Abstract
Obesity is a heritable trait that contributes to substantial global morbidity and mortality. Here, we summarize findings from the past decade of genetic and epigenetic research focused on unravelling the underpinnings of adiposity. More than 140 genetic regions now are known to influence adiposity traits. The genetics of general adiposity, as measured by body mass index, and that of abdominal obesity, as measured by waist-to-hip ratio, have distinct biological backgrounds. Gene expression associated with general adiposity is enriched in the nervous system. In contrast, genes associated with abdominal adiposity function in adipose tissue. Recent population-based epigenetic analyses have highlighted additional distinct loci. We discuss how associated genetic variants can lead to understanding causal mechanisms, and to disentangling reverse causation in epigenetic analyses. Discoveries emerging from population genomics are identifying new disease markers and potential novel drug targets to better define and combat obesity and related diseases.
Keywords: Genetics, Epigenetics, Obesity, Adiposity
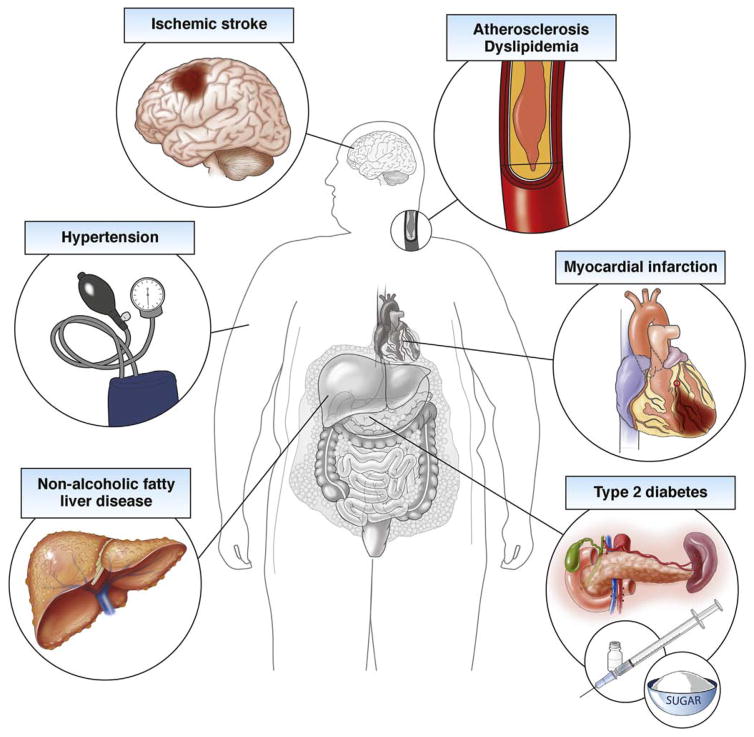
Obesity is a global epidemic and its subsequent health conditions, including nonalcoholic fatty liver disease, type 2 diabetes, and cardiovascular disease, result in extensive morbidity, mortality, and health care expenditures (Figure 1).1 Few effective treatments exist for obesity, in part owing to a limited understanding of its complex and multifactorial etiology. Discoveries emerging from population genomics are identifying biomarkers and relevant causal mechanisms, which enable the development of better preventive, diagnostic, and therapeutic approaches.
Figure 1.
Medical consequences of obesity.
Population Genomics as a Window on Obesity Biology
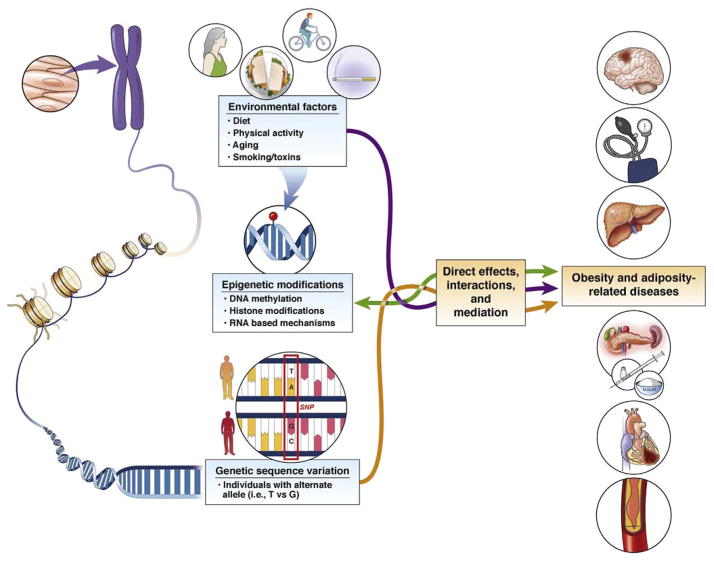
Measures of adiposity are heritable across generations, in part owing to inherited variations in the genetic code.2 In addition to genetic contributions, obesity is the result of a complex interplay of environmental, behavioral, and social factors, which can be relayed from parent to offspring independent of genetic sequence variation. At the molecular level, exposures may influence obesity through lasting epigenetic modifications of DNA. With advances in genomic technologies we can integrate genetic, epigenetic, and environment factors among thousands of individuals to gain insight into the biology of obesity and related traits.
What Genomic Elements Can Be Studied in Relation to Adiposity in a Population?
Although the vast majority of the human DNA code is nonvariable, a typical genome is reported to differ from a reference human genome at 4.1–5.0 million sites.3 The majority of these variations (>99.9%) consist of single-nucleotide polymorphisms (SNPs) (Figure 2) and short insertions or deletions; however, larger structural changes occur and in total affect more base pairs.3 Genetic sequence variants can influence the overall function and the quantity of a gene product. The expression of genes, which varies across tissues and time points, also can be influenced by DNA methylation, an epigenetic modification, through alterations of transcription factor binding and chromatin structure (Figure 2). Modifications to histones, proteins to which DNA coils around to form condensed structures, also are correlated to gene expression. It is not clear whether these modifications have a direct regulatory function or play a stabilizing role after gene expression changes occur.4 Segments of RNA, present in various lengths, which do not code proteins (micro RNAs or long noncoding RNAs), also can influence the quantity and structure of messenger RNA and subsequent protein production. In summary, genetic and epigenetic changes may be silent or alter the quantity, structure, and function of RNA and proteins, which ultimately can influence fat mass and distribution. Both genetic and epigenetic variations across individuals thus can alter adiposity phenotypes and lead to variation in traits across individuals.
