Summary
Human functional MRI (fMRI) research primarily focuses on analyzing data averaged across groups, which limits the detail, specificity, and clinical utility of fMRI resting-state functional connectivity (RSFC) and task activation maps. To push our understanding of functional brain organization to the level of individual humans, we assembled a novel MRI dataset containing five hours of RSFC data, six hours of task fMRI, multiple structural MRIs, and neuropsychological tests from each of ten adults. Using these data, we generated ten high fidelity, individual-specific functional connectomes. This individual connectome approach revealed several new types of spatial and organizational variability in brain networks, including unique network features and topologies that corresponded with structural and task-derived brain features. We are releasing this highly-sampled, individual-focused dataset as a resource for neuroscientists, and we propose precision individual connectomics as a model for future work examining the organization of healthy and diseased individual human brains.
Keywords: fMRI, individual variability, functional connectivity, brain networks, myelin mapping
Introduction
Over the past 30 years, functional magnetic resonance imaging (fMRI) and, more recently, resting-state functional connectivity (RSFC) fMRI studies based on the blood oxygen level-dependent (BOLD) signal, have significantly advanced our knowledge of human brain function and organization. By spatially co-registering and combining data from dozens, hundreds or even thousands of individuals, neuroscientists have been able to reliably identify central tendencies of both task-induced activation patterns (Martin, 2007; Petersen and Posner, 2012; Rugg and Vilberg, 2013; Wager and Smith, 2003) and the large-scale network organization of the brain (Beckmann et al., 2005; Power et al., 2011; Smith et al., 2009; Yeo et al., 2011). However, unlike structural MRI, which demonstrates clear clinical utility by describing the physical structure of individual brains, fMRI and RSFC research has generally shied away from studying individuals, with the notable exception of studies focused on specific regions of the cortex using functional localizers (Kanwisher, 2017) or high-field imaging (Cheng, 2016) (though see Huth et al., 2016 for an alternative approach to studying individual brains). Instead, much of systems neuroscience has focused on examining the group-average brain. While group averaging has revealed many basic principles of functional brain organization, the lack of emphasis on understanding individuals means that clinical applications of fMRI and RSFC have been limited to pre-surgical functional mapping (Mitchell et al., 2013; Sunaert, 2006).
Because the temporal signal-to-noise ratio of BOLD MRI data is relatively low (Welvaert and Rosseel, 2013), results obtained in individuals using small quantities of per-subject fMRI data (5–20 minutes) cannot precisely characterize brain function and organization (Anderson et al., 2011; Laumann et al., 2015; Xu et al., 2016). Hence, neuroimaging researchers have generally chosen to aggregate small amounts of data across many individuals. This approach identifies group-level central tendencies that generalize across individuals but obscures subject-specific features. Similarly, studies describing cross-individual variability in functional brain organization have tended to use large datasets with relatively small amounts of per-individual data (Gordon et al., 2017a, 2017b; Langs et al., 2016; Mueller et al., 2013; Wang et al., 2015), although this quantity has increased very recently (Chen et al., 2015; Xu et al., 2016). Thus, detailed characterization of individual brains has so far been limited (Laumann et al., 2015; Poldrack et al., 2015).
We contend that it is appropriate now to expand the standard human functional neuroimaging approach by developing methods to systematically characterize brain function and organization in single individuals. Individual-specific neuroimaging will be critical for determining whether differences in brain organization are behavior-related, disease-dependent, or epiphenomenal. This approach will also empower the study of individuals with idiosyncratic brain lesions (Fair et al., 2006; Pizoli et al., 2011), rare disorders, or unusual cognitive skills (Dresler et al., 2017) that could deepen our understanding of the fundamental principles of human brain organization. Finally, precise descriptions of brain organization in individual neurological, neurosurgical and psychiatric patients may enable custom treatment approaches beyond basic pre-surgical functional mapping (Hacker et al., 2013).
Recently, Laumann et al. (2015) characterized an individual’s functional brain organization by analyzing many hours of RSFC data collected from a single person across more than a year (Poldrack et al., 2015). This work suggested that 1) with sufficient data, reliable estimates of brain networks can be produced in a single individual; 2) features of an individual’s network map closely correspond with task-driven fMRI activations; and, most notably, 3) individual brain networks are more detailed than group-average networks, as small features often are obscured by averaging across groups. Together, these findings provided a single example of how individual specificity can be achieved using extensive within-individual data collection.
The present work represents a critical next step towards establishing precision individual connectomics as a research paradigm. To advance this nascent area of neuroscience, we collected a large quantity of data from ten individuals (24–34 years; 5F), each of whom underwent many hours of RSFC, task-based fMRI, structural MRI, and a neuropsychological testing battery (Figure 1, Table 1). To standardize time of day effects (Shannon et al., 2013) and reduce data acquisition costs, every scan started at midnight. Therefore, we refer to this dataset as the Midnight Scan Club (MSC) data. The complete MSC dataset is freely available from openfmri.org (Poldrack et al., 2013) and neurovault.org (Gorgolewski et al., 2015) as a resource for neuroscientists.
Figure 1. Per-subject data available in the MSC dataset.
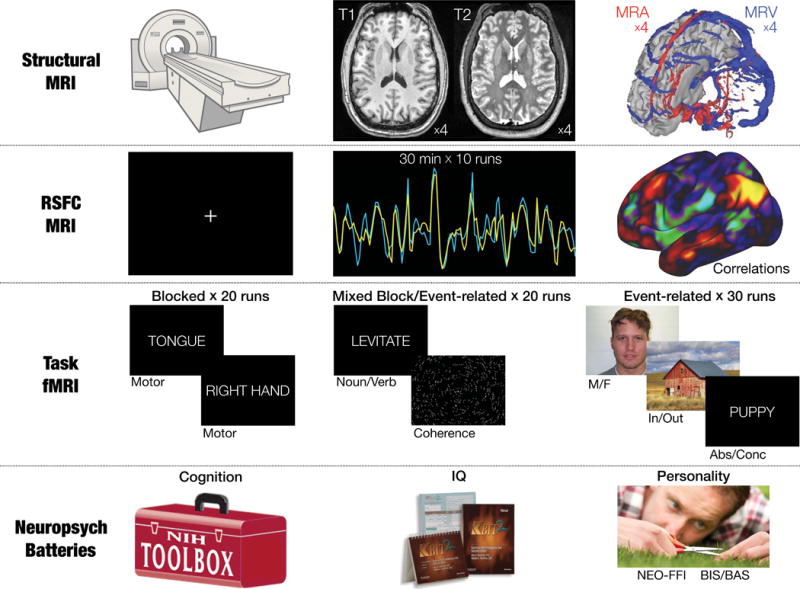
Data includes: four high-resolution T1 scans, four high-resolution T2 scans, four magnetic resonance angiogram (MRA) scans and four pairs of magnetic resonance venogram (MRV) scans, five hours of fMRI RSFC data, six hours of fMRI task data across three different tasks, and four neuropsychological assessments producing 21 individual assessment scores.
Table 1.
Subject demographic information and neuropsychological assessment scores
| 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | AVG | SD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender M=5, F =5 | M | M | F | F | M | F | F | F | M | M | ||
| Age | 34 | 34 | 29 | 28 | 27 | 24 | 31 | 27 | 26 | 31 | 29.1 | 3.3 |
| Education (years) | 22 | 28 | 18 | 22 | 20 | 17.5 | 20 | 21 | 19 | 19 | 20.7 | 3.0 |
| KBIT-2 | ||||||||||||
| Verbal Standard | 129 | 129 | 117 | 127 | 102 | 119 | 127 | 129 | 135 | 135 | 124.9 | 9.9 |
| Nonverbal Standard | 125 | 130 | 112 | 130 | 132 | 125 | 132 | 115 | 115 | 132 | 124.8 | 7.9 |
| IQ | 131 | 134 | 117 | 133 | 120 | 126 | 134 | 126 | 129 | 138 | 128.8 | 6.6 |
| BIS/BAS | ||||||||||||
| BAS Drive | 12 | 15 | 11 | 9 | 15 | 9 | 9 | 8 | 12 | 13 | 11.3 | 2.5 |
| BAS Fun Seeking | 7 | 12 | 10 | 10 | 15 | 11 | 13 | 9 | 10 | 15 | 11.2 | 2.6 |
| BAS Reward Responsiveness | 17 | 17 | 17 | 16 | 19 | 18 | 12 | 15 | 18 | 19 | 16.8 | 2.1 |
| BIS | 23 | 24 | 22 | 26 | 19 | 23 | 26 | 24 | 27 | 26 | 24.0 | 2.4 |
| NEO-FFI | ||||||||||||
| Neuroticism | 35 | 15 | 18 | 30 | 17 | 18 | 35 | 33 | 41 | 28 | 27.0 | 9.3 |
| Extraversion | 17 | 38 | 26 | 27 | 37 | 26 | 30 | 20 | 27 | 30 | 27.8 | 6.5 |
| Openness | 39 | 34 | 30 | 36 | 36 | 27 | 41 | 30 | 32 | 38 | 34.3 | 4.5 |
| Agreeableness | 24 | 31 | 31 | 34 | 29 | 36 | 38 | 30 | 28 | 25 | 30.6 | 4.5 |
| Conscientiousness | 26 | 31 | 31 | 40 | 39 | 25 | 35 | 30 | 38 | 28 | 32.3 | 5.4 |
| NIH Toolbox | ||||||||||||
| Cognition Crystallized Composite | 146.3 | 137.9 | 139.3 | 146.8 | 132.1 | 135.8 | 132.7 | 141.4 | 151.1 | 154.0 | 141.7 | 7.5 |
| 9-Hole Pegboard Dexterity | 112.7 | 105.0 | 99.7 | 102.2 | 98.8 | 102.6 | 100.0 | 110.7 | 92.5 | 104.1 | 102.8 | 5.8 |
| Dimensional Change Card Sort | 121.4 | 109.7 | 99.2 | 99.1 | 95.7 | 91.4 | 115.5 | 93.7 | 113.4 | 115.1 | 105.5 | 10.7 |
| Flanker Inhibitory Control & Attention | 113.8 | 111.7 | 103.2 | 84.8 | 90.8 | 108.2 | 106.0 | 98.5 | 113.4 | 92.8 | 102.3 | 10.2 |
| List Sorting Working Memory | 112.7 | 131.8 | 97.6 | 123.7 | 123.7 | 118.1 | 127.4 | 123.7 | 128.2 | 117.2 | 120.4 | 9.8 |
| Oral Reading Recognition ENG | 136.1 | 130.3 | 128.3 | 131.6 | 120.7 | 126.2 | 126.5 | 130.3 | 131.9 | 130.3 | 129.2 | 4.1 |
| Pattern Comparison Process Speed | 124.4 | 126.2 | 83.2 | 90.9 | 116.5 | 103.1 | 107.9 | 121.7 | 148.9 | 137.7 | 116.0 | 20.3 |
| Picture Sequence Memory | 112.3 | 134.8 | 134.8 | 130.2 | 112.4 | 112.4 | 114.9 | 98.3 | 121.7 | 134.8 | 120.7 | 12.6 |
| Picture Vocabulary | 128.0 | 128.1 | 136.7 | 145.0 | 133.8 | 133.8 | 127.5 | 136.8 | 149.2 | 151.1 | 137.0 | 8.7 |
Here we present a collection of novel and recently developed (Laumann et al., 2015) analyses to characterize the functional brain organization of the ten MSC individuals. This characterization includes 1) assessment of within-subject quality of RSFC data, including the reliability of multiple connectional and graph-theoretical measures of network function, 2) precise description of the brain network organization of each individual MSC subject, including both the physical topography and network topology, and 3) convergence between individuals’ task activation patterns, RSFC-derived brain networks, and distributions of cortical myelin.
Results
Figure 1 shows a graphical summary of the MSC dataset. In each of the 10 subjects, we collected 3.5 hours of structural MRI scanning; five hours of RSFC data; six hours of task-based fMRI data across three different tasks, including a blocked motor task, a mixed blocked/event-related perceptual/language task, and an event-related incidental memory task with multiple stimulus types; and a neuropsychological testing battery (Table 1).
Fidelity of RSFC depends on measure of interest, data quantity, and quality
Averaged activity timecourses were extracted from individual-specific parcels (defined following Laumann et al., 2015) and concatenated across sessions. Iterative split-data reliability analyses (as in Laumann et al., 2015) for multiple FC measures were conducted in each individual. Reliability was assessed for 1) the parcel-to-parcel connectivity matrix and 2) the network membership assigned to each parcel. Previous work has evaluated brain networks using three fundamental graph-theoretic concepts: the brain’s hub structure, efficiency of information transfer, and modular organization (Bassett and Bullmore, 2009; Bullmore and Sporns, 2009). Therefore, we also examined the reliability of three measures commonly employed to evaluate these properties: 3) the participation coefficient (PC), a measure of the diversity of a region’s cross-network connectivity contributing to its hub status (Guimerà et al., 2007; Power et al., 2013); 4) global efficiency, a whole-brain measure of network information transfer (Rubinov and Sporns, 2010); and 5) modularity, a measure of how well the whole-brain graph can be represented as a set of discrete networks (Newman, 2004).
