recombinant protein
For expression of recombinant protein, ORFs for chicken γS and γN were amplified from cDNA templates using primers:
CGSNde1: CATATGTCCAGGGCTGGACCCAAG;
CGSHind3: AAGCTTTACTCAGCGATGCTGCGGAAG;
CGNNde1: CATATGTCTCAGTGCTCAGGAAAAATC;
CGNHind3: AAGCTTCAGCTAGAAGTAGTTGATAAC
and cloned into Nde I/Xho I sites of pET 31b (Novagen).
The plasmid was transformed into E. coli BL21(DE3) pLysS (Novagen). Colonies were grown in LB broth with ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml in ethanol). Protein production was induced by 1mM IPTG with culturing at 30 °C overnight. Bacteria were pelleted by centrifuging at 4 °C for 15 min and resuspended in 50 mM Tris-HCl 7.2, 10 mM EDTA, 300 mM NaCl. Pellets were lysed by repeated sonication with 2 sec burst and 5 sec break for 10 min working time. Cell lysate supernatant was dialyzed to working buffer (50 mM Tris, pH 8.5, 1 mM dithiothreitol, and 1 mM EDTA. Dialyzed protein was loaded on a Mono Q 5/50 GL anionic exchange column using the AKTA Explorer 100 (Amersham Pharmacia Biotech) purification system. Combined fractions were loaded onto a HiLoad 16/60 Superdex 75 size exclusion column (Amersham Pharmacia Biotech). Purified proteins were analyzed by 12% Bis-Tris gels (Novex, San Diego).
For site-specific mutagenesis of residue L16>Q, mismatch double stranded primers CGSmut1: CTATGAAGACAAGAATTTCCAGGGTCGTCGCTACGAGTGC and CGSmut2: GCACTCGTAGCGACGACCCTGGAAATTCTTGTCTTCATAG (PAGE purified; Integrated DNA Technologies, Inc., Coralville, IA) were used to mutate the γS pET plasmid using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). The recombinant chicken γS mutant was expressed and purified by the same protocol as wild type.
Recombinant protein was validated by mass spectroscopy. Purified γS crystallin in 50 mM Tris pH 8.5, 1mM EDTA, 1mM DTT was digested with trypsin gold (Promega), purified by C4 Zip Tip (Millipore) and eluted in 60% acetonitrile/40% 0.1% TFA. Matrix solution was prepared of 10 mg/mL α-cyano-4-hydroxycinnamic acid (CHCA) in 50% actetonitrile/50% 0.1% TFA. CHCA matrix solution and γS crystalline were mixed 1:1 and spotted on a matrix-assisted laser desorption-ionization time (MALDI) plate. MALDI spectra were obtained using an AB Sciex 5800 matrix-assisted laser desorption-ionization time-of-flight (MALDI TOF/TOF) instrument in the reflective positive mode with range m/z 150–4000. Identified MS and MS/MS peaks were entered into MASCOT search engine. γS crystallin was identified by four matching peptides.
CD and Fluorescence spectroscopy
Circular dichroism (CD) spectra were recorded on a Jasco J-715 spectropolarimeter (Jasco International Co., Ltd., Hachioji, Tokyo, Japan) at 20°C. Far UV CD experiments were performed at protein concentration of 0.2mg/ml in buffer using a 2-mm path length cuvette. Data were collected over the wavelength range of 260–200 nm at 1-nm intervals. Ten scans were averaged and normalized by subtracting the base line recorded for the buffer. The results were plotted as ellipticity (mdeg) versus wavelength (nm). For a melting point curve, CD at 200 nm was measured over a temperature range of 20°C to 80°C. Fluorescence emission spectra were measured on a PTI Quantimaster fluorimeter (Photon Technology International, Trenton, NJ) at 20 °C.
Acknowledgments
This work was performed with the support of the NEI Intramural Program and NIH grantsEY010572 and EY007755.
Footnotes
Author Contributions.
Experiments were planned and data analyzed by GW, YC, VS, KP, PW, LD. Experiments were performed by YC,VS, HL,KP, SM, JF, PW,LD. Essential materials were provided by JM. Manuscript was written by GW with contributions from other authors.
