Supplemental Digital Content is Available in the Text.
Key Words: tenofovir microbicide gel, HIV-1, transmission fitness
Abstract
Background:
Women in the CAPRISA 004 trial assigned to use 1% tenofovir (TFV) microbicide gel, who became HIV-1 infected, had higher viral load set-point and slower antibody avidity maturation compared with placebo participants. We investigated whether TFV gel was selected for viruses with altered genetic characteristics.
Setting:
The participants of the CAPRISA 004 trial (n = 28 TFV and 43 placebo) were from KwaZulu-Natal Province, South Africa and were infected with HIV-1 subtype C. After HIV-1 diagnosis, they were recruited into the CAPRISA 002 cohort.
Methods:
We analyzed gag sequences from the earliest time point post infection (within 3 months of estimated time of infection). Transmission index was measured using a model which predicts the likelihood of an amino acid to be transmitted. Phylogenetic distance from a regional consensus sequence was calculated from a maximum likelihood phylogenetic tree.
Results:
Transmission index and distance from the most common (consensus) sequence have been shown to be markers of transmission fitness. We found that viruses infecting TFV gel recipients were closer to the consensus sequence of regional strains (P = 0.003) and had higher transmission index (P = 0.01). The transmission index was weakly correlated with concomitant viral load (Spearman r = 0.22, P = 0.06).
Conclusion:
Decreased acquisition risk may have increased the barrier to infection therefore selecting for fitter, more consensus-like viruses. Such virus fitness effects will need to be considered for future pre-exposure prophylaxis and vaccine trials.
INTRODUCTION
Pre-exposure prophylaxis (PrEP) has been shown to be effective at reducing HIV incidence.1 Microbicides deliver an antiviral compound inside the vagina or rectum to intervene at the site of transmission, and the CAPRISA 004 trial was the first to show that 1% tenofovir (TFV) microbicide gel was effective in reducing HIV acquisition in heterosexually active women.1 CAPRISA 004 found that women who used the gel were 39% less likely to become infected, although high adherence was likely required for this approach to be effective.1 As TFV raised the barrier to infection, it is important to determine whether this affected the characteristics of viruses which overcame this barrier.
Although mutations which confer resistance to TFV have been reported in oral TFV PrEP regimens,2 none were identified in the CAPRISA 004 trial1 suggesting that the TFV gel does not confer a prolonged selective pressure on the virus and is not disadvantageous for postinfection therapy. Despite the gel acting as a barrier at point of first exposure to HIV during infection, no difference in the number of transmitted variants was observed between viruses infecting the TFV and placebo recipients.3 Similarly, gag-protease replication capacity was the same between the 2 arms of the study.4 However, immunological differences were identified, with individuals in the TFV group having a higher postinfection CD4+ T-cell responses,5 and slower antibody avidity maturation,6 compared with those from the placebo. Moreover, TFV recipients had significantly higher viral loads than placebo recipients at 12 months post infection, although CD4 counts were similar between study groups.7
Recent evidence suggests that the transmission bottleneck favors viruses with higher transmission fitness.8 Measures of sequence conservation have been used as surrogates of transmission fitness, including distance from consensus and a transmission index.8,9 Theoretically, such transmission selection bias will increase when per-exposure infection risk decreases. This effect has been observed when comparing men to women, and healthy men to men with genital ulcers or inflammation, in the context of heterosexual transmission,8 and between heterosexual men and men who have sex with men.9 A key parameter that correlates with transmission fitness is the similarity of the sequence to circulating viruses in the population. Viruses that are closer to the consensus sequence may have a transmission advantage as illustrated by the findings in transmission pairs that have shown that viruses closer to the ancestor in the donor are preferentially transmitted, and there is selective transmission of viruses closer to the subtype consensus sequence.10,11 This feature has also been linked to viral load and posttransmission reversion rates,8 within-host fitness,12 and in vitro replicative capacity,13 suggesting that founder viruses with high transmission potential are likely to be linked with higher viral load post transmission, though host-specific immune responses that target common variants will weaken this signal.14,15
We therefore hypothesized that partial protection conferred by TFV would result in increased selection at the time of transmission. Furthermore, such selection would result in founder viruses with higher transmission potential in the TFV arm and may explain the increased viral load observed among these women.7
METHODS
Women who seroconverted while participating in the CAPRISA 004 study were recruited into the CAPRISA 002 acute infection cohort. The study was reviewed by University of KwaZulu-Natal Biomedical Research Ethics Committee, and University of Cape Town Human Research Ethic Committee.7 Written consent was obtained from all participants.