Figure 2A–E illustrates that all measures exhibited relatively poor reliability given “typical” quantities of data (< 10 minutes retained after motion censoring). Reliability of many measures sharply increased with inclusion of additional data. In one individual (MSC08), parcel-to-parcel connectivity similarity was low (Fig. 2A,H), most likely owing to residual motion effects (Fig. S2). However, MSC08′s data was much less reliable than the overall relationship between residual motion effects and reliability would predict. This subject repeatedly reported falling asleep, exhibited frequent prolonged eye closures, and systematic increases in head motion over the course of each run (See Figure S1). Thus low reliability likely is attributable to high levels of drowsiness (Laumann et al., 2016; Tagliazucchi and Laufs, 2014).
Figure 2. Reliability, bias, and similarity of RSFC measures.
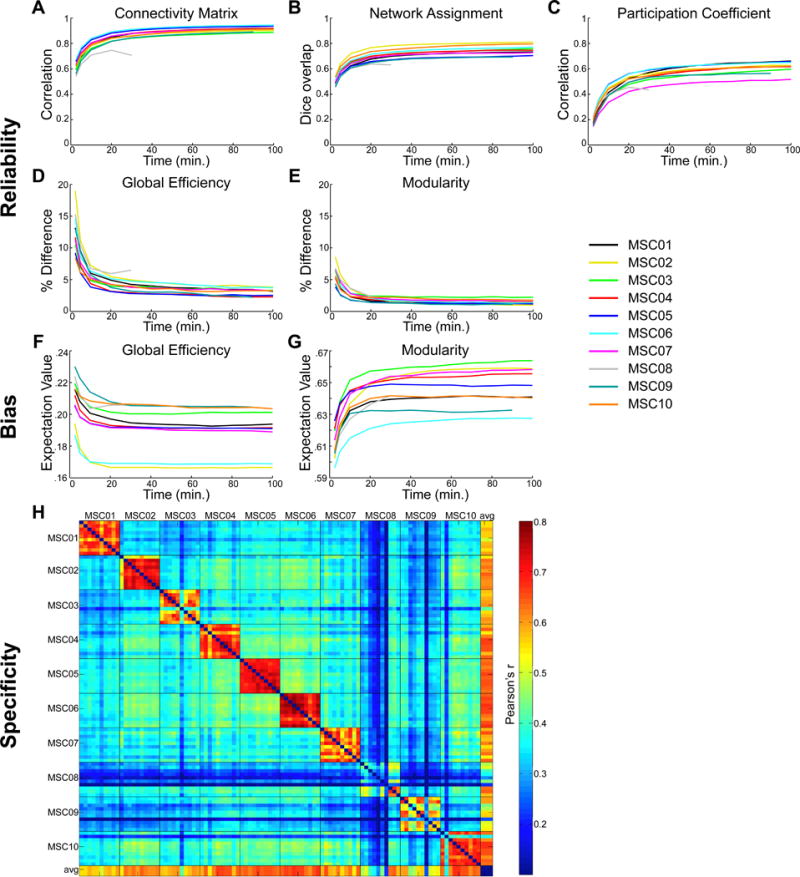
A–E: The reliability of various brain network measures increases with quantity of analyzed data. A given amount of motion-censored data (x-axes) was randomly selected and compared to a random independent sample of 70 minutes of data from the same subject; this was repeated 1000 times. A: Correlations of connectivity matrix upper triangles. B: Dice coefficients representing overlap of the node-wise categorical assignments. C: Correlation of node-wise participation coefficients. D: Percent difference in global efficiency. E: Percent difference in modularity. F–G: The expectation value of graph-theoretic measures depends on quantity of analyzed data. F: Expectation value of global efficiency. G: Expectation value of modularity. H: Pairwise similarity of correlation matrices between all individual subject sessions, as well as the group average (last row/column).
The quantity of data required for reliable estimation depended on the measure of interest. Excluding subject MSC08, we observed that 30 minutes of motion-censored data was required to achieve an average (across subjects) correlation matrix reliability of r > .85 (Figure 2A). Network assignment required even more data (90 minutes) to achieve an average Dice coefficient > .75 (Figure 2B). Modularity achieved an average difference of <3% from the split-data sample (Figure 2E) with only 10 minutes of data, while global efficiency required 90 minutes to achieve the same reliability (Figure 2D). The PC measure never achieved a high degree of split-data similarity in most subjects (maximum r = .5–.65; Figure 2C). Reliabilities calculated at each graph density are shown in Figure S2A–C.
Regarding global efficiency and modularity, increasing the amount of data not only increased reliability, but also altered expectation values, suggesting that these metrics systematically depend on data quantity. This effect is illustrated in Figure 2F–G. Across all subjects, global efficiency calculated from less than 20 minutes of motion-censored data was systematically elevated (Figure 2F) while modularity calculated from less than 20 minutes of data was systematically depressed (Figure 2G).
Session-level RSFC matrix similarity was examined both within and across subjects using an a priori group-level parcellation (Gordon et al., 2016). Within-subject similarity was high across sessions (diagonal blocks in Fig. 2H; worst in MSC08), with inter-session variability most prominent in primary somatosensory/motor cortex (see Figure S2E–F). For each session, correlation matrix similarity was greater within-subject than across subjects (all t>3.34; all p<=.001), meaning that individual RSFC patterns were distinct. In the following sections, we report detailed topographical and topological characterization of these individually distinct networks.
High fidelity individual RSFC mapping reveals individual-specific brain network features obscured by group averaging
To compare network topographies across individuals, we used a graph theory-based community detection approach (Power et al., 2011). Networks were defined in each individual by collapsing across density thresholds (Laumann et al., 2015) and assigning identities based on similarity to a set of template networks (Figure S3A). Identified networks included the Default Mode (DMN), Medial and Lateral Visual (mVis, IVis), Cingulo-Opercular (CON), Salience, Fronto-Parietal (FPN), Dorsal Attention (DAN), Ventral Attention (VAN), Hand, Face, and Leg Somatomotor (hSMN, fSMN, lSMN), Auditory, Premotor, Parietal Memory (PMN), Contextual Association (CAN), and anterior and posterior medial temporal networks (aMTL, pMTL). Network topographies were consistent across density thresholds (Figure S3B,C). With the exception of a small region in parieto-occipital sulcus, cross-subject variability in RSFC patterns was not related to cortical folding (Figure S3D).
We observed broad consistencies in network topographies across the ten individual datasets (Fig. 3A,B). For example, all network maps showed hallmark DMN features in medial parietal cortex, medial prefrontal cortex, and bilateral angular gyri that are consistently seen in group-average data (Raichle and Snyder, 2007). Similarly, all subjects showed canonical FPN features in lateral prefrontal cortex, lateral parietal cortex, dorsomedial prefrontal cortex, and lateral temporal cortex.
Figure 3. Brain network maps from highly-sampled subjects.
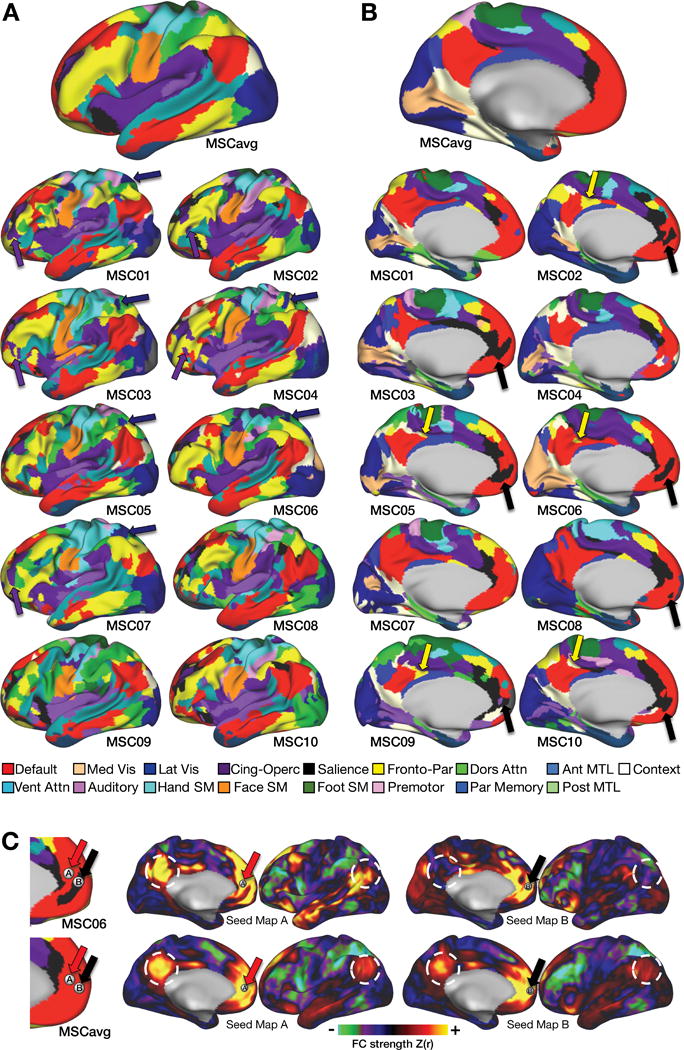
A, B: Lateral and medial views of brain networks identified in group average data (top) and in ten individuals. Several network “pieces” (sets of contiguous vertices in the same network) are highlighted that are observed across individuals but absent from the group average. Purple arrows: Cingulo-Opercular network pieces in anterior middle/inferior frontal gyrus. Dark blue arrows: Lateral Visual network pieces in superior parietal cortex. Yellow arrows: Fronto-Parietal network pieces in posterior cingulate cortex. Black arrows: Salience network pieces in ventromedial prefrontal cortex. C: Individual-specific features of brain networks reflect strong differences in functional connectivity. Adjacent seeds in ventromedial prefrontal cortex (red arrows: seed A; black arrows: seed B) are in Default and Salience networks, respectively, in subject MSC06. Middle and right: The Default seed (seed A) demonstrates strong positive connectivity with posterior cingulate and angular gyrus (white circles), but the Salience seed (seed B) demonstrates weak or negative connectivity with these regions. Only minimal differences are observed in group average data (bottom).
However, certain network features were observed that were absent from the group average. For example, five of ten MSC subjects had a region of CON in anterior inferior/middle frontal gyrus (Figure 3A, purple arrows); six subjects had a IVis feature in lateral parietal cortex (Figure 3A, dark blue arrows); seven subjects had a Salience feature in ventromedial prefrontal cortex (Figure 3B, black arrows); and five subjects had a FPN feature in middle cingulate cortex (Figure 3B, yellow arrows). None of these features were present in the group-average network map. As an example, we highlight the connectivity of the Salience network feature in ventromedial prefrontal cortex in one subject. Although this region has long been considered a core node of the DMN (Raichle et al., 2001), here we show that in subject MSC06, a region in the medial prefrontal cortex (Figure 3C: seed B, black arrow) was strongly correlated with pregenual cingulate, frontal pole, ventral insula, and temporal-parietal junction, but was un- or anti-correlated with canonical DMN regions (white circles). An adjacent seed in a portion of the vmPFC identified as DMN network (Figure 3C, seed A, red arrow) showed the expected strong medial parietal/angular gyrus connectivity. By contrast, the same two seeds showed almost no connectivity differences in the MSC group average (Figure 3C).
Global network organization of individual connectomes differs from the group average
Characterizing brain networks requires understanding the brain’s global network structure, especially the relationships within and between networks. To pursue this analysis in a well-defined, individual-specific network space (Wig et al., 2011), we identified parcel-level brain networks using the individual-specific cortical parcellations employed in the RSFC reliability analyses (above). The topographies of individual-specific brain networks were nearly identical vertexwise (Figure 3A) and parcel-wise (Figure S4), suggesting that the network topographies are consistent across these two spatial scales. Network structures for each subject and for the group-average were visualized using spring-embedded plots (Dosenbach et al., 2008; Power et al., 2011) across a wide range of thresholds (.5% to 5%; see Supplementary Movie). Figure 4A–B shows spring embedded plots from the center of the threshold range (2.5% edge density).
Figure 4. Graph analysis of brain networks.
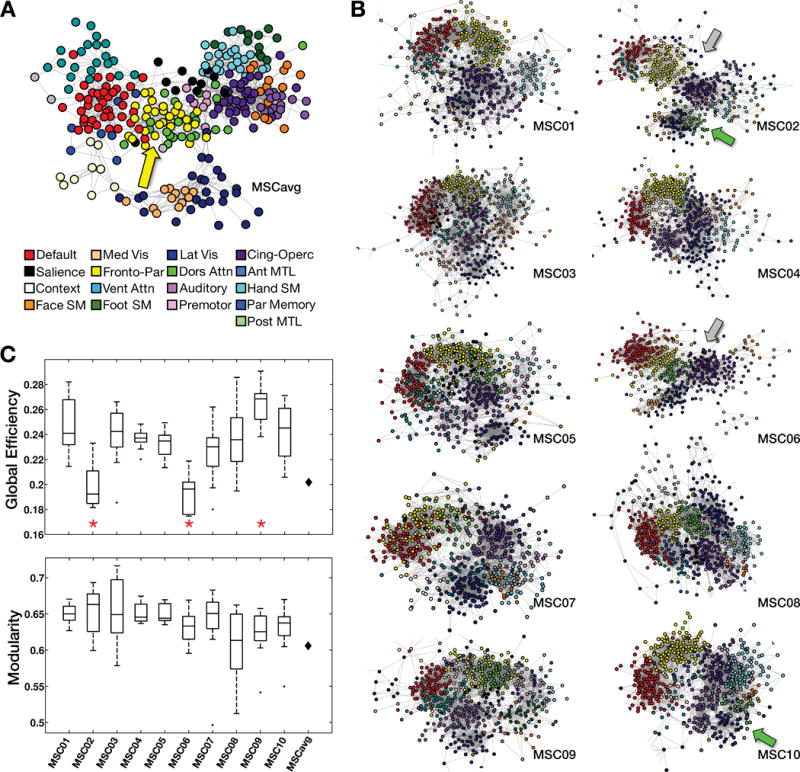
A,B: “Spring embedding” plots visualize networks such that well-connected groups of nodes are pulled together. Note that a few sparsely-connected peripheral nodes are not visualized here. A: In the group average, FrontoParietal (yellow) and Dorsal Attention (green) networks are central (yellow arrow). B: Many individual graphs exhibit a broadly circular organization without a central feature. Two individuals (gray arrows) exhibited a more linear organization. In addition, in two individuals, the Dorsal Attention and Fronto-Parietal networks were not adjacent (green arrows). C: ANCOVAs tested for the effect of subject identity on global efficiency (top) and modularity (bottom) while controlling for data quality and quantity. Significant effects (red asterisks) of subject identity were observed for global efficiency but not modularity. Black diamonds represent group average modularity and global efficiency.