Figure 2.
Genetic and epigenetic variation influences gene expression.
How Are Obesity-Related, Population-Based Genomic and Epigenomic Associations Identified?
Up to the end of the past century, identification of genetic variants associated with obesity relied heavily on linkage studies in rodents with obesity caused by a mutation in a single gene (monogenic obesity) and candidate-gene–based approaches in severely obese human beings. During this time period, several loss-of-function mutations causing monogenic obesity were identified in the appetite-regulating leptin-melanocortin pathway, including leptin,5 leptin receptor,6 pro-opiomelanocortin (POMC), and melanocortin 4 receptor (MC4R).7,8 Children with MC4R mutations experience mealtime hyperphagia and more frequent snacking compared with other obese children.9 In addition, children with leptin deficiency have intense hyperphagia with food-seeking behavior and show aggressive behavior when food is denied.10 This behavior can be normalized with leptin supplementation. Such disrupting mutations, however, explain only a small part of the etiology of early onset severe obesity and very little of the etiology of adiposity in the general population. Although these early studies suggested that appetite regulation was important in genetic susceptibility to obesity, hypothesis-free unbiased approaches in recent years have implicated a wide range of obesogenic mechanisms.
In the past 15 years, technical advancements in molecular biology and genomics have enabled researchers to study variation at millions of sites in the human genome in relation to adiposity. These genome-wide association studies (GWAS) require large numbers of individuals to distinguish real associations from background variation, combining individuals from many smaller studies to increase sample sizes. With the availability of larger biobanks such as the UK Biobank,11 the US Million Veterans Program,12 and the Kadooire Biobank,13 comprising approximately half a million to a million participants each, there likely will be larger single-study reports and massive meta-analyses in the near future.
DNA methylation is the most frequently studied epigenetic modification in population-based studies, largely because of its relative stability and ease of measurement in high-throughput, array-based assays. Methylation of DNA involves the addition of a methyl group to the 5′ position of a cytosine residue of the DNA, specifically at cytosine-phosphate-guanine dinucleotides (CpG) (Figure 2). DNA methylation modulates gene expression and may influence an individual’s susceptibility to obesity or its downstream consequences. Unlike genotype, DNA methylation is cell-and tissue-specific,14 and the procurement of relevant tissues remains a major limitation in population-based epigenome studies of adiposity traits. Second, unlike genotypes, DNA methylation and other epigenetic modifications are malleable and may change in response to environmental exposures or disease states.15,16 Third, DNA methylation and other epigenetic modifications also partly are under genetic control and influenced by nearby genetic sequence variants,17–21 and thus are not entirely independent of genetic effects.
Overall and Abdominal Obesity
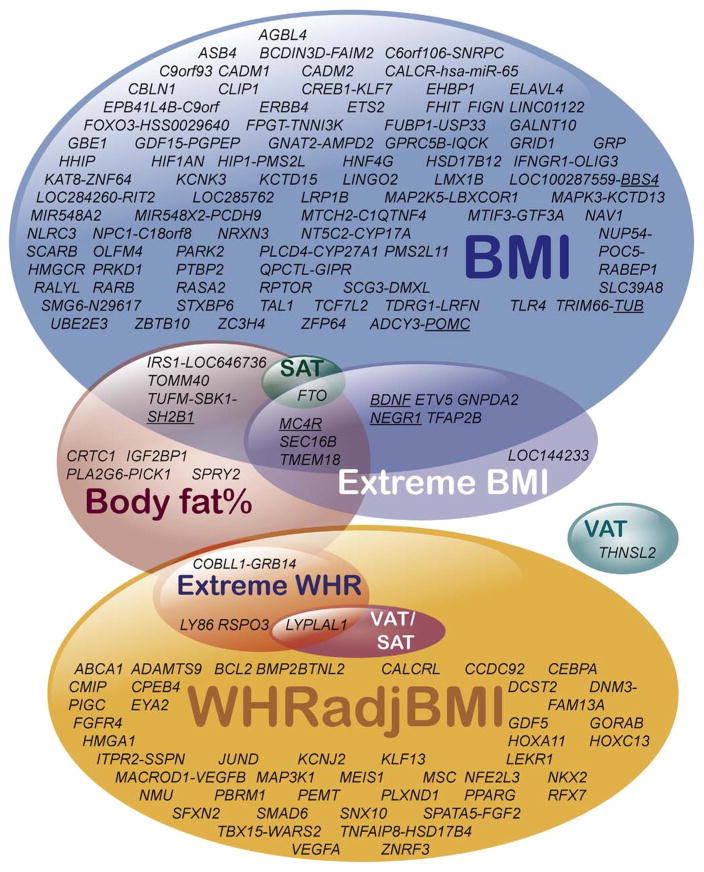
Body mass index (BMI) and waist-to-hip ratio controlled for BMI (WHRadjBMI) are simple measures of overall and abdominal adiposity, respectively. Both adiposity measures are associated with the development of metabolic complications of obesity. In 2007, Frayling et al22 showed that the FTO locus was associated with both BMI and diabetes, identifying the first common genetic locus for an adiposity measure and a cardiometabolic disease trait. It should be noted that for this locus and others, the closest gene often has been used to denote the locus, even when the causal gene is not known. By increasing sample sizes and power, international collaborative efforts performed successively larger meta-analyses of approximately 339,000 individuals for overall obesity, as measured using BMI,23–27 and approximately 224,000 individuals for central obesity, as measured using WHRadjBMI.28–30 These studies have greatly expanded the number of implicated loci to 98 for BMI and 49 for WHRadjBMI, with no overlapping loci between the 2 adiposity traits (Figure 3 and Supplementary Table 1). Examination of the identified loci showed common variants in or near genes previously found in monogenic obesity studies in human beings or mice. Examples include mutations in POMC31 and MC4R.7,8 Specifically, these findings showed that common variants with subtle effects on the function or expression of these genes can influence population BMI, whereas severe rare mutations can lead to early onset monogenic obesity. A study assessing genetic associations with the tails of the BMI and WHR distribution in the general population of adults32 highlighted the same loci as the top findings in the population-based approaches (Figure 3). Once again, this highlighted the observation that similar genes can influence common and extremes of obesity in the general population.
