Abstract
Mature Xenopus oocytes are highly enriched for mitochondria. The organelles are stored and partitioned to newly-arising cells during embryogenesis, when there is little mitochondrial DNA (mtDNA) replication or transcription. A previously described member of the high mobility group (HMG) family of proteins, mtTFA, has been suggested to play a role in control of mtDNA copy number. mtTFA serves as a mitochondrial transcription factor in humans and Xenopus and as an abundant mtDNA packaging protein in yeast, like its prokaryotic histone-like counterpart, HU protein. Northern blot analysis demonstrated that expression of the gene was regulated during Xenopus oogenesis and specifically peaked at stage II. Western and Southern blotting were used to quantify amounts of the protein and mtDNA, respectively, in each stage of oogenesis. mtTFA:mtDNA ratios were found to be relatively low in previtellogenic oocytes while the ratios increased markedly in mature oocytes.
INTRODUCTION
Replication of mitochondrial DNA (mtDNA) is under relaxed temporal control and occurs throughout the cell cycle in cultured mammalian cells (1). Mitochondria have limited autonomy, requiring that the genome must act in concert with the nuclear genome for proper maintenance and biogenesis. Nuclear genes encode all the enzymes required for replication and transcription of mtDNA. These two processes are intimately linked because the control region contains strand-specific promoters and information for the initiation of mtDNA replication. While the overall mechanism by which the cell regulates mtDNA copy number is still poorly understood (2), it is widely held that the generation or utilization of primers for leading-strand replication is a prime candidate for a major control point in replication. Because transcripts initiated from the light strand promoter are used as primers for leading-strand replication (3), a number of studies have attempted to correlate the abundance of the human mitochondrial transcription factor mtTFA with mtDNA copy number.
mtTFA is a member of the high mobility group (HMG) family of proteins, a class of proteins that participate in the regulation of gene expression and the structural organization of chromatin. HMG proteins can bend DNA structure to facilitate the formation of higher-order nucleoprotein complexes (4,5). Some HMG-box proteins, such as mammalian HMG1 and HMG2 (6) and Saccharomyces cerevisiae NHP6 (7), bind DNA with little or no sequence specificity. Others, such as the human RNA polymerase I upstream binding factor (8) and sex determining factor SRY (9), recognize specific sequences. Although mtTFA can bind DNA non-specifically, its binding to the control region of mtDNA is non-random and its binding to preferred sites has been documented by footprinting (10,11). Under some circumstances, levels of mtTFA have been shown to vary concomitantly with levels of mtDNA. Low levels of the protein are seen in human cells depleted transiently or permanently of mtDNA (12–14). Patients with mitochondrial myopathies have increased levels of mtTFA in ragged-red fibers with elevated levels of mtDNA. In addition, in one study, heterozygous mtTFA knockout mice exhibited reduced mtDNA copy numbers while homozygous tissue-specific knockout embryos lacked mtDNA and died (15). Overexpression of mtTFA mRNA in HeLa cells has been shown to increase the abundance of mitochondrial transcripts in vivo (16). However, this study did not determine the increase in the mtTFA:mtDNA ratio. A recent study has shown that Drosophila mtTFA can be reduced markedly by RNA interference without significantly inhibiting mtDNA transcription for a period of several days (17). It is unknown whether this unexpected result may reflect species-specific effects or methodological differences.
The homolog of vertebrate mtTFA in S.cerevisiae appears to play a somewhat different role. Yeast mtTFA was originally termed ABF2 for ARS-binding factor 2 because it bound specifically to the autonomous replicating sequence ARS1 (18). However, it was later shown that mtTFA is not a nuclear protein in yeast and is not involved in chromosomal replication. Disruption of the gene in yeast leads to loss of mtDNA when cells are grown on glucose (18). A deficiency in the yeast mtTFA can be complemented by human mtTFA (19), but also by the bacterial histone-like protein HU (20), by nuclear NHP6A (21) or by overexpression of other yeast mitochondrial proteins (22,23). This suggests that the essential role of S.cerevisiae mtTFA is for general DNA packaging, rather than transcription. In fact, S.cerevisiae mtTFA has little ability to stimulate transcription from yeast promoters (24), although adding the C-terminal tail of human mtTFA to yeast mtTFA does permit the yeast factor to stimulate in vitro transcription from human promoters (25).