The overall network structure of the MSC-average data (Figure 4A) closely matched previous reports (Power et al., 2011). In the group result, sensory, motor, and association networks were arranged around centrally located FPN (yellow) and DAN (green) networks (Figure 4A, yellow arrow). By contrast, none of the individual brain networks exhibited this organization (Figure 4B). In eight of ten MSC subjects, the association networks were arranged in a circular pattern. Within the circle, networks were pairwise connected as follows: DMN↔FPN↔DAN↔CON↔VAN↔DMN. A variety of single nodes were centrally positioned in this circle, but they were not preferentially FPN or DAN nodes. Individual sensory and motor networks tended to be linked to the CON and DAN networks, as in the group results.
In two individuals (MSC02 and MSC06, Figure 4B, gray arrows), the networks were linearly as opposed to circularly connected. Although spring-embedding layouts may change with graph density, the observed topologic distinctions (linear vs circular arrangement) were generally consistent across all but the sparsest or densest connection thresholds (Supplementary Movie). Post-hoc analyses indicated that these two subjects lack strong links between VAN regions and other regions outside the VAN or DMN (Figure S5).
We also observed a second pattern of topological distinction. In the MSC-average graph, as well as in most individuals, the DAN and FPN networks were adjacent or intermixed. However, in two individuals (MSC02 and MSC10; Figure 4B, green arrows) links between the DAN and FPN were absent, and the DAN was adjacent to the IVis network. Again, this observation was consistent across thresholds (Supplementary Movie).
We investigated whether these visually apparent differences in network organization were reflected in quantitative differences in global efficiency or modularity, which are whole-brain measures of network organization, computed for each session in each subject. Analysis of covariance (ANCOVA) tests for the effect of subject identity were computed separately for each measure at each density threshold, controlling for data quantity and quality (see Methods). After correction for multiple comparisons, we found a significant main effect of subject at every density threshold for global efficiency (at 2.5% density: F(9,87)=7.81, corrected p<.001), but not for modularity (at 2.5% density: F(9,87) = .84, p > .5). Figure 4C shows these effects at the 2.5% density threshold; Figure S6A–B illustrates mean (cross-session) values for each subject at every density threshold. Post-hoc pairwise comparisons revealed that the global efficiency effect was driven primarily by the two subjects with more linear graphs (MSC02 and MSC06) in whom global efficiency was low compared to all other subjects. We also observed significant global efficiency increases in MSC09. See Figure S6C for complete pairwise comparisons.
High fidelity mapping of individual fMRI task activations
Task-based fMRI allows functional localization at the individual level. The topography of task-fMRI responses in each individual was examined using data averaged over scanning sessions. Motor task responses are shown in Figure 5. Subject-specific somatotopy is evident in responses to movements of the tongue (ventrolateral somatomotor strip), hand (dorsolateral somatomotor strip), and foot (dorsomedial somatomotor strip). Including all data from all subjects produced group-average somatotopic maps that were statistically robust but spatially nonspecific, incorporating large swaths of white matter. By contrast, including only one session from each subject (i.e., using the same total amount of data) produced average maps with very few statistically robust responses. These results indicate that fMRI task responses become blurred in group averaged data, even when the individuals have been mutually co-registered using advanced nonlinear methods.
Figure 5. Highly-sampled task fMRI data enables precise localization of responses in individual subjects.
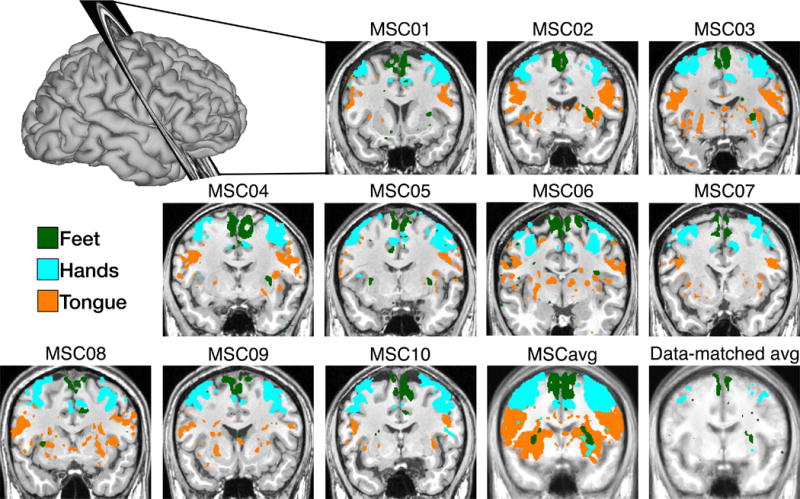
Regions near the central sulcus show BOLD responses to foot, hand and tongue movements. The “Hands” and “Feet” activation patterns represent contrasts between left- and right-sided movement, and are thresholded at |t|>5 and |t|>3, respectively. “Tongue” activations represent a tongue movement vs baseline contrast, and are thresholded at |t|>10. Responses are robust but poorly localized when all data from all subjects are tested (MSCavg). Average responses are sparse when data quantity is matched to the individual contrasts (data-matched avg).
Task activation patterns closely conform to RSFC network maps in individuals
Previous work has demonstrated individual-specific correspondence between RSFC and task fMRI in one highly-sampled subject (Laumann et al., 2015) and for multiple lower-data subjects (Tavor et al., 2016). Here, we extend this approach to demonstrate correspondence between RSFC networks and task responses across multiple tasks for ten highly-sampled subjects (Figure 6). Strong activations in response to hand and tongue movements (hand > tongue contrast) tightly respected individual-specific RSFC-defined hSMN and fSMN network boundaries (Fig. 6A,B; cyan and orange outlines, respectively). Similarly, responses to visual scenes (scene > face contrast) respected the boundaries of each individual’s resting state-defined CAN network (Fig 6D,E). By contrast, individual motor and scene-related responses did not respect network boundaries derived from other subjects or the group average, thus demonstrating individual topographic specificity (Figure 6C,F). Task/rest convergences can be seen for all pairwise subject comparisons in Figure S7.
Figure 6. Task-evoked BOLD responses align closely with individual-specific networks derived from resting-state data.
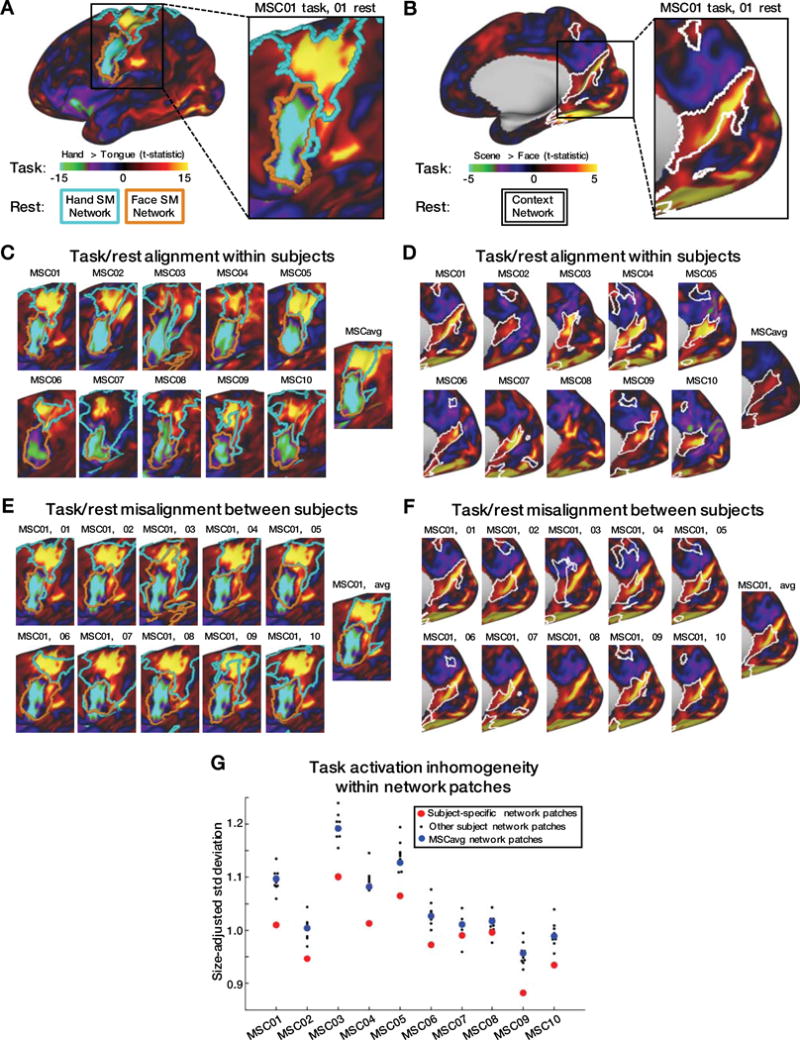
A,B: Hand > Tongue (A) and Scene > Face (B) task contrast maps for a single subject. Boundaries of this subject’s hand Somatomotor (cyan), face Somatomotor (orange) (A), and Contextual Association (B) RSFC networks are shown on the same surface. C,D: Hand > Tongue (C) and Scene > Face (D) task contrasts and RSFC network boundaries for each subject. E,F: Hand > Tongue (E) and Scene > Face (F) task contrasts from MSC01, compared to network boundaries of every subject. G: In each subject, the t-map inhomogeneities across many task contrasts was lower within all pieces of the subject’s own RSFC networks (red) than within pieces of other subjects’ networks (black) or group-average networks (blue). Seven task contrasts were tested: 1) Tongue motion > baseline; 2) Left Hand motion > Right Hand motion; 3) Left Leg motion > Right Leg motion; 4) Face stimulus > Word stimulus; 5) Scene stimulus > Face stimulus; 6) Glass dot pattern > baseline; 7) Noun/Verb stimulus > baseline.
Quantitative assessment of topographic correspondence between task responses and resting-state networks was evaluated in terms of response magnitude inhomogeneity within all discrete network regions. Inhomogeneity was defined as the standard deviation of all t-score values within a given region, adjusted for the size of that region and summed across regions (see Methods for details). Low inhomogeneity indicates that network boundaries accurately predict the spatial pattern of task-evoked activity. Individual topographic specificity was quantified by contrasting within- vs. across-individual inhomogeneity (Fig. 6D). In 8 of 10 subjects, the task vs. network topographic correspondence was better within as opposed to across subjects in every instance. In two subjects (MSC07 and MSC08) this result was not strictly obtained (one other subject demonstrated lower inhomogeneity). However, in aggregate, these results demonstrate the tight correspondence between task-evoked responses and network boundaries in individuals.
Spatially variable myelin features closely conform to RSFC network maps in individuals
Cortical myelin content can be derived from the ratio between T1- and T2-weighted images (Glasser and Van Essen, 2011; Glasser et al., 2016). A region of high cortical myelin in lateral occipito-temporal cortex, previously identified as the human MT+ complex (Glasser and Van Essen, 2011), was found in each MSC subject. The spatial location of this highly myelinated region varied across subjects (Table S1). Here, we tested whether anatomical differences in putative MT+ correspond to differences in RSFC network topography.
In all subjects, the entire putative MT+ region fell within a single RSFC-derived network, often requiring the network to exhibit extrusions encompassing the region (Figure 7A,B). Further, MT+ regions did not respect network boundaries derived from other subjects or the group average. These results demonstrate convergence of topographic specificity between RSFC networks and cortical myelin distributions in individuals (Figure 7C). Notably, the MT+ region was assigned to the IVis network in seven subjects, but the DAN in three other subjects (MSC02, MSC08, MSC10). Seed maps of the MT+ region were grossly similar across subjects (Figure S8). Thus, these individual differences likely reflect network assignment rather than network topography.
Figure 7. Regions of elevated cortical myelin density align closely with individual-specific networks derived from resting-state data.