Figure 3.
Venn diagram for loci associated with BMI (Locke 201524), body fat percentage (Lu 201635), waist-hip-ratio adjusted for BMI (Shungin, 201529), VAT, SAT, and their ratio (VAT/SAT) (Fox, 201237), and extremes of body mass index and waist-hip-ratio (Berndt, 201332). Genes implicated in monogenic obesity are underlined. Loci are named according to nearby genes by the original authors, but are not necessarily proven to be the causal gene.
The remarkable overlap between genes identified from studies in the general population, extremes of distribution, and severe early onset monogenic disorders suggest that targeting the genes identified in adiposity GWAS with small effect sizes could have strong effects on preventing obesity. As proof of principle, for example, the common SNP at the HMG-CoA reductase locus has a small effect size in relation to increased low-density lipoprotein (LDL) cholesterol, where each effect allele increases LDL cholesterol by only 2.5 mg/dL.33 However, statin therapy, which directly inhibits the HMG-CoA reductase enzyme, has a large effect on LDL cholesterol levels. Statins routinely lower LDL cholesterol by 30%–50%,34 and reductions of more than 100 mg/dL can occur for some individuals, which is an approximately 40 times greater effect than that observed with naturally occurring genetic variation.
Furthermore, the adiposity GWAS shows that many of the identified common variants do not fall in the coding regions of genes. This suggests that most SNPs identified in adiposity GWAS may regulate the magnitude of gene expression, rather than cause severe changes to the coding part of the gene that can precipitate syndromic changes. After identifying adiposity-related loci, it is possible to examine the effects of genetic variants identified in adiposity GWAS on the development of obesity-related metabolic disorders. As expected, there was enrichment for variants that increase both obesity and metabolic diseases, such as type 2 diabetes, dyslipidemia, and cardiovascular disease, although some variants have the opposite effect (see the Pleiotropic Effects section).
From Genetic Variants to Function to Better Understand Obesity Biology
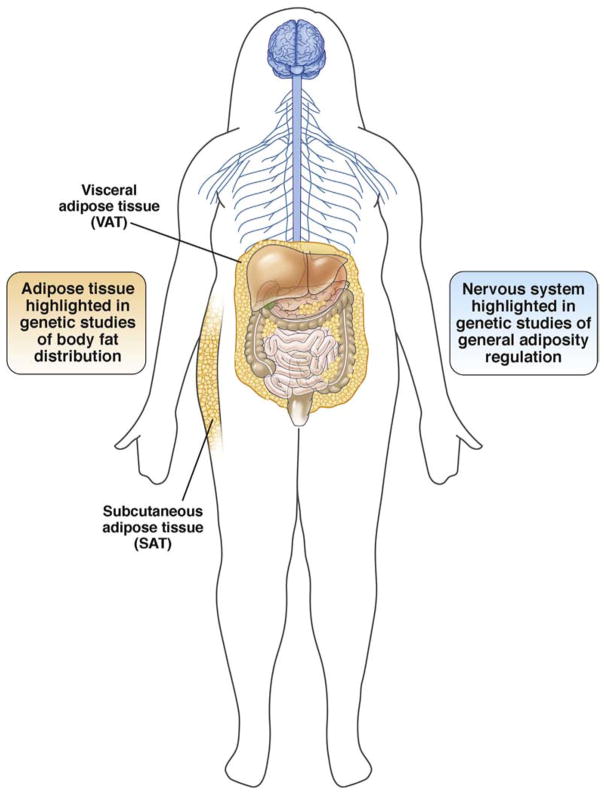
To identify tissues, genes, and pathways that influence adiposity traits, investigators integrated genetic sequence variants with gene expression across tissues and grouped genes by related pathways. These integrative analyses showed that the nervous system, in general, and the hypothalamus, hippocampus, and limbic system, specifically, play prominent roles for regulating BMI (Figure 4).24 Further analyses highlighted genes related to synaptic function and neurotransmitter signaling (ELAVL4, GRID1, CADM2, NRXN3, NEGR1, and SCG3), and energy homeostasis (HNF4G, TLR4, BDNF, POMC, MC4R, and ETV5) as important for influencing overall obesity. For abdominal obesity, in contrast, integrative analyses highlighted effects in abdominal and subcutaneous adipose tissues (Figure 4).29 Highlighted genes function in adiponectin signaling, insulin sensitivity and regulation of glucose levels, skeletal growth, angiogenesis (VEGFA, VEGFB, RSPO3, STAB1, WARS2, PLXND1, MEIS1, FGF2, SMAD6, and CALCRL), transcriptional regulation (CEBPA, PPARG, MSC, SMAD6, HOXA, HOXC, ZBTB7B, JUND, KLF13, MEIS1, RFX7, NKX2-6, and HMGA1), and adipose tissue development (CEBPA, PPARG, BMP2, HOXC-mir196, SPRY1, TBX15, and PEMT).29 Taken together, integrating genetic variation with multitissue gene expression helps to dissect the causal biological susceptibility to identify new targets for therapeutic interventions.