In this paper, we explore the role of mtTFA in the dramatic accumulation of mtDNA that occurs in amphibian oocytes. Xenopus laevis has been a particularly useful model system for studies of mitochondrial biogenesis because mature oocytes are 105-fold enriched for mitochondria compared with somatic cells (26). Consequently, mtDNA is the major DNA component in oocytes. It is estimated that 16–17 rounds of mtDNA replication occur in the absence of nuclear DNA replication to provide an egg with a sufficient supply of mtDNA (27). Accumulation of mtDNA is rapid in previtellogenic oocytes but slows considerably as the oocyte matures (28,29). We quantified mtTFA protein and mtDNA to permit calculation of the stoichiometry of protein:DNA at each stage of oogenesis. Our findings indicate that mtTFA is an abundant protein that may help regulate mtDNA copy number during Xenopus oogenesis as it packages mtDNA.
MATERIALS AND METHODS
Oocytes
Sexually mature X.laevis females were obtained from Nasco (Fort Atkinson, WI) and anesthetized by treatment with 0.1% Tricaine and hypothermia prior to decapitation and removal of ovaries. Oocytes were isolated by treating ovaries with 0.2% collagenase (Type II; Sigma) in DNOM (60 mM NaCl, 2 mM KCl, 1 mM Na2HPO4 pH 7.4, 5 mM HEPES pH 7.4, 1 mM MgCl2), staged (30) and stored at –80°C in buffers appropriate for RNA, protein or for mtDNA determination.
Northern blotting
Total RNA from known numbers of staged oocytes was obtained by homogenization in 0.5 ml RNA homogenization buffer (0.3 M NaCl, 2% SDS, 50 mM Tris pH 7.5, 1 mM EDTA) (31). The sample was then extracted with 1:1 phenol:chloroform, precipitated with ethanol and resuspended in TE, 1% SDS at a concentration of 0.5 µl/oocyte for stages I and II, 0.1 µl/oocyte for stages III and IV and 2 µl/oocyte for stage IV and combined stages V and VI (V+VI). RNA was subjected to electrophoresis on a 1% formaldehyde-containing agarose gel (32) and passively transferred to Hybond–N+ nylon membrane (Amersham Life Sciences). A fragment containing nearly the entire coding region (nucleotides 234–2003) (10) of the Xenopus mtTFA cDNA was 32P-labeled by random priming using a Prime-It II kit (Stratagene). Hybridization and washes were carried out at 65°C and bands were detected and quantified by PhosphorImager (Molecular Dynamics).
Quantitative immunoblotting of protein from immature oocytes
Specific numbers of stage I–IV oocytes or stages V+VI were collected and homogenized in sample loading buffer in microfuge tubes before storing at –80°C. Recombinant mtTFA (rmtTFA) was purified as described (10) and the concentration determined using a molar extinction coefficient of 28 590 calculated for the mature protein. Samples were thawed as required, then boiled for 2 min before loading onto a 12% denaturing polyacrylamide gel. In parallel, appropriate amounts of rmtTFA were loaded to generate a standard curve for each gel. Proteins were transferred onto nitrocellulose membrane (Schleicher and Schuell), which was then incubated with primary antibodies directed against purified recombinant protein at a dilution of 1:10 000 in Tween–PBS. Blots were incubated with 125I-labeled Protein A (Amersham Life Sciences) diluted to 1:2000 and washed with an immunoprecipitation wash buffer (20 mM Tris pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.02% SDS) (33). Signals were quantified with a PhosphorImager (Molecular Dynamics). Data that were within the concentration range of the standard curve were used for calculations. The mean value (ng/cell) and standard deviation were calculated for each stage.
Quantitative Southern blot analysis of mtDNA from immature oocytes
After staging and collecting a specific number of oocytes per tube, each sample was homogenized in whole cell homogenizing buffer (30 mM Tris pH 8, 2 mM DTT, 0.5 mM EDTA, 2% SDS) before storing at –80°C. Samples were then thawed as required. Varying amounts of pXLM32, a plasmid that contains the entire X.laevis mtDNA genome, were added as an internal standard (34). The samples were extracted twice with 1:1 phenol:chloroform. The nucleic acids were ethanol precipitated and digested with BglII and ScaI (New England Biolabs) in the presence of 20 µg/ml RNase A. This resulted in several bands, including a 4670 bp fragment from sample mtDNA and a 3809 bp fragment from pXLM32. Samples were re-extracted then electrophoresed on a 0.7% agarose gel. DNA in the gel was depurinated, denatured and passively transferred onto Hybond–N+ as described (32). A 1775 bp mtDNA fragment generated by HindIII digestion of pXLM32 was used as template to make a random primed probe (Prime-It II kit). Bands from sample mtDNA and pXLM32 both contained this entire 1.8 kb region. After several washes with 2× SSC, 0.1% SDS and 0.1× SSC, 0.1% SDS at 65°C, the signals were quantified by PhosphorImager (Molecular Dynamics) and corrected for their molecular weight. For each stage, the mean value (ng/cell) and standard deviation were calculated as for mtTFA. The mean amount of mtTFA or mtDNA in each stage was divided by their respective molecular weights of 28 000 or 11.7 × 106. The resulting quotients were further divided to obtain protein:mtDNA ratios.