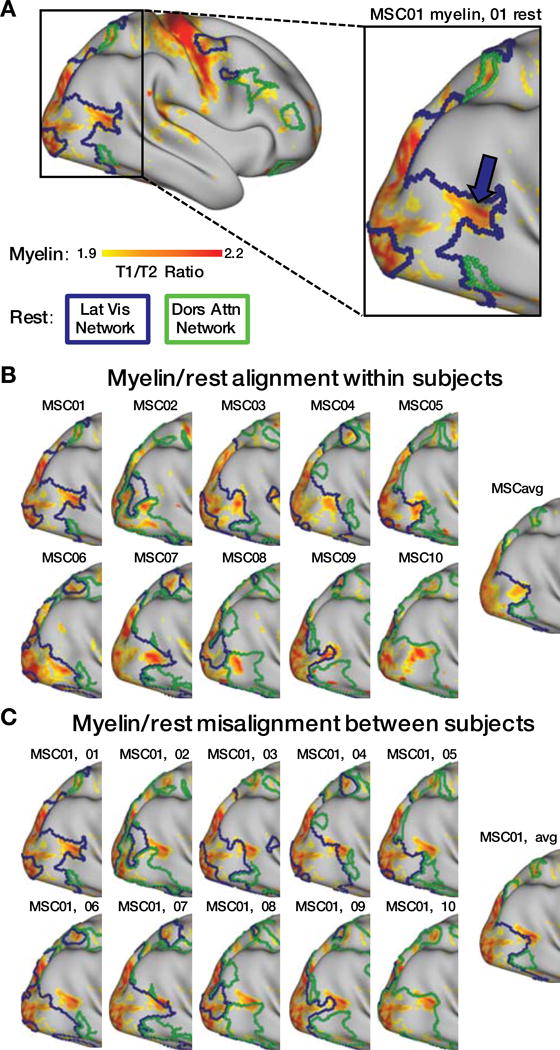
A: Cortical myelin density map for a single subject, thresholded at T1/T2 ratio>1.9 for visual purposes. A magenta arrow indicates the putative MT+ complex. Boundaries of this subject’s Lateral Visual (blue) and Dorsal Attention (green) RSFC networks are shown. B: Myelin maps showing the location of MT+ and RSFC network boundaries for each subject and the MSC average. Network boundaries align well with myelin maps on an individual-specific basis. C: Myelin maps showing the location of MT+ in MSC01, compared to the network boundaries of every subject and the MSC average, demonstrating that myelin maps do not align well with RSFC boundaries from other subjects.
Discussion
Descriptors of brain organization such as RSFC, task-based fMRI, and cortical myelin content have enhanced our scientific understanding of human brain function, and have the potential to improve the clinical care of neurological, neurosurgical, and psychiatric patients beyond current pre-surgical planning applications. However, functional neuroimaging may not achieve its full potential until very accurate individual-level brain network estimates can be achieved (Gordon et al., 2017a, 2017b; Harrison et al., 2015; Laumann et al., 2015; Wang et al., 2015).
To advance the goal of characterizing brain networks in individuals, we are providing this highly-sampled ten-subject dataset as a public resource for neuroscientists. In addition to 300 minutes of resting state data, each subject also contributed 350 minutes of task fMRI, 200 minutes of high-resolution structural and vascular imaging, and extensive neuropsychological testing (Figure 1, Table 1). This quantity of data represents at least a threefold increase in per-subject data for each of the resting state, task, and structural modalities over any other currently publicly available dataset (e.g., Chen et al., 2015; Van Essen et al., 2013). This greater quantity of data allowed us to develop procedures to assess data reliability, as well as to conduct extensive individualized delineation of task responses, connectome profiling, and cross-modal comparisons against task activations and myelin maps with greater precision than would be possible with any other dataset.
Highly-sampled fMRI data allow reliable, individual-specific, and externally valid descriptions of brain organization
Descriptions of an individual’s brain network organization are of greatest value if they are both reliable and externally valid. That is, we must be able to demonstrate that 1) repeated characterizations of the network organization produce the same answer in the same individual; and 2) the individual’s brain network organization corresponds to external measures of brain function or structure.
Reliability
We observed that for most subjects, several RSFC-derived measures converged to stable estimates given sufficient data. Convergence of correlation matrices, network assignments, global efficiency estimates, and modularity estimates are illustrated in Figure 2A–E. These findings extend previous work illustrating single-subject reliability of RSFC correlations and network assignments (Anderson et al., 2011; Laumann et al., 2015; Xu et al., 2016). The quantity of data needed for reliable characterization of individual-specific measures varied from 10 minutes to 80 minutes, depending on the specific measure. Less than 10 minutes of data (retained after motion correction) yielded low reliability estimates for all measures, as well as systematically biased graph-theoretic measures (see below).
Although modularity and global efficiency measures exhibited reasonable reliability with sufficient data (<4% error), the PC measure was poorly reliable (similarity < r=.65), even when large quantities of data were included in the comparisons (Figure 2C). A possible explanation for this result is that evaluating PC requires nonlinear transforms of the correlation matrix including thresholding and community assignment. Requiring precision from both measures simultaneously may account for the instability of PC estimates.
Of note, one subject exhibited substantially lower reliability of connectivity matrices (MSC08). As data quantity was matched across subjects for this analysis, this poor reliability most likely reflects drowsiness (Laumann et al., 2016; Tagliazucchi and Laufs, 2014). Previous work suggests that subjects who are drowsy may repeatedly transition into sleep states, resulting in less reliable RSFC data (Laumann et al., 2016; Yeo et al., 2015a). We believe that drowsiness did not significantly contaminate the data obtained in the other subjects, as they 1) self-reported continued wakefulness; 2) did not exhibit prolonged eye closures (as measured with eye-tracking); and 3) did not display the gradual within-session head motion increases characteristic of drifting off to sleep (see Figure S1).
We also observed that RSFC measures were highly subject-specific: session-to-session similarities were high within subject but much lower across subjects (Figure 2H). Similar findings have been reported by others as the basis for connectivity-based ‘fingerprinting’ of individuals (Anderson et al., 2011; Finn et al., 2015; Miranda-Dominguez et al., 2014; Pannunzi et al., 2017; Xu et al., 2016). However, the ability to distinguish subjects may at least in part be explained by differences in the spatial locations of brain networks across individuals (Gordon et al., 2017a; Laumann et al., 2015; Satterthwaite and Davatzikos, 2015). Additionally, systematic differences in other unconstrained biological (e.g., brain size, gyral folding patterns) or non-biological factors (e.g., data quality) may contribute to subject discrimination. We suggest that discriminating between individuals is a less important objective than accurately characterizing each individual’s functional connectome, which requires large quantities of high-quality data.
Bias
For two graph-theoretic measures of network organization—global efficiency and modularity—insufficient data not only resulted in unreliable measures, but also produced systematically biased values that were independent from the increased variance (Figure 2F–G). This effect converges with theoretical predictions that noise may subtly bias network measures (Sporns, 2014). Here, the effect may be driven by retained noise in limited data inducing the appearance of factitious cross-network connections that artificially shorten paths between nodes, increasing the apparent global efficiency and reducing modular structure.
These findings raise important issues to be considered when conducting RSFC studies using short scan times. Limited quantities of data increase noise in single-subject RSFC estimates. Additionally, for some measures, short scan times introduce systematic bias, which potentially compromises group comparisons. Theoretically, limited-data scans could be used in group comparisons, as long as the groups are both large enough to overcome the noise and strictly matched for quantity of data retained after motion censoring to equalize the bias. Future work should explore the nature of these biases to determine the feasibility of such approaches.
Validity
We observed precise, subject-specific correspondences between the spatial topography of RSFC-derived brain networks and the spatial extent of activations for motor and perceptual tasks (Figure 6). The scene-related activation elicited activity in regions consistent with previous reports that localized contextual processing to retrosplenial cortex (Bar and Aminoff, 2003), but did so in a highly subject-specific fashion. Broadly, these observations are consistent with previous task/rest correspondences observed in group average (Smith et al., 2009) and individual-specific data (Laumann et al., 2015; Tavor et al., 2016).
Individual-specific RSFC network topography also co-localized with a highly myelinated region in lateral occipito-temporal cortex (Figure 7) that likely represents the human MT+ complex (Glasser and Van Essen, 2011), one of the most spatially variable cortical areas described to date (Van Essen et al., 2012). Interestingly, this area was usually (7/10 subjects) within the IVis network, despite previous arguments that area MT+ may be a primary node of the DAN (Fox et al., 2006). Indeed, these seven subjects had no strong connections between putative MT+ and DAN (Figure S8).
Together, these findings reinforce the notion that 1) individual differences in the topography of resting state networks correspond to individual differences in the representation of function as determined by task fMRI, and that 2) this correspondence emerges because both task activations and RSFC network maps represent anatomically variable cortical regions such as the MT+ complex.
Precision mapping of individual brains reveals phenomenology obscured by group-averaging
Group-level analyses commonly assume that the organization of resting state networks is constant when evaluating variable correlation magnitude across individuals. The present results demonstrate how individual-specific brain networks violate this assumption. Network pieces anatomically varied across individuals, even after non-linear co-registration of the cortical surface. It is for this reason that group-averaging obscures the small, spatially variable network pieces that characterize individual brains (Figure 3). This result extends previous reports of small, individual-specific network pieces that are absent from group-average networks (Gordon et al., 2017b; Harrison et al., 2015; Laumann et al., 2015; Wang et al., 2015).
Interestingly, individual-specific network features were frequently observed across the ten MSC subjects. For example, seven of the subjects had a piece of the Salience network (Seeley et al., 2007) in ventromedial prefrontal cortex, a region generally assigned to the DMN (Buckner et al., 2008; Raichle and Snyder, 2007). Similarly, six subjects had an FPN network piece in posterior cingulate, another classical DMN region; six subjects had Visual network pieces in superior parietal cortex; and seven subjects had Somatomotor network pieces in middle opercular cortex. Comparable observations previously were made on the basis of less extensively sampled data (Gordon et al., 2017b; Tavor et al., 2016; Sepulcre et al., 2012; Eickhoff et al., 2010).
The functional significance of these individual-specific network features is not yet clear. Task fMRI data that is spatially convergent with RSFC networks (Figure 6) will be critical for understanding the function of these regions. In future work, comparing activated regions to RSFC networks (as has been done previously at the group level; Smith et al., 2009; Yeo et al., 2015b) will allow a functional annotation of the network features in each individual. Such data will aid not only in understanding the function of these features, but also in matching them across individuals.
Global network topology
The global network topology of individual-specific functional connectomes fundamentally differs from the network topology of group-average data (Figure 4A–B). Specifically, in the spring-embedding representation of the group-average network, the FPN and DAN occupy a central position (yellow arrow), as previously reported (Power et al., 2011). This observation has been interpreted as suggesting that these networks integrate information across more peripheral processing systems (Cole et al., 2013; Power and Petersen, 2013; Power et al., 2011). However, while many of the sensory/motor processing systems remain peripheral in individual data, there is no consistent ‘central’ network in the spring-embedding representation. Instead, in individual network topologies, “primary” networks (i.e., Visual, Auditory, Motor) tend to be connected to the CON (Dosenbach et al., 2006, 2007, 2008), likely via individual-specific network nodes in lateral parietal and posterior temporal cortex (Driver and Noesselt, 2008; Sepulcre et al., 2012). Meanwhile, “association” networks (i.e., CON, VAN, DMN, FPN, DAN) tend to be arrayed in a broadly circular formation (though two subjects did not share this arrangement—see below). Rather than a central integrating network, individual nodes from many networks were observed in the structure’s center.
It is plausible that the central position of the FPN in group-average data reflects mixing of signals from different networks. As we have shown (Figure 3), different networks exist at the same anatomical location across subjects. Additionally, group average parcels may not precisely fit a given individual’s brain organization. These factors contribute to signal mixing from multiple networks within and across subjects. Signal mixing is likely to be prominent in FPN regions, which are among the most individually variable in the brain (Mueller et al., 2013).
Eight of the ten MSC individuals exhibited a “circular” network structure, but two individuals exhibited a more linear organization (gray arrows, Figure 4B). This linearity was the result of the VAN being more weakly linked to non-DMN regions (Figure S5). This difference in network organization was associated with significantly reduced global efficiency (Figure 4C), indicating that longer “paths” are required to traverse the graph. Caution is required in interpreting the global efficiency metric in functional connectivity data, as the “paths” we observe represent correlated activity rather than physical links (Power et al., 2013).
We also observed a difference across individuals in the topology of the DAN. In eight of ten subjects, as well as in the group-average, the DAN was closely linked to the FPN. However, in two subjects, this network was not strongly connected to the FPN, but was closely tied to the IVis network (green arrows, Figure 4B). Interestingly, the highly myelinated putative MT+ region in these two subjects was also part of the DAN instead of the IVis network (the network containing MT+ in the majority of subjects) (Figure 7; Figure S8).
Further investigation of these apparent DAN variants is needed. We currently favor the possibility that variable inter- and intra-network connections of DAN regions may explain different sub-network configurations in different individuals. Specifically, it appears that the “DAN” identified in these two subjects is lacking superior parietal regions (possibly because they are incorporated into Premotor or FPN networks), but has instead incorporated lateral occipitotemporal regions (including MT+). These alternate DAN configurations may represent networks with only partially overlapping functions that nonetheless are assigned the same network label. This idea is supported by the observation that the MT+ region showed relatively consistent functional connectivity across individuals, despite being assigned to a different sub-network by the community detection algorithm (Figure S8).
