Abstract
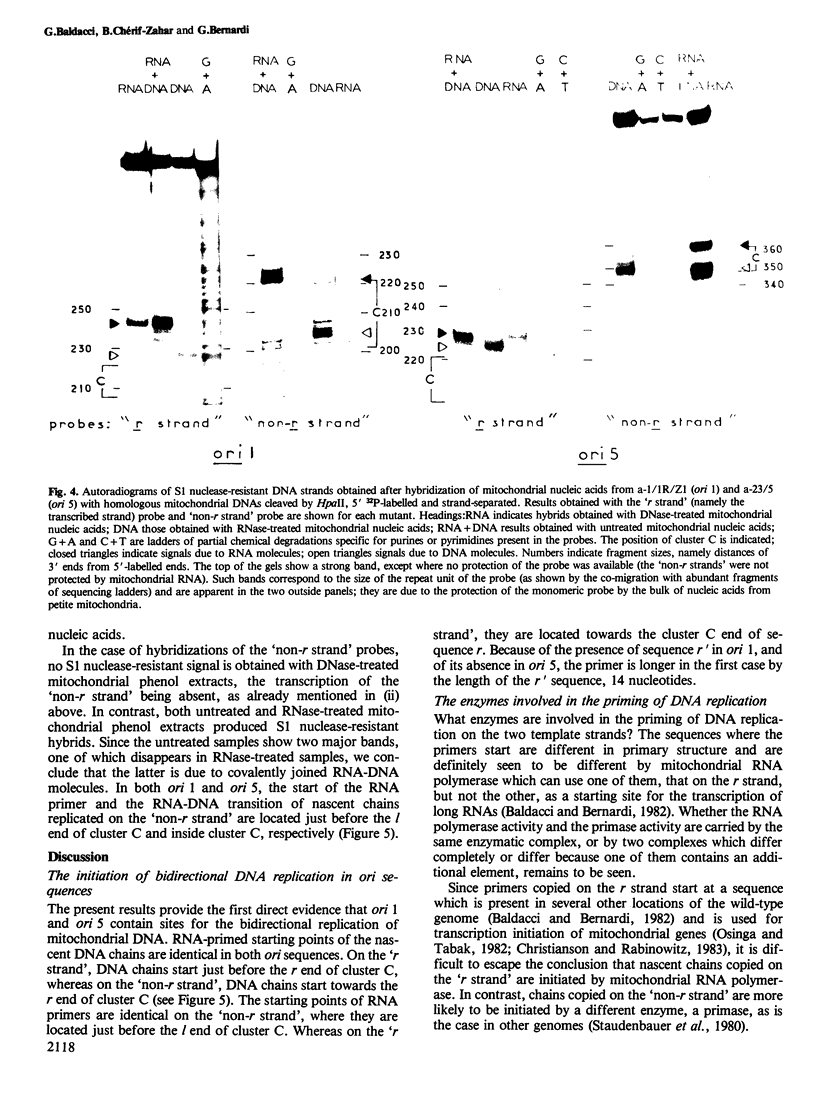
We report here the first direct demonstration that the active ori sequences of the mitochondrial genome of Saccharomyces cerevisiae are indeed origins of DNA replication, as previously postulated on the basis of compelling but indirect evidence. Basically, such sequences are formed by four regions: (i) GC clusters A and B, which are separated by a 29-bp AT stretch; (ii) a central 200-bp AT stretch, l; (iii) GC cluster C; (iv) a 16-bp AT stretch r, which comprises a site for transcription initiation. The ori sequences investigated, ori 1 and ori 5, have opposite orientations on the parental wild-type genome; ori 1 has but ori 5 does not have an additional 14-bp AT stretch r', between cluster C and sequence r; they were carried by the genomes of two spontaneous petites. In both ori sequences, nascent DNA chains using as template the strand containing sequence r (the 'r strand') start at the r end of cluster C, are elongated towards sequence l, and follow an RNA primer starting at sequence r. Nascent DNA chains copied on the 'non-r strand' start within cluster C, are elongated towards sequence r, and follow an RNA primer starting in sequence l just before cluster C. Ori 1 and 5 are, therefore, used as sites for RNA-primed bidirectional replication of mitochondrial DNA. Several aspects of this process are discussed.
