Abstract
Cell-autonomous immunity relies on the ubiquitin coat surrounding cytosol-invading bacteria functioning as an 'eat-me' signal for xenophagy. The origin, composition, and precise mode of action of the ubiquitin coat remain incompletely understood. Here, by studying Salmonella Typhimurium, we show the E3 ligase LUBAC to generate linear (M1-linked) polyubiquitin patches in the ubiquitin coat that serve as anti-bacterial and pro-inflammatory signaling platforms. LUBAC is recruited via its subunit HOIP to bacterial surfaces that are no longer shielded by host membranes and already displaying ubiquitin, suggesting LUBAC amplifies and refashions the ubiquitin coat. LUBAC-synthesized polyubiquitin recruits Optineurin and Nemo for xenophagy and local activation of NF-κB, respectively, which independently restrict bacterial proliferation. In contrast, the professional cytosol-dwelling Shigella flexneri escapes from LUBAC-mediated restriction through the antagonizing effects of the effector E3 ligase IpaH1.4 on the deposition of M1-linked poly-ubiquitin and the subsequent recruitment of Nemo and Optineurin. We conclude that LUBAC-synthesized M1-linked ubiquitin transforms bacterial surfaces into signaling platforms for anti-bacterial immunity reminiscent of anti-viral assemblies on mitochondria.
Mammalian cells maintain a sterile cytosol by deploying galectins to survey endomembrane damage and subsequently coat invading bacteria as well as damaged membranes with poly-ubiquitin 1. The bacterial ubiquitin coat comprises multiple linkage types, synthesized by several E3 ligases such as LRSAM1, Parkin, Smurf1 and RNF166 2–5. Galectin-8 and the poly-ubiquitin coat provide ligands ('eat-me' signals) for NDP52, Optineurin and other cargo receptors that induce anti-bacterial autophagy ('xenophagy') and restrict bacterial proliferation 1,6–10. Autophagy cargo receptors are not evenly dispersed around cytosol-invading bacteria 7,9,11. Such biased distribution of cargo receptors, sometimes referred to as 'microdomains', reflects the availability of their ligands, suggesting that recruitment signals and substrate specificity of individual E3 ligases are crucial to successful xenophagy.
Results
LUBAC synthesizes M1-linked ubiquitin chains on cytosolic S.Typhimurium to restrict bacterial proliferation
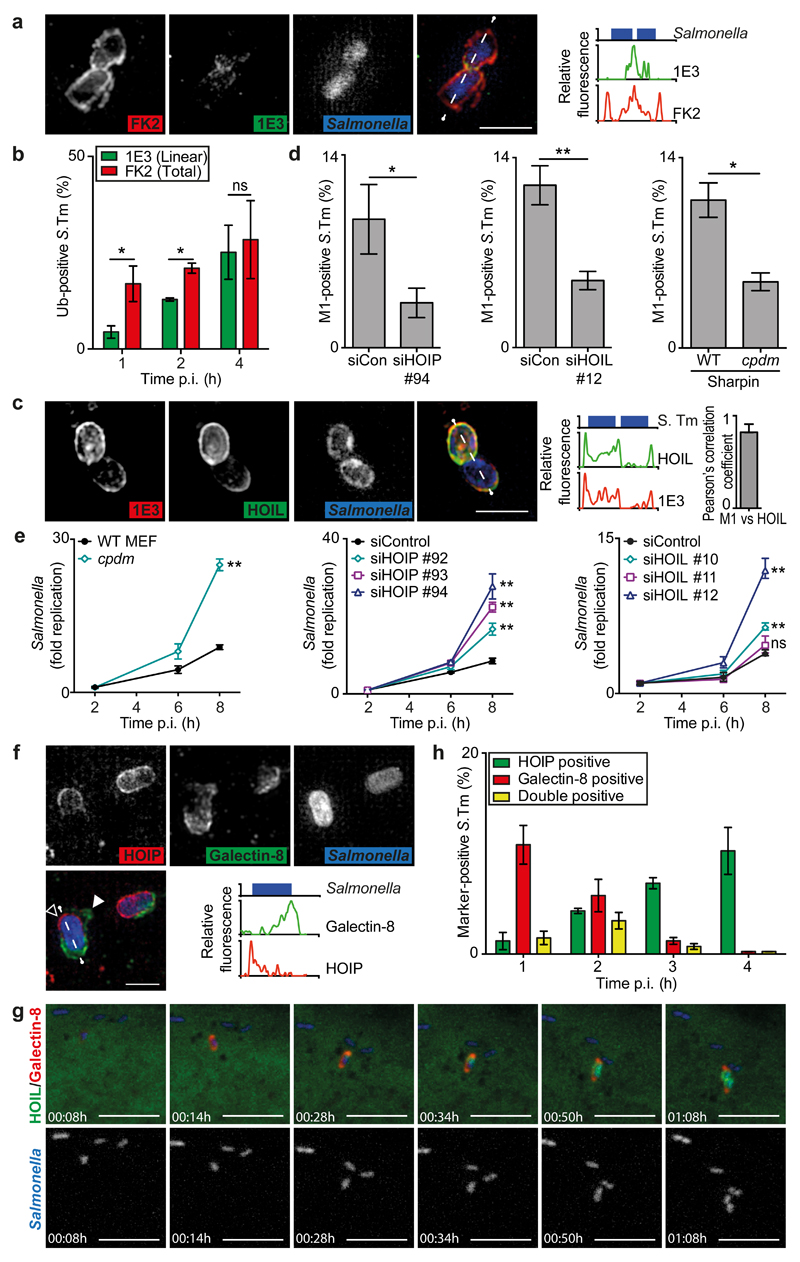
To understand microdomain origin we investigated the detailed topology of the ubiquitin coat of Salmonella enterica serovar Typhimurium (S.Typhimurium) in infected murine embryonic fibroblasts (MEFs) using Structured Illumination Microscopy (SIM), a superresolution technique. Co-staining with antibodies 1E3 and FK2, which recognize M1-linked ubiquitin or all linkage types except K6, respectively (Fig.S1), revealed that M1-linked ubiquitin is not homogenously distributed throughout the bacterial ubiquitin coat (Fig.1a) and that it is synthesized later than other chain types (Fig.1b,S2a). The existence of M1-linked ubiquitin patches suggests that specific E3 ligases, the availability of their substrates, and/or deubiquitinating enzymes ultimately determine the distribution of the ubiquitin-sensing cargo receptors. We next tested whether the synthesis of M1-linked ubiquitin chains on S.Typhimurium requires LUBAC, a multimeric E3 ubiquitin ligase comprised of HOIP, HOIL-1, and Sharpin, that is known to specifically form this linkage type12–15. Superresolution microscopy revealed extensive co-localization of M1-linked polyubiquitin with LUBAC (Fig.1c). Depletion of HOIP or HOIL-1 or a defect in Sharpin due to a spontaneous mutation (Sharpincpdm) substantially reduced the fraction of S.Typhimurium coated with M1-linked ubiquitin chains while not affecting the fraction of FK2-labelled bacteria (Fig.1d,S2b,S3a,b,c). We conclude that LUBAC contributes to the ubiquitin-coat of cytosol-invading S.Typhimurium by synthesizing M1-linked ubiquitin chains in a manner requiring all three LUBAC subunits.
