Abstract
The first rudimentary evidence that the human body harbors a microbiota hinted at the complexity of host-associated microbial ecosystems. Now, almost 400 years later, a renaissance in the study of microbiota spatial organization, driven by coincident revolutions in imaging and sequencing technologies, is revealing functional relationships between biogeography and health, particularly in the vertebrate gut. In this review, we present our current understanding of principles governing the localization of intestinal bacteria, and spatial relationships between bacteria and their hosts. We further discuss important emerging directions that will enable progressing from the inherently descriptive nature of localization and –omics technologies to provide functional, quantitative, and mechanistic insight into this complex ecosystem.
Introduction
The complex consortium of microbes that inhabits the distal human gut has become a topic of intense scientific study over the past decade, resulting in huge gains of sequence-based, molecular, and functional understanding. Still, many of the governing principles by which the human body acts as a home to microbial life were established by pioneering research from several hundred years ago. In the 17th century, Antonie van Leeuwenhoek created lenses powerful enough to observe bacteria for the first time, and quickly focused on his own body. In scrapings from the plaque between his teeth, he found a diverse, abundant array of “animalcules”. Nevertheless, it was not until the 20th century that Elie Metchnikoff elucidated that humans are born sterile and then populated immediately after birth by numerous microbes (Metchnikoff, 1901), and laid out an atlas for the gastrointestinal tract that still serves as the framework of our current understanding. His microscopy studies revealed that the density of bacteria increases along the length of the gastrointestinal tract, that different diets foster the growth of distinct microbial communities, and that bacteria are most abundant in the large intestine (Metchnikoff, 1901). However, it was uncertain at that time whether this community performed any valuable function(s). Instead, there was largely consensus that the majority of the gastrointestinal tract and its microbial community were evolutionary relics (Metchnikoff, 1901).
Recent insight into the gut microbiota’s tremendous impact on health and development has largely been driven by technological advances. Continually decreasing sequencing costs have helped with establishing a foundational understanding of the natural variation of microbiota composition among healthy subjects, and variation due to numerous factors, ranging from diet to lifestyle to delivery method. Although cataloging membership of the gut microbiota has become routine, understanding the functionality of individual members in the context of their ecosystem remains challenging. To that end, high-throughput assays focused on analysis of DNA (metagenomics), proteins (proteomics), and small molecules (metabolomics) are providing key tools to interrogate the elaborate crosstalk among gut microbes and between microbes and their host. Nonetheless, these techniques are typically applied to homogenized samples and neglect the spatial heterogeneity that underlies operations within this complex ecosystem.
While these high-throughput –omics techniques can be applied to virtually any biological sample (at least in principle), most analyses rely on stool, which has both advantages and shortcomings. Fecal material can be obtained non-invasively, is readily available, and allows for repeated sampling of an individual over time. However, sensitivity to change or decay of microbes and the molecules they produce (e.g., transcripts, metabolites) means that what is detected in stool may not be representative of what is occurring within the host. Perhaps most importantly, the snapshot of the gastrointestinal system provided by a single fecal sample is unable to capture the variation in bacterial localization and function along the length of the digestive tract.
Efforts have been made to quantify microbiota composition using 16S rRNA sequencing from samples derived invasively from distinct locations along the gastrointestinal tract, either by biopsy (Eckburg et al., 2005; Wang et al., 2005), laser-capture microdissection (Nava et al., 2011), or manual dissection (Turnbaugh et al., 2009; Yasuda et al., 2015). However, reliance upon bulk material for these measurements still results in loss of spatial resolution at the level of individual cells or even large consortia, thus failing to capture relationships within multi-cellular structures known to play important roles in complex microbial ecosystems (Dekas et al., 2014; Sekiguchi et al., 1999). To determine which bacteria are where, how their gene expression and function is connected to their local environment, and how global spatial organization varies in health and disease, we must return to where the field began – the microscope.
Biogeography of the gut microbiota
The gut contains a number of distinct habitats that select for heterogeneous spatial organization of the microbes housed within (reviewed in detail in (Donaldson et al., 2016; Fung et al., 2014; Kamada et al., 2013; Mowat and Agace, 2014). Variation in microbial localization and density within the digestive tract has been described along both the length of the gut (from mouth to rectum) and the cross-section (from mucosa-associated into the lumen). Chemical gradients (e.g. pH) (O’May et al., 2005), oxygen levels (Albenberg et al., 2014), nutrient availability (Berry et al., 2013), and immune effectors (Vaishnava et al., 2011) are among the many factors hypothesized to drive heterogeneity along the longitudinal and transverse axes. Experimental models such as mice have led to a general understanding of gut biogeography, including epithelial topography, mucus architecture, and the spatial distribution of various phyla (Figure 1).
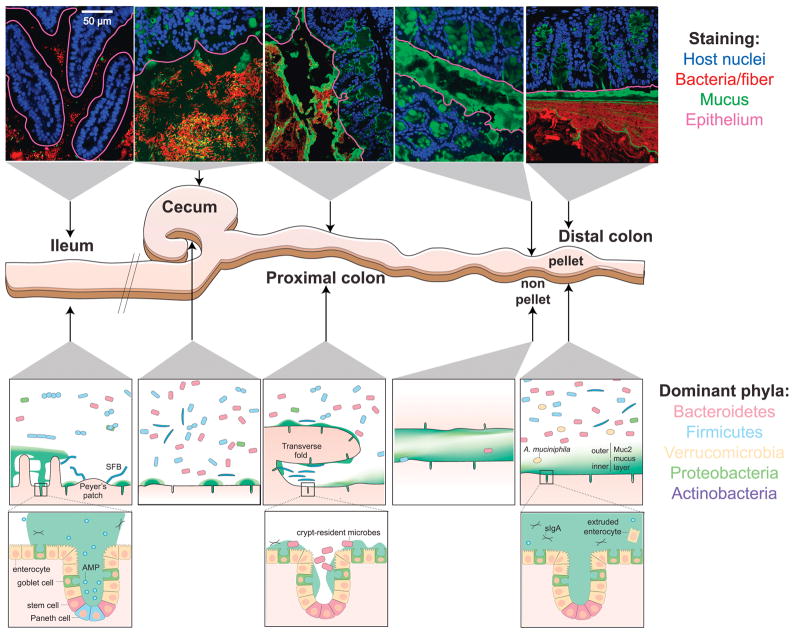
Figure 1. Biogeography of the mouse gastrointestinal microbiota.