Future work must develop approaches to identify and validate the types of network variants discussed above, quantify how they may be distributed in different populations, and explore what their functional consequences might be. It is tempting to hypothesize that presently observed network variants may relate to differences in demographics, cognitive abilities, or personality measures. Demographic information and neuropsychological batteries were collected to ensure that our subjects were relatively homogenous across a wide variety of measures (Table 1). However, the MSC dataset was not designed to examine brain-behavior correspondences, as reliable brain-behavior correlations cannot be computed in a sample of only 10 subjects. A power analysis shows that, to be 80% powered in ten subjects, such correlations would have to be extremely strong (uncorrected for multiple comparisons: r = .71; corrected: r=.87). Exploratory testing did not discover any effects of that magnitude (all rs < .60, all ps > .05). Thus, the relationship between global network variants discovered in the MSC data and behavior will need to be tested in other datasets. However, given the homogenous and normal-to-supernormal neuropsychological measurements across subjects, the variants we describe are unlikely to induce medically significant harm to cognitive function.
Future benefits of precision functional mapping of individual human brains
For systems neuroscience
The observed differences between individual and group-average network descriptions carry important implications for future brain network analyses (Dubois and Adolphs, 2016; Satterthwaite and Davatzikos, 2015). The majority of extant RSFC-based network analyses define nodes based on regions of interest (ROIs) derived from group-averaged data, applied identically in every individual. However, the present findings indicate that group-averaged ROIs may encompass multiple individual-specific network features, resulting in time series with mixed signals. This effect is illustrated by our finding that individual task responses only aligned well with each subject’s own resting-state networks (Figure 6). Thus, the precision of task responses and network measures can be improved by delineating brain features in an individual-specific fashion. Alternative techniques for individualized network analyses include machine learning techniques (Glasser et al., 2016; Hacker et al., 2013), mapping of semantic representations (Huth et al., 2016), and projection of individual data into an abstract representational space (Guntupalli et al., 2016; Langs et al., 2016). The convergence between RSFC, task activation, and myelin measures also indicates that task activation and/or myelin mapping may be useful as priors to help identify RSFC networks in circumstances where network identity/function is unclear—e.g., when brain damage has caused networks to reorganize into a nonstandard configuration that cannot be matched against a template.
For medicine
For over 25 years, functional MRI has tantalized us with the largely unfulfilled promise of clinical relevance—of the ability to non-invasively evaluate a patient’s brain function in order to predict a clinical outcome or influence treatment. So far, the quantity of RSFC data typically collected in patients (5–15 min) is adequate for pre-surgical planning around a few brain networks, but insufficient for precisely mapping function. To make predictions about individuals, machine learning algorithms can classify subjects as patients or controls using small quantities of data (Fair et al., 2013; Greene et al., 2016). However, these classification approaches have yet to be translated into routine clinical use.
Collecting hours of RSFC or task fMRI data per subject may help to fulfill the clinical promise of functional neuroimaging. Unlike neurodiagnostic techniques such as CT and PET, which expose patients to ionizing radiation, or invasive electrophysiology (electrocorticography, depth electrodes), fMRI is without significant medical risk. Thus, intensive fMRI scanning could be utilized in diagnostic workups, especially when accurate individualized assessment is critical. The risks and costs of even multiple hours of fMRI scanning compare favorably to a brain biopsy, or to the implantation of electrical grids for recordings in neurosurgical patients.
Conclusions
We present preliminary analyses of the multi-modal, highly-sampled Midnight Scan Club dataset. The primary purpose of this work is to describe the dataset, which is now publicly available as a general resource for neuroscientists. We have also presented several novel findings that emerged from our analyses of these data, including the reliability of, and potential biases within, multiple functional connectivity and graph-theoretic measures; the identification of variants in global network architecture; and the demonstration of individual-specific correspondences between task-evoked brain activity, resting state-derived networks, and the distribution of cortical myelin content. Together, these analyses underscore the scientific value of the MSC dataset for for precision functional mapping of individual brains.
We observed interesting differences in global network organization across ten individuals. However, additional individuals must be characterized to understand the prevalence of brain network variants in the general population. An important next step will be to study many more highly-sampled individuals in order to investigate which differences are linked to behavioral measures, and which are degenerate or epiphenomenal. Currently, the most significant impediment to high fidelity individual brain mapping is cost per subject. We aim to reduce this cost by making the MSC dataset publicly available to other researchers, so that they may utilize it to further describe individual-specific aspects of brain function and organization.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Dr. Nico Dosenbach (ndosenbach@wustl.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Data were collected from ten healthy, right-handed, young adult subjects (5 females; age: 24–34). Two of the subjects are authors (NUFD and SMN), and the remaining subjects were recruited from the Washington University community. Informed consent was obtained from all participants. The study was approved by the Washington University School of Medicine Human Studies Committee and Institutional Review Board. Other findings using these participants have been previously reported in Laumann et al. (2016).
METHOD DETAILS
Neuropsychological evaluation
The following behavioral assessments were administered to each subject:
The Kaufman Brief Intelligence Test, Second Edition (KBIT-2). This tool assesses overall intelligence in verbal and nonverbal domains (Kaufman and Kaufman, 2013).
The Behavioral Inhibition/Approach System (BIS/BAS) scale. This tool assesses the strength of three motivational drives and one aversive drive (Carver and White, 1994).
The NEO Five-Factor Inventory (NEO-FFI). This tool assesses personality traits within five independent factors (Costa and MacCrae, 1992).
The NIH Toolbox. This is a standardized set of tools to assess neurobehavioral function across a broad set of domains (www.nihtoolbox.org).
MRI image acquisition
Imaging for each subject was performed on a Siemens TRIO 3T MRI scanner over the course of 12 sessions conducted on separate days, each beginning at midnight. Structural MRI was conducted across two separate days. In total, four T1-weighted images (sagittal, 224 slices, 0.8 mm isotropic resolution, TE=3.74 ms, TR=2400 ms, TI=1000 ms, flip angle = 8 degrees), four T2-weighted images (sagittal, 224 slices, 0.8 mm isotropic resolution, TE=479 ms, TR=3200 ms), four MRA (transverse, 0.6 × 0.6, × 1.0mm, 44 slices, TR=25ms, TE=3.34ms) and eight MRVs, including four in coronal and four in sagittal orientations (sagittal: 0.8 × 0.8 × 2.0mm thickness, 120 slices, TR=27ms, TE=7.05ms; coronal: 0.7 × 0.7 × 2.5mm thickness, 128 slices, TR=28ms TE= 7.18ms), were obtained for each subject. Analyses of the MRA and MRV scans are not reported here.
On ten subsequent days, each subject underwent 1.5 hour functional MRI scanning beginning at midnight. In each session, we first collected thirty contiguous minutes of resting state fMRI data, in which subjects visually fixated on a white crosshair presented against a black background. Each subject was then scanned during performance of three separate tasks: motor (2 runs per session, 7.8 minutes combined), incidental memory (3 runs per session, 13.1 minutes combined), mixed design (2 runs per session, 14.2 minutes combined). Across all sessions, each subject was scanned for 300 total minutes during the resting state and approximately 350 total minutes during task performance. All functional imaging was performed using a gradient-echo EPI sequence (TR = 2.2 s, TE = 27 ms, flip angle = 90° voxel size = 4 mm × 4 mm × 4 mm, 36 slices). In each session, one gradient echo field map sequence was acquired with the same prescription as the functional images. An EyeLink 1000 eye-tracking system (http://www.sr-research.com) allowed continuous monitoring of subjects’ eyes in order to check for periods of prolonged eye closure, potentially indicating sleep. Only one subject (MSC08) demonstrated prolonged eye closures.
Task design
Motor task design
The motor task was adapted from that used in the Human Connectome Project (Barch et al., 2013). Subjects were presented with visual cues that directed them to close and relax their hands, flex and relax their toes, or wiggle their tongue. Each block started with a 2.2 s cue indicating which movement was to be made. After this cue, a centrally-presented caret replaced the instruction and flickered once every 1.1 s (without temporal jittering). Each time the caret flickered, subjects executed the proper movement. 12 movements were made per block. Each task run consisted of 2 blocks of each type of movement as well as 3 blocks of resting fixation, which lasted 15.4 s.
Incidental memory task
In this task, subjects made binary decisions about scenes, faces, and words, each of which was presented multiple times. Within each session, a single scan run was collected of each type of stimulus. In each task run, subjects viewed 24 stimuli, 3 times each. Stimuli were presented for 1.7 s with a jittered inter-stimulus interval ranging from 500–4900 ms. For scenes, subjects indicated whether an indoor or outdoor picture was presented. For faces, male/female judgments were made. For words, subjects made abstract/concrete judgments. Subjects were instructed to make their decisions as quickly as possible irrespective of the number of times a given stimulus had been presented. Subjects made their responses using a fiber-optic response box, and the finger used for each response type was counterbalanced within participants across sessions. Face stimuli were taken from several publicly available databases: The Psychological Image Collection at Stirling’s 2D face set (http://pics.psych.stir.ac.uk/); the CNBC Tarrlab “Face Place” repository (wiki.cnbc.cmu.edu/Face_Place; Righi et al., 2012); the Park Aging Mind Laboratory Face Database (agingmind.utdallas.edu/facedb; Minear and Park, 2004); and Libor Spacek’s Facial Imaging Database (cmp.felk.cvut.cz/~spacelib/faces/). Scene stimuli were drawn from a larger set reported by Chen et al. (2017). Lexical properties used to create the abstract and concrete word lists were sourced from the English Lexicon Project (Balota et al., 2007).
Mixed block/event-related design task
This task was adapted from experimental conditions reported by (Dubis et al., 2016). One task was a spatial coherence discrimination task, which used concentric dot patterns (Glass, 1969) that were either 0% or 50% coherent. During this task, subjects had to identify each pattern as concentric or random. The other task was a verbal discrimination task. Subjects were presented with nouns and verbs, and had to identify which type of word was being presented on the screen. Task blocks began with a 2.2 s cue screen indicating which task was to be conducted in the following block. Blocks consisted of 30 trials (half concentric/half non-concentric for coherence, half noun/half verb for verbal). Stimuli were presented for 0.5 s with a variable 1.7–8.3 s ISI. A stop cue displayed for 2.2 s signaled the end of each task block. Each scan run consisted of two blocks of each task. Task blocks were separated by 44 s periods of rest. For each task, the finger used for each response was counterbalanced within participants across sessions. No words overlapped between this task and the implicit memory task.
QUANTIFICATION AND STATISTICAL ANALYSIS
Cortical surface generation
Generation of cortical surfaces from the MRI data followed a procedure similar to that previously described in (Glasser et al., 2013) and (Laumann et al., 2015). First, anatomical surfaces were generated from the subject’s average T1-weighted image in native volumetric space using FreeSurfer’s default recon-all processing pipeline (version 5.3). This pipeline first conducted brain extraction and segmentation. After this step, segmentations were hand-edited to maximize accuracy. Subsequently, the remainder of the recon-all pipeline was conducted on the hand-edited segmentations, including generation of white matter and pial surfaces, inflation of the surfaces to a sphere, and surface shape-based spherical registration of the subject’s original surface to the fsaverage surface (Dale & Sereno, 1993; Dale et al., 1999; Fischl et al., 1999; Segonne et al., 2004). The fsaverage-registered left and right hemisphere surfaces were brought into register with each other using deformation maps from a landmark-based registration of left and right fsaverage surfaces to a hybrid left-right fsaverage surface (‘fs_LR’; (Van Essen et al., 2012)) and resampled to a resolution of 164,000 vertices (164k fs_LR) using Caret tools (Van Essen et al., 2001). Finally, each subject’s 164k fs_LR surface was down-sampled to a 32,492 vertex surface (fs_LR 32k). The various deformations from the original surfaces to the fs_LR 32k surface were composed into a single deformation map allowing for one step resampling. A script for this procedure is available on the Van Essen laboratory website (Freesurfer_to_fs_LR Pipeline, http://brainvis.wustl.edu). These various surfaces in native volumetric space were then transformed into atlas volumetric space by applying the previously calculated T1-to-atlas transformation.
Myelin Mapping
Myelin mapping was performed following methods described in (Glasser and Van Essen, 2011) and (Glasser et al., 2013). This procedure is based on the insight that myelin content positively covaries with T1-weighted image intensity, but negatively covaries with T2-weighted image intensity. Thus, dividing the T1 image by the T2 image will enhance image contrast for myelin content. At the same time, taking the ratio of T1/T2 eliminates the intensity bias field, as it is the same in both the T1 and T2 images. To perform this procedure, the T2 image was registered to the T1 image in native 0.8 mm resolution scanner space using in-house 4dfp tools for intensity gradient-based cross-modal registration (Rowland et al., 2005). The resulting T1/T2 ratio image was then sampled to the surface using a specialized algorithm available in the Connectome Workbench suite of tools (‘-volume-to-surface-mapping –myelin-style’; available at http://humanconnectome.org). The algorithm maps volumetric data to the midthickness surface by selectively sampling voxels delimited by the white matter and pial surfaces, i.e. the cortical ribbon. To remove voxels presumed to predominantly reflect large blood vessels, voxels with T1/T2 values exceeding ±1 SD of all T1/T2 values within the cortical ribbon were excluded (Glasser and Van Essen, 2011). The final value at each surface vertex was computed by averaging the remaining voxels around that position using a Gaussian-weighted function (FWHM = 5 mm). Finally, to correct for variable residual bias across subjects, we used the average MSC myelin map to estimate the expected low spatial frequency content of T1/T2 image intensity, following (Glasser et al., 2013). The difference between the average myelin map and each subject’s myelin map was heavily smoothed (FWHM = 14 mm) and then subtracted from each subject’s myelin map, leaving the low spatial frequency content of the group map while preserving the higher spatial frequency content specific to each individual.
fMRI Preprocessing
Functional data were preprocessed to reduce artifact and to maximize cross-session registration. All sessions underwent correction of odd vs. even slice intensity differences attributable to interleaved acquisition, intensity normalization to a whole brain mode value of 1000, and within run correction for head movement. Atlas transformation was computed by registering the mean intensity image from a single BOLD session to Talairach atlas space (Talairach and Tournoux, 1988) via the average high-resolution T2-weighted image and average high-resolution T1-weighted image. All subsequent BOLD sessions were linearly registered to this first session. This atlas transformation, mean field distortion correction (see below), and resampling to 3-mm isotropic atlas space were combined into a single interpolation using FSL’s applywarp tool (Smith et al., 2004). All subsequent operations were performed on the atlas-transformed volumetric time series.