HIV full-length gag was sequenced from plasma collected at the earliest time point post infection (enrollment) as previously described4 from the CAPRISA 004 1% TFV gel microbicide trial participants (n = 71) who seroconverted during the trial.1 An alignment of 929 sequences from antiretroviral naive, chronically infected individuals presenting at antenatal and outpatient clinics in Durban, South Africa,15 was used as the reference. This alignment represents the ancestral population of circulating viral strains from the same source population where the participants of this study were sampled. Therefore, the consensus sequence of this alignment was used as the most recent ancestral virus. HIV-1 Gag was selected for analysis because of its importance in determining viral replication and early set-point viral load.16,17
A maximum likelihood phylogenetic tree, based on DNA of all CAPRISA 004 sequences was generated (see Supplemental Digital Content 1A, http://links.lww.com/QAI/B43) using PhyML with 100 bootstraps.18 The tree was rooted on the reference consensus. To generate the phylogeny, a GTR nucleotide substitution model was used, equilibrium frequencies were estimated using maximum likelihood while the transition/transversion ratio, proportion of invariable sites, and gamma shape parameter were estimated from the data. The best of nearest neighbor interchange and sub-tree pruning and re-grafting was used to optimize the tree topology and branch lengths. The total branch length for each terminal sequence from the root was measured using the “branchlength” function available within the TreeRate tool in Los Alamos National Laboratory databases,19,20 and used to assess phylogenetic divergence of each sequence. Phylogenetic divergence from the reference consensus was compared between viral sequences from TFV and placebo recipients. We also estimated phylogenetic divergence by rooting the tree with the 2004 HIV-1 subtype C ancestral sequence (https://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html), and again with a most recent common ancestor of the combined alignment of the reference sequences and the study sequences (generated using the “ancestral reconstruction tool” function in HyPhy21,22 with default settings for a codon alignment). Genetic distances were also determined from the number of substitutions per site between each CAPRISA 004 sequence and the reference consensus using a phylogenetic-based composite log-likelihood approach.23 This analyses was conducted in MEGA v624 using a gamma distribution model for the rate variation among sites and allowing for substitution heterogeneity among lineages.
The transmission index of a sequence was calculated as the mean of the expected log-odds of transmission for each site in the sequence, as estimated by a logistic regression model that included a second-order polynomial of cohort frequency, the number of co-varying sites, and offsets and cohort–frequency interactions for each protein domain. We used published model weights that were previously trained using linked transmission pairs from Zambia.8 The expected position-specific amino acid frequencies were taken from the reference alignment.25 The resulting transmission index is unitless, and was standardized to zero mean and unit variance across all samples.
Statistical tests were implemented in Prism 5.0 (GraphPad Software, Inc., San Diego, CA) and R-Statistical package version 3.1.0. Spearman correlation was used to test association between transmission index and plasma viral load.