Quantitative immunoblotting and Southern blotting of material from mature oocyte mitochondria
Due to a large accumulation of proteins in stage V and VI oocytes, whole cell lysate analyses were not possible. For these stages, crude mitochondrial lysates were used. To control for the use of two different methods, data were collected for stage IV oocytes using both whole cell preparations and mitochondrial lysates. Oocytes were homogenized in STE (0.25 M sucrose, 30 mM Tris pH 8, 0.5 mM EDTA, 2 mM DTT, 0.5 mM PMSF) and centrifuged at 1500 g at 4°C to pellet debris and yolk. The supernatant was the crude mitochondrial fraction used for analyses. Loading buffer was added to a portion of the mitochondria and frozen at –80°C for future protein analysis. The remainder of the mitochondrial suspension was stored frozen in aliquots for future mtDNA determination. Known volumes of the lysates were analyzed on 12% SDS–PAGE. The immunoblotting procedure for mitochondrial lysates was the same as described above except that washes were performed using PBS, 0.5% Tween. Aliquots of the mitochondrial suspensions were thawed as required for mtDNA determination. An equal volume of whole cell homogenizing buffer was added along with Proteinase K (0.5 mg/ml) for a 1 h incubation at 37°C. The treatment breaks open mitochondria and facilitates subsequent phenol/chloroform extractions. Known amounts of plasmid pXLM32 DNA were added after the Proteinase K incubation and before phenol/chloroform digestion. Restriction enzyme digestions, electrophoresis and hybridization were the same as described for immature oocytes.
RESULTS
Expression of Xenopus mtTFA mRNA
RNA blot hybridization was performed to examine the expression pattern of Xenopus mtTFA during oogenesis (Fig. 1A). RNA isolated from defined numbers of oocytes was subjected to analysis. Stages I–IV were collected as four separate samples, but the mature oocytes (stages V and VI) were grouped as one (V+VI). Fewer mature oocytes were used in this analysis due to the large quantity of rRNA that accumulates at these stages. Signals from the 2.2 kb major transcript were quantified, normalized for the number of cells and graphed relative to each other (Fig. 1B). Transcript levels increased 5-fold from stage I to stage II. After relative expression peaked at stage II, there was a slow decline throughout the remainder of oogenesis to ∼50% of the maximal level. Early peak expression has also been observed for another nuclear-encoded mitochondrial product, which is required for replication, the catalytic subunit of DNA polymerase γ (35). We did not attempt to determine whether the declining abundance of mRNA during vitellogenesis was due to mRNA turnover or to regulation of gene expression.
Figure 1.