Distortion correction
A mean field map was generated based on the field maps collected in each subject (Laumann et al., 2015). This mean field map was then applied to all sessions for distortion correction. To generate the mean field map the following procedure was used: (1) Field map magnitude images were mutually co-registered. (2) Transforms between all sessions were resolved. Transform resolution reconstructs the n-1 transforms between all images using the n·(n-1)/2 computed transform pairs. (3) The resolved transforms were applied to generate a mean magnitude image. (4) The mean magnitude image was registered to an atlas representative template. (5) Individual session magnitude image to atlas space transforms were computed by composing the session-to-mean and mean-to-atlas transforms. (6) Phase images were then transformed to atlas space using the composed transforms, and a mean phase image in atlas space was computed.
Application of mean field map to individual fMRI sessions: (1) For each session, field map uncorrected data was registered to atlas space, as above. (2) The generated transformation matrix was then inverted and applied to the mean field map to bring the mean field map into the session space. (3) The mean field map was used to correct distortion in each native-space run of resting state and task data in the session. (4) The undistorted data was then re-registered to atlas space. (5) This new transformation matrix and the mean field map then were applied together to resample each run of resting state and task data in the session to undistorted atlas space in a single step.
RSFC preprocessing
Additional preprocessing steps to reduce spurious variance unlikely to reflect neuronal activity were executed as recommended in (Ciric et al., 2017; Power et al., 2014). First, temporal masks were created to flag motion-contaminated frames. We observed that two subjects (MSC03 and MSC10) had a high-frequency artifact in the motion estimates calculated in the phase encode (anterior-posterior) direction that did not appear to reflect biological movement. We thus filtered the motion estimate timecourses in this direction only to retain effects occurring below 0.1 Hz, in these subjects only. Motion contaminated volumes were then identified by frame-by-frame displacement (FD, described in (Power et al., 2012)), calculated as the sum of absolute values of the differentials of the 3 translational motion parameters (including one filtered parameter) and 3 rotational motion parameters. Frames with FD > .2mm were flagged as motion-contaminated. Across all subjects, these masks censored 28% ± 18% (range: 6% – 67%) of the data; on average, subjects retained 5929 ± 1508 volumes (range: 2733 – 7667). Note that in this paradigm, even the worst subject retained almost two hours of data. See Figure S1 for motion parameter estimates in each subject.
After computing the temporal masks for high motion frame censoring, the data were processed with the following steps: (i) demeaning and detrending, (ii), multiple regression including: whole brain, ventricular and white matter signals, and motion regressors derived by Volterra expansion (Friston et al., 1996), with censored data ignored during beta estimation, (iii) interpolation across censored frames using least squares spectral estimation of the values at censored frames (Power et al., 2014) so that continuous data can be passed through (iv) a band-pass filter (0.009 Hz < f < 0.08 Hz) without re-introducing nuisance signals (Hallquist et al., 2013) or contaminating frames near high motion frames (Carp, 2013; Power et al., 2012). Censored frames were then excised from the data for all subsequent analyses.
Surface processing and CIFTI generation of BOLD data
Surface processing of the RSFC BOLD data proceeded through the following steps. First, the BOLD fMRI volumetric timeseries (both resting-state and task) are sampled to each subject’s original mid-thickness left and right-hemisphere surfaces (generated as the average of the white and pial surfaces) using the ribbon-constrained sampling procedure available in Connectome Workbench 1.0. This procedure samples data from voxels within the gray matter ribbon (i.e., between the white and pial surfaces) that lie in a cylinder orthogonal to the local mid-thickness surface weighted by the extent to which the voxel falls within the ribbon (Glasser & Van Essen, 2011). Voxels with a timeseries coefficient of variation 0.5 standard deviations higher than the mean coefficient of variation of nearby voxels (within a 5 mm sigma Gaussian neighborhood) were excluded from the volume to surface sampling, as described in (Glasser et al., 2013). Once sampled to the surface, timecourses were deformed and resampled from the individual’s original surface to the 32k fs_LR surface in a single step using the deformation map generated above (in “Cortical surface generation”). This resampling allows point-to-point comparison between each individual registered to this surface space.
These surfaces were then combined with volumetric subcortical and cerebellar data into the CIFTI format using Connectome Workbench (Marcus et al., 2011), creating full brain timecourses excluding non-gray matter tissue. Subcortical (including accumbens, amygdala, caudate, hippocampus, pallidum, putamen, and thalamus) and cerebellar voxels were selected based on the FreeSurfer segmentation of the individual subject’s native-space average T1, transformed into atlas space. Finally, the resting-state timecourses were smoothed with geodesic 2D (for surface data) and Euclidean 3D (for volumetric data) Gaussian kernels (σ = 2.55 mm).
Within-subject reliability of RSFC and derived measures
First, each subject’s RSFC data was parcellated into discrete homogenous parcels using the gradient-based parcellation technique (Cohen et al., 2008; Gordon et al., 2016; Laumann et al., 2015; Nelson et al., 2010; Wig et al., 2014). Briefly, RSFC data from all sessions were concatenated. Timecourses from all points in the brain were correlated against each other to generate a correlation map seeded from every point, and then these maps were correlated against each other to calculate the similarity of RSFC maps between each pair of points in the brain. A map of spatial gradients was then calculated on each column of the resulting similarity matrix using Workbench tools. Edges were identified in each of the resulting gradient maps using the watershed edge detection technique (Beucher and Lantuejoul, 1979), and all resulting edge maps were summed. Parcels were built from the resulting summed edge map by again applying the watershed edge detection technique. Neighboring parcels with edge counts less than a predefined threshold (here, the 50th percentile of edge count values) were merged. The average timecourse within each resulting parcel was then calculated.
We examined the reliability of five measures derived from these individual-specific parcel timecourses:
Parcel-to-parcel RSFC matrix. All parcel timecourses were correlated against each other to generate a parcel-to-parcel RSFC matrix for each subject. Values in the resulting matrix were Fisher-transformed to improve normality. Values taken from the upper triangle of this matrix represent all pairwise parcel relationships.
Parcel network identity. Network identification closely followed the procedures outlined in the “ Vertex-wise network mapping” section below, except that the parcel-to-parcel matrix was used instead of the vertex-to-vertex matrix. Briefly, correlations between parcels with centroids within 30mm of each other were set to zero in the matrix. The matrix was thresholded at a range of graph densities ranging from .3% to 5%, and the Infomap algorithm (Rosvall and Bergstrom, 2008) was applied to each thresholded matrix. The resulting communities were assigned network identities based on similarities to known group-average networks, and a consensus network assignment was computed by collapsing across thresholds.
Participation coefficient. Participation coefficient (PC) is a measure of the degree to which a graph node is connected to multiple networks. PC for each parcel was calculated as in (Guimerà et al., 2007). This calculation was done separately at each density threshold using the Infomap-derived communities identified at that threshold, and these threshold-specific PC values were then averaged across thresholds.
Global Efficiency. Global efficiency is a whole-graph measure of how efficiently information can be transferred over a graph. We used the Brain Connectivity Toolbox (http://www.brain-connectivity-toolbox.net; Rubinov and Sporns, 2010) to calculate Global Efficiency at each density threshold, and we then averaged these threshold-specific Global Efficiency measures across thresholds.
Modularity. Modularity is a whole-graph measure representing the degree to which the graph can be well represented as a set of discrete modules (Newman, 2004). We calculated modularity at each density threshold using the Infomap-derived communities identified at that threshold, and we then averaged these threshold-specific Modularity measures across thresholds.
The within-subject reliability of each of these measures was assessed in each subject using an iterative comparison of random data subsets, as in (Laumann et al., 2015). For each subject, the 10 sessions were split into two equal-sized, randomly selected subsets of sessions. Seventy minutes of (post-motion censoring) data was randomly selected from one of the two subsets. A varying amount of data (ranging from 2.5 minutes to 100 minutes, when possible) was randomly selected from the other subset. This data was contiguous within sessions but did not necessarily include temporally adjacent sessions. Each of the six above measures was calculated in each subset, and measures from the two subsets were compared. The comparisons used were as follows: 1) Parcel RSFC matrices were compared by correlating the upper triangles of the matrix. 2) Parcel network identities were compared via dice overlap. 3) Parcel PC values were compared by correlation. 4–6) The whole-graph measures were compared by calculating the absolute difference between the values from the two subsets, expressed as a percentage of the value from the 70-minute data subset.
To obtain robust estimates of the reliabilities of these measures for each subject, this procedure was iterated 1000 times for each quantity of data tested, with a different random selection of data in each iteration.
Finally, to examine whether reliability of the three graph metrics depends on the density threshold used, we conducted this iterated reliability estimation procedure separately for each density threshold, calculated using the greater of 100 minutes or the maximum amount of data available from each subject.
Within- and across-subject similarity of RSFC
To evaluate the within- and across-subject similarity of RSFC measures, we calculated the pairwise similarity between all subject sessions. First, for each subject in each session, we generated a parcel-to-parcel RSFC matrix, as above, using a common set of parcels (from (Gordon et al., 2016). Although these group-derived parcels lack the individual specificity of the subject-derived parcels, they allow direct comparisons of data between subjects. We then calculated a “similarity matrix” by correlating the upper triangle of each session’s RSFC matrix against the upper triangle of all other sessions in all other subjects. We then examined whether the similarities of sessions were higher within a subject than across subjects. Finally, we averaged the connectivity matrices across individuals and computed the similarity between each individual session matrix and this group average matrix.
Localization of within-subject variability in RSFC
To determine which regions of the brain tend to exhibit more within-subject variability in RSFC correlation patterns across sessions, we generated a parcel-to-parcel connectivity matrix for each session in each subject. For each subject, we calculated the standard deviation of each parcel-to-parcel connectivity value across sessions. We then averaged across the rows of this matrix in order to determine the average standard deviation in connection strength for a given parcel. To identify regions that were commonly variable in many subjects, we averaged these parcel-wise standard deviations spatially (i.e. at each vertex) across all subjects (except for subject MSC08, who exhibited variability out-of-scale with the other subjects), excluding zero-values in border vertices.
Vertex-wise network mapping
The network organization of each subject’s brain was derived using the graph-theory-based Infomap algorithm for community detection (Rosvall and Bergstrom, 2008), following (Power et al., 2011). In this approach, we calculated the cross-correlation matrix of the time courses from all brain vertices (on the cortical surfaces) and voxels (in subcortical structures), concatenated across sessions. Correlations between vertices/voxels within 30 mm of each other were set to zero in this matrix to avoid basing network membership on correlations attributable to spatial smoothing. Geodesic distance was used for within-hemisphere surface connections and Euclidean distance for sub-cortical connections. Inter-hemispheric connections between the cortical surfaces were retained, as smoothing was not performed across the mid-sagittal plane.
This matrix was then thresholded at a range of values calculated based on the resulting density of the matrix; the density thresholds ranged from .3% to 5%. These thresholded matrices were used as inputs for the Infomap algorithm, which calculated community assignments (representing brain networks) separately for each threshold. Small networks with 400 or fewer vertices/voxels were considered unassigned and removed from further consideration. The above analysis was conducted in each individual subject, and in data averaged across all subjects.
To identify putative networks we may find in each subject, we conducted a re-analysis of our previous work on group-average data (Gordon et al., 2017a; Laumann et al., 2015), in which the Infomap algorithm was conducted in data averaged across a large, independent group of 120 individuals with low amounts of per-subject data. Analysis procedures were identical to the analyses conducted on this group in (Gordon et al., 2017a; Laumann et al., 2015), except that the minimum density threshold tested was reduced to .3%. As a result, we were able to identify group-average networks corresponding to a) early visual cortex, b) somatomotor cortex associated with the foot, and c) to a strip along postcentral gyrus corresponding closely to “premotor” activation in the Neurosynth platform (Yarkoni et al., 2011). See Figure S2A for these independent group-average networks.
Putative network identities were then assigned to each subject’s communities (and to the communities from the average of the individual subjects) by matching them at each threshold to the above independent group networks. This matching approach proceeded as follows. At each density threshold, all identified communities within an individual were compared (using spatial overlap, quantified with the Jaccard index) to each one of the independent group networks in turn. The best-matching (highest-overlap) community was assigned that network identity; that community was not considered for comparison with other networks within that threshold. Matches lower than Jaccard = .1 were not considered (to avoid matching based on only a few vertices). Matches were first made with the large, well-known networks (Default, Lateral Visual, Motor, Cingulo-Opercular, Fronto-Parietal, Dorsal Attention), and then to the smaller, less well-known networks (Ventral Attention, Salience, Parietal Memory, Contextual Association, Medial Visual, Motor Foot). See Figure S2B for “block” diagrams showing networks present at each threshold, following Power et al. (2011). In each individual and in the average, a “consensus” network assignment was derived by collapsing assignments across thresholds, giving each node the assignment it had at the sparsest possible threshold at which it was successfully assigned to one of the known group networks.