Figure 4.
Tissues highlighted in genetic studies of general and central adiposity.
More Detailed Adiposity Measures: Fat Mass and Visceral and Subcutaneous Adipose Tissue
Although BMI and WHR are easy to measure in the population, they do not directly quantify fat mass or fat depots, which are likely more relevant in disease development. GWAS analysis in approximately 100,000 individuals identified 12 loci associated with body fat percentage (BF %), as measured by bioimpedance analysis or dual-energy X-ray absorptiometry.35 The majority of BF% loci overlap with BMI findings, and one, COBLL1/GRB14, with WHRadjBMI (Figure 3). The BF% increasing allele at COBLL1/GRB14 was associated with lower WHRadjBMI, suggesting that the risk variant promotes gluteal rather than abdominal fat storage. Seven loci showed a larger effect on BF% than on BMI, suggestive of a primary association with adiposity in particular. Five loci showed larger effects on BMI than on BF%, which may be explained by an association with increases in both fat and lean mass.
The ratio of visceral adipose tissue to subcutaneous adipose tissue (VAT/SAT) is an alternative measure of body fat distribution with cardiometabolic disease relevancy because a higher VAT/SAT is correlated with the development of metabolic complications.36 Fox et al37 performed a GWAS analysis of VAT/SAT in approximately 10,000 individuals. They found that a variant at the LYPLAL1 locus, previously identified in association with WHR, also was associated with a VAT/SAT ratio with larger effect sizes in women as compared with men. The strongest association for SAT was within the FTO locus. Unlike SAT, the largest effect sizes for VAT were seen at a variant near THNSL2, also with greater effects in women as compared with men. Thus, examining the genetic associations of complementary measures of adiposity (BMI, %BF, SAT, VAT) can begin to show the underlying biology by which specific fat deposition may contribute to disease development.
FTO/IRX3, Variant to Function Studies Identifies Novel Obesity Biology
SNPs within the first intron (ie, the noncoding part) of the FTO gene have been reported to be associated robustly with common obesity since 2007. Recent studies have suggested a recessive effect, in which individuals carrying zero vs one BMI-raising allele have a similar BMI, however, individuals with zero vs two BMI-raising alleles at the FTO locus had an average BMI of 27.3 vs 28.1 kg/m2, respectively.38 The strongest effect of the FTO locus was observed in individuals younger than 50 years of age.39 There is some evidence from observational studies in human beings that the FTO SNP effect on adiposity is modified by physical activity level40,41 and may affect dietary preferences.42 However, the association of FTO locus variation and obesity is not completely explained by interactions or mediation by lifestyle factors. So how do these FTO variants actually exert their effect on BMI?
Smemo et al43 found that obesity-increasing noncoding variants within the first FTO intron are associated with increased expression of the neighboring gene IRX3, but not with FTO in human brains. Furthermore, Irx3-deficient mice had a reduction in body weight of 25%–30%, primarily through the loss of fat mass and increase in basal metabolic rate, with browning of white adipose tissue. Previous work has shown that IRX3 and other homeobox transcription factors were overexpressed in adipocytes after weight loss, indicating that changes in these genes were important in weight regulation.44 Claussnitzer et al45 combined information on regulatory elements and methylation patterns to show that the FTO SNP rs1421085 disrupts the binding site of the ARID5B repressor. The disruption of the repressor led to increased expression of IRX3 and IRX5 during adipocyte differentiation, causing energy-consuming beige adipocytes to become energy-storing white adipocytes. This decade-long effort to understand the biology of one of the earliest and most robust association signals for adiposity highlights the challenges and rewards in translating genomic discoveries to validated, potentially actionable, biology.
Pleiotropic Effects
Although obesity is known to increase the risk of metabolic diseases, some extremely obese individuals do not develop adiposity-related metabolic diseases. Why this is the case is not completely understood. This phenomenon also has been observed in genetic studies, in which a few recently identified genetic variants that increase adiposity also protect against the development of metabolic diseases.24,46 This observation suggests that genetics can dissociate epidemiologically correlated traits and identify mechanisms by which metabolic disease can be abrogated even in the obese. For example, the minor allele rs2943650 near the IRS1 (encoding the insulin-receptor substrate 1) gene was associated with a higher BF% (especially in men),46 however, paradoxically, it also was associated with a favorable lipid profile, and a decreased risk of type 2 diabetes and coronary heart disease.46 Gene expression profiles have shown that the variant was related to a higher expression of the IRS1 gene in adipose tissue, but not in liver, brain, or blood, suggesting that the adipose tissue may be the tissue in which it acts to have its effect. Furthermore, they found that the allele was associated with a decreased VAT/SAT ratio and higher adiponectin levels in men, suggesting that the protective effects for insulin resistance and dyslipidemia could be explained partly by increased deposition of fat in subcutaneous tissue and decreased ectopic fat deposition in men. These findings are in line with the adipose tissue expandability hypothesis, which states that when adipose tissue cannot expand by cell size or number, lipid will accumulate preferentially in ectopic sites such as cardiac, liver, and pancreatic β-cells. Ectopic fat depots are associated with the development of cardiometabolic disease.47
Interestingly, 10 additional loci increase body fat but protect from a lipodystrophy-like phenotype and insulin resistance, in line with what is described for IRS1.48 Furthermore, multiple cross-trait analyses have shown complex patterns of effects and begins to delineate metabolic disease subtypes that are discernable by shared genetic etiology.24,29 Genomic discoveries thus may enable the identification of patients at high risk of developing metabolic complications and help predict the complications they will develop to target early tailored treatment based on their individual risk rather than the population risk. Such tailoring of care would increase the effectiveness of treatments and decrease the medical and human costs of disease.
