Abstract
Electronic excited molecular oxygen (singlet oxygen, 1O2) is known to damage DNA, yielding mutations. In this work, the mutagenicity induced by 1O2 in a defined sequence of DNA was investigated after replication in Escherichia coli mutants deficient for nucleotide and base excision DNA repair pathways. For this purpose a plasmid containing a 1O2-damaged 14 base oligonucleotide was introduced into E.coli by transfection and mutations were screened by hybridization with an oligonucleotide with the original sequence. Mutagenesis was observed in all strains tested, but it was especially high in the BH20 (fpg), AYM57 (fpg mutY) and AYM84 (fpg mutY uvrC) strains. The frequency of mutants in the fpg mutY strain was higher than in the triple mutant fpg mutY uvrC, suggesting that activity of the UvrABC excinuclease can favor the mutagenesis of these lesions. Additionally, most of the mutations were G→T and G→C transversions, but this was dependent on the position of the guanine in the sequence and on repair deficiency in the host bacteria. Thus, the kind of repair and the mutagenesis associated with 1O2-induced DNA damage are linked to the context of the damaged sequence.
INTRODUCTION
Oxidative DNA damage and mutagenesis have been proposed to be directly involved as possible causes of the aging process, neurological degenerative diseases and cancer (1). Reactive oxygen species, such as hydroxyl radicals, superoxide, hydrogen peroxide and singlet oxygen (1O2), are products of cell metabolism which may reach the genetic material yielding DNA damage. Cells have evolved different DNA repair systems that deal with these lesions, in order to maintain the integrity of this molecule. In some instances, however, mutations create a permanent sequel of oxidative DNA damage (2). The DNA damage induced by 1O2 is mainly at guanine sites and several DNA alterations have been identified when free 2′-deoxyguanosine (dG) or DNA is treated with 1O2. The major type of DNA lesion induced by this oxidative agent is 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), but other mutagenic and/or lethal lesions are also formed (3,4).
Many DNA repair pathways have been characterized as implicated in the removal of DNA damage induced by 1O2. In Escherichia coli, formamidopyrimidine-DNA N-glycosylase (FPG) is probably the most important protein involved in oxidative DNA damage repair: this enzyme excises 8-oxodG, when paired with cytosine, and also recognizes several imidazole ring-opened purines. After removing the altered base by its glycosylase activity, it cleaves the phosphodiester bound at the abasic site formed, acting as an AP lyase (5,6). On the other hand, MutY glycosylase removes the unmodified base adenine when mispaired with 8-oxodG (7,8). That UvrABC-mediated nucleotide excision repair participates in removal of DNA oxidative lesions was first suggested after analysis of the transformation efficiency of plasmids treated with 1O2. It was observed that plasmid inactivation by oxidative damage was more pronounced in bacteria deficient in the fpg and uvrA genes, compared to wild-type or single mutant strains, suggesting that FPG and UvrABC excinuclease complement each other in the repair of lethal damage induced by 1O2 (9,10). Similar experiments indicated that exonuclease III (the xth gene) and endonuclease IV (the nfo gene) also participate in the repair of DNA damage induced by 1O2 in independent and complementary pathways, probably acting on abasic sites and on deoxyriboses modified at the 3′-OH end causing blocks to DNA polymerase action (11).
The mutagenicity of 1O2-induced damage has been demonstrated in bacteria (12–14) and mammalian cells (15–18). As expected, mutations are mainly base substitutions affecting G:C base pairs, implicating mutations targeted to base damage. G:C→T:A transversions are interpreted as being due to the ability of 8-oxodG to mispair with dG, yielding these mutations after one round of DNA replication (19). The second class of common mutations are G:C→C:G transversions, but the lesion involved in induction of this kind of mutation is unknown (14,15,20,21).
In this work, the frequency and spectra of mutations induced by 1O2 were analyzed by damaging a specific DNA sequence within a plasmid, which was then introduced by transfection in E.coli bacterial strains with different DNA repair deficiencies. A sequence of 14 nt was treated with 1O2, generated by thermodissociation of disodium 3,3′-(1,4-naphthylidene) diproprionate endoperoxide (NDPO2), and used to construct the vector pUC3GN. After vector replication in bacteria, mutations were screened by hybridization. The results indicate that both G→T and G→C transversions are frequent mutations in the 1O2-damaged oligonucleotide, with clear differences in the position within the sequence. As expected, DNA repair deficiencies (fpg, mutY and uvrC) affect 1O2-induced mutagenesis but, surprisingly, MutY also reduces G→C mutations. Moreover, the activity of UvrABC may increase mutagenesis in fpg mutY strains. Thus, 1O2-induced DNA mutagenesis is affected by these DNA repair pathways and is dependent on the sequence context neighboring the lesion.