Fig.1. LUBAC synthesizes M1-linked ubiquitin chains on cytosolic S.Typhimurium and restricts its proliferation.
(a,c,f) Structured illumination micrographs. MEFs infected with BFP-expressing S.Typhimurium, PFA-fixed at (a,c) 90min or (f) 1h post infection (p.i.) and stained for (a) total (FK2, red) and M1-linked (1E3, green) polyubiquitin, (c) co-expressing GFP:HOIL-1, Flag:HOIP and Flag:Sharpin and stained for M1-linked (1E3, red) polyubiquitin, or (f) expressing mCherry:HOIP and stained for Galectin-8 (false-coloured green); empty arrowhead indicates region with no galectin-8 and filled arrowhead indicates retracting Galectin-8+ve membrane. Line graphs show fluorescence plots along indicated lines. Data are representative of n>5 images from at least three independent experiments. Scale bar 2μm. (c) Far right graph Pearsons’s correlation coefficient of M1-linked polyubiquitin and HOIL. Mean±SD of n=5 images
(b,d,h) Percentage of marker-positive S. Typhimurium as counted by eye using widefield microscopy at 90 min p.i. or at the indicated time points in PFA-fixed MEFs (b,d) stained with antibodies 1E3 for M1-linked and FK2 for total polyubiquitin or (h) expressing GFP:HOIP and mCherry:galectin-8. Mean±SEM of triplicate coverslips from three independent repeats, n>100 bacteria per coverslip. *p<0.05, **p<0.01, Student’s t-test.
(e) Fold replication of S.Typhimurium in siRNA-treated or Sharpincpdm MEFs. Bacteria were counted based on their ability to grow on agar plates. Mean±SD of triplicate MEF cultures and duplicate colony counts, representing three independent repeats. ns=non-significant, **p<0.01, one-way ANOVA with Dunnett’s multiple comparisons test (siRNA-treated MEFs) or Student’s t-test (cpdm MEFs).
(g) Selected frames from live imaging on a confocal spinning disk microscope (see Supplementary Video 1) of MEFs co-expressing mCherry:Galectin-8, GFP:HOIL-1, Flag:HOIP and Flag:Sharpin, infected with BFP-expressing S.Typhimurium and imaged every 2 min. The shown event is representative of >9 videos from three independent experiments. Scale bar 10μm.
Since the ubiquitin coat of cytosol-invading bacteria activates anti-bacterial autophagy, we tested whether LUBAC is required to restrict the proliferation of S.Typhimurium (Fig.1e,S3a,b,c,S4). Bacteria proliferated significantly more in cells derived from Sharpincpdm mice, in MEFs depleted of HOIP or HOIL-1, as well as in human HOIP-/- epithelial cells, suggesting that all three subunits are required for LUBAC to protect cells against invading bacteria.
LUBAC is recruited to the surface of S. Typhimurium after escape from SCVs
Since the bacterial ubiquitin coat comprises both ubiquitylated bacterial surface proteins as well as host proteins on damaged Salmonella-containing vacuoles 16–18, we used galectin-8 as a marker of damaged endomembranes 7 to investigate the localization of LUBAC on cytosol-invading S.Typhimurium relative to the vacuolar membrane remnant (Fig.1f). Superresolution microscopy revealed that LUBAC is recruited to the bacterial surface and that galectin-8+ve membrane remnants 1,7, rather than attracting LUBAC, shield bacteria from it. Only where the membrane has been destroyed (open arrowhead) or where the membrane remnants retract (filled arrowhead) can LUBAC gain access to the bacterial surface.