RESULTS
A total of 71 gag sequences (28 TFV and 43 placebo) were generated at enrollment from all available samples from participants who were recruited within 3 months of HIV-1 infection. The median [interquartile range (IQR)] days post infection at which the earliest sequence data were obtained were 84 (84–91) for TFV group and 84 (77–84) for placebo. We found that viruses within the TFV arm were less diverse from each other compared with those from the placebo group (P = 0.02; randomization test; TFV mean 0.026 (95% CI: 0.023 to 0.029) and placebo median 0.030 (95% CI: 0.028 to 0.032), see Supplemental Digital Content 1B, http://links.lww.com/QAI/B43). This constrained diversity of viruses infecting the TFV arm was not due to founder effects as sequences from the 2 groups (TFV and placebo) of the trial were interspersed on the phylogenetic tree, showing no lineage effects on the viruses infecting the 2 groups (see Supplemental Digital Content 1A, http://links.lww.com/QAI/B43). Instead minor subclustering of sequence pairs on the extant branches alone was evident in the tree and confirmed by a weak tree-based likelihood for clustering of −46.1 (P = 0.04).
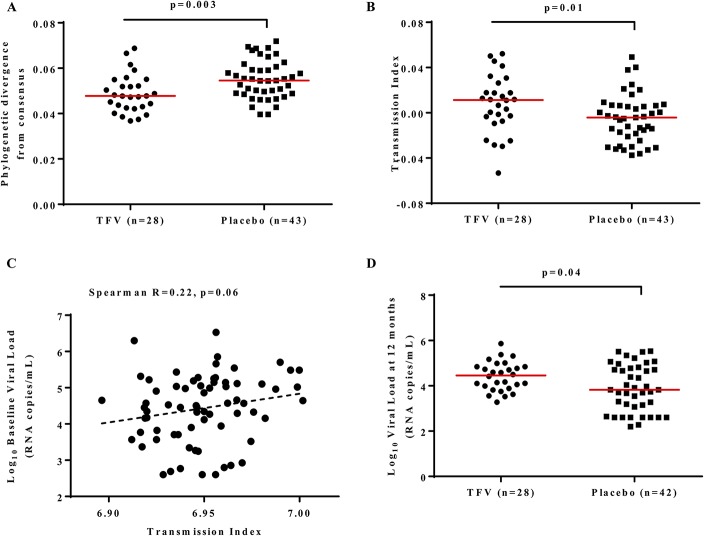
To assess whether the 1% TFV gel microbicide resulted in selection of viruses with altered transmission potential, we first generated a consensus sequence of subtype C viruses from the same geographical region. We then compared the phylogenetic divergence from the consensus sequence between the TFV and placebo viruses. HIV gag sequences from the TFV group were more similar to the consensus [phylogenetic distance median of 0.048 (IQR 0.043–0.054) compared with 0.055 (IQR 0.049–0.061) in placebo, P = 0.003; Mann-Whitney U test, Fig. 1A], suggesting preferential transmission of more consensus-like viruses in women who were assigned to the TFV arm. Similar results to the subtype C reference were found when we used the most recent common ancestor [Mann-Whitney U test P = 0.009 with TFV median of 0.097 (IQR 0.077–0.112) and placebo median of 0.108 (IQR 0.092–0.131)]. However we lost the signal when we used a subtype C global reference set from earlier in the epidemic (the 2004 HIV-1 subtype C ancestral sequence) as reference [Mann-Whitney U test for distance from the P = 0.528 with TFV median of 0.117 (IQR 0.102–0.130) and placebo median of 0.120 (IQR 0.108–0.128)].
FIGURE 1.
Comparison of phylogenetic divergence, transmission fitness and viral load between placebo and TFV arms. A, Comparison of phylogenetic divergence from the consensus subtype C reference sequence from the same geographic region. The phylogenetic distance median for TFV = 0.048 (IQR 0.043–0.054) and for placebo = 0.055 (IQR 0.049–0.061). B, Transmission index comparison between gag sequences from the TFV [median = 0.011 (IQR −0.006 to 0.024)] and those from the placebo [median = −0.004 (IQR −0.021 to 0.006)] recipients. C, Correlation between the baseline viral load and transmission index. D, Comparison of plasma viral loads at 12 months post infection between participants from the TFV [median viral load = 4.46 (IQR 4.06–4.78)] and placebo groups [median viral load = 3.83 (IQR 3.30–4.65)] (one participant from the placebo group did not have viral load data at or close to 12 months post infection).