MATERIALS AND METHODS
Oligonucleotide treatment with singlet oxygen
The target oligonucleotide (5′-CATACCCGGGACAT-3′, 2 µg) was incubated with 50 mM NDPO2. Pure 1O2 is generated through thermal decomposition of NDPO2 for 1 h at 37°C (22). During thermal decomposition of the water-soluble NDPO2 to 3,3′-(1,4-naphthylidene) dipropionate (NDP) and molecular oxygen, half of the oxygen is in the excited state (23). The maximal rate of 1O2 generation was calculated by measuring NDP formation (24). For example, at 6 min after addition of 10 mM endoperoxide the maximal rate of 1O2 generation was 38 µM/min, decreasing to 10 µM after 30 min. Thus, the steady-state of 1O2 in the incubation medium was calculated to be 31 × 10–12 M at 6 min and 8 × 10–12 M at 30 min after addition of NDPO2 (24). The oligonucleotide was purified by ethanol precipitation before pUC3GN construction.
Plasmids and bacteria
pUC3GN is a derivative of pUC8, containing a sequence of 14 nt treated with NDPO2 before insertion into the vector, through the gapped duplex technique, described by Koehl et al. (25) and Lambert et al. (26). The oligonucleotide contains only three contiguous dG, which are the major target for 1O2 treatment. Plasmid pUC3G contains the untreated oligonucleotide sequence and was used as a control. The plasmids were used to transform wild-type and repair-deficient E.coli strains. The bacterial strains used were AB1157 (F– thr-1 leuB6 proA2 his-4 thi-1 argE2 lacY1, galK2 rpsL supE44 ara-14 xyl-15 mtl-1 tsx-33), as wild-type, and its derivatives BH20 (fpg), AB1886 (uvrA), BW9101 (xth), AYM57 (fpg mutY) and AYM84 (fpg mutY uvrC).
Mutagenesis assay
After DNA transfection, the bacterial cells were spread on LB–agar plates supplemented with antibiotics for plasmid selection (27). The colonies were grown overnight at 37°C and potential mutants were screened by colony differential hybridization, using an oligonucleotide (20 bases) which includes the wild-type target sequence as probe. The probe was 5′-labeled with T4 polynucleotide kinase employing [γ-32P]ATP (Amersham Pharmacia Biotech). After autoradiography, colonies with decreased radioactive signal were selected and plasmid DNA purified (28). For each strain tested approximately 10 independent transfections and hybridization assays (each with around 100 colonies tested) were performed. The potentially mutated plasmids were sequenced by the DNA polymerase termination method (29).
RESULTS
A plasmid containing 1O2-induced damage in a specific sequence (pUC3GN) was transfected into different DNA repair-deficient strains and potentially mutated colonies were sequenced. The mutation frequency in each strain is shown in Table 1. There was an increase in mutagenesis with the damaged vector (pUC3GN) in all strains tested, when compared with the unmodified vector (pUC3G). This confirms the mutagenic consequences of 1O2-induced DNA damage. Although UvrABC (9,10) and exonuclease III, the product of the xth gene (14), have been implicated in the repair of 1O2-induced damage, the mutation frequencies in the single mutant strains BW9109 (xth) and AB1886 (uvrA) were similar to that found in the wild-type strain, AB1157. Thus, it is likely that these oxidative damage repair pathways are redundant. The BH20 (fpg), AYM57 (fpg mutY) and AYM84 (fpg mutY uvrC) strains exhibited mutation frequencies for the treated vector significantly higher than the wild-type strain. Curiously, the triple mutant AYM84 had a lower mutation frequency when compared to the double mutant AYM57. These data suggest that, in the fpg mutY background, UvrABC activity contributes to the increase in mutagenesis.