Live microscopy directly confirmed that membrane damage and galectin-8 accumulation preceded LUBAC recruitment, which initially was confined to a discrete area in the gap of the galectin coat before spreading around the bacterium (Fig.1g, Video1). Notably, after bacterial division both daughter bacteria retained their LUBAC+ve status, suggesting that the bacterial ubiquitin coat provides a more durable signal than galectin-8+ve membranes. Consistent with this notion, the fraction of LUBAC+ve bacteria increased between 1h and 4h post infection, while the fraction of galectin-8+ve bacteria decreased (Fig.1h). Membrane remnants and LUBAC therefore provide two complementary signals to the cell — an immediate but relatively short lived cue that arises from the membrane remnant and a delayed but more sustainable one that originates from the bacterial surface, which are characterized by galectin-8 and M1-linked polyubiquitin, respectively.
HOIP recruits the LUBAC complex to pre-existing ubiquitin on cytosol-invading Salmonella
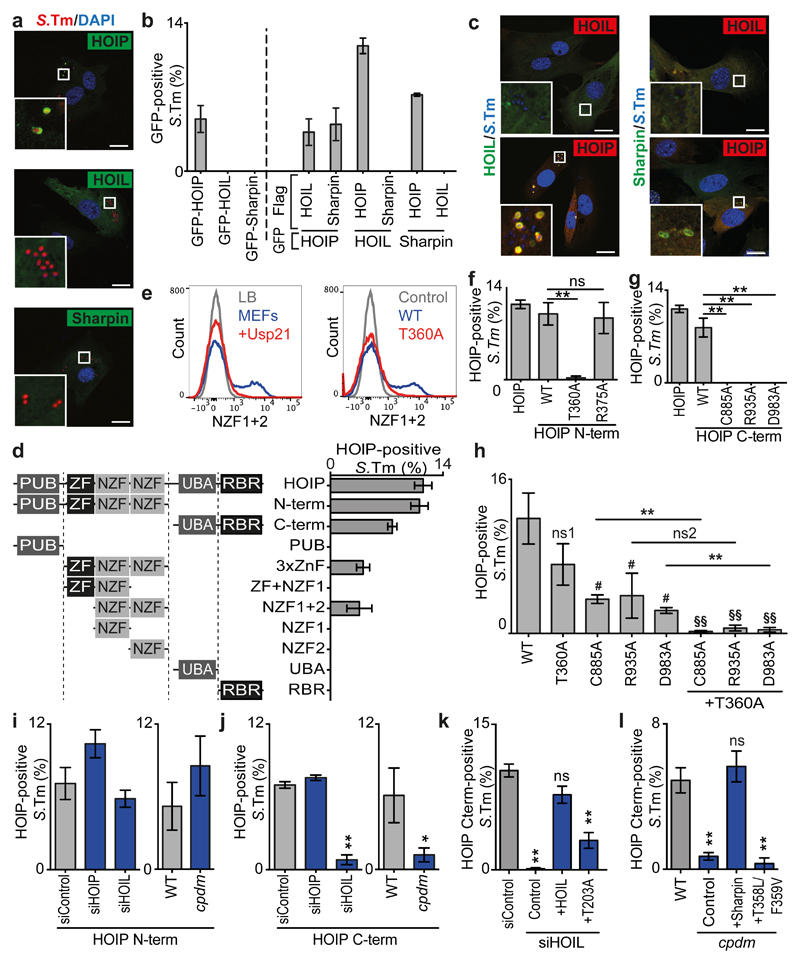
To understand how LUBAC is recruited to cytosol-invading S.Typhimurium, we expressed individual LUBAC subunits. Only GFP-HOIP, not GFP-HOIL-1 or GFP-Sharpin, accumulated on S.Typhimurium (Fig.2a,b,S5a). Recruitment of GFP-HOIP required the presence of endogenous HOIL-1, consistent with the need for HOIL-1 to stabilize HOIP (Fig.S5b) 13,14. Since GFP-HOIL-1 and GFP-Sharpin were recruited to S.Typhimurium only if co-expressed with FLAG-HOIP (Fig.2b,c,S5c), we conclude that HOIP recruits HOIL-1 and Sharpin to cytosol-invading S. Typhimurium.
Fig.2. HOIP senses and amplifies the ubiquitin coat of S.Typhimurium.
(a,c) Confocal micrographs of PFA-fixed MEFs infected with (a) mCherry-expressing or (c) DAPI-stained S.Typhimurium (blue) and expressing (a) GFP-tagged LUBAC subunits only or (c) co-expressing Flag-tagged LUBAC subunits (red) at 1h p.i.. Data are representative of three independent experiments. Scale bar 20μM. Split channels displayed in Fig.S2A,C.
(b,d,f-l) Percentage of marker-positive S. Typhimurium as counted by eye using widefield microscopy at 1h p.i. in PFA-fixed MEFs expressing (b) the indicated GFP- and FLAG-tagged LUBAC subunits or (d,f-l) GFP-tagged HOIP alleles. (i-l) MEFs from wild type or Sharpincpdm mice were treated with control siRNA or siRNA against the indicated murine LUBAC components and complemented with Flag-tagged human HOIP, HOIL-1 or Sharpin alleles as indicated. (b) Mean±SD of triplicate coverslips, representing two independent repeats. (d,f-l) Mean±SEM of triplicate coverslips from three independent repeats, n>100 bacteria per coverslip. *p<0.05, **p<0.01, (f,g,h,l) one-way ANOVA with Dunnett’s multiple comparisons test or (h,j) Student’s t-test. (h) # or ns1; compared to WT, §§; compared to T360A.