Full text
PDF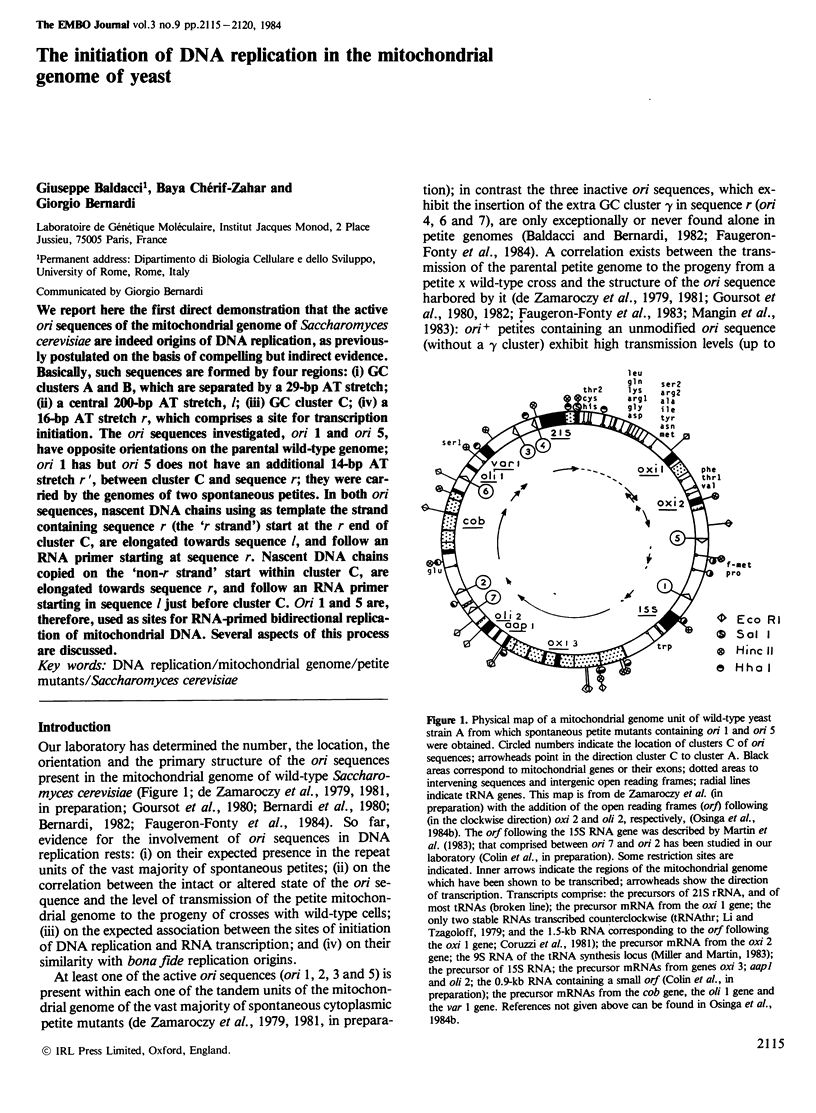




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldacci G., Bernardi G. Replication origins are associated with transcription initiation sequences in the mitochondrial genome of yeast. EMBO J. 1982;1(8):987–994. doi: 10.1002/j.1460-2075.1982.tb01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc H., Dujon B. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3942–3946. doi: 10.1073/pnas.77.7.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T., Rabinowitz M. Identification of multiple transcriptional initiation sites on the yeast mitochondrial genome by in vitro capping with guanylyltransferase. J Biol Chem. 1983 Nov 25;258(22):14025–14033. [PubMed] [Google Scholar]
- Clayton D. A. Replication of animal mitochondrial DNA. Cell. 1982 Apr;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Coruzzi G., Bonitz S. G., Thalenfeld B. E., Tzagoloff A. Assembly of the mitochondrial membrane system. Analysis of the nucleotide sequence and transcripts in the oxi1 region of yeast mitochondrial DNA. J Biol Chem. 1981 Dec 25;256(24):12780–12787. [PubMed] [Google Scholar]
- Faugeron-Fonty G., Culard F., Baldacci G., Goursot R., Prunell A., Bernardi G. The mitochondrial genome of wild-type yeast cells. VIII. The spontaneous cytoplasmic "petite" mutation. J Mol Biol. 1979 Nov 5;134(3):493–457. doi: 10.1016/0022-2836(79)90365-6. [DOI] [PubMed] [Google Scholar]
- Faugeron-Fonty G., Mangin M., Huyard A., Bernardi G. The mitochondrial genomes of spontaneous orir petite mutants of yeast have rearranged repeat units organized as inverted tandem dimers. Gene. 1983 Sep;24(1):61–71. doi: 10.1016/0378-1119(83)90131-2. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Gaillard C., Bernardi G. The nucleotide sequence of the mitochondrial genome of a spontaneous "petite" mutant of yeast. Mol Gen Genet. 1979 Jul 24;174(3):335–337. doi: 10.1007/BF00267807. [DOI] [PubMed] [Google Scholar]
- Goursot R., Mangin M., Bernardi G. Surrogate origins of replication in the mitochondrial genomes of ori-zero petite mutants of yeast. EMBO J. 1982;1(6):705–711. doi: 10.1002/j.1460-2075.1982.tb01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobom G. Replication signals in prokaryotic DNA. Curr Top Microbiol Immunol. 1981;94-95:93–142. doi: 10.1007/978-3-642-68120-2_3. [DOI] [PubMed] [Google Scholar]
- Li M., Tzagoloff A. Assembly of the mitochondrial membrane system: sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell. 1979 Sep;18(1):47–53. doi: 10.1016/0092-8674(79)90352-0. [DOI] [PubMed] [Google Scholar]