Top: confocal micrographs of intestinal sections stained with UEA-1 (green) and DAPI (blue in epithelium, red in lumen). The UEA-1 glycan epitope is most abundant in the mouse distal colon and less so in the cecum and proximal colon. The epithelial boundary is overlaid (maroon). Middle: schematic of the distal mouse gastrointestinal tract. Bottom: schematic of key characteristics of each intestinal location after fixation with dry Carnoy’s fixative. Mucus structure and bacterial localization are heterogeneous along the longitudinal and transverse axes of the murine gastrointestinal tract. The MUC2-dependent layer becomes increasingly dense and impenetrable along the length of the intestines; the non-continuous appearance of the mucus in the ileum and proximal colon is potentially due to artefacts during mucus preparation. The density and diversity of bacteria increase along the longitudinal axis, with the small intestine favoring facultative anaerobic, proteolytic bacteria, and the colon favoring anaerobic, saccharolytic bacteria. Along the transverse axis, most bacteria are spatially segregated from the host tissue by immunological and physical barriers (Johansson et al., 2008; Vaishnava et al., 2011), with a few notable exceptions (Ivanov et al., 2009; Lee et al., 2013; Sonnenberg et al., 2012). Mucus structure in live animals is reviewed in (Pelaseyed et al., 2014). AMP, antimicrobial peptide; sIgA, secretory IgA; SFB, segmented filamentous bacteria.
Bacteria are not uniformly distributed throughout the lumen
The vast majority of gut bacteria are found within the transient digesta passing through the lumen of the lower gastrointestinal tract. Although the lumen is a continuous space, properties of the local microenvironment drive variation in both identity and abundance of taxa. In the small intestine, where transit is faster, and simple sugar and amino acid metabolism is favored, the community is dominated by rapidly dividing facultative anaerobes such as Proteobacteria and Lactobacillales (Gu et al., 2013). By contrast, in the large intestine the flow is slower, and metabolism favors fermentation of complex polysaccharides derived either from undigested plant material (fiber), or from the host mucus. This results in greater species richness (Faith et al., 2013; Seedorf et al., 2014; Turnbaugh et al., 2009) and dominance of the saccharolytic Bacteroidales and Clostridiales orders.
Within the colon, bacteria are also organized along the transverse axis, from the middle of the lumen to the mucosa. Moving toward the epithelium, mucin utilizers, such as Akkermansia muciniphila (Berry et al., 2013) and certain Bacteroides spp. (Berry et al., 2013; Yasuda et al., 2015), are thought to be enriched in the loose outer mucus layer that in humans extends several hundred microns into the lumen. Closer to the mucosa, an oxygen gradient selects for aerotolerant taxa, such as Proteobacteria and Actinobacteria, that express higher levels of oxygen-detoxifying catalases than other lumen residents (Albenberg et al., 2014).
Most commensal organisms are kept segregated from the epithelium by the mucosa
The inflammatory risk of housing trillions of microbes within our gut is minimized by immunological and physical barriers that limit bacterial colonization of the intestinal mucosa (reviewed in detail in (Johansson and Hansson, 2016)). The best studied physical barrier to the intestinal epithelium is the gastrointestinal gel-forming mucin MUC2, which polymerizes to form a mesh-like microbial sieve (Johansson et al., 2011). In mice, the mucus is thickest and densest in the distal colon, where it is organized into two layers: the striated, attached inner layer that is generally devoid of bacteria, and the outer loose layer, which provides a habitat and scaffold for bacterial attachment and nourishment (Johansson et al., 2008). The inner layer physically excludes bacteria (Atuma et al., 2001; Ermund et al., 2013b; Swidsinski et al., 2007b) and is enriched in innate and adaptive immune effectors targeting the microbiota, thereby providing a biochemical barrier as well (Barr et al., 2013; Bergstrom et al., 2016; Macpherson et al., 2015; Rogier et al., 2014).
Like the microbiota composition, the mucus varies throughout the colon, becoming denser and more continuous along the length toward the rectum, in part due to decreased water content. Interestingly, mucus thickness also fluctuates depending on circadian rhythms, affecting the taxonomic composition of the mucosal-associated ecosystem (Thaiss et al., 2016). In the small intestine, where most nutrient absorption occurs, the mucus is loose and penetrable (Ermund et al., 2013a). Nonetheless, the spaces between villi are largely free of bacteria, due to secretion of antimicrobial peptides (Salzman et al., 2010; Vaishnava et al., 2011).
In mice, close association of bacteria with the epithelial surface and within crypts appear to be common features of the proximal colon and cecum (Pedron et al., 2012), where the inner mucus layer is less dense and more penetrable by bacteria than the distal colon (Figure 1) (Ermund et al., 2013a; Swidsinski et al., 2007b). The bacteria residing in crypts are distinct from the luminal population and enriched in members from the phyla Proteobacteria (Lee et al., 2013; Pedron et al., 2012) and Firmicutes (Swidsinski et al., 2005a). Though the Bacteroidetes are less abundant in crypts, the best-studied resident is Bacteroides fragilis, which is able to bind to gastrointestinal mucin (Huang et al., 2011). Stable association of B. fragilis in crypts has been attributed to a species-specific locus predicted to target polysaccharides (Lee et al., 2013). Whether these results reflect immature mucus present in gnotobiotic mice colonized for short periods of time ((Johansson et al., 2015) or are representative of a fully developed barrier will require additional studies.
Similar to crypts, the transverse folds of the proximal colon of mice provide an epithelium-associated microhabitat for a specific subset of the mouse gut microbiota. 16S sequencing of sections isolated through laser capture microdissection showed that these interfold regions are enriched in the Firmicutes families Lachnospiraceae and Ruminococcaceae (Nava et al., 2011). Crypts and transverse folds provide a protected habitat separated from the flow that transports luminal digesta. Thus, microbes localized to such safe havens likely have an advantage in reseeding the colon during normal transit of gut contents and after disruption by antibiotics and infection (Lee et al., 2013).