Finally, contiguous network pieces that were smaller than 30 mm2 were removed, following (Gordon et al., 2017a) as such regions are smaller than the approximate effective resolution of our data (originally 4mm isotropic voxels, smoothed 6mm FWHM on the cortical surface). This procedure serves as a spatial filter, eliminating implausibly small objects without smoothing the underlying data. Neighboring network identities were then dilated into the removed regions one vertex at a time until the region was filled.
Across network density thresholds, we observed that the locations of borders between individual-specific networks were largely consistent. However, this was not universally true; in some cases, the borders shifted substantially from threshold to threshold. This shifting represents an uncertainty in the spatial topography of the networks that is not well captured by the consensus network map. To characterize this uncertainty, in each subject we identified all cortical vertices that had inconsistent network assignments across the above density thresholds. A vertex was deemed inconsistent if it changed network assignment from one density threshold to the next, and both networks were present at both thresholds. This uncertainty can be seen in Figure S3.
Relationship between anatomical variability and functional connectivity variability
To ensure that anatomical variability across subjects did not drive RSFC variability in this data, we used spatial correlation to compare the RSFC patterns of every cortical point between each pair of subjects. We then determined the differences in anatomy between each pair of subjects at each cortical point by calculating the absolute difference in areal distortion—the amount of cortical shape distortion needed to conduct the surface-based registration procedure (an index of how different the individual’s anatomy was from the template)—between the subjects at each point. We then correlated the RSFC similarities with the anatomical differences at each cortical point, across all subject pairings. Finally, these correlations were corrected for multiple comparisons by employing a two dimensional surface-based permutation test (described in Gordon et al., 2017a), in which the identities of the anatomical difference maps are randomly permuted and then correlated with the unpermuted RSFC similarity maps in order to generate a null distribution of variously-sized patches. This approach established the correction threshold at R>.474 (corresponding to p<.001 uncorrected) with a cluster extent threshold of 25 mm2; this corresponded to an overall corrected level of p<.05.
Parcel-wise network structure
Network structure for each subject and for the group average was visualized using spring-embedded plots, following (Power et al., 2011). The parcel-wise RSFC matrix was thresholded at a variety of densities from .3% to 5% after removing local connections within 30mm, as above. The surviving connections act as “springs” in the spring-embedded plot in order to position nodes (in this case, parcels) in space such that well-connected groups of nodes are pulled together. For visualization purposes, nodes of the spring-embedded plot are assigned a network identity in the same fashion as the vertex-wise networks (described above): by conducting Infomap on the parcel-wise RSFC matrix across density thresholds, matching resulting networks to known group-average networks, and generating a consensus assignment. This procedure allows easy visualization of each subject’s brain network structure. To quantify the visualized network structures, we calculated modularity and global efficiency in each individual. To determine whether subjects differed from one another in these measures, we repeated these calculations for each individual session at each density and calculated a one-way ANCOVA testing for a main effect of subject. As data quantity/quality can affect these measures, we used the following three metrics as continuous covariates of no interest: number of frames retained after motion scrubbing in each session; mean FD in retained framed in each session; and mean DVARS (Power et al., 2012) in retained frames in each session.
Task Analysis
Task evoked activations were modeled individually for each voxel with a general linear model (GLM) (Miezin et al., 2000), using in-house image analysis software written in IDL (Research Systems, Inc.). First level analyses were conducted separately for each session in a given subject, and second level analyses grouped data across the ten sessions of a single subject. The tasks had three different types of designs (motor = block design, coherence/semantic = mixed block/event-related design, incidental memory = event-related design). In the block design motor task, a block regressor was convolved with a canonical hemodynamic response to model the five experimental conditions: tongue, left hand, right hand, left foot, right foot. In the event-related incidental memory design, a finite impulse response (FIR) approach was used to model the timecourse of activations for each event, with a single delta function used to model each of 8 timepoints after a response; this approach has the advantage of making no assumptions regarding the shape of the hemodynamic response function (Ollinger et al., 2001). Separate regressors were included for each stimulus type (male, female, indoor, outdoor, abstract, concrete) as well as the number of presentations of a given item (first, second, or third presentation). The coherence and semantic judgment tasks were jointly modeled in a mixed block-event-related design. Events were modeled with an FIR model (as above, with 8 timepoints for each event); separate event regressors were included for the start and end cues in each task, and for the different trial types (noun, verb, 50% coherence, 0% coherence). The block (sustained activity) was modeled with a square block regressor, with separate regressors for sustained activity in the semantic and coherence task. Given the low number of error trials, errors were not modeled in any task. In addition to these terms, constant and linear effects were modeled for each run to remove any influences of baseline or linear signal drift.
Evaluating convergence between task activation patterns and vertex-wise RSFC topography
If RSFC-derived vertex-wise networks represent the individual-specific organization of the brain, then the physical shapes of these networks should converge well with the physical shapes of task-derived activation patterns, and task activations should be relatively homogenous within pieces of the networks. First, for each subject, we visually compared RSFC-derived network shapes against the shapes of activation patterns of all of the task contrasts.
Second, we tested the inhomogeneity of task activation in each contiguous network piece. The following seven task contrasts were specifically examined: Motor task – 1) Tongue motion > baseline; 2) Left Hand motion > Right Hand motion; 3) Left Leg motion > Right Leg motion; Incidental Memory task – 4) Face stimulus > Word stimulus; 5) Scene stimulus > Face stimulus; Mixed task – 6) Glass pattern > baseline; 7) Noun/Verb stimulus > baseline. To assess the functional inhomogeneity of these contrasts within a set of networks, we computed the standard deviation of z-transformed t-values for each contiguous piece of the networks. A lower standard deviation indicates higher functional homogeneity within the network piece. As (in)homogeneity depends critically on the size of the region in which it is tested (Gordon et al., 2016), the standard deviations were averaged over all network pieces while accounting for the size of those pieces, following Schaefer et al. (in press):
where sdl is the standard deviation of task activation z-values for network piece l and |l| is the number of vertices in network piece l. This inhomogeneity value was calculated separately for each task contrast and averaged across contrasts for each subject.
Third, we tested whether the calculated inhomogeneity of an individual’s task activation within networks was lower than if network maps were used that were not specific to that individual. For each subject, we repeated the above inhomogeneity calculation across task contrasts; however, instead of using the subject’s individual-specific network map, we used a) the network maps from all other subjects, and b) the group average network map.
DATA AND SOFTWARE AVAILABILITY
Raw MRI data, as well as segmented cortical surfaces, preprocessed volumetric and ciftispace RSFC timecourses, myelin maps, and subject-specific parcellations and networks, have been deposited in the Openfmri data repository (https://openfmri.org/) under the label “Midnight Scan Club”. Session- and subject-specific volumetric task responses have been deposited in the Neurovault repository (http://neurovault.org/) under the label “Midnight Scan Club task data”.
Code to perform preprocessing and analysis is available at https://github.com/MidnightScanClub.
Supplementary Material
Spring-embedded network configurations across all tested density thresholds for all subjects.
Acknowledgments
This work was supported by National Institutes of Health Grants NS088590, TR000448 (NUFD), MH104592 (DJG), 1P30NS098577 (to the Neuroimaging Informatics and Analysis Center), and HD087011 (to the Intellectual and Developmental Disabilities Research Center at Washington University); the Jacobs Foundation (NUFD); the Child Neurology Foundation (NUFD); the McDonnell Center for Systems Neuroscience (NUFD, BLS); the Mallinckrodt Institute of Radiology (NUFD); the Hope Center for Neurological Disorders (NUFD, BLS, SEP); an American Psychological Association dissertation research award (AWG); and Dart Neuroscience LLC. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the U.S. government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
TOL, AWG, SMN and NUFD initiated the study. TOL, AWG, KBM, JSS, AZS, BLS, SEP, SMN and NUFD designed the protocol. AWG, DJG, JJB, CHD, AN, SMN and NUFD collected the data. EMG, TOL, AZS, AWG, DJN, DJG, JJB, MO, CHD, CG, JMH, RSC, and AN processed the data. EMG, TOL, AWG, DJN, MO, CG, HS, RSC, and AZS analyzed the data. EMG, TOL, AWG, DJN, AZS, SEP, SMN, and NUFD wrote the paper with input from all the authors.
References
- Anderson JS, Ferguson MA, Lopez-Larson M, Yurgelun-Todd D. Reproducibility of single-subject functional connectivity measurements. Am J Neuroradiol. 2011;32:548–555. doi: 10.3174/ajnr.A2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balota DA, Yap MJ, Cortese MJ, Hutchison KA, Kessler B, Loftis B, Neely JH, Nelson DL, Simpson GB, Treiman R. The English Lexicon Project. Behav Res Methods. 2007;39:445–459. doi: 10.3758/bf03193014. [DOI] [PubMed] [Google Scholar]
- Bar M, Aminoff E. Cortical analysis of visual context. Neuron. 2003;38:347–358. doi: 10.1016/s0896-6273(03)00167-3. [DOI] [PubMed] [Google Scholar]
- Barch DM, Burgess GC, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, Glasser MF, Curtiss S, Dixit S, Feldt C, et al. Function in the human connectome: Task-fMRI and individual differences in behavior. NeuroImage. 2013;80:169–189. doi: 10.1016/j.neuroimage.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET. Human brain networks in health and disease. Curr Opin Neurol. 2009;22:340–347. doi: 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher S, Lantuejoul C. Use of Watersheds in Contour Detection In International Workshop on Image Processing: Real-Time Edge and Motion Detection/Estimation. Rennes, France: 1979. [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The Brain’s Default Network: Anatomy, Function, and Relevance to Disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Carp J. Optimizing the order of operations for movement scrubbing: Comment on Power et al. NeuroImage. 2013;76:436–438. doi: 10.1016/j.neuroimage.2011.12.061. [DOI] [PubMed] [Google Scholar]
- Carver CS, White TL. Behavioral inhibition, behavioral activation, and affective responses to impending reward and punishment: The BIS/BAS Scales. J Pers Soc Psychol. 1994;67:319–333. [Google Scholar]
- Chen B, Xu T, Zhou C, Wang L, Yang N, Wang Z, Dong HM, Yang Z, Zang YF, Zuo XN, et al. Individual Variability and Test-Retest Reliability Revealed by Ten Repeated Resting-State Brain Scans over One Month. PLoS ONE. 2015;10:e0144963. doi: 10.1371/journal.pone.0144963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Gilmore AW, Nelson SM, McDermott KB. Are there Multiple Kinds of Episodic Memory? An fMRI Investigation Comparing Autobiographical and Recognition Memory Tasks. J Neurosci. 2017:1534–16. doi: 10.1523/JNEUROSCI.1534-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. Exploration of Human Visual Cortex Using High Spatial Resolution Functional Magnetic Resonance Imaging. NeuroImage. 2016 doi: 10.1016/j.neuroimage.2016.11.018. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Ciric R, Wolf DH, Power JD, Roalf DR, Baum G, Ruparel K, Shinohara RT, Elliott MA, Eickhoff SB, Davatzikos C, et al. Benchmarking of participant-level confound regression strategies for the control of motion artifact in studies of functional connectivity. NeuroImage. 2017 doi: 10.1016/j.neuroimage.2017.03.020. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NUF, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. Defining functional areas in individual human brains using resting functional connectivity MRI. NeuroImage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Reynolds JR, Power JD, Repovs G, Anticevic A, Braver TS. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat Neurosci. 2013;16:1348–1355. doi: 10.1038/nn.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PT, MacCrae RR. Revised NEO personality inventory (NEO PI-R) and NEO five-factor inventory (NEO-FFI): Professional manual. Psychological Assessment Resources Incorporated; 1992. [Google Scholar]