Table 1. Mutation frequency in the pUC3GN vector after replication in wild-type and DNA repair-deficient E.coli strains.
| Strain (plasmid)a |
Genotype |
Number of colonies |
Number of mutants |
Frequency
(%) |
| AB1157 (pUC3G) | wt | 1152 | 0 | <0.087 |
| AYM84 (pUC3G) | fpg mutY uvrC | 1110 | 0 | <0.09 |
| AYM57 (pUC3G) | fpg mutY | 984 | 0 | <0.1 |
| AB1157 (pUC3GN) | wt | 1300 | 8 | 0.6 |
| BW9109 (pUC3GN) | xthA | 1846 | 9 | 0.5 |
| AB1886 (pUC3GN) | uvrA | 661 | 2 | 0.3 |
| BH20 (pUC3GN) | fpg | 1041 | 15 | 1.4b |
| AYM84 (pUC3GN) | fpg mutY uvrC | 1052 | 19 | 1.8b |
| AYM57 (pUC3GN) | fpg mutY | 1059 | 35 | 3.1b,c |
apUC3G and pUC3GN correspond to control and 1O2-damaged plasmids, respectively.
bValues statistically significant (P < 0.05) when compared to wild-type bacteria and c values significantly higher (P < 0.05) when compared with all other strains. Statistics were performed with the Bross test (42), which is used for rare events. Mutant proportions were also assessed by G-tests (log likelihood ratio tests) with Bonferroni’s corrected levels of significance, with identical results (43).
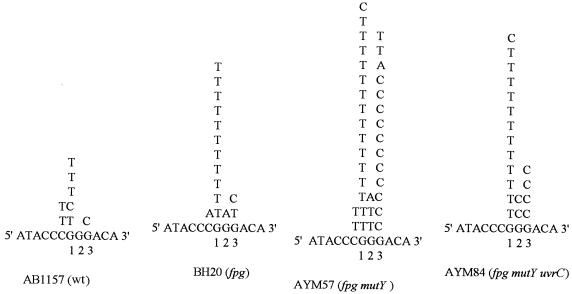
DNA sequencing of the pUC3GN mutants identified only single base substitutions in all strains tested, with no deletions or multiple mutations observed. The distributions of such mutations on the damaged oligonucleotide are presented in Figure 1. Only five mutations of 77 (6.5%) were found at bases other than the three guanines (mutations at G1, G2 and G3 represent 93.5% of the mutations), but all five were at the cytosine flanking (5′) the run of three G nucleotides. The most frequent substitutions were G→T and G→C transversions.
Figure 1.
Mutation spectra in the pUC3GN vector after replication in wild-type and repair-deficient E.coli strains. The DNA sequence presented corresponds to the damaged oligonucleotide inserted into pUC3GN. The mutations are indicated above this sequence. The guanine positions and the bacteria where mutations were generated are also indicated.
Analyzing all the mutations that originated in the different strains, a clear increase in mutagenesis at positions G1 and G3 was observed. Mutations at G1 were very frequent in all tested strains and the great majority of them were G→T (42 of 45, 93.3%). Very few mutations were found at G2 (6 of 77, 7.8%) and mutations in G3 appeared mostly in the AYM57 strain (fpg mutY). Curiously, the distributions of G→T and G→C mutations were quite distinct for G1 and G3: only three G→C mutations were found at G1 (3 of 45, 6.7%), while these mutations were mainly located at G3 (17 of 21, 81.0%).
DISCUSSION
Mutations found at this specific DNA target confirm previous reports on the mutagenic action of 1O2 (12,14,15). G→T transversion is the most frequent type of mutation induced by 1O2 and has been associated with the presence of 8-oxodG, which is able to mispair with adenine (30,31). 8-OxodG is the major DNA lesion caused by 1O2 (32–34). FPG and MutY glycosylase are the main enzymes involved in reduction of the mutagenic potential of 8-oxodG. FPG removes this lesion when it is paired with dC. MutY glycosylase acts by removing dA inserted in front of 8-oxodG. Together, these enzymes reduce the occurrence of G→T transversions. This explains the high induction of this kind of mutation observed in bacteria, which are deficient in the FPG and MutY-mediated DNA repair pathways (Table 1).