(e) Flow cytometry of S.Typhimurium grown in LB as indicated or extracted from MEFs (all other samples), treated with Usp21 as indicated, and stained with recombinant HOIP NZF1+2 (WT, blue), mutant allele (T360A, red) or secondary reagent only (Control, grey).
To further investigate how HOIP senses cytosol-invading S.Typhimurium we generated truncation mutants. HOIPN-term and HOIPC-term accumulated independently around S.Typhimurium, thus revealing the existence of two independent recruitment signals for HOIP (Fig.2d). Within HOIPN-term the double NZF (dNZF) domain was sufficient for recruitment to bacteria in cells. The purified dNZF also bound S.Typhimurium in vitro, but only if bacteria had been extracted from MEFs, not if they had been grown in Luria Bertani (LB) medium (Fig.2e,S6). The dNZF domain may therefore sense a host-derived ligand such as ubiquitin or Nemo, for which dNZF comprises binding sites 19. Treating host cell-extracted S.Typhimurium in vitro with the deubiquitinase USP21 to remove ubiquitin precluded binding of dNZF, as did a mutation in the ubiquitin binding site of NZF1 (T360A) 19, which prevented binding of dNZFT360A and HOIPN-termT360A to bacteria in vitro and in cells, respectively (Fig.2e,f). In contrast, HOIPN-termR375A, deficient in binding to Nemo 19, was recruited at wild type levels (Fig.2f). We therefore conclude that the dNZF domain recruits HOIP to cytosol-invading S.Typhimurium in a ubiquitin-dependent manner. Within the C-terminal fragment both the UBA and the RBR domain are required for recruitment to S.Typhimurium as deletion of either domain prevented accumulation of HOIPC-term around bacteria (Fig.2d). The RBR domain provides E3 ligase activity; it contains the catalytic cysteine C885 and binding sites for the acceptor and donor ubiquitins of the M1-linked polyubiquitin chain under synthesis, which are inactive in HOIPR935A and HOIPD983A, respectively 1,6–9,20. We found that HOIPC-term C885A, R935A, and D983A were not recruited to S.Typhimurium (Fig.2g). Catalytic activity is therefore essential for HOIPC-term to accumulate on cytosol-invading bacteria.
We next tested the effect of the inactivating point mutations on full length HOIP (Fig.2h). Individually inactivating the ubiquitin binding domain (HOIPT360A) or the catalytic domain (HOIPC885A, HOIPR935A, or HOIPD983A) had partial effects on HOIP recruitment, whereas T360A in combination with the catalytic mutations completely abrogated HOIP accumulation on S.Typhimurium. We conclude that the efficient recruitment of HOIP relies on two cooperating modi operandi, namely HOIP’s ability to bind and synthesize ubiquitin chains via its N- and C-terminus, respectively.
The nature of the two recruitment mechanisms for HOIP revealed above suggests the potential existence of a feed-forward loop, which inspired us to investigate whether the accumulation of either of HOIP’s termini is independent of other LUBAC subunits. Using siRNA resistant HOIP fragments, we observed that the recruitment of HOIPN-term occurred equally efficiently in control cells, cells from Sharpincpdm mice, or cells depleted of HOIL-1 or HOIP (Fig.2i). We conclude that the ubiquitin-mediated recruitment of HOIPN-term is not dependent on LUBAC activity, thus revealing the existence of an upstream E3-ubiquitin ligase mediating LUBAC recruitment. Based on the binding specificity of HOIPdNZF to ubiquitin chain types in vitro (Fig.S7), the upstream E3 ligase may produce K63 chains but depletion of ligases previously implicated in LUBAC activation during NF-κB signaling (cIAP1, cIAP2, XIAP, Traf3, Traf6) or ubiquitin-coating of cytosol-invading bacteria (LRSAM1, Parkin) affected neither the recruitment of HOIPN-term nor the deposition of M1-linked ubiquitin chains (Fig.S3d,e,S8). In contrast to HOIPN-term, the recruitment of HOIPC-term required HOIL-1 and Sharpin (Fig.2i,j), as well as catalytic activity in HOIP (Fig.2g). Both HOIL-1 and Sharpin contain ubiquitin-binding domains (UBDs) 13,21,22 that were individually required for the efficient recruitment of HOIPC-term to S.Typhimurium (Fig.2k,l). Taken together, our data reveal two ubiquitin-dependent recruitment mechanisms for LUBAC, namely i) to pre-existing ubiquitin chains via HOIPdNZF and ii) to LUBAC synthesized M1-linked ubiquitin chains via HOIL-1 and Sharpin, thereby stabilizing existing and, importantly, recruiting new LUBAC complexes in a feed-forward mechanism. A ubiquitin-centered feed-forward loop also occurs during mitophagy, where the kinase PINK1 activates the E3 ligase Parkin by phosphorylating Parkin as well as Parkin-synthesized ubiquitin chains, an allosteric activator of Parkin23. However, in contrast to CCCP-treated, depolarized mitochondria, no phospho-Ser65-ubiquitin accumulated on S.Typhimurium (Fig.S9). Taken together, we propose a model in which HOIP is recruited to cytosol-invading S.Typhimurium via two cues; the primary provided by an upstream E3 ligase and sensed by the ubiquitin-binding dNZF domain of HOIP, the secondary comprising a feed-forward loop of LUBAC synthesizing and binding M1-linked polyubiquitin.