Sex and Age Effects
Some studies have assessed the possibility of effect modification by age and sex for the genetic associations with adiposity. In a longitudinal study across the life course, a genetic risk score including 32 BMI-related SNPs was associated with a higher average lifetime BMI. There was a steeper increase in BMI up to 65 years of age with a higher genetic risk score, but not beyond 65 years of age, indicating that some genetic effects might be age-dependent.49 Winkler et al39 from the Genetic Investigation of Anthropometric Traits consortium conducted a genome-wide search for loci that show distinct age- and/or sex-specific effects for BMI and WHRadjBMI in a total of approximately 320,000 adults of European ancestry. Age was dichotomized at 50 years. In this study, 15 loci with age-specific effects were identified for BMI and 44 loci for were identified for WHRadjBMI. Most of these loci were known from previous GWAS, which was expected because relatively larger effect sizes are needed to achieve significance in this type of analysis. Eleven of the 15 age-dependent BMI loci showed larger effects in younger vs older adults. The 4 loci that were more pronounced in older adults had previously been found to be associated with diabetes or coronary heart disease. This may have biased the results because such diseases may affect body weight. Of the 44 sex-specific loci, 11 showed an opposite effect direction in women vs men, and 28 showed a larger effect in women and smaller or no effect in men. Only 4 showed a larger effect in men. It is seemingly paradoxical that the effects generally are more apparent in women than in men given that men have more abdominal obesity than women, perhaps the effect is more noticeable in women who do not usually have this type of distribution. No 3-way interaction (gene × age × sex) was noted in the study, however, power was limited for this type of analysis.
Genetic Studies of Adiposity in Non-European Ancestry
Overall, genetic loci conferring susceptibility for different adiposity traits are similar across ethnic ancestries. The majority of early studies of adiposity genetics were performed in individuals of European ancestry. Smaller sample sizes and lower-powered GWAS studies in African and Asian ancestry individuals confirmed associations for many of the variants identified in the European ancestry studies, therefore suggesting a general shared transethnic susceptibility to adiposity that likely arose before migrations out of Africa.50–52 However, a few novel loci were first identified in non-European populations. For example, GWAS of BMI in more than 70,000 individuals of African ancestry and more than 80,000 individuals of East Asian Ancestry identified 2 new loci (GALNT10 and MIR148A-NFE2L3)50 and 4 new loci (KCNQ1, ALDH2/MYL2, ITIH4, and NT5C2) among African and Asian ancestry groups, respectively.52 Parallel studies in approximately 50,000 East Asian individuals identified 4 novel loci near the EFEMP1, ADAMTSL3, CNPY2, and GNAS genes that were associated with waist circumference adjusted for BMI, and 2 loci near the NID2 and HLA-DRB5 genes associated with WHRadjBMI.51 Many of these variants were found to have similar effects in individuals of European ancestry and likely were not identified previously owing to lower allele frequencies and thus lower power in studies of individuals of European ancestry.
Ancestry-specific variants also have been discovered. These include a GWAS in more than 5000 Samoans that identified a common missense variant in CREBRF with a large effect on BMI.53 This variant is rare in other populations. Salinas et al54 performed a GWAS in fewer than 2000 individuals of 4 different ancestries and found suggestive evidence of variants with differential effects across ancestries. Such heterogeneity may be owing to genetic or environmental modifiers, or more technical causes such as the casual variant not being located on the same haplotype as the associated variant across ancestries. Further work will be required to validate the proposed heterogeneity and to differentiate among these possibilities.
Overall, these studies show that there are both shared and unique genetic contributions to obesity across ancestries.
Epigenetic Studies of Adiposity
Early candidate region and hypothesis-free genome-wide scans of blood-derived DNA methylation in relation to BMI identified numerous differentially methylated loci; however, similar to early GWAS, candidate region and small sample-sized methylation studies often failed to show replicable results. One of the first robust associations of differential DNA methylation with BMI was reported in 2014 by Dick et al55 at the hypoxia-inducible factor 3α (HIF3A) locus. The investigators conducted an epigenome-wide association study of BMI by examining DNA methylation at approximately 480,000 CpGs in leukocyte-derived DNA from 459 individuals, and replicated findings in blood samples from 1789 individuals, and 395 skin and 635 adipose tissue biopsy specimens. They identified a correlation with BMI among Europeans with differential methylation at 3 CpGs in the first intron of HIF3A in blood and adipose tissue, but not skin. HIF3A, a component of the hypoxia-inducible transcription factor, regulates the cellular and physiological response to reduced oxygen. In animal models, HIF3A responds to glucose and insulin, and accelerates adipocyte differentiation. The association of HIF3A locus methylation with BMI and adiposity-related conditions has been replicated further, including for childhood obesity and birth weight. Whether this methylation change causally influences BMI or whether it is the result of increased BMI is controversial. Suggestive evidence of reverse causation in which BMI affects HIF3A locus methylation was found by Richmond et al,56 who examined approximately 1000 mother–offspring pairs using longitudinal methylation data in childhood and bidirectional Mendelian randomization approaches. Mendelian randomization leverages the random distribution of genetic variants free from confounders to infer the causal relation between an exposure (imputed from the genetic variants) and an outcome. These statistical models are powerful tools for observational studies but are subject to numerous caveats and assumptions.
Two recent meta-analyses examining BMI in relation to methylation of blood-derived DNA from separate samples of approximately 8000 and 10,000 participants have expanded the number of replicated regions showing BMI-related differences in epigenetic signaling to more than 200 independent loci.57,58 The association between BMI and DNA methylation was found to be largely independent of nearby genetic variants or known BMI-related SNPs (ie, FTO or MC4R loci), emphasizing the complementarity of genetic and epigenetic approaches. Differential methylation at 1 locus, intronic to sterol regulatory element binding transcription factor 1 (SREBF1), was found in Mendelian randomization analysis to have a causal relation to BMI. In addition, this analysis found evidence suggesting that methylation at the SREBF1 locus was associated causally with SREBF1 gene expression, and additional adiposity-related traits (WHRadjBMI, adiponectin, and birth weight), glycemic traits, obesity-related dyslipidemia, and risk of coronary artery disease. SREBF1 plays a central role in energy homeostasis by promoting glycolysis, lipogenesis, and adipogenesis via induction of the conversion of acetyl-CoA to triglycerides. At least 1 genetic study supports a causal relationship of triglyceride-rich lipoprotein increase and coronary disease.59 Taken together, the results highlight an example in which genomic regulatory mechanisms may promote obesity and adiposity-related disease.