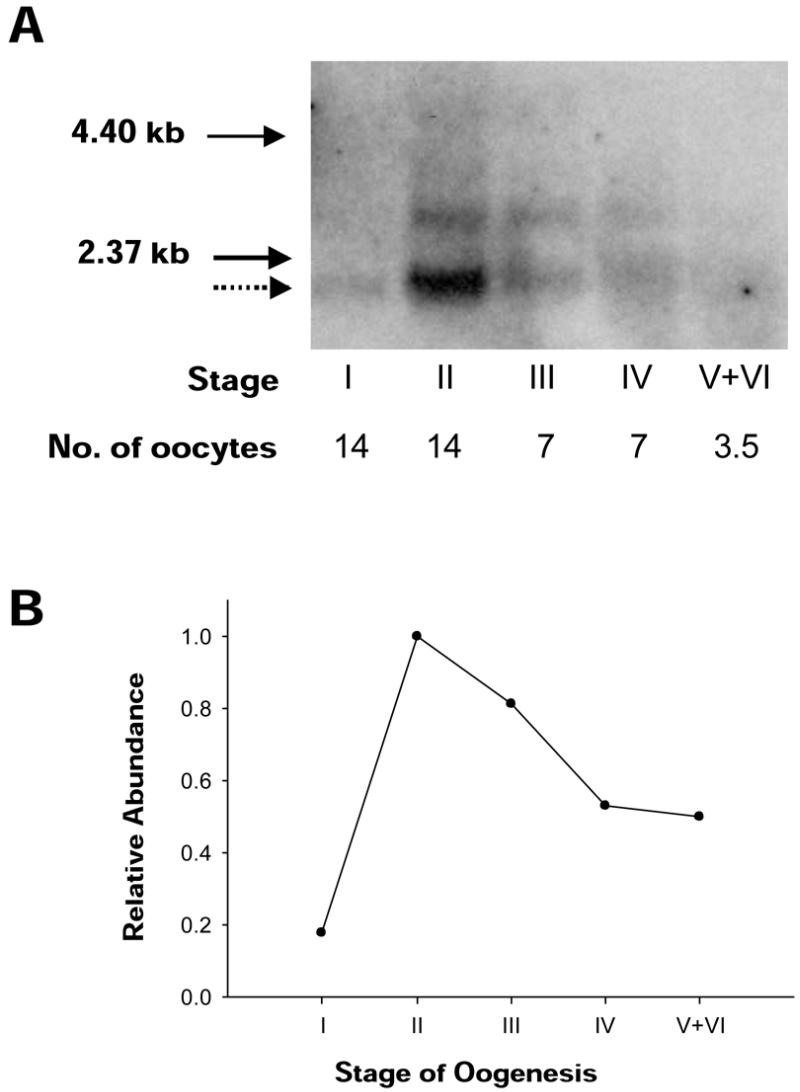
Expression of Xenopus mtTFA mRNA during oogenesis. (A) Northern blot of mtTFA transcripts in all stages of oogenesis, as classified by Dumont (30). Volumes equivalent to a specific number of oocytes for each stage were loaded on the same gel. The dotted arrow indicates the major mRNA species. The longer cross-hybridizing RNA species appears to result from alternative splicing in the 3′ untranslated region. Solid arrows indicate the sizes of RNA ladder markers (Life Technologies). (B) The major mRNA species was quantified with a phosphorimager. After the data were normalized for the number of cells in each stage, the relative abundance of mRNA was graphed.
Accumulation of mtTFA during oogenesis
We measured the ratio of mtTFA:mtDNA directly in total cell lysates and in mitochondrial lysates of defined numbers of staged Xenopus oocytes. The procedure used to quantify mtTFA by western blotting is shown in Figure 2. Known amounts of recombinant mtTFA were used to generate a standard curve to quantify the samples on every blot (Fig. 2E). Data in Figure 2A and B were derived from whole cell homogenates of a known number of staged oocytes. The accumulation of protein and yolk in mature oocytes (stages V and VI) made it difficult to obtain quantifiable western blots with whole cell lysates from these larger oocytes. Therefore, crude mitochondrial lysates from defined numbers of stage IV–VI oocytes were used to complete the analysis (Fig. 2C and D). Although this method might involve some loss of mitochondrial material, this would not affect the mtTFA:mtDNA ratio as the same lysates were used to measure mtDNA as described below. Both whole cell lysates and mitochondrial lysates were analyzed for stage IV oocytes to validate this approach. We observed a slightly higher per cell content of mtTFA when we analyzed mitochondrial extracts as compared with total cell extracts. The reason for this is not clear, although we suspect the increased total content of protein in stage IV oocytes may reduce the efficiency of transfer of protein to the membrane. As shown below, we saw a comparable increase in the signal for mtDNA in mitochondrial extracts compared with whole cell extracts for stage IV oocytes, so that both methods gave a similar estimate of the mtTFA:mtDNA ratio. Results from experiments with linear standard curves were pooled to determine the mean amount of mtTFA per cell for stages I–IV, or the amount per microliter for mitochondrial lysates (stages IV and V+VI). Data from at least three determinations of the mtTFA content at each stage in oogenesis are shown in Figure 3, documenting a dramatic increase in the level of mtTFA during oogenesis.
Figure 2.