In addition, there are some notable exceptions to the theme that commensals are segregated from the epithelium by mucus and immune factors. In the ileum of mice, the Firmicutes segmented filamentous bacteria (SFB) adhere tightly to epithelial cells of villi and Peyer’s patches, and this intimate association has pronounced immunomodulatory effects (Ivanov et al., 2009). SFB strains are host-specific, and detected in a wide range of vertebrates (Klaasen et al., 1993), including humans (Yin et al., 2013). While it is not clear whether SFB, or an analogous taxon, occupy such a privileged physical niche in humans, there is recent evidence that humans harbor other lymphoid (Obata et al., 2010; Sonnenberg et al., 2012) and adipose tissue-resident commensals (Schieber et al., 2015). In the latter case, it was shown that in mice, a commensal member of Enterobacteriaceae can be detected in host adipose tissue and promotes a metabolic blockade to muscle wasting (Schieber et al., 2015). However, the mechanisms by which specific taxa colonize and are restricted to particular anatomical sites are still largely unknown, and future studies are required to determine how stable colonization of host tissue affects host biology.
Spatial redistribution of commensals during disease
Historically, research into the impact of the microbiota on human health has focused on either compositional changes associated with disease, or the emergence of pathogens that can colonize or breach the epithelium. Now, it is becoming clear that spatial reprogramming of the microbiota may be a common and functionally relevant feature of chronic inflammatory diseases. Recently, specific microbiota members, such as the mucus degrader A. muciniphila, have been implicated in wound healing by stimulating enterocyte proliferation and migration (Alam et al., 2016). Conversely, inflammatory bowel diseases (IBD), which include ulcerative colitis and Crohn’s disease, are characterized by a compromised mucosal barrier and inappropriate immune activation by commensals mislocalized to the mucosa (Bergstrom et al., 2017; Swidsinski et al., 2009). Biopsies of patients with IBD have revealed biofilms of adherent B. fragilis covering the mucosa at inflamed sites (Swidsinski et al., 2005b), although it is still unclear whether this localization is a cause or an effect of IBD. Furthermore, there is evidence that mucus-consuming bacteria with increased prevalence in IBD patients are also better mucus utilizers in vitro (Png et al., 2010), indicating an important role in mucus utilization, mucosal proximity and disease. It has also been shown that bacterial relocalization during colitis promotes immune stimulation in genetically susceptible murine hosts via outer membrane vesicles (Hickey et al., 2015). Host genetics may also play an important role in digestive tract diseases. Recently, a specific host molecular chaperone, Cosmc, was shown to be associated with IBD in humans, and deletion of Cosmc in mice led to loss of gut microbiota diversity (Kudelka et al., 2016). Furthermore, the structure of the fecal microbiota is affected in patients with chronic idiopathic diarrhea, suggesting that gut homeostasis is an important feature in establishing commensal organization (Swidsinski et al., 2008).
Polymicrobial biofilms of otherwise commensal organisms are also a spatial feature of colorectal tumors (Dejea et al., 2014). The lack of a consensus bacterial composition at tumor sites suggests that the organization of commensals may be secondary to initiation and potentiation of the tumor by a narrower subset of phylotypes may be the relevant characteristic of this pathology (Hajishengallis, 2014). Unsurprisingly, the best understood examples of inappropriate microbiota localization involve diseases for which biopsies are a routine diagnostic, suggesting the possibility of ascertainment bias. Further investment in microscopy and spatial imaging technology, and their use in disease models in transparent organisms such as Caenorhabditis elegans or zebrafish, are needed to determine the dynamics of mislocalization during, and its contribution to, disease processes.
Overcoming imaging challenges in the gut
Intestinal tissues have long been stained and imaged for diagnostic purposes. Hematoxylin and eosin (H&E) stain differentially colors basophilic and acidophilic substances, respectively, and is commonly used in histology. Other stains target broad categories of molecules (e.g., Gram, targeting the cell wall) and hence can be used to generally define bacterial localization, but do not permit analysis of specific species or structures such as biofilms. Recent advances have combined multiple labeling approaches, including general stains, specific antibodies, and/or fluorescent probes, to dissect spatial heterogeneity in taxonomic composition and host environment across gut locations.
Mucus preservation and visualization
The gastrointestinal tract and its microbial inhabitants are difficult to observe intact. Key morphological features, including mucus and digesta where the vast majority of gut microbes reside, are difficult to preserve during routine histological preparation.
MUC2 is a large 5,197-amino acid protein core containing two mucin domains rich in proline, threonine, and serine repeats that become heavily O-glycosylated during processing in the Golgi apparatus (Johansson et al., 2011). These glycans account for nearly 80% of the mass of secreted MUC2 and result in the formation of a highly hydrated mucus layer upon secretion from the secretory vacuoles of goblet cells (Ambort et al., 2012). These properties render mucus difficult to preserve and observe, although some recent methodological advances have allowed the concurrent staining of mucus, epithelium and microbiota (Figure 2, Box 1). However, due to the fact that hydration status can alter mucus thickness, fixed sections are not necessarily representative of the in vivo landscape, but rather should be used in comparative studies with appropriate, similarly treated controls. To assay in vivo characteristics, live tissue from mouse intestines can be mounted in perfusion chambers or as colonic explants ((Bergstrom et al., 2016; Birchenough et al., 2016), although the use of buffers and hydrating reagents must be carefully considered as mucus will readily absorb water, potentially altering thickness.
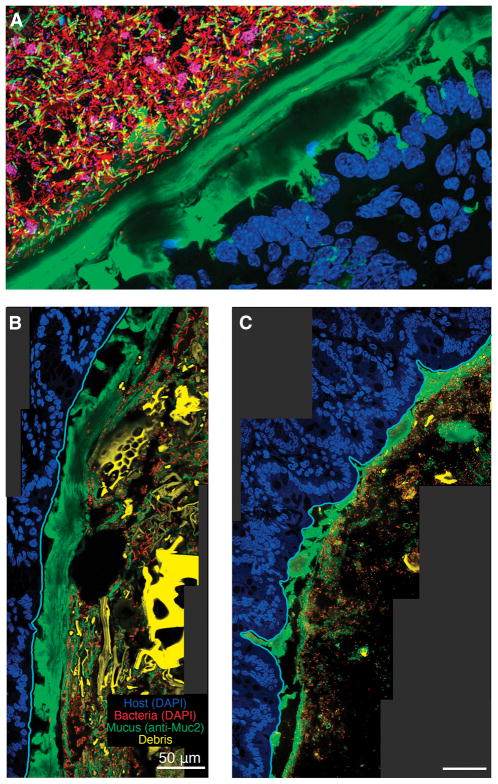
Figure 2. Improved histological methods allow visualization of mucus heterogeneity in the mouse colon.