G→C transversion is also common in DNA damaged by 1O2, but the lesion responsible for this kind of mutation is unknown. There is an increase in G→C transversion frequency in the fpg mutY strain transformed with pUC3GN. The data are consistent with a report of Zhang et al. (35), who observed an increase in G→C transversions in a mutY strain. Moreover, using in vitro assays, these authors observed that MutY glycosylase is able to remove dG when paired with 8-oxodG. They proposed that this enzyme could act on a 8-oxodG:dG mispair, reducing the occurrence of G→C transversions. However, other reports, involving in vitro replication and in vivo mutagenesis assays with 8-oxodG, indicated that this lesion is mainly used as a template for dC and dA insertion (17,30,31,36), which would result only in G→T transversions. Other authors have also observed increased levels of mutagenesis, including G:C→C:G transversions, on γ-irradiated M13 DNA transfected in a MutY- and FPG-deficient E.coli strain (37). Recently, Duarte et al. (21), using in vitro systems, observed that the 8-oxodG is more vulnerable to oxidation than dG and identified some products of 8-oxodG oxidation. One of them, guanidinohydantoin, is used by the Klenow fragment of DNA polymerase as the template for dA and dG insertion through in vitro replication assays. The authors proposed that lesions produced by 8-oxodG oxidation could be involved in G→T as well as G→C transversions induced by oxidative agents such as 1O2. These findings are consistent with the increase in G→C transversions observed in the fpg mutY strains reported here. In fact, treatment of the specific oligonucleotide with 1O2 increased the possibility of a second oxidation of 8-oxodG. The resulting base could mispair with dG. As G→C mutations occur in MutY-deficient strains, the data indicate that this protein may in fact remove the dG in this mispair, reducing its mutagenic potential, in agreement with the data of Zhang et al. (35). Therefore, it is reasonable to suppose that this enzyme plays a role in the removal of damaged guanine mispaired with dG. Schulz et al., treating plasmid DNA with 1O2 generated by NDPO2 or with the photosensitizer riboflavin, also observed G:C→C:G transversions after replication in E.coli, which they interpreted as due to lesions other than 8-oxodG (38).
Another striking observation is the significantly lower mutation frequency detected in strain AYM84 (fpg mutY uvrC), when compared with AYM57 (fpg mutY) (Tables 1 and 2). The overall decrease in mutagenesis was 1.7-fold, but the reduction was more pronounced at the third dG base (3.5-fold). Involvement of the UvrABC enzymes in the repair of 1O2-induced DNA damage has been proposed, but the lesion repaired by this system is unknown (9). The results shown in this work, however, indicate that UvrABC activity in strain AYM57 (fpg mutY) contributes to mutation fixation. Similar data were obtained for ionizing radiation-induced mutagenesis in double-stranded M13 DNA (39), i.e. G:C→C:G transversions were observed in DNA repair-proficient E.coli and not in uvrA-deficient cells. The authors also suggest that the nucleotide excision repair (UvrABC) pathway is implicated in the generation of this particular kind of mutation, by removing the damaged base when mispaired with an undamaged dG. The final product would be a transversion mutation due to excision of the damaged base. This model can also account for the G→C transversions at position G3 observed in the fpg mutY strain, described in this work. In this case, a deficiency in the ability to recognize the damaged dG (dG*) by FPG or the mispair dG*:dG by MutY would enrich substrates for nucleotide excision repair, which originates the mutation. Otherwise, in the triple mutant other DNA repair pathway(s) would remove the mispaired dG or a second round of replication would allow correct insertion of a dC pairing with the damaged base and the final result would be a decreased frequency of mutations in the absence of the UvrABC pathway.
Table 2. Frequency of G→T and G→C in pUC3GN after replication in wild-type and DNA repair-deficient E.coli strains.
| Strain |
Number of colonies |
Number of G→T |
G→T
frequency (%) |
Number of G→C |
G→C frequency (%) |
| AB1157 (wt) | 1300 | 4 | 0.3 | 2 | 0.15 |
| BW9109 (xth) | 1846 | 5 | 0.27 | 1 | 0.05 |
| BH20 (fpg) | 1041 | 12 | 1.1 | 1 | 0.1 |
| AYM84 (fpg mutY uvrC) | 1059 | 12 | 1.1 | 7 | 0.66 |
| AYM57 (fpg mutY) | 1052 | 19 | 1.8 | 12 | 1.1 |
The importance of sequence context to the mutagenesis induced by 1O2 is clear from the results presented here. In all strains studied the mutations occurred preferentially in guanine sites, as expected for 1O2 action (14). There is a distinct mutation pattern at the three guanines present in the target. Mutations were much more frequent at G1 and G3, with only 6 (of 77) mutations found at G2. Another interesting observation is the occurrence of G→T and G→C transversions predominantly at the G1 and G3 positions, respectively. These data are consistent with the idea that DNA structure at specific sequence contexts may influence the DNA repair/tolerance pathways that deal with damaged bases. Alternatively, different lesions may be produced at different positions of the DNA target. The lower frequency of mutations at G2 may simply be due to the induction of a lower frequency of damage at this position. The heterogeneous distribution of this damage in DNA has already been observed in ssDNA treated with NDPO2 (40). However, the ability to deal with lesions at G2 by a more error-free DNA repair or replication mechanism may also account for the low frequency of mutagenesis at this position.