Supporting Information
Shotgun proteomics analysis of chicken lens. This PDF flat file contains complete instrument settings, search parameters, and pipeline processing steps along with complete lists of identified peptides, identified proteins, and quantitative information. The PDF file is a text version of an Excel spreadsheet that is available upon request.
References
- 1.Bloemendal H, De Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–85. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Wistow GJ, Piatigorsky J. Lens crystallins: the evolution and expression of proteins for a highly specialized tissue. Annu Rev Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 3.Wistow G. Molecular Biology and Evolution of Crystallins: Gene Recruitment and Multifunctional Proteins in the Eye Lens. R.G. Landes Company; Austin, TX: 1995. [Google Scholar]
- 4.Slingsby C, Wistow GJ, Clark AR. Evolution of crystallins for a role in the vertebrate eye lens. Protein Sci. 2013;22:367–80. doi: 10.1002/pro.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark AR, Lubsen NH, Slingsby C. sHSP in the eye lens: Crystallin mutations, cataract and proteostasis. Int J Biochem Cell Biol. 2012;44:1687–97. doi: 10.1016/j.biocel.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Ray ME, Wistow G, Su YA, Meltzer PS, Trent JM. AIM1, a novel non-lens member of the betagamma-crystallin superfamily, is associated with the control of tumorigenicity in human malignant melanoma. Proc Natl Acad Sci U S A. 1997;94:3229–34. doi: 10.1073/pnas.94.7.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wistow G, Summers L, Blundell T. Myxococcus xanthus spore coat protein S may have a similar structure to vertebrate lens beta gamma-crystallins. Nature. 1985;315:771–3. doi: 10.1038/315771a0. [DOI] [PubMed] [Google Scholar]
- 8.Shimeld SM, Purkiss AG, Dirks RP, Bateman OA, Slingsby C, Lubsen NH. Urochordate betagamma-crystallin and the evolutionary origin of the vertebrate eye lens. Curr Biol. 2005;15:1684–9. doi: 10.1016/j.cub.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 9.Kappe G, Purkiss AG, van Genesen ST, Slingsby C, Lubsen NH. Explosive expansion of betagamma-crystallin genes in the ancestral vertebrate. J Mol Evol. 2010;71:219–30. doi: 10.1007/s00239-010-9379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wistow G, Wyatt K, David L, Gao C, Bateman O, Bernstein S, Tomarev S, Segovia L, Slingsby C, Vihtelic T. gamma N-crystallin and the evolution of the betagamma-crystallin superfamily in vertebrates. FEBS J. 2005;272:2276–91. doi: 10.1111/j.1742-4658.2005.04655.x. [DOI] [PubMed] [Google Scholar]
- 11.Brakenhoff RH, Aarts HJ, Reek FH, Lubsen NH, Schoenmakers JG. Human gamma-crystallin genes. A gene family on its way to extinction. Journal of Molecular Biology. 1990;216:519–532. doi: 10.1016/0022-2836(90)90380-5. [DOI] [PubMed] [Google Scholar]
- 12.Simpanya MF, Wistow G, Gao J, David LL, Giblin FJ, Mitton KP. Expressed sequence tag analysis of guinea pig (Cavia porcellus) eye tissues for NEIBank. Mol Vis. 2008;14:2413–27. [PMC free article] [PubMed] [Google Scholar]
- 13.Harding JJ, Crabbe MJC. The lens: Development, proteins, metabolism and cataract. In: Davson H, editor. The Eye. Academic Press; New York: 1984. pp. 207–492. [Google Scholar]
- 14.de Jong WW, Bloemendal H. Molecular and Cellular Biology of the Eye Lens. Wiley-Interscience; New York: 1981. Evolution of lens and crystallins; pp. 221–278. [Google Scholar]
- 15.Wilmarth PA, Taube JR, Riviere MA, Duncan MK, David LL. Proteomic and sequence analysis of chicken lens crystallins reveals alternate splicing and translational forms of beta B2 and beta A2 crystallins. Invest Ophthalmol Vis Sci. 2004;45:2705–15. doi: 10.1167/iovs.04-0131. [DOI] [PubMed] [Google Scholar]
- 16.Wistow G. Lens crystallins: gene recruitment and evolutionary dynamism. Trends Biochem Sci. 1993;18:301–6. doi: 10.1016/0968-0004(93)90041-k. [DOI] [PubMed] [Google Scholar]