LUBAC-synthesized M1-linked polyubiquitin recruits Nemo and Optineurin to cytosol-invading bacteria
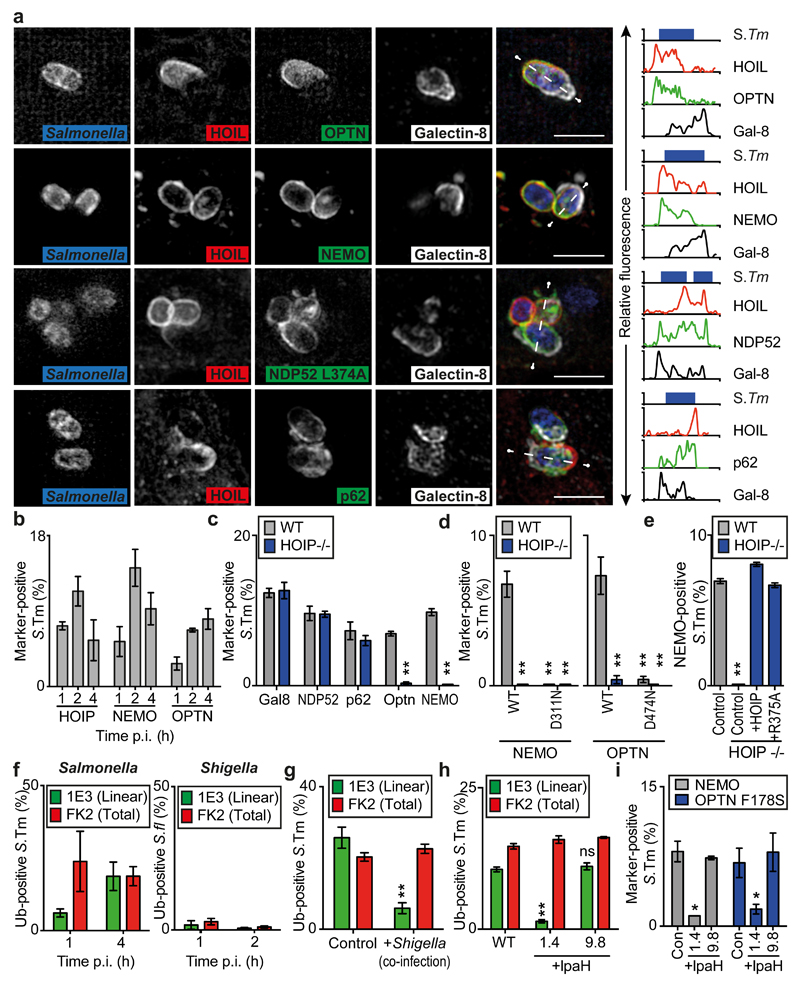
To investigate how LUBAC restricts bacterial proliferation and affects host cell behavior, we investigated the LUBAC-dependent recruitment of cytosolic ubiquitin receptors. Superresolution microscopy revealed that NDP52 and p62 only partially overlapped with LUBAC on bacteria and were also recruited to LUBAC-ve, galectin-8+ve areas (Fig.3a,S10a). In contrast, the linear ubiquitin binder Nemo 24 and the related ubiquitin binding protein Optineurin coincided selectively with LUBAC and were, like LUBAC, excluded from galectin-8+ve areas. Nemo, Optineurin and HOIP were present continuously at S.Typhimurium over the course of an infection (Fig.3b). Using HOIP-/- human epithelial cells we found that Nemo and Optineurin, but not galectin-8, NDP52, and p62, required LUBAC for their recruitment to S. Typhimurium (Fig.3c,S3f). Nemo and Optineurin are recruited via ubiquitin, since alleles deficient in ubiquitin binding (NemoD311N, OptnD474N)25 failed to accumulate on S. Typhimurium (Fig.3d). In contrast, direct contacts of Nemo with HOIP are not essential, since HOIPR375A, an allele deficient in Nemo binding19, complemented HOIP-/- cells (Fig.3e). In murine cells the conjugation of LC3 to the membrane of the damaged Salmonella-containing vacuole (SCV) occurs independently of macro-autophagy, thus providing an alternative ligand for Optn26. Revealing the essential contribution of LUBAC to the recruitment of Optn therefore required inactivation of its LC3-binding LIR motif (OptnF178S)9, while p62 and NDP52 recruitment was independent of LUBAC even if their LIR motifs and the NDP52 galectin-8 binding site were destroyed (p62DDDW335-338AAAA, NDP52V136S,L374A)27–29 (Fig.S10b-e). We conclude that Optineurin and Nemo are effector proteins of LUBAC, which via M1-linked ubiquitin chains provides the critical signal for their recruitment to cytosol-invading S.Typhimurium.
Fig.3. LUBAC recruits Optineurin and Nemo to S.Typhimurium.
(a) Structured illumination micrographs. MEFs infected with BFP-expressing S.Typhimurium co-expressing the indicated GFP-tagged proteins and mCherry:HOIL-1, Flag:HOIP and Flag:Sharpin were PFA-fixed at 1h p.i. and stained for Galectin-8 (white). NDP52 constructs lack SKICH domain to prevent aggregation. Graphs show fluorescence plots along indicated line. Data are representative of n>5 images from three independent experiments. Scale bar, 2μM
(b-e) Percentage of marker-positive S. Typhimurium as counted by eye using widefield microscopy at 1h p.i. in PFA-fixed (b) MEFs or (c-e) wild type or HOIP−/− HCT116 cells expressing GFP-tagged alleles of HOIP, Nemo, Optn or antibody-stained for Galectin-8, NDP52 or p62. (e) HOIP expression was complemented with Flag-tagged HOIP alleles as indicated. Mean±SEM of triplicate coverslips from three independent repeats, n>100 bacteria per coverslip. *p<0.05, Student’s t-test.