Quantitation of mtTFA by western blotting. An example is shown of quantitation of mtTFA on western blots using specific antiserum and I125-protein A with phosphorimager detection. (A and B) Standards of recombinant mtTFA samples are shown on the left and oocyte samples on the right for manually staged and counted oocytes from stage I–III (A) or stage IV (B). (C and D) Data are similar to (A) and (B), except that crude mitochondrial fractions were used for analysis of stage IV (C) or stage V+VI oocytes (D). (E) A representative standard curve showing the relationship between phosphorimager intensity and the quantity (in ng) of rmtTFA. The line drawn through the data points is a linear regression.
Figure 3.
mtTFA content of staged oocytes. At least three independent experiments similar to that shown in Figure 2 were used to calculate the mean amount of mtTFA and standard deviation for each oocyte stage. The values are indicated above each bar in the graph. Asterisks denote that the analyzed samples were mitochondrial lysates, while all others were whole cell lysates.
Accumulation of mtDNA during oogenesis
Our approach to directly determine mtTFA:mtDNA ratios required quantitation of mtDNA from the same lysates used for mtTFA quantitation. Figure 4 shows the blot hybridization strategy used to quantify mtDNA in the presence of internal standards of pXLM32 DNA. For each experiment, known quantities of pXLM32 were added to duplicate or triplicate collections of oocytes prior to homogenization. The DNA mixtures were organically extracted and digested with restriction endonucleases BglII and ScaI. The DNA fragments were separated by electrophoresis, passively transferred to nylon membranes and probed with a labeled 1.8 kb fragment that recognizes a 4.7 kb band in the mtDNA digest and a 3.8 kb band in the internal plasmid standard (Fig. 4A). Every mtDNA quantitation required a separate internal standard curve on the same blot (Fig. 4B). Figure 5A and B shows representative blots from each stage. Data that were within the range bracketed by the internal standards were pooled to calculate the average mtDNA content in each stage (Fig. 6). Our data can be compared with those from two previous studies that suggested different patterns for the accumulation of mtDNA. Webb and Smith (28) used complementary RNA hybridization to measure mtDNA levels, with no internal control to account for losses during extractions. This work suggested that mtDNA amplification was essentially complete in stage IV oocytes, with very little additional accumulation of mtDNA in pigmented oocytes. In contrast, Callen et al. (27) employed two independent techniques to suggest a more gradual accumulation of mtDNA. Our results closely parallel those of Callen et al. (27), confirming the observation that a few rounds of mtDNA replication occur in vitellogenic oocytes.
Figure 4.
Strategy for quantitation of mtDNA by Southern blot analysis. (A) The ScaI and BglII maps of mtDNA and pXLM32 are shown to indicate how these two DNAs yield fragments of different sizes that contain the entire extent of the 1.8 kb HindIII fragment used as a hybridization probe. The pBR322 vector in pXLM32 is shown as a thick line. The HindIII sites are shown only in the pXLM32 diagram. (B) A representative standard curve showing the relationship between phosphorimager intensity and quantity of pXLM32 DNA. The line drawn through the data points is a linear regression.
Figure 5.
Quantitation of mtDNA by Southern blot analysis. (A) Variable amounts of plasmid pXLM32 were added to mitochondrial lysates prior to extraction and processing. Representative Southern blot results are shown for oocyte stages I–IV. (B) Mitochondrial lysates (indicated by asterisks) from stages IV and V+VI were used to quantify the amount of mtDNA on one Southern blot. As with data for mtTFA, mtDNA was quantified for whole stage IV oocytes (panel A) as well as for lysates.
Figure 6.

Accumulation of mtDNA during Xenopus oogenesis. At least three independent experiments were used to calculate the mean amount of mtDNA and standard deviation in each stage. These values are indicated above each bar in the graph. Asterisks denote data derived from mitochondrial lysates, while all others were whole cell lysates.
DISCUSSION
The mtTFA:mtDNA ratio varies during Xenopus oocyte development
Quantitative methods were used to study the abundance of Xenopus mtTFA in relation to mtDNA during oogenesis. The mean values from Figures 3 and 6A were used to calculate the number of mtTFA molecules per mtDNA molecule, as shown in Table 1. We found that mtTFA accumulates to very high levels in mature oocytes, exceeding 2000 copies per mtDNA. We have shown that the large majority of mtTFA remains bound to mtDNA in nucleoids in low salt lysates of Xenopus oocyte mitochondria, ruling out the possibility that mature oocytes contain a large pool of mtTFA not bound to mtDNA (data not shown). mtTFA is likely to be a major component of the Xenopus mtDNA nucleoprotein complexes observed using electron microscopy (36). Since Xenopus mtTFA binds as a tetramer with a 35 bp footprint (10,37), we can estimate that 2000 copies per mtDNA molecule are sufficient to cover the entire mtDNA genome. This calculation assumes that mtTFA is always a tetramer with a stable footprint. It is unknown whether the packing density may change as the ratio of mtTFA:mtDNA changes.