The transmission index predicts the likelihood of a given amino acid sequence to be transmitted and is based primarily on how conserved observed amino acids are in a reference alignment.8 We found that viruses from the TFV recipients had higher transmission indexes, median 0.011 (IQR 0.006–0.024) compared with those from the placebo recipients—0.004 (IQR 0.021–0.006) (P = 0.01; Mann-Whitney U test, Fig. 1B). After 12 months post infection, the median transmission index of viruses in the placebo increased to a level indistinguishable from those in the TFV group (P = 0.9; see Supplemental Digital Content 1C, http://links.lww.com/QAI/B43). The transmission indexes of placebo viruses increased by a mean of 0.020 from early to 12 months post infection compared with a mean increase of 0.007 in TFV viruses (P = 0.004; Mann-Whitney U test).
We investigated whether the transmission index and phylogenetic divergence of the viruses were associated with markers of disease progression. There were no clear relationships between transmission index or phylogenetic divergence and viral load set-point or CD4 count (data not shown). However, we found a modest correlation between the transmission index and the baseline concomitant viral load (Spearman r = 0.22; P = 0.06, Fig. 1C). Similar to published data,7 the median viral loads for this subset of assessed samples were higher in the TFV group compared with the placebo group at 12 months post infection (Fig. 1D median log viral loads: TFV = 4.46 (IQR 4.06–4.78), placebo = 3.83 (IQR 3.30–4.65); P = 0.04; Mann-Whitney U).
Antibody avidity index was used to measure antibody maturation, with women assigned to the TFV group having slower antibody avidity maturation. We found a positive correlation between the transmission index and antibody avidity index (Spearman r = 0.36; P = 0.018) but only among the placebo group.
DISCUSSIONS
Theoretical models predict that increasing the barrier to infection will increase the likelihood that infection is established by a virus with relatively high transmission fitness.8,9 To the extent that transmission fitness is related to in vivo replication fitness, such increased selection pressure will result in higher viral loads (on average) among individuals at lower per-exposure risk of infection. In our cross-sectional study comparing viruses from recently infected women in the TFV and placebo groups of the CAPRISA 004 1% TFV gel microbicide trial, we report that viruses from TFV recipients were more consensus-like with a higher transmission index, suggesting that TFV increased selection pressure at the transmission barrier. However, the 2004 HIV-1 subtype C ancestral sequence is too far in the ancestral lineage (as seen from the large branch lengths) to be able to pick up these differences. Interestingly, there was a possible correlation between transmission index and the baseline viral load, consistent with previous reports linking within-host fitness to the similarity of host virus to circulating viruses.8,9 Thus the selection of viruses with higher transmission fitness may explain our previously reported finding of increased acute viremia among women in the TFV arm of the trial.7 Unfortunately data for adherence to TFV were not available for these study participants, to confirm differences in viral load between different adherence levels. It is interesting however that antibody avidity maturation was positively correlated with transmission fitness among the placebo. In the original study of antibody avidity,6 TFV was found to delay antibody avidity maturation and this study further confirms that it masks the natural relationship between transmission fitness and sensitivity to host antibodies.
These results are consistent with recent studies reporting differences in transmission selection bias based on per-exposure risk of infection, using models that estimate transmission fitness based on sequence features alone. In the context of heterosexual transmission in Zambia, less selection was observed among women and men with genital ulcers or inflammation (group with higher infection risk) compared with those without genital ulcers or inflammation.8 Similarly, founder viruses isolated from heterosexual men from North America had higher transmission indexes than those isolated from men who have sex with men, indicative of lower per-exposure risk of infection among the heterosexual men in that study.9 Here, because TFV is a nucleotide analog, it is effective only after viral entry through inhibition of HIV-1 reverse transcription.26 It has been shown that multiple infections per cell, as occurs during cell–cell transfer, can overcome the intracellular drug levels in the absence of drug resistance.27 It is possible that TFV exerted a “sieve” effect allowing for infection after exposure to a high number of infectious units that may occur in semen with a high infectious dose, donors with sexually transmitted infections, or through cell–cell transfer.