A) Distal colon of a conventional mouse stained with UEA-1 (green), DAPI (blue in epithelium, red in lumen), and FISH probes specific to Firmicutes (yellow) and Bacteroidales (maroon). The thick, continuous, laminated inner layer of mucus adheres to the epithelium and excludes bacteria.
B–C) Distal colon of a gnotobiotic mouse colonized with Bacteorides thetaiotaomicron fed a high-fiber diet (B) or polysaccharide-free diet (C) (Earle et al., 2015). The sections are stained with anti-MUC2 antibody and DAPI; images show the epithelium (blue), mucus (green), bacteria (red), and plant matter (yellow) in the lumen. The epithelial border (cyan) was identified using the software BacSpace (Earle et al., 2015). The mucus layer in (C) is thinner than in (B), likely due to its consumption by B. thetaiotaomicron.
Box 1. Technical challenges of mucus and microbiota imaging.
Recently developed histological methods for improved mucus preservation enable the observation of bacteria and mucus in a relatively intact state (Johansson and Hansson, 2012; Matsuo et al., 1997). Fixation of tissue with fresh chloroform, dry methanol, and glacial acetic acid (called Carnoy’s fixative, or methacarn), and processing in water-free solutions before embedding in paraffin, results in a mucus layer that is structurally comparable to in vivo samples (Johansson et al., 2011). The presence of luminal contents further stabilizes the mucus and allows for the study of resident bacteria. Properly preserved, the mucus layer can be visualized using antibodies to MUC2 (Johansson and Hansson, 2012) or lectins (Essner et al., 1978) such as Ulex europaeus agglutinin I (UEA-1) for mouse colon (Gouyer et al., 2011) or wheat germ agglutinin (WGA) (Goto et al., 2014), which label specific carbohydrate structures attached to the protein core (Figure 1–3). However, even this fixation can cause decreases in apparent mucus thickness due to dehydration. The effects of this dehydration are most severe in the small intestine and in the proximal colon, due to the high water content of these intestinal regions. One limitation of this preparation method is the incompatibility of alcohol-based fixatives with actin stains, such as phalloidin. Nonetheless, this methodology has opened the door for systematic studies of the spatial organization of the gut microbiota within the colon environment, and has laid the foundation for understanding the bacteria-mucus interface (Chassaing et al., 2015; Loonen et al., 2014; Martinez-Medina et al., 2014; Nava et al., 2011).
Imaging the microbiota alongside mucus presents additional challenges. Preservation of endogenous fluorescence is not compatible with denaturing fixatives required for maintenance of mucus. Cross-linking fixatives such as 10% formalin or 4% paraformaldehyde are compatible with fluorescent reporters, and coupled with sucrose immersion and OCT embedding and cryosectioning, high-resolution images of bacteria in morphologically intact tissue with luminal contents have been achieved (Arena et al., 2015; Muller et al., 2012). In cases where mucus preservation is desired, methacarn fixation coupled with immunofluorescence against cloned or native bacterial proteins enables the simultaneous visualization of mucus and gut bacteria. Immunofluorescence-based bacterial detection methods are compatible with commercially available host-specific antibodies, and are widely used with a variety of tissue preparation strategies (Barthel et al., 2003; Deng et al., 2003; Lee et al., 2013; Sigal et al., 2015).
Mucus staining can be coupled to bacterial detection using fluorescence in situ hybridization (FISH) probes that target individual microbiota members (Chassaing et al., 2015; Huang et al., 2011; Johansson et al., 2008; Loonen et al., 2014; Pedron et al., 2012; Swidsinski et al., 2005a; Swidsinski et al., 2007a; Swidsinski et al., 2005b; Vaishnava et al., 2011). Despite its general utility, there are some limitations that must be considered when using FISH. Although probes have been designed to generally label bacteria, certain taxa are less permeable to nucleic acid oligomers or have lower ribosomal contents (Wagner et al., 2003), and hence general probes are unable to detect every bacterium in a given sample (Daims et al., 1999). Conversely, general stains such as the DNA dye 4′,6-diamidino-2-phenylindole (DAPI) present other challenges, for example autofluorescence from tissue and plant material is particularly bright in the blue emission range, and hence generates substantial noise that is computationally challenging to separate from the relatively weak bacterial signal (Earle et al., 2015). FISH can also be challenging when trying to distinguish between closely related strains (Wagner et al., 2003). Finally, the contents of lysed bacteria, which may be particularly prevalent during perturbations that challenge viability, can generate background signals that reduce signal to noise. Nonetheless, these recent advances indicate a bright future for targeted imaging of microbiota members and their mucosal environment.
Using the techniques highlighted in Box 1, multiple groups have shown that diet is an important modulator of mucus thickness (Desai et al., 2016; Earle et al., 2015). Specifically, during fiber deprivation, microbes that would otherwise consume microbiota-accessible carbohydrates (MACs) degrade the mucus barrier, reducing its thickness (Figure 2B,C). Furthermore, during dietary MAC starvation, mice were shown to be more susceptible to the bacterial pathogen, Citrobacter rhodentium, highlighting the importance of the mucus layer as a mechanical barrier against bacterial invasion (Desai et al., 2016). Given the difficulties of preserving mucus in its original in vivo state, it is imperative that studies are comparative in nature and include proper controls, rather than make absolute claims of mucus thickness and structure.