The association of DNA methylation with BMI generally is stronger and of greater magnitude as compared with the association of BMI with genetic sequence variants. This likely is owing to changes in DNA methylation that occur in response to increasing adiposity. Indeed, further causal inference modeling has indicated that at least 20%, and likely more, of identified differential methylation is a consequence of differences in BMI.57 DNA methylation findings largely were replicated across Caucasian and African American ancestries,60,61 indicating that shared exposures across ancestries may contribute to adiposity-related differences in DNA methylation. This suggests that epigenetic discoveries may help to identify transethnic targets to alter adiposity-related disease outcomes in the future.
Some of these downstream methylation changes may be useful as biomarkers for subsequent obesity-related disease development. The carnitine palmitoyl-transferase 1A (CPT1A) locus was associated with BMI and, in other studies, with metabolic syndrome, low high-density lipoprotein levels, and hypertriglyceridemia.62–68 CPT1A is a major rate-limiting enzyme for long-chain fatty acid oxidation, transporting acetyl-CoA into the mitochondria for reduction in the tricarboxylic acid cycle. Severe homozygous mutations in CPT1A in Northern populations increase susceptibility to fatal decompensation as a result of hypoketotic hypoglycemia. Preservation of heterozygous mutations in CPT1A in Arctic populations may confer an energy metabolism advantage. Thus, genetic variants may have been evolutionarily selected in some groups for their role in energy metabolism, and, independently, epigenetic mechanisms at the same loci may influence energy metabolism in the general population.
Methylation at the adenosine triphosphate binding cassette G1 (ABCG1) locus also was shown to be linked to adiposity-related disease because it was associated with triglycerides, high-density lipoprotein cholesterol, and predictive of incident coronary heart disease events. DNA methylation differences at the ABCG1 locus were found to be a consequence of increased BMI based on Mendelian randomization analysis. ABCG1 is a cell-membrane lipid transporter that has an established role in reverse-cholesterol transport, and its role in obesity has been supported in previous animal and human studies.69,70 Each standard deviation difference in methylation in the population at the ABCG1 locus was associated with a 38% increased risk of future coronary heart disease events.71
Taken together, the earlier-described results highlight examples in which epigenomic changes may be useful not only in identifying genes causally related to the development of obesity, but also the development of obesity-related metabolic diseases.
Transgenerational and Lifetime Exposures on Epigenetic Modifications and Obesity Susceptibility
Accumulating evidence has suggested that the roots of adult obesity and cardiometabolic disease may begin in utero through fetal exposure to maternal health conditions. Numerous epidemiologic studies have shown a robust association between maternal and offspring obesity. Because the epigenome is thought to be more pliable during fetal development, studies of human fetal programming support epigenetic inheritance as a potential mechanism. For example, offspring born with nutritional or emotional distress during natural disasters, such as the Dutch Hunger Strike or a major ice storm, show marked differences in DNA methylation signatures. Children born before and after maternal bariatric surgery show differences in DNA methylation in glucoregulatory and inflammatory pathways. Women in rural Gambia, during periods of hunger and differing maternal methyl-donor intake, have offspring that show differences in DNA methylation. Transgenerational effects can be substantial; for example, maternal pre-pregnancy obesity was associated with 30% and 35% increased risk in the offspring of cardiovascular events and all-cause mortality, respectively. It is important to note that the transgenerational epidemiologic associations do not prove a causal role of epigenetic inheritance in adiposity because there may be alternate pathways or other factors that confound this relationship (eg, learned behaviors, inherited socioeconomic status, microbiome transfer). Whether epigenetic modifications can be passed on directly between generations is also unclear. Animal studies have provided supportive evidence, for example, paternal epigenetic inheritance via sperm through ribosomal DNA methylation and RNA-based mechanisms. Whether this also is true in human beings remains an area of active research and debate.
Road to Precision Medicine?
From GWAS and epigenome-wide association studies of adiposity, we have learned much about the biology of obesity. We also have identified many potential therapeutic drug targets. These studies provide the basis for Mendelian randomization studies to provide evidence to support future clinical trials, in which the causal determinants of adiposity can be evaluated for effects on a number of traits ranging from cardiovascular disease72 to multiple sclerosis.73 Thereby potentially avoiding an expensive clinical trial focused on an adiposity target that is not causal for the disease of interest in a human population.
To date, identified variants have small effect sizes and therefore by themselves are unsuitable for prediction of polygenic obesity compared with using family history of obesity or information on childhood obesity.74 The area under the receiver operating characteristic curve, which estimates the predictive performance of a test, is substantially higher when using parental obesity as opposed to the strongest 32 BMI-related SNPs to predict an individual’s risk of obesity (0.75 as compared with 0.57).74 Current genetic risk scores to guide personalized diets or prevention regimens have shown some promise,75,76 but further research showing substantial benefits and cost benefits is warranted before widespread use.77
Despite poor predictive abilities, many non–genome-wide significant genetic loci are likely to be true BMI influencing variants.24,29 As we go beyond the genome-wide significant associations, the predictive power of using these variants (alone or in combination with other -omic markers) in predictive models likely will improve. Use of circulating epigenetic markers as liquid biopsies from blood also could replace more traditional tissue biopsies to diagnose tissue-specific disease and reduce patient pain and risk of complications associated with traditional biopsies, as have been proposed for liver fibrosis epigenetic markers. The US National Institute of Health has recognized the remarkable potential of these developments in guiding efforts to reduce obesity and improve the health of the population and is setting research priorities to take these new discoveries and translate them into effective treatments for obesity.