Reducing the barrier to infection allows establishment of infection by viruses with lower in vivo fitness. Consequently, such viruses will be under increased pressure to revert to higher fitness variants. Consistent with this model, faster reversion rates were previously observed among recently infected Zambian women compared with men.8 Similarly, in this study, transmission index increased at a faster rate among the placebo recipients, such that by 12 months post infection it was similar between the 2 groups. Such rapid reversion suggests that any negative effects of increased transmission selection bias will be short lived and is consistent with the convergence of viral load measured in these 2 arms over the 2 years after infection.7
As transmission fitness cannot be directly measured, we used 2 measures of sequence conservation as surrogates, which have previously shown positive correlation with transmission fitness.11,28 Similarly, conservation of circulating viruses have been observed to positively correlate with within-host fitness.12 It seems likely that conservation among circulating viruses is a result of evolutionary constraints during within-host evolution and on transmission, making sequence conservation a useful (though noisy) estimator for both transmission and within-host fitness.
Taken together, these and previous results support a model in which a side effect of incompletely reducing the per-exposure risk of infection is to increase the virulence among observed infections. Although theoretically this effect could lead to both higher viral load and faster disease progression, we have only observed a modest and transient increase in viral load. Although this modest impact may be due to incomplete adherence,1 it is also possible that increasing transmission selection bias, while measurable, has a subclinical impact that is largely mitigated by elevated reversion rates among high-risk subjects. However, even a subclinical impact on viral load may affect downstream transmission rates,29–31 which would reduce (but not eliminate) community-wide effectiveness of microbicide rollout. Monitoring and modeling these effects will be important for future PrEP and vaccine trials.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the CAPRISA 004 trial participants and the CAPRISA laboratory personnel. They also thank Dr Oliver Laeyendecker for providing the antibody avidity index data. Some computations were performed using computer cluster facilities provided by the University of Cape Town's ICTS High Performance Computing team (see http://hpc.uct.ac.za).
Footnotes
Supported by grants from India-South Africa collaborative grant provided through the South African Technology Innovation Agency (TIA), and the Centre for the AIDS Program of Research in South Africa (CAPRISA) to C.W. The CAPRISA 004/CAPRISA Tenofovir gel Research for AIDS Prevention Science (TRAPS) program was supported by the United States Agency for International Development and Family Health International (co-operative agreement #GPO-A-00-05-00022-00 contract #132119); the South African Department of Science & Technology (DST) and CONRAD (co-operative Grant #GP00-08-00005-00, subproject PPA-09-046). D.R.C. is a recipient of the South African National Research Foundation (NRF) Research Career Advancement Fellowship. N.K.N. was partially supported by the South African Medical Research Council.
Presented at the AIDS 2016 conference; July 18–22, 2016; Durban, South Africa.
The authors have no funding or conflicts of interest to disclose.
The views expressed by the authors are entirely their own and do not necessarily reflect those of their funding sources.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.jaids.com).