- Mangin M., Faugeron-Fonty G., Bernardi G. The orir to ori+ mutation in spontaneous yeast petites is accompanied by a drastic change in mitochondrial genome replication. Gene. 1983 Sep;24(1):73–81. doi: 10.1016/0378-1119(83)90132-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller D. L., Martin N. C. Characterization of the yeast mitochondrial locus necessary for tRNA biosynthesis: DNA sequence analysis and identification of a new transcript. Cell. 1983 Oct;34(3):911–917. doi: 10.1016/0092-8674(83)90548-2. [DOI] [PubMed] [Google Scholar]
- Osinga K. A., De Vries E., Van der Horst G. T., Tabak H. F. Initiation of transcription in yeast mitochondria: analysis of origins of replication and of genes coding for a messenger RNA and a transfer RNA. Nucleic Acids Res. 1984 Feb 24;12(4):1889–1900. doi: 10.1093/nar/12.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osinga K. A., De Vries E., Van der Horst G., Tabak H. F. Processing of yeast mitochondrial messenger RNAs at a conserved dodecamer sequence. EMBO J. 1984 Apr;3(4):829–834. doi: 10.1002/j.1460-2075.1984.tb01892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osinga K. A., Tabak H. F. Initiation of transcription of genes for mitochondrial ribosomal RNA in yeast: comparison of the nucleotide sequence around the 5'-ends of both genes reveals a homologous stretch of 17 nucleotides. Nucleic Acids Res. 1982 Jun 25;10(12):3617–3626. doi: 10.1093/nar/10.12.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudenbauer W. L., Scherzinger E., Lanka E. Replication of the colicin E1 plasmid in extracts of Escherichia coli: uncoupling of leading strand from lagging strand synthesis. Mol Gen Genet. 1979;177(1):113–120. doi: 10.1007/BF00267260. [DOI] [PubMed] [Google Scholar]
- Tapper D. P., Clayton D. A. Precise nucleotide location of the 5' ends of RNA-primed nascent light strands of mouse mitochondrial DNA. J Mol Biol. 1982 Nov 25;162(1):1–16. doi: 10.1016/0022-2836(82)90159-0. [DOI] [PubMed] [Google Scholar]
- Wong J. F., Ma D. P., Wilson R. K., Roe B. A. DNA sequence of the Xenopus laevis mitochondrial heavy and light strand replication origins and flanking tRNA genes. Nucleic Acids Res. 1983 Jul 25;11(14):4977–4995. doi: 10.1093/nar/11.14.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Baldacci G., Bernardi G. Putative origins of replication in the mitochondrial genome of yeast. FEBS Lett. 1979 Dec 15;108(2):429–432. doi: 10.1016/0014-5793(79)80580-3. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Faugeron-Fonty G., Bernardi G. Excision sequences in the mitochondrial genome of yeast. Gene. 1983 Mar;21(3):193–202. doi: 10.1016/0378-1119(83)90002-1. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Marotta R., Faugeron-Fonty G., Goursot R., Mangin M., Baldacci G., Bernardi G. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature. 1981 Jul 2;292(5818):75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]