Mutations at the cytosine preceding G1 were also observed at low frequency (5 of 77, 6.5%). The presence of 8-oxodG in the G1 position could facilitate the occurrence of mutagenesis in the 5′-flanking nucleotide. In fact, experiments reporting in vitro replication assays, using an oligonucleotide with a single 8-oxodG as template, indicated that DNA polymerases could make an error at the 5′ neighbor pyrimidine (41). Mutations at the 5′-flanking sites were also observed when a monomodified vector carrying 8-oxodG was transfected and replicated in mammalian cells (37). Thus, 8-oxodG at position G1 may be responsible not only for the targeted G→T transversions but also for these mutations at the 5′-flanking pyrimidine.
The effects of DNA sequence context on mutation induction by 1O2 indicate a strong role of DNA repair and replication mechanisms that confront DNA lesions. According to the data reported in this work, the participation of base excision repair, with FPG and MutY glycosylase, and nucleotide excision repair, with UvrABC, in 1O2-induced lesion repair may depend upon the position of the damage. A DNA sequence effect may be one of the explanations for the high frequency of G→C mutations found in several 1O2-induced (and other oxidative damage) mutational spectra studies (14,15,20). As analogous and/or homologous genes to these bacterial proteins have been found in human cells, it would be expected that the related mammalian proteins would also have their activity influenced by DNA sequence context, influencing mutational fingerprinting of tumor-inducing agents.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are indebted to Dr S. R. Matioli for his support in the statistical analysis. This work was supported by FAPESP (São Paulo, Brazil, 98/11119-7) and CNPq (Brasília, Brazil). L.F.A-L. was the recipient of a fellowship from CAPES (Brasília, Brazil). This work was part of the cooperative project USP/COFECUB (a Brazil–France agreement).
References
- 1.Briviba K., Klotz,L.O. and Sies,H. (1997) Toxic and signaling effects of chemically or photochemically generated singlet oxygen in biological systems. Biol. Chem., 378, 1259–1265. [PubMed] [Google Scholar]
- 2.Sies H. and Menck,C.F.M. (1992) Singlet oxygen induced DNA damage. Mutat. Res., 275, 367–375. [DOI] [PubMed] [Google Scholar]
- 3.Ravanat J.L. and Cadet,J. (1995) Reaction of singlet oxygen with 2′-deoxyguanosine and DNA. Isolation and characterization of the main oxidation products. Chem. Res. Toxicol., 8, 379–388. [DOI] [PubMed] [Google Scholar]
- 4.Cadet J., Berger,M., Douki,T., Morin,B., Raoul,S., Ravanat,J.L. and Spinelli,S. (1997) Effects of UV and visible radiation on DNA—final base damage. Biol. Chem., 378, 1275–1286. [PubMed] [Google Scholar]
- 5.Boiteux S., O’Connor,T.R., Lederer,F., Gouyett,A. and Laval,J. (1990) Homogenous Escherichia coli FPG protein. J. Biol. Chem., 265, 3916–3922. [PubMed] [Google Scholar]
- 6.Boiteux S.; Gajewski,E., Laval,J. and Dizdaroglu,M. (1992) Substrate specificity of Eschericha coli FPG protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry, 31, 106–110. [DOI] [PubMed] [Google Scholar]
- 7.Michaels M.L., Cruz,C., Grollman,A.P. and Miller,J.H. (1992) Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl Acad. Sci. USA, 89, 7022–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulychev N.V., Varaprasad,C.V., Dorman,G., Miller,J.H., Eisenberg,M., Grollman,A.P. and Johnson,F. (1996) Substrate specificity of Escherichia coli MutY protein. Biochemistry, 35, 13147–13156. [DOI] [PubMed] [Google Scholar]