Bacterial detection
The lumen of the gastrointestinal tract is one of the densest and most diverse ecosystems described, with hundreds of bacterial phylotypes that can collectively reach densities exceeding 1011 bacteria per gram of content. Visualizing both individual bacterial cells and the breadth of diversity of these bacteria via microscopy presents a substantial technical challenge. Early efforts to visualize the gut microbiota within the colon relied on electron microscopy. Scanning and transmission electron microscopy of intestinal tissue, while limited in scale and bacterial identification strategies, rendered stunning, high-resolution images of bacteria associated with food particles and host tissue (Bollard et al., 1986; Croucher et al., 1983; Savage and Blumershine, 1974; Savage et al., 1971) and produced vivid depictions of morphological diversity in the gut (Croucher et al., 1983; Savage and Blumershine, 1974) (Figure 3A,B). However, these methods require fixation and processing that result in the collapse of colonic mucus, and hence loss of biologically relevant spatial information at the mucus interface.
Figure 3. Microscopic visualization of the gut microbiota.
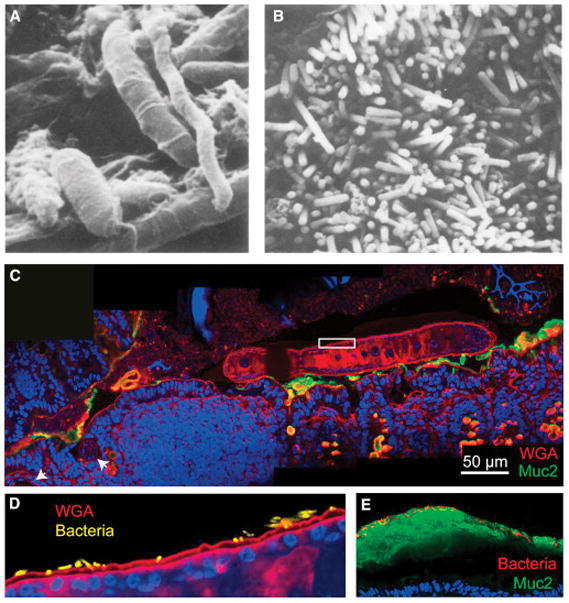
A) Scanning electron micrograph (Savage and Blumershine, 1974) from the mouse distal gut. Rod-, fusiform- and spiral-shaped bacteria are present, illustrating the morphological diversity of the gut microbiota.
B) Scanning electron micrograph from the mouse colon (Savage and Blumershine, 1974), highlighting the high density of bacteria in the gut.
C) Inter-kingdom spatial interactions between helminths and bacteria have been poorly studied to date. Trichuris muris, a mouse model of whipworm, was visualized by labeling with WGA (red) in the proximal colon of a conventional Swiss-Webster mouse 14 days after inoculation with ~200 T. muris ova. The anterior end is embedded in the epithelium near gastrointestinal lymphoid tissue (white arrowheads), while the posterior end is free in the lumen. MUC2 is labeled in green, and the mouse and worm nuclei are labeled with DAPI in blue.
D) Magnification of white box in (C) with bacterial DAPI signal segmented from the worm DAPI signal and false-colored yellow. Bacteria can be seen embedded in the cuticle of the worm.
E) Much of what we know about localization in the gut is derived from mouse studies. Further studies of human biopsies are needed, and in particular, on samples with preserved mucus. Here, a biopsy from a healthy patient has been fixed in methacarn, processed, sectioned and stained as described in Box 1. Bacteria, which are labeled with DAPI and false-colored red, are visible on the luminal side of the inner mucus layer.
Optical microscopy is a versatile and powerful method to visualize the spatial organization of the gut microbiota, based on its ease of use, diverse colorimetric bacterial labeling strategies, and applicability to larger tissue sections. Early applications of light microscopy to Gram-stained histological sections revealed that bacteria colonize the mucosa (Savage et al., 1968; Tannock, 1987), providing some of the first evidence for distinct gut habitats that select for particular phylotypes. However, reliance on general stains and cell morphology to identify bacteria limits insight into this spatial partitioning.
Fluorescence microscopy provides several options for visualizing particular bacterial taxa in vivo: (1) direct identification of bacteria expressing fluorescent proteins, (2) immunofluorescence applied to an epitope of interest, and (3) fluorescence in situ hybridization (FISH) to RNA (particularly 16S rRNA). Fluorescent protein expression has been used successfully to study several pathogens (Arena et al., 2015; Muller et al., 2012) and commensals (Amar et al., 2011; Jemielita et al., 2014), although such in vivo bacterial localization studies require some persistence of the strain within the community. Moreover, to engineer protein expression within such a strain requires that the species be culturable and genetically tractable. While there have been advances in cultivation methods for gut microbes (Browne et al., 2016; Lagier et al., 2016; Schnupf et al., 2015), genetic tools remain limited. Furthermore, the incompatibility of fixation methods with either fluorescent proteins or mucus preservation currently prohibits direct visualization of both fluorescent proteins and mucus within the same tissue section (Box 1).
Application of FISH to 16S rRNA has provided a flexible method of bacterial identification that does not require culturing of taxa. The sequencing data required to design probes is readily available, and validated probes already exist for many abundant gut commensals (Loy et al., 2007). Furthermore, FISH protocols are compatible with nearly every fixation and processing method, and combinatorial labeling strategies enable detection of at least 15 different phylotypes in a single sample (Valm et al., 2011). Using multi-label FISH, the presence of colon tumors was shown to be correlated with biofilms and bacterial tissue invasion (Dejea et al., 2014). Imaging of the oral microbiota revealed the development of plaque microbes into spatially differentiated structures from a few to hundreds of microns in size (Welch et al., 2016). 16S rRNA FISH has become a widely employed method for detection of commensal gut microbes in vivo. The convenience and reliability of 16S rRNA FISH far outweigh its limitations (Box 1), making it an essential tool for studying microbiota biogeography (Chassaing et al., 2015; Huang et al., 2011; Johansson et al., 2008; Loonen et al., 2014; Pedron et al., 2012; Swidsinski et al., 2005a; Swidsinski et al., 2007a; Swidsinski et al., 2005b; Vaishnava et al., 2011).