- 17.Lee S, Mahler B, Toward J, Jones B, Wyatt K, Dong L, Wistow G, Wu Z. A single destabilizing mutation (F9S) promotes concerted unfolding of an entire globular domain in gammaS-crystallin. J Mol Biol. 2010;399:320–30. doi: 10.1016/j.jmb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandilands A, Hutcheson AM, Long HA, Prescott AR, Vrensen G, Loster J, Klopp N, Lutz RB, Graw J, Masaki S, Dobson CM, MacPhee CE, Quinlan RA. Altered aggregation properties of mutant gamma-crystallins cause inherited cataract. Embo J. 2002;21:6005–14. doi: 10.1093/emboj/cdf609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wistow G. The human crystallin gene families. Hum Genomics. 2012;6:26. doi: 10.1186/1479-7364-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian J, Ishibashi K, Reiser K, Grebe R, Biswal S, Gehlbach P, Handa JT. Advanced glycation endproduct-induced aging of the retinal pigment epithelium and choroid: a comprehensive transcriptional response. Proc Natl Acad Sci U S A. 2005;102:11846–51. doi: 10.1073/pnas.0504759102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinha D, Esumi N, Jaworski C, Kozak CA, Pierce E, Wistow G. Cloning and mapping the mouse Crygs gene and non-lens expression of [gamma]S-crystallin. Mol Vis. 1998;4:8. [PubMed] [Google Scholar]
- 22.Organisciak D, Darrow R, Gu X, Barsalou L, Crabb JW. Genetic, age and light mediated effects on crystallin protein expression in the retina. Photochem Photobiol. 2006;82:1088–96. doi: 10.1562/2005-06-30-RA-599. [DOI] [PubMed] [Google Scholar]
- 23.Wistow G, Bernstein SL, Wyatt MK, Behal A, Touchman JW, Bouffard G, Smith D, Peterson K. Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants. Mol Vis. 2002;8:171–84. [PubMed] [Google Scholar]
- 24.Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99:14682–7. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wistow G. The NEIBank project for ocular genomics: data-mining gene expression in human and rodent eye tissues. Prog Retin Eye Res. 2006;25:43–77. doi: 10.1016/j.preteyeres.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Wistow G, Peterson K, Gao J, Buchoff P, Jaworski C, Bowes-Rickman C, Ebright JN, Hauser MA, Hoover D. NEIBank: genomics and bioinformatics resources for vision research. Mol Vis. 2008;14:1327–37. [PMC free article] [PubMed] [Google Scholar]
- 27.Goncalves J, Nolasco S, Nascimento R, Lopez Fanarraga M, Zabala JC, Soares H. TBCCD1, a new centrosomal protein, is required for centrosome and Golgi apparatus positioning. EMBO Rep. 2010;11:194–200. doi: 10.1038/embor.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duran RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13:121–8. doi: 10.1038/embor.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boardman PE, Sanz-Ezquerro J, Overton IM, Burt DW, Bosch E, Fong WT, Tickle C, Brown WR, Wilson SA, Hubbard SJ. A comprehensive collection of chicken cDNAs. Curr Biol. 2002;12:1965–9. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
- 30.Cvekl A, Yang Y, Chauhan BK, Cveklova K. Regulation of gene expression by Pax6 inocular cells: a case of tissue -preferred expression of crystallins in lens. Int J Dev Biol. 2004;48:829–44. doi: 10.1387/ijdb.041866ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klement JF, Wawrousek EF, Piatigorsky J. Tissue-specific expression of the chicken àA-crystallin gene in cultured lens epithelia and transgenic mice. Journal of Biological Chemistry. 1989;264:19837–19844. [PubMed] [Google Scholar]
- 32.Carosa E, Kozmik Z, Rall JE, Piatigorsky J. Structure and expression of the scallop Omega-crystallin gene. Evidence for convergent evolution of promoter sequences. J Biol Chem. 2002;277:656–64. doi: 10.1074/jbc.M107004200. [DOI] [PubMed] [Google Scholar]
- 33.Lee DC, Gonzalez P, Wistow G. Zeta-crystallin: a lens-specific promoter and the gene recruitment of an enzyme as a crystallin. J Mol Biol. 1994;236:669–78. doi: 10.1006/jmbi.1994.1178. [DOI] [PubMed] [Google Scholar]