- Dosenbach NUF, Visscher KM, Palmer ED, Miezin FM, Wenger KK, Kang HC, Burgund ED, Grimes AL, Schlaggar BL, Petersen SE. A core system for the implementation of task sets. Neuron. 2006;50:799–812. doi: 10.1016/j.neuron.2006.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NUF, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RAT, Fox MD, Snyder AZ, Vincent JL, Raichle ME, et al. Distinct brain networks for adaptive and stable task control in humans. Proc Natl Acad Sci. 2007;104:11073–11078. doi: 10.1073/pnas.0704320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NUF, Fair DA, Cohen AL, Schlaggar BL, Petersen SE. A dualnetworks architecture of top-down control. Trends Cogn Sci. 2008;12:99–105. doi: 10.1016/j.tics.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresler M, Shirer WR, Konrad BN, Müller NCJ, Wagner IC, Fernández G, Czisch M, Greicius MD. Mnemonic Training Reshapes Brain Networks to Support Superior Memory. Neuron. 2017;93:1227–1235.e6. doi: 10.1016/j.neuron.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver J, Noesselt T. Multisensory Interplay Reveals Crossmodal Influences on ‘Sensory-Specific’ Brain Regions, Neural Responses, and Judgments. Neuron. 2008;57:11–23. doi: 10.1016/j.neuron.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubis JW, Siegel JS, Neta M, Visscher KM, Petersen SE. Tasks Driven by Perceptual Information Do Not Recruit Sustained BOLD Activity in Cingulo-Opercular Regions. Cereb Cortex. 2016;26:192–201. doi: 10.1093/cercor/bhu187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J, Adolphs R. Building a Science of Individual Differences from fMRI. Trends Cogn Sci. 2016;20:425–443. doi: 10.1016/j.tics.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Jbabdi S, Caspers S, Laird AR, Fox PT, Zilles K, Behrens TEJ. Anatomical and Functional Connectivity of Cytoarchitectonic Areas within the Human Parietal Operculum. J Neurosci. 2010;30:6409–6421. doi: 10.1523/JNEUROSCI.5664-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Brown TT, Petersen SE, Schlaggar BL. fMRI reveals novel functional neuroanatomy in a child with perinatal stroke. Neurology. 2006;67:2246–2249. doi: 10.1212/01.wnl.0000249348.84045.0e. [DOI] [PubMed] [Google Scholar]
- Fair DA, Nigg JT, Iyer S, Bathula D, Mills KL, Dosenbach NUF, Schlaggar BL, Mennes M, Dickstein DP, Martino AD, et al. Distinct neural signatures detected for ADHD subtypes after controlling for micro-movements in resting state functional connectivity MRI data. Front Syst Neurosci. 2013;6:80. doi: 10.3389/fnsys.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci. 2006;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RSJ, Turner R. Movement-Related effects in fMRI time-series. Magn Reson Med. 1996;35:346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Glass L. Moiré effect from random dots. Nature. 1969;223:578–580. doi: 10.1038/223578a0. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. Mapping Human Cortical Areas In Vivo Based on Myelin Content as Revealed by T1- and T2-Weighted MRI. J Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb Cortex. 2016;26:288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Petersen SE. Individual Variability of the System-Level Organization of the Human Brain. Cereb Cortex. 2017a;27:386–399. doi: 10.1093/cercor/bhv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Gilmore AW, Nelson SM, Dosenbach NUF, Petersen SE. Individual-specific features of brain systems identified with resting state functional correlations. NeuroImage. 2017b;146:918–939. doi: 10.1016/j.neuroimage.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski KJ, Varoquaux G, Rivera G, Schwarz Y, Ghosh SS, Maumet C, Sochat VV, Nichols TE, Poldrack RA, Poline JB, et al. NeuroVault.org: a web-based repository for collecting and sharing unthresholded statistical maps of the human brain. Front Neuroinformatics. 2015;9 doi: 10.3389/fninf.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene DJ, Church JA, Dosenbach NUF, Nielsen AN, Adeyemo B, Nardos B, Petersen SE, Black KJ, Schlaggar BL. Multivariate pattern classification of pediatric Tourette syndrome using functional connectivity MRI. Dev Sci. 2016;19:581–598. doi: 10.1111/desc.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimerà R, Sales-Pardo M, Amaral LAN. Classes of complex networks defined by role-to-role connectivity profiles. Nat Phys. 2007;3:63–69. doi: 10.1038/nphys489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntupalli JS, Hanke M, Halchenko YO, Connolly AC, Ramadge PJ, Haxby JV. A Model of Representational Spaces in Human Cortex. Cereb Cortex. 2016;26:2919–2934. doi: 10.1093/cercor/bhw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker CD, Laumann TO, Szrama NP, Baldassarre A, Snyder AZ, Leuthardt EC, Corbetta M. Resting state network estimation in individual subjects. Neuroimage. 2013;82:616–633. doi: 10.1016/j.neuroimage.2013.05.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallquist MN, Hwang K, Luna B. The nuisance of nuisance regression: Spectral misspecification in a common approach to resting-state fMRI preprocessing reintroduces noise and obscures functional connectivity. NeuroImage. 2013;82:208–225. doi: 10.1016/j.neuroimage.2013.05.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SJ, Woolrich MW, Robinson EC, Glasser MF, Beckmann CF, Jenkinson M, Smith SM. Large-scale Probabilistic Functional Modes from resting state fMRI. NeuroImage. 2015;109:217–231. doi: 10.1016/j.neuroimage.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth AG, de Heer WA, Griffiths TL, Theunissen FE, Gallant JL. Natural speech reveals the semantic maps that tile human cerebral cortex. Nature. 2016;532:453–458. doi: 10.1038/nature17637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwisher N. The Quest for the FFA and Where It Led. J Neurosci. 2017;37:1056–1061. doi: 10.1523/JNEUROSCI.1706-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman AS, Kaufman NL. Encyclopedia of Special Education. Second. John Wiley & Sons, Inc.; 2013. Kaufman Brief Intelligence Test. [Google Scholar]
- Langs G, Wang D, Golland P, Mueller S, Pan R, Sabuncu MR, Sun W, Li K, Liu H. Identifying Shared Brain Networks in Individuals by Decoupling Functional and Anatomical Variability. Cereb Cortex. 2016;26:4004–4014. doi: 10.1093/cercor/bhv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NUF, et al. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron. 2015;87:657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Snyder AZ, Mitra A, Gordon EM, Gratton C, Adeyemo B, Gilmore AW, Nelson SM, Berg JJ, Greene DJ, et al. On the Stability of BOLD fMRI Correlations. Cereb Cortex. 2016:1–14. doi: 10.1093/cercor/bhw265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus D, Harwell J, Olsen T, Hodge M, Glasser M, Prior F, Jenkinson M, Laumann T, Curtiss S, Van Essen D. Informatics and Data Mining Tools and Strategies for the Human Connectome Project. Front Neuroinformatics. 2011;5 doi: 10.3389/fninf.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. The representation of object concepts in the brain. Annu Rev Psychol. 2007;58:25–45. doi: 10.1146/annurev.psych.57.102904.190143. [DOI] [PubMed] [Google Scholar]
- Miezin FM, Maccotta L, Ollinger JM, Petersen SE, Buckner RL. Characterizing the hemodynamic response: effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing. NeuroImage. 2000;11:735–759. doi: 10.1006/nimg.2000.0568. [DOI] [PubMed] [Google Scholar]
- Minear M, Park DC. A lifespan database of adult facial stimuli. Behav Res Methods Instrum Comput. 2004;36:630–633. doi: 10.3758/bf03206543. [DOI] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Mills BD, Carpenter SD, Grant KA, Kroenke CD, Nigg JT, Fair DA. Connectotyping: Model Based Fingerprinting of the Functional Connectome. PLOS ONE. 2014;9:e111048. doi: 10.1371/journal.pone.0111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TJ, Hacker CD, Breshears JD, Szrama NP, Sharma M, Bundy DT, Pahwa M, Corbetta M, Snyder AZ, Shimony JS, et al. A Novel Data-Driven Approach to Preoperative Mapping of Functional Cortex Using Resting-State Functional Magnetic Resonance Imaging. Neurosurgery. 2013;73:969–983. doi: 10.1227/NEU.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BTT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H. Individual Variability in Functional Connectivity Architecture of the Human Brain. Neuron. 2013;77:586–595. doi: 10.1016/j.neuron.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, Velanova K, Donaldson DI, Phillips JS, Schlaggar BL, et al. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010;67:156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Fast algorithm for detecting community structure in networks. Phys Rev E. 2004;69:066133. doi: 10.1103/PhysRevE.69.066133. [DOI] [PubMed] [Google Scholar]
- Ollinger JM, Corbetta M, Shulman GL. Separating Processes within a Trial in Event-Related Functional MRI: II. Analysis. NeuroImage. 2001;13:218–229. doi: 10.1006/nimg.2000.0711. [DOI] [PubMed] [Google Scholar]
- Pannunzi M, Hindriks R, Bettinardi RG, Wenger E, Lisofsky N, Martensson J, Butler O, Filevich E, Becker M, Lochstet M, et al. Resting-state fMRI correlations: From link-wise unreliability to whole brain stability. NeuroImage. 2017;157:250–262. doi: 10.1016/j.neuroimage.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Petersen SE, Posner MI. The Attention System of the Human Brain: 20 Years After. Annu Rev Neurosci. 2012;35:73–89. doi: 10.1146/annurev-neuro-062111-150525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizoli CE, Shah MN, Snyder AZ, Shimony JS, Limbrick DD, Raichle ME, Schlaggar BL, Smyth MD. Resting-state activity in development and maintenance of normal brain function. Proc Natl Acad Sci U S A. 2011;108:11638–11643. doi: 10.1073/pnas.1109144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Barch DM, Mitchell J, Wager T, Wagner AD, Devlin JT, Cumba C, Koyejo O, Milham M. Toward open sharing of task-based fMRI data: the OpenfMRI project. Front Neuroinformatics. 2013;7 doi: 10.3389/fninf.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen MY, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, et al. Long-term neural and physiological phenotyping of a single human. Nat Commun. 2015;6:8885. doi: 10.1038/ncomms9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Petersen SE. Control-related systems in the human brain. Curr Opin Neurobiol. 2013;23:223–228. doi: 10.1016/j.conb.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, et al. Functional Network Organization of the Human Brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Lessov-Schlaggar CN, Petersen SE. Evidence for Hubs in Human Functional Brain Networks. Neuron. 2013;79:798–813. doi: 10.1016/j.neuron.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Snyder AZ. A default mode of brain function: A brief history of an evolving idea. NeuroImage. 2007;37:1083–1090. doi: 10.1016/j.neuroimage.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righi G, Peissig JJ, Tarr MJ. Recognizing disguised faces. Vis Cogn. 2012;20:143–169. [Google Scholar]
- Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland DJ, Garbow JR, Laforest R, Snyder AZ. Registration of [18F]FDG microPET and small-animal MRI. Nucl Med Biol. 2005;32:567–572. doi: 10.1016/j.nucmedbio.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Rugg MD, Vilberg KL. Brain networks underlying episodic memory retrieval. Curr Opin Neurobiol. 2013;23:255–260. doi: 10.1016/j.conb.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Davatzikos C. Towards an Individualized Delineation of Functional Neuroanatomy. Neuron. 2015;87:471–473. doi: 10.1016/j.neuron.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo XN, Holmes AJ, Eickhoff SB, Yeo BTT. Local-Global Parcellation of the Human Cerebral Cortex From Intrinsic Functional Connectivity MRI. Cereb Cortex. doi: 10.1093/cercor/bhx179. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J, Sabuncu MR, Yeo TB, Liu H, Johnson KA. Stepwise Connectivity of the Modal Cortex Reveals the Multimodal Organization of the Human Brain. J Neurosci. 2012;32:10649–10661. doi: 10.1523/JNEUROSCI.0759-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon BJ, Dosenbach RA, Su Y, Vlassenko AG, Larson-Prior LJ, Nolan TS, Snyder AZ, Raichle ME. Morning-evening variation in human brain metabolism and memory circuits. J Neurophysiol. 2013;109:1444–1456. doi: 10.1152/jn.00651.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014;17:652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- Sunaert S. Presurgical planning for tumor resectioning. J Magn Reson Imaging JMRI. 2006;23:887–905. doi: 10.1002/jmri.20582. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H. Decoding Wakefulness Levels from Typical fMRI Resting-State Data Reveals Reliable Drifts between Wakefulness and Sleep. Neuron. 2014;82:695–708. doi: 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. New York: Thieme Medical Publishers, Inc; 1988. [Google Scholar]
- Tavor I, Jones OP, Mars RB, Smith SM, Behrens TE, Jbabdi S. Task-free MRI predicts individual differences in brain activity during task performance. Science. 2016;352:216–220. doi: 10.1126/science.aad8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J, Coalson T. Parcellations and Hemispheric Asymmetries of Human Cerebral Cortex Analyzed on Surface-Based Atlases. Cereb Cortex. 2012;22:2241–2262. doi: 10.1093/cercor/bhr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, WU-Minn HCP Consortium The WU-Minn Human Connectome Project: an overview. NeuroImage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Smith EE. Neuroimaging studies of working memory: a meta-analysis. Cogn Affect Behav Neurosci. 2003;3:255–274. doi: 10.3758/cabn.3.4.255. [DOI] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, Langs G, Pan R, Qian T, Li K, et al. Parcellating cortical functional networks in individuals. Nat Neurosci. 2015;18:1853–1860. doi: 10.1038/nn.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welvaert M, Rosseel Y. On the Definition of Signal-To-Noise Ratio and Contrast-To-Noise Ratio for fMRI Data. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0077089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Schlaggar BL, Petersen SE. Concepts and principles in the analysis of brain networks. Ann N Y Acad Sci. 2011;1224:126–146. doi: 10.1111/j.1749-6632.2010.05947.x. [DOI] [PubMed] [Google Scholar]
- Wig GS, Laumann TO, Petersen SE. An approach for parcellating human cortical areas using resting-state correlations. NeuroImage. 2014;93:276–291. doi: 10.1016/j.neuroimage.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Opitz A, Craddock RC, Wright MJ, Zuo XN, Milham MP. Assessing Variations in Areal Organization for the Intrinsic Brain: From Fingerprints to Reliability. Cereb Cortex. 2016;26:4192–4211. doi: 10.1093/cercor/bhw241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8:665–670. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Tandi J, Chee MWL. Functional connectivity during rested wakefulness predicts vulnerability to sleep deprivation. NeuroImage. 2015a;111:147–158. doi: 10.1016/j.neuroimage.2015.02.018. [DOI] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Eickhoff SB, Yaakub SN, Fox PT, Buckner RL, Asplund CL, Chee MWL. Functional Specialization and Flexibility in Human Association Cortex. Cereb Cortex. 2015b;25:3654–3672. doi: 10.1093/cercor/bhu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spring-embedded network configurations across all tested density thresholds for all subjects.