Table 1. mtTFA/mtDNA ratios during Xenopus oogenesis.
| Stage |
mtTFA/mtDNAa (mol/mol) |
| I | 394 ± 14 |
| II | 396 ± 112 |
| III | 756 ± 244 |
| IV | 1480 ± 163 |
| IVb | 1585 ± 642 |
| V+VIb | 2658 ± 756 |
aMean amounts of the protein and DNA (Figs 3 and 6A) were used to generate molar ratios.
bMitochondrial lysates were used in the analysis.
It is interesting to compare the Xenopus mitochondrial system with those of yeast and human mtDNA. Diffley and Stillman (18) estimated that yeast contain ∼250 000 copies of mtTFA (Abf2p) per cell, which translates to roughly one protein per 15 bp based on published estimates of mtDNA content. Using a simplified purification procedure, Fisher et al. (38) found that human mtTFA was more abundant than had previously been thought. By comparing the levels of purified mtTFA to literature values for the cellular mtDNA content, they suggested a minimal estimate of ∼15 molecules per mtDNA in cultured KB cells. Thus, Xenopus mtTFA is remarkably abundant, like yeast mtTFA, despite the fact that it resembles human mtTFA in the ability to stimulate transcription.
What are the consequences of mtTFA accumulation in Xenopus oocytes?
Stimulation of in vitro transcription in Xenopus depends on the binding of mtTFA to a site between two promoters in the control region (10). In vitro experiments have shown that concentrations of mtTFA 2–3-fold higher than optimal stimulatory concentrations are sufficient to shut down transcription (10). Because the HMG-box protein mtTFA is known to wrap, bend and compact DNA (37,39), the protein accumulated in mature oocytes may package the mtDNA in complexes that prevent access by other regulatory proteins. Unfortunately, the absolute rate of transcription per mtDNA template has not been studied in staged oocytes. Transcription is still detected by radiolabeling in mature oocytes (40), but is essentially inactive in unfertilized eggs and early embryos (41,42). It is possible that the accumulation of high levels of mtTFA may be part of the developmental down-regulation of mitochondrial transcription. Accumulation of high levels of mtTFA coincides with the reduced rate of mtDNA replication (28,29) and the reduced frequency of partially replicated D-loop molecules as oocytes mature (43). Similarly, the state of mtDNA nucleoprotein may be a factor in the block to mtDNA replication in eggs and early oocytes (26).
To our knowledge, this work is the first to show a significant variation in the mtTFA:mtDNA ratio in animal tissues. Studies of mtTFA in other systems have rarely attempted to measure this critical ratio directly. Our results illustrate the need to measure the mtTFA:mtDNA ratio in various cell types under different physiological conditions. Xenopus laevis oocytes are particularly well suited for the in vitro investigation of the relative abundance of mtTFA. The amplification of mtDNA is more extreme in amphibian oocytes but also occurs in mammals. Mouse and bovine oocytes have been estimated to contain 100 times as much mtDNA as a somatic cell. Approximately one-third of the total cellular DNA is mitochondrial in mouse eggs (44), but this is still nearly 2000 times less than what is found in frog eggs. Developing bovine oocytes also contain 1000 times less mtDNA than mature frog oocyte (45). It is possible that mtTFA:mtDNA ratios vary in mammalian oocytes as in frogs. However, it would be more difficult to obtain biochemical quantities of mammalian oocytes required to measure the ratio directly, particularly if the mtTFA:mtDNA ratio is lower in mammalian cells as suggested by Fisher et al. (38).
This investigation has concentrated on mtTFA as this is the only well characterized non-enzymatic protein known to bind double-stranded mtDNA in vivo. Other binding proteins that help make up mitochondrial chromatin or are partners of mtTFA may play a role in regulating mitochondrial nucleic acid synthesis. Characterizing other components of the nucleoid could contribute significantly to the underlying mechanisms of mtDNA maintenance.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Grant GM29681 from the NIH to D.F.B.