- 34.Wilmarth PA, Riviere MA, David LL. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J Ocul Biol Dis Infor. 2009;2:223–234. doi: 10.1007/s12177-009-9042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krey JF, Wilmarth PA, Shin JB, Klimek J, Sherman NE, Jeffery ED, Choi D, David LL, Barr-Gillespie PG. Accurate label-free protein quantitation with high-and low -resolution mass spectrometers. J Proteome Res. 2014;13:1034–44. doi: 10.1021/pr401017h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Z, Delaglio F, Wyatt K, Wistow G, Bax A. Solution structure of (gamma)S -crystallin by molecular fragment replacement NMR. Protein Sci. 2005;14:3101–14. doi: 10.1110/ps.051635205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hackett SJ, Kimball RT, Reddy S, Bowie RC, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han KL, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–8. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 38.Sinha D, Wyatt MK, Sarra R, Jaworski C, Slingsby C, Thaung C, Pannell L, Robison WG, Favor J, Lyon M, Wistow G. A temperature-sensitive mutation of Crygs in the murine Opj cataract. J Biol Chem. 2001;276:9308–15. doi: 10.1074/jbc.M010583200. [DOI] [PubMed] [Google Scholar]
- 39.Purkiss AG, Bateman OA, Wyatt K, Wilmarth PA, David LL, Wistow GJ, Slingsby C. Biophysical properties of gammaC-crystallin in human and mouse eye lens: the role of molecular dipoles. J Mol Biol. 2007;372:205–22. doi: 10.1016/j.jmb.2007.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahler B, Chen Y, Ford J, Thiel C, Wistow G, Wu Z. Structure and Dynamics of the Fish Eye Lens Protein, gammaM7-Crystallin. Biochemistry. 2013;50:3579–3587. doi: 10.1021/bi400151c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Callis PR, King J. Mechanism of the very efficient quenching of tryptophan fluorescence in human gamma D-and gamma S -crystallins: the gamma-crystallin fold may have evolved to protect tryptophan residues from ultraviolet photodamage. Biochemistry. 2009;48:3708–16. doi: 10.1021/bi802177g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Organisciak D, Darrow R, Barsalou L, Rapp C, McDonald B, Wong P. Light induced and circadian effects on retinal photoreceptor cell crystallins. Photochem Photobiol. 2011;87:151–9. doi: 10.1111/j.1751-1097.2010.00844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xi J, Farjo R, Yoshida S, Kern TS, Swaroop A, Andley UP. A comprehensive analysis of the expression of crystallins in mouse retina. Mol Vis. 2003;9:410–9. [PubMed] [Google Scholar]
- 44.Fan J, Dong L, Mishra S, Chen Y, Fitzgerald P, Wistow G. A role for gammaS-crystallin in the organization of actin and fiber cell maturation in the mouse lens. FEBS J. 2012;279:2892–904. doi: 10.1111/j.1742-4658.2012.08669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wistow GJ, Mulders JW, de Jong WW. The enzyme lactate dehydrogenase as a structural protein in avian and crocodilian lenses. Nature. 1987;326:622–624. doi: 10.1038/326622a0. [DOI] [PubMed] [Google Scholar]
- 46.Wistow G, Anderson A, Piatigorsky J. Evidence for neutral and selective processes in the recruitment of enzyme-crystallins inavian lenses. Proc Natl Acad Sci U S A. 1990;87:6277–80. doi: 10.1073/pnas.87.16.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wistow G, Sardarian L, Gan W, Wyatt MK. The human gene for gammaS-crystallin: alternative transcripts and expressed sequences from the first intron. Mol Vis. 2000;6:79–84. [PubMed] [Google Scholar]
- 48.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–82. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 52.Wilmarth PA, Tanner S, Dasari S, Nagalla SR, Riviere MA, Bafna V, Pevzner PA, David LL. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to crystallin insolubility? J Proteome Res. 2006;5:2554–66. doi: 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., 3rd MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid communications in mass spectrometry: RCM. 2004;18:2162–8. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 54.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak MY, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nature biotechnology. 2012;30:918–20. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–4. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 56.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 57.Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]