A specific challenge associated with bacterial detection in the gut is the cell density present within the colon. Even in thinly cut paraffin sections, the cellular cross-sections of maximal area lie in different planes, and rarely are perpendicular to the imaging axis. Due to the large spatial extent of point spread functions, the out-of-focus signal arising from cells a few microns away from the focal plane pollutes standard epifluorescence images, making detection of single cells extremely difficult. Thus, a smaller depth of focus, which is achieved by confocal microscopy, is necessary to distinguish individual cells. Even then, the close contact between bacteria makes automated segmentation and cell counting computationally challenging. Several tools have emerged to facilitate user-friendly manual and semi-automated curation of cell boundaries (Ducret et al., 2016; Van Valen et al., 2016). For large-scale measurements, where manual curation is prohibitively time intensive, pixel-based quantitative measurements that ignore cell boundaries can be used (Earle et al., 2015) as long as cellular resolution is not required. New methods for imaging of thick sections, thereby allowing full visualization of colonic crypts and biopsy samples (Liu et al., 2015), should motivate the development of three-dimensional segmentation algorithms.
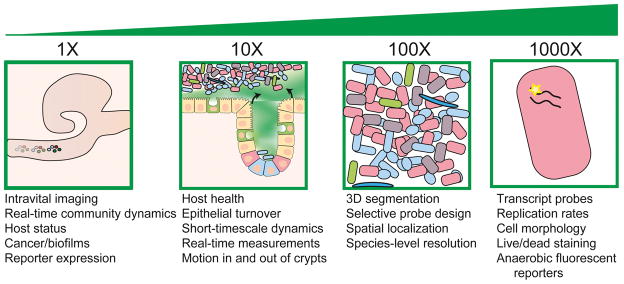
Given the limitations of existing methods, and the difficulties inherent in preserving morphological features of the gut, both imaging and computational advances have great potential to empower systematic study of the organization and spatial dynamics of the gut microbial community (Figure 4).
Figure 4. Future directions and challenges for localization studies of the gut microbiota.
Biogeography studies require technological and methodological development from the organismal/organ scale (left) to the cellular and sub-cellular scales (right). Highlighted are some of the potential challenges and improvements that will advance the field of microbiota organization.
Future directions
In the era of high throughput –omics research, technological advances combined with user-friendly data analysis platforms have made many approaches to microbiota investigation highly accessible. As we continue to unravel the complex microbial communities and the interactions that underlie microbial localization in the gut, several emerging technologies stand to contribute substantially. Recent advances in microbiota imaging approaches have pushed the field forward (Earle et al., 2015; Geva-Zatorsky et al., 2015), and promise to provide spatial information in tandem with functional interrogations through other approaches. Bioorthogonal click chemistry labeling of gut-resident bacteria is a powerful addition to the current set of visualization tools. Transparent vertebrates such as zebrafish have enabled real-time imaging of microbes within a host (Jemielita et al., 2014; Wiles et al., 2016), and imaging windows that have potentiated real-time tracking of tumor cells may provide the capacity for measuring the dynamics of the colonic community in mice (Ritsma et al., 2013).
Synthetic biology, the application of engineering principles to design biological systems, has had limited focus on abundant members of the gut microbiota (Khalil and Collins, 2010). Developing tools for gut commensals has the potential to provide biosensors and memory devices that can address basic questions about factors that individual bacteria sense and respond to in the gut (Mimee et al., 2015). Paired with a quantitative imaging pipeline, these tools can be used to discover the metabolic interactions and niche properties that shape the gut community and host health, including the spatial extent to which the effects of local inflammation are experienced by the microbiota, whether certain microhabitats are protected from antibiotic exposure, and how colonization order affects spatial partitioning or the ability to stably introduce a new strain. Imaging mass spectrometry (Berry et al., 2013; Rath et al., 2012) provides a complementary and valuable culture- and isolation-independent method to determine which bacteria are responsible for producing bioactive small molecules and proteins, and how these products drive spatial organization.
Finally, microbiota imaging must focus on expanding our biological scope (Figure 3,4). Nearly everything we know about spatial organization in the gut is limited to bacteria in mice. Few studies have examined how gut-resident viruses (Barr et al., 2013; Virgin, 2014), fungi (Iliev et al., 2012; Underhill and Iliev, 2014), protozoans (Berrilli et al., 2012) and helminths (parasitic worms; Figure 3C, D) (Broadhurst et al., 2012; Hasnain et al., 2011) remodel their environment and interact with the bacterial community. These groups include many important commensals and pathogens, and understanding how our physiology is affected by their presence (or in some cases, absence) is critical.
On the host side, while mice, and gnotobiotic mice in particular, are essential model organisms, there are clear differences physiologically (e.g., immune system), anatomically (Treuting and Dintzis, 2012), and microbially (Spor et al., 2011) between mice and humans. Further studies of human tissue, from biopsies (Johansson et al., 2014; Swidsinski et al., 2007a; Swidsinski et al., 2005b) and colectomies (Dejea et al., 2014), will be critical for elucidating these differences.
The localization of pathogens within their respective hosts is an essential aspect of their biology and a key determinant of the pathology that ensues. Only recently has it been recognized that the localization of commensals within their vertebrate hosts is equally important, and that mislocalization of these otherwise benign organisms is a feature of chronic inflammatory diseases. Now, with imaging and computational tools in hand, we are poised to answer many more questions by quantifying the often subtle spatial factors that have profound effects on our health.
Acknowledgments
The authors thank Dr. Joel Weinstock (Tufts University’s Sackler School) and Mr. Joseph Urban (USDA) for their assistance in conceptualizing and performing helminth experiments. Funding was provided by NSF CAREER Award MCB-1149328 (to K.C.H.). CT is supported by a James S. McDonnell Studying Complex Systems Postdoctoral Fellowship. This work was funded by grants from the National Institutes of Health NIDDK (R01-DK101674, R01-DK085025) to J.L.S.
Footnotes
Author Contributions
J.S., K.C.H, C.T., and K.E. contributed to the conceptualization and production of the manuscript text. K.E. and C.T. provided images for Figures 1–4.