REFERENCES
- 1.Abdool Karim Q, Abdool Karim SS, Frohlich JA, et al. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329:1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehman DA, Baeten JM, McCoy CO, et al. Risk of drug resistance among persons acquiring HIV within a randomized clinical trial of single- or dual-agent preexposure prophylaxis. J Infect Dis. 2015;211:1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valley-Omar Z, Sibeko S, Anderson J, et al. CAPRISA 004 tenofovir microbicide trial: no impact of tenofovir gel on the HIV transmission bottleneck. J Infect Dis. 2012;206:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopera DR, Mann JK, Mwimanzi P, et al. No evidence for selection of HIV-1 with enhanced gag-protease or Nef function among breakthrough infections in the CAPRISA 004 tenofovir microbicide trial. PLoS One. 2013;8:e71758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mureithi MW, Poole D, Naranbhai V, et al. Preservation HIV-1-specific IFNgamma+ CD4+ T-cell responses in breakthrough infections after exposure to tenofovir gel in the CAPRISA 004 microbicide trial. J Acquir Immune Defic Syndr. 2012;60:124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laeyendecker O, Redd AD, Nason M, et al. Antibody maturation in women who acquire HIV infection while using antiretroviral preexposure prophylaxis. J Infect Dis. 2015;212:754–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrett NJ, Werner L, Naicker N, et al. HIV disease progression in seroconvertors from the CAPRISA 004 tenofovir gel pre-exposure prophylaxis trial. J Acquir Immune Defic Syndr. 2015;68:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson JM, Schaefer M, Monaco DC, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345:1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tully DC, Ogilvie CB, Batorsky RE, et al. Differences in the selection bottleneck between modes of sexual transmission influence the genetic composition of the HIV-1 founder virus. PLoS Pathog. 2016;12:e1005619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deymier MJ, Ende Z, Fenton-May AE, et al. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-alpha resistance. PLoS Pathog. 2015;11:e1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redd AD, Collinson-Streng AN, Chatziandreou N, et al. Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J Infect Dis. 2012;206:1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanini F, Brodin J, Thebo L, et al. Population genomics of intrapatient HIV-1 evolution. Elife. 2015;4:e11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann JK, Barton JP, Ferguson AL, et al. The fitness landscape of HIV-1 gag: advanced modeling approaches and validation of model predictions by in vitro testing. PLoS Comput Biol. 2014;10:e1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson AL, Mann JK, Omarjee S, et al. Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity. 2013;38:606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlson JM, Du VY, Pfeifer N, et al. Impact of pre-adapted HIV transmission. Nat Med. 2016;22:606–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prince JL, Claiborne DT, Carlson JM, et al. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog. 2012;8:e1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brockman MA, Brumme ZL, Brumme CJ, et al. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J Virol. 2010;84:11937–11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. [DOI] [PubMed] [Google Scholar]
- 19.Maljkovic Berry I, Athreya G, Kothari M, et al. The evolutionary rate dynamically tracks changes in HIV-1 epidemics: application of a simple method for optimizing the evolutionary rate in phylogenetic trees with longitudinal data. Epidemics. 2009;1:230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maljkovic Berry I, Ribeiro R, Kothari M, et al. Unequal evolutionary rates in the human immunodeficiency virus type 1 (HIV-1) pandemic: the evolutionary rate of HIV-1 slows down when the epidemic rate increases. J Virol. 2007;81:10625–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosakovsky Pond SL, Frost DW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. [DOI] [PubMed] [Google Scholar]
- 26.Rohan LC, Moncla BJ, Kunjara Na Ayudhya RP, et al. In vitro and ex vivo testing of tenofovir shows it is effective as an HIV-1 microbicide. PLoS One. 2010;5:e9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigal A, Kim JT, Balazs AB, et al. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature. 2011;477:95–98. [DOI] [PubMed] [Google Scholar]
- 28.Sagar M, Laeyendecker O, Lee S, et al. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis. 2009;199:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen MS, Chen YQ, McCauley M, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes JP, Baeten JM, Lingappa JR, et al. Determinants of per-coital-act HIV-1 infectivity among African HIV-1-serodiscordant couples. J Infect Dis. 2012;205:358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baeten JM, Kahle E, Lingappa JR, et al. Genital HIV-1 RNA predicts risk of heterosexual HIV-1 transmission. Sci Transl Med. 2011;3:77ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.