(f-i) Percentage of marker-positive S.flexneri or S.Typhimurium as counted by eye using widefield microscopy at 1h p.i. in PFA-fixed MEFs (f-h) stained with antibodies 1E3 for M1-linked and FK2 for total polyubiquitin or (i) expressing the indicated GFP-tagged genes.. Stable expression of Flag-tagged IpaH genes as indicated. In co-infections S.Typhimurium were identified by expression of mCherry. Mean±SEM of triplicate coverslips from three independent repeats, n>100 bacteria per coverslip. *p<0.05, **p<0.01, Student’s t-test.
The Shigella-encoded E3 ligase IpaH1.4 antagonizes LUBAC-mediated recruitment of Nemo and Optineurin to cytosol-invading bacteria
Next we tested whether Shigella flexneri, a Gram-negative bacterium highly adapted to the cytosolic environment, has evolved countermeasures against LUBAC-mediated deposition of M1-linked ubiquitin chains and restriction of proliferation. Similar to S.Typhimurium, S.flexneri recruited LUBAC in a HOIP-dependent manner (Fig.S11a,b). However, in contrast to S.Typhimurium, depletion of HOIP did not significantly affect proliferation of S.flexneri (Fig.S11c), suggesting that S.flexneri indeed antagonizes LUBAC-mediated restriction. We therefore compared the deposition of M1-linked ubiquitin chains on both bacteria and found that a much smaller fraction of S.flexneri was labeled with M1-linked ubiquitin chains (Fig.3f). Note that S.flexneri enters the cytosol almost quantitatively, while most S.Typhimurium remain membrane enclosed. To distinguish whether S.flexneri avoids recognition by the ubiquitylation machinery or whether it actively antagonizes ubiquitylation, we performed co-infection experiments. The deposition of M1-linked ubiquitin chains on S.Typhimurium was lower in cells co-infected with S.flexneri, revealing that S.flexneri actively antagonizes the accumulation of M1-linked ubiquitin chains even in trans on cytosol-invading S.Typhimurium (Fig.3g), possibly by secreting LUBAC-targeting effector proteins. Consistent with a recent report on S.flexneri suppressing NF-κB signaling by targeting LUBAC for degradation via IpaH1.4, a secreted E3 ubiquitin ligase 30, we found that in cells transduced with IpaH1.4 but not IpaH9.8 fewer S.Typhimurium became positive for M1-linked ubiquitin chains and recruited Nemo or Optineurin (Fig.3h,i). Active antagonism of M1-linked polyubiquitin coating of bacterial surfaces by S.flexneri indicates the evolutionary importance of the LUBAC pathway for the cell-autonomous defense against cytosol-invading bacteria.
Bacteria coated with M1-linked polyubiquitin activate NF-κB
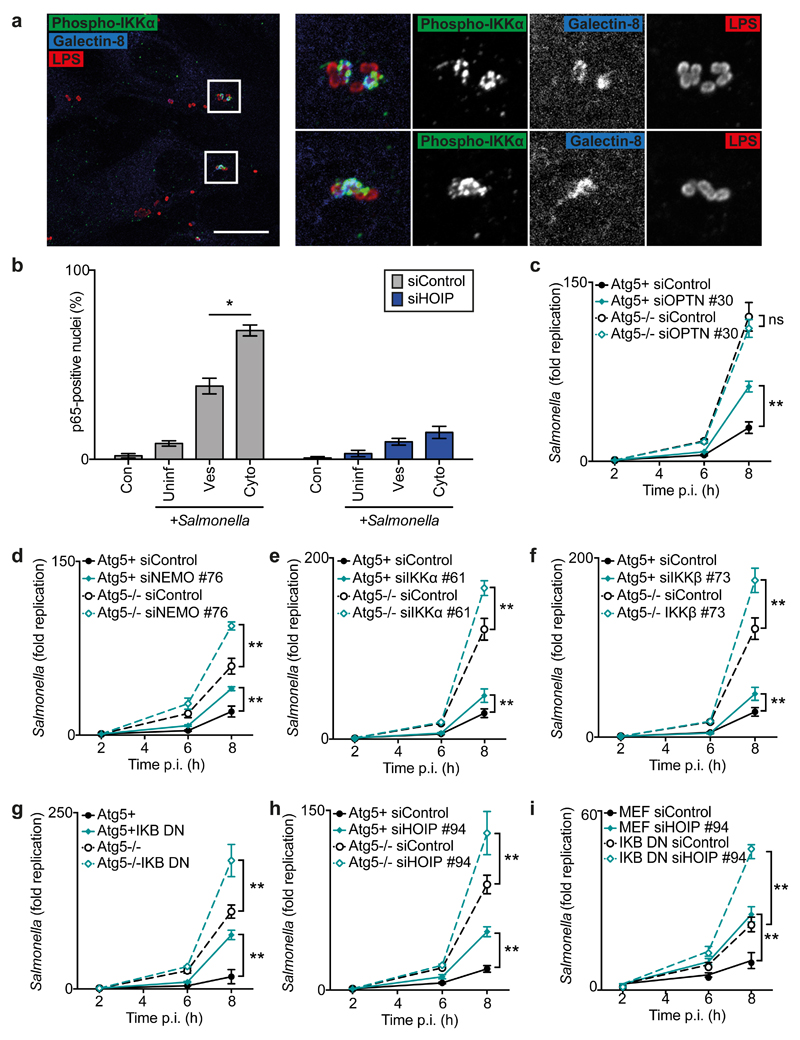
The generation of M1-linked polyubiquitin on cytosol-invading bacteria by LUBAC and the subsequent recruitment of Nemo suggest that such bacteria may directly induce NF-κB. To test whether the IκB kinase (IKK) complex is recruited to and activated on cytosol-invading bacteria, we stained Salmonella-infected cells with phospho-Ser176/180-IKKα, a marker of activated IKKα. Phospho-IKKα was specifically observed on galectin-8+ve bacteria, i.e. those exposed to the cytosol, and labelled 16.6±1.4% of intracellular S.Typhimurium at 1h post infection (p.i.) (Fig.4a). To investigate whether S.Typhimurium induces NF-κB specifically upon entering the host cytosol, we evaluated the nuclear translocation of the NF-κB subunit p65 in cells exposed to extracellular, vacuole-contained, or cytosol-invading bacteria. At 1h p.i. 68±5% of cells containing at least one galectin-8+ve (i.e. cytosolic) bacterium had activated NF-κB, compared to only 39±7.2% of cells where bacteria had remained in SCVs, and 8±2.4% of cells without intracellular bacteria (Fig.4b). Treatment with siRNA against HOIP abrogated the induction of NF-κB. In contrast, lack of RipK2, an essential component of the peptidoglycan-sensing NOD pathway31, had no effect on the recruitment of HOIP, the deposition of M1-linked ubiquitin chains, the recruitment of phospho-Ser176/180-IKKα, or the proliferation of S.Typhimurium (Fig.S3g,h,S12). We conclude that by depositing M1-linked polyubiquitin, LUBAC transforms the bacterial surface into a pro-inflammatory signaling platform for the local activation of NF-κB.