References
- 1.Bogenhagen D. and Clayton,D.A. (1977) Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell, 11, 719–727. [DOI] [PubMed] [Google Scholar]
- 2.Moraes C.T. (2001) What regulates mitochondrial DNA copy number in animal cells? Trends Genet., 17, 199–205. [DOI] [PubMed] [Google Scholar]
- 3.Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- 4.Grosschedl R., Giese,K. and Pagel,J. (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet., 10, 94–99. [DOI] [PubMed] [Google Scholar]
- 5.Wolffe A.P. (1994) Architectural transcription factors. Science, 264, 1100–1103. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi M., Beltrame,M. and Paonessa,G. (1989) Specific recognition of cruciform DNA by nuclear protein HMG 1. Science, 243, 1056–1059. [DOI] [PubMed] [Google Scholar]
- 7.Kolodrubetz D. and Burgum,A. (1990) Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J. Biol. Chem., 265, 3234–3239. [PubMed] [Google Scholar]
- 8.Jantzen H., Admon,A., Bell,S. and Tjian,R. (1990) Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature, 344, 830–836. [DOI] [PubMed] [Google Scholar]
- 9.Sinclair A.H., Berta,P., Palmer,M.S., Hawkins,J.R., Griffiths,B.L., Smith,M.J., Foster,J.W., Frischauf,A.M., Lovell-Badge,R. and Goodfellow,P.N. (1990) A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature, 346, 240–244. [DOI] [PubMed] [Google Scholar]
- 10.Antoshechkin I. and Bogenhagen,D.F. (1995) Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol. Cell. Biol., 15, 7032–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher R.P., Lisowsky,T., Parisi,M.A. and Clayton,D.A. (1992) DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem., 267, 3358–3367. [PubMed] [Google Scholar]
- 12.Davis A.F., Ropp,P.A., Clayton,D.A. and Copeland,W.C. (1996) Mitochondrial DNA polymerase γ is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res., 24, 2753–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson N.-G., Oldfors,A., Holme,E. and Clayton,D.A. (1994) Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem. Biophys. Res. Commun., 200, 1374–1381. [DOI] [PubMed] [Google Scholar]
- 14.Poulton J., Morten,K., Freman-Emmerson,C., Potter,C., Sewry,C., Dubowitz,V., Kidd,H., Stephenson,J., Whitehouse,W., Hansen,F. et al. (1994) Deficiency of the human mitochondrial transcription factor h-mtTFA in infantile mitochondrial myopathy is associated with mtDNA depletion. Hum. Mol. Genet., 3, 1763–1769. [DOI] [PubMed] [Google Scholar]
- 15.Larsson N.-G., Wang,J., Wilhelmsson,H., Oldfors,A., Rustin,P., Lewandoski,M., Barsh,G.S. and Clayton,D.A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet., 18, 231–236. [DOI] [PubMed] [Google Scholar]
- 16.Montoya J., Perez-Martos,A., Garstka,H.L. and Wiesner,R.J. (1997) Regulation of mitochondrial transcription by mitochondrial transcription factor A. Mol. Cell. Biochem., 174, 227–230. [PubMed] [Google Scholar]
- 17.Goto A., Matsushima,Y., Kadowaki,T. and Kitagawa,Y. (2001) Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. Biochem. J., 354, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diffley J.F. and Stillman,B. (1991) A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA, 88, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parisi M.A., Xu,B. and Clayton,D.A. (1993) A human mitochondrial activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol., 13, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Megraw T.L. and Chae,C.-B. (1993) Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem., 268, 12758–12763. [PubMed] [Google Scholar]
- 21.Kao L., Megraw,T. and Chae,C.-B. (1993) Essential role of the HMG domain in the function of yeast mitochondrial histone HM: functional complementation of HM by the nuclear nonhistone protein NHP6A. Proc. Natl Acad. Sci. USA, 90, 5598–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho J.H., Lin,S.J., Kao,L.R., Megraw,T.L. and Chae,C.-B. (1998) A novel DNA-binding protein bound to the mitochondrial inner membrane restores the null mutation of mitochondrial histone Abf2p in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 5712–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zelenaya-Troitskaya O., Perlman,P.S. and Butow,R.A. (1995) An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J., 14, 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu B. and Clayton,D.A. (1992) Assignment of a yeast protein necessary for mitochondrial transcription initiation. Nucleic Acids Res., 20, 1053–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dairaghi D.J., Shadel,G.S. and Clayton,D.A. (1995) Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol., 249, 11–28. [DOI] [PubMed] [Google Scholar]