Supplementary Material
Acknowledgments
Funding
Supported by grants from the Swedish Research Council (2015-03477), Swedish Heart-Lung Foundation (20150429), Swedish Research Council Formas (221-2013-1673) (T.F.); Göran Gustafssons Stiftelse; and by National Institutes of Health RO1DK107904 and RO1DK106621 grants as well as the University of Michigan Department of Internal Medicine and University of Michigan Biological Sciences Scholars Program (E.K.S.). The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the US Department of Health and Human Services.
Abbreviations used in this paper
- BF%
body fat percentage
- BMI
body mass index
- CpG
cytosine-phosphate-guanine dinucleotides
- GWAS
genome-wide association study
- LDL
low-density lipoprotein
- SNP
single nucleotide polymorphism
- SREBF1
sterol regulatory element binding transcription factor 1
- VAT/SAT
visceral adipose tissue to subcutaneous adipose tissue ratio
- WHRadjBMI
waist-to-hip ratio controlled for body mass index
Footnotes
Conflicts of interest
The authors disclose no conflicts.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2017.01.054.
References
- 1.Lette M, Bemelmans WJ, Breda J, et al. Health care costs attributable to overweight calculated in a standardized way for three European countries. Eur J Health Econ. 2016;17:61–69. doi: 10.1007/s10198-014-0655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nan C, Guo B, Warner C, et al. Heritability of body mass index in pre-adolescence, young adulthood and late adulthood. Eur J Epidemiol. 2012;27:247–253. doi: 10.1007/s10654-012-9678-6. [DOI] [PubMed] [Google Scholar]
- 3.1000 Genomes Project Consortium. Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benveniste D, Sonntag HJ, Sanguinetti G, et al. Transcription factor binding predicts histone modifications in human cell lines. Proc Natl Acad Sci U S A. 2014;111:13367–13372. doi: 10.1073/pnas.1412081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montague CT, Farooqi IS, Whitehead JP, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 6.Clement K, Vaisse C, Lahlou N, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 7.Vaisse C, Clement K, Guy-Grand B, et al. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20:113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 8.Yeo GS, Farooqi IS, Aminian S, et al. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet. 1998;20:111–112. doi: 10.1038/2404. [DOI] [PubMed] [Google Scholar]
- 9.Valette M, Bellisle F, Carette C, et al. Eating behaviour in obese patients with melanocortin-4 receptor mutations: a literature review. Int J Obes (Lond) 2013;37:1027–1035. doi: 10.1038/ijo.2012.169. [DOI] [PubMed] [Google Scholar]
- 10.Farooqi IS, O’Rahilly S. 20 years of leptin: human disorders of leptin action. J Endocrinol. 2014;223:T63–T70. doi: 10.1530/JOE-14-0480. [DOI] [PubMed] [Google Scholar]
- 11.Ganna A, Ingelsson E. 5 year mortality predictors in 498, 103 UK Biobank participants: a prospective population-based study. Lancet. 2015;386:533–540. doi: 10.1016/S0140-6736(15)60175-1. [DOI] [PubMed] [Google Scholar]
- 12.Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Chen J, Collins R, et al. China Kadoorie Biobank of 0. 5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011;40:1652–1666. doi: 10.1093/ije/dyr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimas AS, Deutsch S, Stranger BE, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busche S, Shao X, Caron M, et al. Population whole-genome bisulfite sequencing across two tissues highlights the environment as the principal source of human methylome variation. Genome Biol. 2015;16:290. doi: 10.1186/s13059-015-0856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell JT, Pai AA, Pickrell JK, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drong AW, Nicholson G, Hedman AK, et al. The presence of methylation quantitative trait loci indicates a direct genetic influence on the level of DNA methylation in adipose tissue. PLoS One. 2013;8:e55923. doi: 10.1371/journal.pone.0055923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaunt TR, Shihab HA, Hemani G, et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;17:61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shakhbazov K, Powell JE, Hemani G, et al. Shared genetic control of expression and methylation in peripheral blood. BMC Genomics. 2016;17:278. doi: 10.1186/s12864-016-2498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 27.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindgren CM, Heid IM, Randall JC, et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2011;43:1164. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krude H, Biebermann H, Luck W, et al. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat Genet. 1998;19:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 32.Berndt SI, Gustafsson S, Magi R, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins R, Reith C, Emberson J, et al. Interpretation of the evidence for the efficacy and safety of statin therapy. Lancet. 2016;388:2532–2561. doi: 10.1016/S0140-6736(16)31357-5. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y, Day FR, Gustafsson S, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Commun. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaess BM, Pedley A, Massaro JM, et al. The ratio of visceral to subcutaneous fat, a metric of body fat distribution, is a unique correlate of cardiometabolic risk. Diabetologia. 2012;55:2622–2630. doi: 10.1007/s00125-012-2639-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox CS, Liu Y, White CC, et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood AR, Tyrrell J, Beaumont R, et al. Variants in the FTO and CDKAL1 loci have recessive effects on risk of obesity and type 2 diabetes, respectively. Diabetologia. 2016;59:1214–1221. doi: 10.1007/s00125-016-3908-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkler TW, Justice AE, Graff M, et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11:e1005378. doi: 10.1371/journal.pgen.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kilpelainen TO, Qi L, Brage S, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8:e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young AI, Wauthier F, Donnelly P. Multiple novel gene-by-environment interactions modify the effect of FTO variants on body mass index. Nat Commun. 2016;7:12724. doi: 10.1038/ncomms12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi Q, Kilpelainen TO, Downer MK, et al. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014;23:6961–6972. doi: 10.1093/hmg/ddu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smemo S, Tena JJ, Kim KH, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dankel SN, Fadnes DJ, Stavrum AK, et al. Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss. PLoS One. 2010;5:e11033. doi: 10.1371/journal.pone.0011033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Claussnitzer M, Dankel SN, Kim KH, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilpelainen TO, Zillikens MC, Stancakova A, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Virtue S, Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the metabolic syndrome–an allostatic perspective. Biochim Biophys Acta. 2010;1801:338–349. doi: 10.1016/j.bbalip.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Lotta LA, Gulati P, Day FR, et al. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2017;49:17–26. doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahl AK, Reynolds CA, Fall T, et al. Multifactorial analysis of changes in body mass index across the adult life course: a study with 65 years of follow-up. Int J Obes (Lond) 2014;38:1133–1141. doi: 10.1038/ijo.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monda KL, Chen GK, Taylor KC, et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45:690–696. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen W, Kato N, Hwang JY, et al. Genome-wide association studies in East Asians identify new loci for waist-hip ratio and waist circumference. Sci Rep. 2016;6:17958. doi: 10.1038/srep17958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen W, Zheng W, Okada Y, et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23:5492–5504. doi: 10.1093/hmg/ddu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minster RL, Hawley NL, Su CT, et al. A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat Genet. 2016;48:1049–1054. doi: 10.1038/ng.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salinas YD, Wang L, DeWan AT. Multiethnic genome-wide association study identifies ethnic-specific associations with body mass index in Hispanics and African Americans. BMC Genet. 2016;17:78. doi: 10.1186/s12863-016-0387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 56.Richmond RC, Sharp GC, Ward ME, et al. DNA methylation and BMI: investigating identified methylation sites at HIF3A in a causal framework. Diabetes. 2016;65:1231–1244. doi: 10.2337/db15-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendelson MM, Marioni RE, Joehanes R, et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a mendelian randomization approach. PLoS Med. 2017;14:e1002215. doi: 10.1371/journal.pmed.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wahl S, Drong A, Lehne B, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Do R, Willer CJ, Schmidt EM, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Demerath EW, Guan W, Grove ML, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24:4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aslibekyan S, Demerath EW, Mendelson M, et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity (Silver Spring) 2015;23:1493–1501. doi: 10.1002/oby.21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frazier-Wood AC, Aslibekyan S, Absher DM, et al. Methylation at CPT1A locus is associated with lipoprotein subfraction profiles. J Lipid Res. 2014;55:1324–1330. doi: 10.1194/jlr.M048504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gagnon F, Aissi D, Carrie A, et al. Robust validation of methylation levels association at CPT1A locus with lipid plasma levels. J Lipid Res. 2014;55:1189–1191. doi: 10.1194/jlr.E051276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Irvin MR, Zhi D, Joehanes R, et al. Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid-lowering Drugs and Diet Network study. Circulation. 2014;130:565–572. doi: 10.1161/CIRCULATIONAHA.114.009158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dekkers KF, van Iterson M, Slieker RC, et al. Blood lipids influence DNA methylation in circulating cells. Genome Biol. 2016;17:138. doi: 10.1186/s13059-016-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mamtani M, Kulkarni H, Dyer TD, et al. Genome- and epigenome-wide association study of hypertriglyceridemic waist in Mexican American families. Clin Epigenetics. 2016;8:6. doi: 10.1186/s13148-016-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sayols-Baixeras S, Subirana I, Lluis-Ganella C, et al. Identification and validation of seven new loci showing differential DNA methylation related to serum lipid profile: an epigenome-wide approach. The REGICOR study. Hum Mol Genet. 2016;25:4556–4565. doi: 10.1093/hmg/ddw285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das M, Sha J, Hidalgo B, et al. Association of DNA methylation at CPT1A locus with metabolic syndrome in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study. PLoS One. 2016;11:e0145789. doi: 10.1371/journal.pone.0145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buchmann J, Meyer C, Neschen S, et al. Ablation of the cholesterol transporter adenosine triphosphate-binding cassette transporter G1 reduces adipose cell size and protects against diet-induced obesity. Endocrinology. 2007;148:1561–1573. doi: 10.1210/en.2006-1244. [DOI] [PubMed] [Google Scholar]
- 70.Frisdal E, Lay SL, Hooton H, et al. Adipocyte Atp-binding cassette G1 promotes triglyceride storage, fat mass growth and human obesity. Diabetes. 2015;64:840–855. doi: 10.2337/db14-0245. [DOI] [PubMed] [Google Scholar]
- 71.Hedman ÅK, Mendelson MM, Marioni RE, et al. Epigenetic patterns in blood associated with lipid traits predict incident coronary heart disease events and are enriched for results from genome-wide association studies. Circ Cardiovasc Genet. 2017;10:e001487. doi: 10.1161/CIRCGENETICS.116.001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fall T, Hagg S, Magi R, et al. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med. 2013;10:e1001474. doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mokry LE, Ross S, Timpson NJ, et al. Obesity and multiple sclerosis: a Mendelian randomization study. PloS Med. 2016;13:e1002053. doi: 10.1371/journal.pmed.1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loos RJ. Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab. 2012;26:211–226. doi: 10.1016/j.beem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 75.Wang C, Gordon ES, Norkunas T, et al. A randomized trial examining the impact of communicating genetic and lifestyle risks for obesity. Obesity (Silver Spring) 2016;24:2481–2490. doi: 10.1002/oby.21661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kullo IJ, Jouni H, Austin EE, et al. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial) Circulation. 2016;133:1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Godino JG, van Sluijs EM, Marteau TM, et al. Lifestyle advice combined with personalized estimates of genetic or phenotypic risk of type 2 diabetes, and objectively measured physical activity: a randomized controlled trial. PLoS Med. 2016;13:e1002185. doi: 10.1371/journal.pmed.1002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.