Fig.4. LUBAC activates autophagy and NF-κB, which independently restrict cytosolic S.Typhimurium.
(a) Confocal micrographs of methanol-fixed MEF cells at 1h p.i. with S.Typhimurium, stained with antibodies against LPS (red), phosphorylated IKKα (green) and Galectin-8 (blue). Data are representative of three independent experiments. Scale bar, 20μM.
(b) Percentage of p65-positive nuclei as counted by eye using widefield microscopy at 1h p.i. in siRNA-treated PFA-fixed MEFs infected with GFP-expressing S.Typhimurium and stained for p65 and Galectin-8. Cells in infected sample (+Salmonella) were classified by bacterial status: no bacteria (Uninfected, ‘Uninf’), Galectin-8-ve bacteria only (Vesicular, ‘Ves’) or ≥1 Galectin-8+ve bacteria (Cytosolic, ‘Cyto’). Mean±SEM of triplicate coverslips from three independent repeats. n>100 bacteria counted per coverslip.
(c-i) Fold replication of S.Typhimurium in MEFs treated with indicated siRNAs against (c) Optn, (d) Nemo, (e) IKKα, (f) IKKβ, (h-i) HOIP or (g,i) expressing I-κBαSS32,36AA (I-κBα DN). Bacteria were counted based on their ability to form colonies on agar plates. Mean±S.D. of triplicate MEF cultures and duplicate colony counts, representing two (e) or three (c-d,f-i) independent repeats. **p<0.01, Student’s t-test, (c-i) calculated for 8h timepoint.
LUBAC activates autophagy and NF-κB, which independently restrict proliferation of cytosolic S. Typhimurium
We next investigated the contribution of the LUBAC effectors Optineurin and Nemo to LUBAC-mediated restriction of Salmonella proliferation. We found that Optineurin, as reported previously 9, is required to antagonize S.Typhimurium and that, unexpectedly, cells depleted of Nemo also failed to restrict bacterial growth (Fig.S3i,j,S13a,b). We therefore tested whether Nemo-mediated restriction of bacterial growth depends on the other subunits of the IKK complex and found that in cells depleted of IKKα or IKKβ S.Typhimurium also hyperproliferated (Fig.S3k,l,S13c,d). We therefore conclude that Optineurin and Nemo, as well as IKKα and IKKβ, protect cells against bacterial proliferation. To investigate whether the IKK complex, like Optineurin9, restricts bacterial proliferation in an autophagy-dependent manner, the IKK subunits were depleted in autophagy-proficient and -deficient cells. Consistent with Optineurin protecting cells via autophagy, depleting Optineurin increased bacterial proliferation only in wild type cells, not in autophagy-deficient ATG5-/- cells (Fig.4c). In contrast, depleting Nemo, IKKα, or IKKβ enhanced bacterial proliferation in both wild type and autophagy-deficient ATG5-/- cells (Fig.4d-f), revealing that the IKK complex protects cells against S.Typhimurium in an autophagy-independent fashion. To test whether the anti-bacterial activity of the IKK complex is manifested via NF-κB we deployed I-κBαSS32,36AA, a dominant negative inhibitor of NF-κB (Fig.S14). Expression of I-κBαSS32,36AA enhanced proliferation of S.Typhimurium in autophagy-proficient and -deficient ATG5-/- cells (Fig.4g), revealing that NF-κB, like the IKK complex, protects cells in an autophagy-independent fashion. Taken together, our data suggests that LUBAC coordinates two pathways of cell-autonomous anti-bacterial defense, namely Optineurin-induced anti-bacterial autophagy and Nemo-controlled NF-κB activation. If correct, LUBAC should appear non-epistatic to either pathway. To test this hypothesis, we depleted HOIP from wild-type, autophagy-deficient ATG5-/-, and I-κBαSS32,36AA-expressing cells (Fig.4h,i). HOIP antagonized bacterial proliferation in either scenario, consistent with its proposed role at the branching point of Optineurin-induced anti-bacterial autophagy and Nemo-controlled NF-κB activation.