- 9.Czeczot H., Tudek,B., Lambert,B., Laval,J. and Boiteux,S. (1991) Escherichia coli FPG protein and UvrABC endonuclease repair DNA damage induced by methylene blue plus light in vivo and in vitro. J. Bacteriol., 173, 3419–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tudek B., Laval,J. and Boiteux,S. (1992) SOS-independent mutagenesis in lacZ induced by methylene blue plus visible light. Mol. Gen. Genet., 236, 433–439. [DOI] [PubMed] [Google Scholar]
- 11.Agnez L.F., Costa de Oliveira,R.L., Di Mascio,P. and Menck,C.F.M. (1996) Involvement of Escherichia coli exonuclease III and endonuclease IV on the repair of singlet oxygen-induced DNA damage. Carcinogenesis, 17, 1183–1185. [DOI] [PubMed] [Google Scholar]
- 12.Decuyper-Debergh D., Piette,J. and Van den Vorst,A. (1987) Singlet oxygen-induced mutations in M13 lacZ phage DNA. EMBO J., 6, 3155–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Retél J., Hoebee,B., Braun,J.E.F., Lutgerink,J.T., Van den Akker,E., Wanamarta,A.H., Joenje,H. and Lafleur,M.V.M. (1993) Mutational specificity of oxidative DNA damage. Mutat. Res., 299, 165–182. [DOI] [PubMed] [Google Scholar]
- 14.Agnez-Lima L.F., Di Mascio,P., Napolitano,R.L., Fuchs,R.P.P. and Menck,C.F.M. (1999) Mutation spectrum induced by singlet oxygen in Escherichia coli deficient in exonuclease III. Photochem. Photobiol., 70, 505–511. [PubMed] [Google Scholar]
- 15.Costa de Oliveira R., Ribeiro,D.T., Nigro,R.G., Di Mascio,P. and Menck,C.F.M. (1992) Singlet oxygen induced mutation spectrum in mammalian cells. Nucleic Acids Res., 20, 4319–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro D.T., Madzak,C., Sarasin,A., Di Mascio,P., Sies,H. and Menck,C.F.M. (1992) Singlet oxygen induced DNA damage and mutagenicity in a single-strand SV40-based shuttle vector. Photochem. Photobiol., 55, 39–45. [DOI] [PubMed] [Google Scholar]
- 17.Moriya M. (1993) Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G:C→T:A transversions in simian kidney cells. Proc. Natl Acad. Sci. USA, 90, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribeiro D.T., Costa de Oliveira,R., Di Mascio,P. and Menck,C.F.M. (1994) Singlet oxygen induces predominantly G to T transversions on a single-stranded shuttle vector replicated in monkey cells. Free Radic. Res. Commun., 21, 75–83. [DOI] [PubMed] [Google Scholar]
- 19.Menck C.F.M., Di Mascio,P., Agnez,L.F., Ribeiro,D.T. and Costa de Oliveira,R. (1993) Genetic deleterious effects of singlet oxygen. Química Nova, 16, 328–336. [Google Scholar]
- 20.McBride T.J., Schneider,J.E., Floyd,R.A. and Loeb,L.A. (1992) Mutations induced by methylene blue plus light in single-stranded M13mp2. Proc. Natl Acad. Sci. USA, 89, 6866–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duarte V., Muller,J.G. and Burrows,C.J. (1999) Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res., 27, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierlot C., Aubry,J.-M., Briviba,K., Sies,H. and Di Mascio,P. (2000) Naphthalene endoperoxides as generators of singlet oxygen in biological media. Methods Enzymol., 319, 3–20. [DOI] [PubMed] [Google Scholar]
- 23.Di Mascio P. and Sies,H. (1989) Quantification of singlet oxygen generated by thermolysis of 3,3′-(1,4-naphthylidene) dipropionate. Monomol and dimol photoemission and effects of 1,4-diazabicyclo[2.2.2]octane. J. Am. Chem. Soc., 111, 2909–2914. [Google Scholar]
- 24.Di Mascio P., Wefers,H., Do-Thi,H.P., Lafleur,M.V.H. and Sies,H. (1989) Singlet molecular oxygen causes loss of biological activity in plasmid and bacteriophage DNA and induces single-strand breaks. Biochim. Biophys. Acta, 1007, 151–157. [DOI] [PubMed] [Google Scholar]