- 26.Chase J.W. and Dawid,I.B. (1972) Biogenesis of mitochondria during Xenopus laevis development. Dev. Biol., 27, 504–518. [DOI] [PubMed] [Google Scholar]
- 27.Callen J.C., Dennebouy,N. and Mounolou,J.-C. (1980) Development of the mitochondrial mass and accumulation of mtDNA in previtellogenic stages of Xenopus oocytes. J. Cell Sci., 41, 307–320. [DOI] [PubMed] [Google Scholar]
- 28.Webb A.C. and Smith,L.D. (1977) Accumulation of mitochondrial DNA during oogenesis in Xenopus laevis. Dev. Biol., 56, 219–225. [DOI] [PubMed] [Google Scholar]
- 29.Callen J.C., Tourte,M., Dennebouy,N. and Mounolou,J.-C. (1980) Mitochondrial development in oocytes of Xenopus laevis. Biol. Cellulaire, 38, 13–18. [Google Scholar]
- 30.Dumont J.N. (1972) Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J. Morphol., 136, 153–180. [DOI] [PubMed] [Google Scholar]
- 31.Gurdon J.B. and Wickens,M.P. (1983) The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol., 101, 370–386. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Stuurman N., Maus,N. and Fisher,P.A. (1995) Interphase phosphorylation of the Drosophila nuclear lamin: site-mapping using a monoclonal antibody. J. Cell Sci., 108, 3137–3144. [DOI] [PubMed] [Google Scholar]
- 34.Ramirez J.L. and Dawid,I.B. (1978) Mapping of mitochondrial DNA in Xenopus laevis and X. borealis: the positions of ribosomal genes and D-loops. J. Mol. Biol., 119, 133–146. [DOI] [PubMed] [Google Scholar]
- 35.Ye F., Carrodeguas,J.A. and Bogenhagen,D.F. (1996) The γ subfamily of DNA polymerases: cloning of a developmentally regulated cDNA encoding Xenopus laevis mitochondrial DNA polymerase γ. Nucleic Acids Res., 24, 1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinon H., Barat,M., Tourte,M., Dufresne,C. and Mounolou,J.-C. (1978) Evidence for a mitochondrial chromosome in Xenopus laevis oocytes. Chromosoma, 65, 383–389. [Google Scholar]
- 37.Antoshechkin I., Bogenhagen,D.F. and Mastrangelo,I.A. (1997) The HMG-box mitochondrial transcription factor xl-mtTFA binds DNA as a tetramer to activate bidirectional transcription. EMBO J., 16, 3198–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher R.P., Lisowsky,T., Breen,G.A. and Clayton,D.A. (1991) A rapid, efficient method for purifying DNA-binding proteins. J. Biol. Chem., 266, 9153–9160. [PubMed] [Google Scholar]
- 39.Diffley J.F. and Stillman,B. (1992) DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem., 267, 3368–3374. [PubMed] [Google Scholar]
- 40.Webb A., LaMarca,M. and Smith,L. (1975) Synthesis of mitochondrial RNA by full-grown and maturing oocytes of Rana pipiens and Xenopus laevis. Dev. Biol., 45, 44–55. [DOI] [PubMed] [Google Scholar]
- 41.El Meziane A., Callen,J.C. and Mounolou,J.-C. (1989) Mitochondrial gene expression during Xenopus laevis development: a molecular study. EMBO J ., 8, 1649–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ammini C. and Hauswirth,W. (1999) Mitochondrial gene expression is regulated at the level of transcription during early embryogenesis of Xenopus laevis. J. Biol. Chem., 274, 6265–6271. [DOI] [PubMed] [Google Scholar]
- 43.Callen J.C., Tourte,M., Dennebouy,N. and Mounolou,J.-C. (1983) Changes in D-loop frequency and superhelicity among the mitochondrial DNA molecules in relation to organelle biogenesis in oocytes of Xenopus laevis. Exp. Cell Res., 143, 115–125. [DOI] [PubMed] [Google Scholar]
- 44.Pikó L. and Matsumoto,L. (1976) Number of mitochondria and some properties of mitochondrial DNA in the mouse egg. Dev. Biol., 49, 1–10. [DOI] [PubMed] [Google Scholar]
- 45.Michaels G.S., Hauswirth,W.W. and Laipis,P.J. (1982) Mitochondrial DNA copy number in bovine oocytes and somatic cells. Dev. Biol., 94, 246–251. [DOI] [PubMed] [Google Scholar]