Competing Financial Interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H, Jones RM, Nusrat A, Neish AS. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nature Microbiology. 2016;1:15021. doi: 10.1038/nmicrobiol.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063. e1058. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermudez-Humaran LG, Smirnova N, Berge M, Sulpice T, Lahtinen S, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO molecular medicine. 2011;3:559–572. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambort D, Johansson ME, Gustafsson JK, Nilsson HE, Ermund A, Johansson BR, Koeck PJ, Hebert H, Hansson GC. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5645–5650. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arena ET, Campbell-Valois FX, Tinevez JY, Nigro G, Sachse M, Moya-Nilges M, Nothelfer K, Marteyn B, Shorte SL, Sansonetti PJ. Bioimage analysis of Shigella infection reveals targeting of colonic crypts. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3282–3290. doi: 10.1073/pnas.1509091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2001;280:G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom JH, Birchenough GM, Katona G, Schroeder BO, Schutte A, Ermund A, Johansson ME, Hansson GC. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc Natl Acad Sci U S A. 2016;113:13833–13838. doi: 10.1073/pnas.1611400113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom K, Fu J, Johansson ME, Liu X, Gao N, Wu Q, Song J, McDaniel JM, McGee S, Chen W, et al. Core 1- and 3-derived O-glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal Immunol. 2017;10:91–103. doi: 10.1038/mi.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrilli F, Di Cave D, Cavallero S, D’Amelio S. Interactions between parasites and microbial communities in the human gut. Frontiers in cellular and infection microbiology. 2012;2:141. doi: 10.3389/fcimb.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, Wanek W, Richter A, Rauch I, Decker T, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchenough GM, Nystrom EE, Johansson ME, Hansson GC. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535–1542. doi: 10.1126/science.aaf7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollard JE, Vanderwee MA, Smith GW, Tasman-Jones C, Gavin JB, Lee SP. Location of bacteria in the mid-colon of the rat. Applied and environmental microbiology. 1986;51:604–608. doi: 10.1128/aem.51.3.604-608.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst MJ, Ardeshir A, Kanwar B, Mirpuri J, Gundra UM, Leung JM, Wiens KE, Vujkovic-Cvijin I, Kim CC, Yarovinsky F, et al. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS pathogens. 2012;8:e1003000. doi: 10.1371/journal.ppat.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher SC, Houston AP, Bayliss CE, Turner RJ. Bacterial populations associated with different regions of the human colon wall. Applied and environmental microbiology. 1983;45:1025–1033. doi: 10.1128/aem.45.3.1025-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Systematic and applied microbiology. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekas AE, Chadwick GL, Bowles MW, Joye SB, Orphan VJ. Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME archaea. Environmental microbiology. 2014;16:3012–3029. doi: 10.1111/1462-2920.12247. [DOI] [PubMed] [Google Scholar]
- Deng W, Vallance BA, Li Y, Puente JL, Finlay BB. Citrobacter rodentium translocated intimin receptor (Tir) is an essential virulence factor needed for actin condensation, intestinal colonization and colonic hyperplasia in mice. Molecular microbiology. 2003;48:95–115. doi: 10.1046/j.1365-2958.2003.03429.x. [DOI] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353. e1321. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nature reviews Microbiology. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature microbiology. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell host & microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermund A, Gustafsson JK, Hansson GC, Keita AV. Mucus properties and goblet cell quantification in mouse, rat and human ileal Peyer’s patches. PloS one. 2013a;8:e83688. doi: 10.1371/journal.pone.0083688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermund A, Schutte A, Johansson MEV, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2013b;305:G341–G347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner E, Schreiber J, Griewski RA. Localization of carbohydrate components in rat colon with fluoresceinated lectins. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1978;26:452–458. doi: 10.1177/26.6.670681. [DOI] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TC, Artis D, Sonnenberg GF. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunological reviews. 2014;260:35–49. doi: 10.1111/imr.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Alvarez D, Hudak JE, Reading NC, Erturk-Hasdemir D, Dasgupta S, von Andrian UH, Kasper DL. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nature medicine. 2015;21:1091–1100. doi: 10.1038/nm.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouyer V, Gottrand F, Desseyn JL. The extraordinarily complex but highly structured organization of intestinal mucus-gel unveiled in multicolor images. PloS one. 2011;6:e18761. doi: 10.1371/journal.pone.0018761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Chen D, Zhang JN, Lv X, Wang K, Duan LP, Nie Y, Wu XL. Bacterial community mapping of the mouse gastrointestinal tract. PloS one. 2013;8:e74957. doi: 10.1371/journal.pone.0074957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nature Reviews Immunology. 2014;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain SZ, Evans CM, Roy M, Gallagher AL, Kindrachuk KN, Barron L, Dickey BF, Wilson MS, Wynn TA, Grencis RK, et al. Muc5ac: a critical component mediating the rejection of enteric nematodes. The Journal of experimental medicine. 2011;208:893–900. doi: 10.1084/jem.20102057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, et al. Colitogenic Bacteroides thetaiotaomicron Antigens Access Host Immune Cells in a Sulfatase-Dependent Manner via Outer Membrane Vesicles. Cell host & microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Lee SM, Mazmanian SK. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe. 2011;17:137–141. doi: 10.1016/j.anaerobe.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielita M, Taormina MJ, Burns AR, Hampton JS, Rolig AS, Guillemin K, Parthasarathy R. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio. 2014;5 doi: 10.1128/mBio.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Hansson GC. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods in molecular biology. 2012;842:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nature Reviews Immunology. 2016 doi: 10.1038/nri.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Jakobsson HE, Holmén-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, Arike L, Wising C, Svensson F, Bäckhed F, et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host and Microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nature immunology. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nature reviews Genetics. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Laboratory animals. 1993;27:141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- Kudelka MR, Hinrichs BH, Darby T, Moreno CS, Nishio H, Cutler CE, Wang J, Wu H, Zeng J, Wang Y, et al. Cosmc is an X-linked inflammatory bowel disease risk gene that spatially regulates gut microbiota and contributes to sex-specific risk. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14787–14792. doi: 10.1073/pnas.1612158114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nature microbiology. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Dube PE, Girish N, Reddy AT, Polk DB. Optical reconstruction of murine colorectal mucosa at cellular resolution. American journal of physiology Gastrointestinal and liver physiology. 2015;308:G721–735. doi: 10.1152/ajpgi.00310.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonen LM, Stolte EH, Jaklofsky MT, Meijerink M, Dekker J, van Baarlen P, Wells JM. REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal immunology. 2014;7:939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- Loy A, Maixner F, Wagner M, Horn M. probeBase--an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic acids research. 2007;35:D800–804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Koller Y, McCoy KD. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol. 2015;36:460–470. doi: 10.1016/j.it.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, Darfeuille-Michaud A, Barnich N. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63:116–124. doi: 10.1136/gutjnl-2012-304119. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Histochemistry of the surface mucous gel layer of the human colon. Gut. 1997;40:782–789. doi: 10.1136/gut.40.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metchnikoff E. The Wilde Medal and Lecture of the Manchester Literary and Philosophical Society. Br Med J. 1901;1:1027–1028. [PMC free article] [PubMed] [Google Scholar]
- Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell systems. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nature reviews Immunology. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- Muller AJ, Kaiser P, Dittmar KE, Weber TC, Haueter S, Endt K, Songhet P, Zellweger C, Kremer M, Fehling HJ, et al. Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell host & microbe. 2012;11:19–32. doi: 10.1016/j.chom.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. The ISME journal. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’May GA, Reynolds N, Smith AR, Kennedy A, Macfarlane GT. Effect of pH and antibiotics on microbial overgrowth in the stomachs and duodena of patients undergoing percutaneous endoscopic gastrostomy feeding. Journal of clinical microbiology. 2005;43:3059–3065. doi: 10.1128/JCM.43.7.3059-3065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Goto Y, Kunisawa J, Sato S, Sakamoto M, Setoyama H, Matsuki T, Nonaka K, Shibata N, Gohda M, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, Sansonetti PJ. A crypt-specific core microbiota resides in the mouse colon. mBio. 2012;3 doi: 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaseyed T, Bergstrom JH, Gustafsson JK, Ermund A, Birchenough GM, Schutte A, van der Post S, Svensson F, Rodriguez-Pineiro AM, Nystrom EE, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. The American journal of gastroenterology. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Rath CM, Alexandrov T, Higginbottom SK, Song J, Milla ME, Fischbach MA, Sonnenburg JL, Dorrestein PC. Molecular Analysis of Model Gut Microbiotas by Imaging Mass Spectrometry and Nanodesorption Electrospray Ionization Reveals Dietary Metabolite Transformations. Analytical chemistry. 2012;84:9259–9267. doi: 10.1021/ac302039u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsma L, Steller EJA, Ellenbroek SIJ, Kranenburg O, Borel Rinkes IHM, van Rheenen J. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat Protocols. 2013;8:583–594. doi: 10.1038/nprot.2013.026. [DOI] [PubMed] [Google Scholar]
- Rogier EW, Frantz AL, Bruno ME, Kaetzel CS. Secretory IgA is Concentrated in the Outer Layer of Colonic Mucus along with Gut Bacteria. Pathogens. 2014;3:390–403. doi: 10.3390/pathogens3020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nature immunology. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC, Blumershine RV. Surface-surface associations in microbial communities populating epithelial habitats in the murine gastrointestinal ecosystem: scanning electron microscopy. Infection and immunity. 1974;10:240–250. doi: 10.1128/iai.10.1.240-250.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. The Journal of experimental medicine. 1968;127:67–76. doi: 10.1084/jem.127.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC, McAllister JS, Davis CP. Anaerobic bacteria on the mucosal epithelium of the murine large bowel. Infection and immunity. 1971;4:492–502. doi: 10.1128/iai.4.4.492-502.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber AM, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, Evans RM, Ayres JS. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science. 2015;350:558–563. doi: 10.1126/science.aac6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015;520:99–103. doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, Smith MI, Simon GM, Scheffrahn RH, Woebken D, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Applied and environmental microbiology. 1999;65:1280–1288. doi: 10.1128/aem.65.3.1280-1288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal M, Rothenberg ME, Logan CY, Lee JY, Honaker RW, Cooper RL, Passarelli B, Camorlinga M, Bouley DM, Alvarez G, et al. Helicobacter pylori Activates and Expands Lgr5(+) Stem Cells Through Direct Colonization of the Gastric Glands. Gastroenterology. 2015;148:1392–1404. e1321. doi: 10.1053/j.gastro.2015.02.049. [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature reviews Microbiology. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Herber A. Mucosal flora in Crohn’s disease and ulcerative colitis - an overview. Journal of physiology and pharmacology : an official journal of the Polish Physiological Society. 2009;60(Suppl 6):61–71. [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World journal of gastroenterology. 2005a;11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S, Lochs H, Dorffel Y. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007a;56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology. 2008;135:568–579. doi: 10.1053/j.gastro.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Sydora BC, Doerffel Y, Loening-Baucke V, Vaneechoutte M, Lupicki M, Scholze J, Lochs H, Dieleman LA. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflammatory bowel diseases. 2007b;13:963–970. doi: 10.1002/ibd.20163. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. Journal of clinical microbiology. 2005b;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW. Demonstration of mucosa-associated microbial populations in the colons of mice. Applied and environmental microbiology. 1987;53:1965–1968. doi: 10.1128/aem.53.8.1965-1968.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell. 2016;167:1495–1510. e1412. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Treuting P, Dintzis SM. Lower gastrointestinal tract. Paper presented at: Elsevier Inc.2012. [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science translational medicine. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nature reviews Immunology. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen DA, Kudo T, Lane KM, Macklin DN, Quach NT, DeFelice MM, Maayan I, Tanouchi Y, Ashley EA, Covert MW. Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments. PLoS computational biology. 2016;12:e1005177. doi: 10.1371/journal.pcbi.1005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Horn M, Daims H. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Current opinion in microbiology. 2003;6:302–309. doi: 10.1016/s1369-5274(03)00054-7. [DOI] [PubMed] [Google Scholar]
- Wang M, Ahrne S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS microbiology ecology. 2005;54:219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Welch JLM, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proceedings of the National Academy of Sciences. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles TJ, Jemielita M, Baker RP, Schlomann BH, Logan SL, Ganz J, Melancon E, Eisen JS, Guillemin K, Parthasarathy R. Host Gut Motility Promotes Competitive Exclusion within a Model Intestinal Microbiota. PLoS biology. 2016;14:e1002517. doi: 10.1371/journal.pbio.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell host & microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang Y, Zhu L, Liu W, Liao N, Jiang M, Zhu B, Hongwei DY, Xiang C, Wang X. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. The ISME journal. 2013;7:615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]