- 25.Koehl P., Burnouf,D. and Fuchs,R.P.P. (1989) Construction of plasmids containing a unique acetylaminofluorene adduct located within a mutation hot spot: a new probe for frameshift mutagenesis. J. Mol. Biol., 207, 355–364. [DOI] [PubMed] [Google Scholar]
- 26.Lambert I.B., Napolitano,R.L. and Fuchs,R.P.P. (1992) Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc. Natl Acad. Sci. USA, 89, 1310–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Birboim H.C. and Doly,J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 31.Wood M.L., Dizdaroglu,M., Gajewski,E. and Essigmann,J. (1990) Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry, 29, 7024–7032. [DOI] [PubMed] [Google Scholar]
- 32.Floyd R.A., West,M.S., Eneff,K.L. and Schneider,J.E. (1989) Methylene blue plus light mediates 8-hydroxyguanine formation in DNA. Arch. Biochem. Biophys., 273, 106–111. [DOI] [PubMed] [Google Scholar]
- 33.Schneider J.E., Price,S., Maldt,L., Gutteridge,J.M.C. and Floyd,R.A. (1990) Methylene blue plus light mediates 8-hydroxy-2′-deoxyguanosine formation in DNA preferentially over strand breakage. Nucleic Acids Res., 18, 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devasagayam T.P.A., Steenken,S., Obendorf,M.S.W., Schulz,W.A. and Sies,H. (1991) Formation of 8-hydroxy-deoxyguanine and generation of strand breaks at guanine residues in DNA by singlet oxygen. Biochemistry, 30, 6283–6289. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q.-M., Ishikawa,N., Nakahara,T. and Yonei,S. (1998) Escherichia coli MutY protein has a guanine-DNA glycosylase that acts on 7,8-dihydro-8-oxoguanine:guanine mispair to prevent spontaneous G:C to C:G transversions. Nucleic Acids Res., 26, 4669–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamiya H., Murata-Kamiya,N., Koizume,S., Inoue,H., Nishimura,S. and Ohtsuka,E. (1995) 8-Hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene: effects of sequence contexts on mutation spectra. Carcinogenesis, 16, 883–889. [DOI] [PubMed] [Google Scholar]
- 37.Kuipers G.K., Slotman,B.J., Poldevaart,H.A., Reitsma-Wijker,C.A. and Lafleur,V.M. (2000) The influence of combined FPG- and MutY-deficiency on the spontaneous and γ-radiation-induced mutation spectrum in the lacZα gene of M13mp10. Mutat. Res., 461, 189–195. [DOI] [PubMed] [Google Scholar]
- 38.Schulz I., Mahler,H.C., Boiteux,S. and Epe,B. (2000) Oxidative DNA base damage induced by singlet oxygen and photosensitization: recognition by repair endonucleases and mutagenicity. Mutat. Res., 461, 145–156. [DOI] [PubMed] [Google Scholar]
- 39.Braun J.E., Wanamarta,A.H., Westmijze,E.J., Wientjes,N.M., Wijker,C.A., Lafleur,M.V.H. and Retél,J. (1997) Influence of nucleotide excision repair of Escherichia coli on radiation-induced mutagenesis of double-stranded M13 DNA. Mutat. Res., 384, 45–53. [DOI] [PubMed] [Google Scholar]
- 40.Ribeiro D.T., Bourre,F., Sarasin,A., Di Mascio,P. and Menck,C.F.M. (1992) DNA synthesis blocking lesions induced by singlet oxygen are targeted to deoxyguanosines. Nucleic Acids Res., 20, 2465–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuchino Y., Mori,F., Kasai,H., Inoue,H., Iwai,S., Miura,K., Ohtsuka,E. and Nishimura,S. (1987) Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature, 327, 77–79. [DOI] [PubMed] [Google Scholar]
- 42.Bross I. (1954) A confidence interval for a percentage increase. Biometrics, 10, 245–250. [Google Scholar]
- 43.Sokal R.R. and Rohlf,F.J. (1995) Biometry: the Principles and Practice of Statistics in Biological Research, 3rd Edn. W.H. Freeman & Co., New York, NY.