Discussion
Our data reveal an essential role for LUBAC in coordinating the cell-autonomous defense against cytosol-invading S.Typhimurium. By synthesizing M1-linked polyubiquitin on bacteria LUBAC transforms the bacterial surface into a multivalent signaling platform that coordinates two cellular defense pathways, namely anti-bacterial autophagy and NF-κB signaling, for which we demonstrate a direct anti-bacterial function independent of autophagy.
The poly-ubiquitin coat on the surface of cytosol-invading bacteria, first described in 20046, has so far been considered a homogenous entity. Although different linkage types are known to occur within the bacterial ubiquitin coat 4,5,32,33, information on the spatial distribution of specific linkage types is not available. Our data reveal that M1-linked poly-ubiquitin localizes to the bacterial surface, but not to membrane remnants of the burst SCV, and that M1-linked ubiquitin chains can occur in distinct islands within the poly-ubiquitin coat. The origin of M1-linked poly-ubiquitin islands remains unknown but is most likely caused by the localized recruitment of LUBAC (as seen in Video 1), the accessibility of LUBAC substrates (for example when SCV remnants shield the bacterial surface as seen in Fig.1F), and / or the antagonizing effects of Otulin (see the accompanying manuscript by van Wijk et al.).
LUBAC recruitment to Gram-negative bacteria is mediated by the N-terminal double NZF domain (dNZF) of HOIP. HOIPdNZF binds ubiquitin on the bacterial surface and accumulates on cytosol-invading bacteria even in cells lacking LUBAC function, indicating that LUBAC refashions an existing ubiquitin coat and that an upstream E3 ligase may function as a novel pattern recognition receptor to direct LUBAC to cytosol-invading bacteria. In addition to recruitment via HOIPdNZF, LUBAC also accumulates in a manner requiring the catalytic activity of HOIP and the ubiquitin-binding domains in Sharpin and HOIL-1, suggesting that M1-linked ubiquitin chains synthesized by LUBAC anchor LUBAC in the bacterial vicinity and, importantly, may recruit further LUBAC molecules. The resulting amplification mechanism is functionally reminiscent of Parkin-mediated ubiquitylation of damaged mitochondria leading to mitophagy 23, although no phospho-Ser65-ubiquitin was detected on cytosol-invading S. Typhimurium.
LUBAC synthesized M1-linked ubiquitin chains on the bacterial surface activate anti-bacterial autophagy and NF-kB signalling. We therefore propose that by synthesizing M1-linked poly-ubiquitin, LUBAC transforms the bacterial surface into an anti-bacterial and pro-inflammatory signaling platform that by its polyvalent nature efficiently recruits adaptor proteins and initiates cellular signaling pathways. M1-linked ubiquitin chains are essential for the recruitment of Optineurin and Nemo, which activate selective autophagy and recruit the IKK complex, respectively. The LUBAC-dependent occurrence of phosphorylated and hence catalytically active IKK on the surface of cytosol-invading bacteria reveals a previously unrecognized principle of IKK activation distinct from receptor-mediated IKK activation. Further work will reveal whether upstream components typically required for receptor-mediated NF-kB activation, for example TAK1, are also recruited and activated on cytosol-invading S. Typhimurium. In light of the evolutionary relationship between bacteria and mitochondria, the functional parallels between signaling platforms on bacteria and anti-viral assemblies on mitochondria seem intriguing and worthy of further comparison 34,35.
The potent anti-bacterial function of LUBAC-synthesized M1-linked ubiquitin chains against cytosol-invading S. Typhimurium suggests that professional cytosol-dwelling bacteria may have evolved efficient means to evade or antagonize the pathway. We observed that S. flexneri, a Gram-negative enterobacterium like S. Typhimurium, is not restricted by LUBAC and that S. flexneri deploys IpaH1.4, a member of the multi-gene family of secreted IpaH E3 ligases, to antagonize the accumulation of M1-linked ubiquitin chains on bacterial surfaces, as well as the recruitment of Optineurin and Nemo. The effect of IpaH1.4 on the coating of bacteria with M1-linked ubiquitin chains is most likely related to its recently discovered ability to target LUBAC for degradation 30. Importantly, protection against the coating of bacterial surfaces with M1-linked ubiquitin provided by IpaH1.4 extends in trans to co-infecting bacteria, as demonstrated here for S. Typhimurium, suggesting that S. flexneri profoundly cripples LUBAC-dependent cellular defense mechanisms with potentially far reaching consequences for the outcome of co-infections.
Methods
Antibodies
Antibodies were from Enzo Life Science (ubiquitin FK2), Millipore (linear ubiquitin 1E3, Phospho-ubiquitin Ser65), R&D Systems (Galectin-8, cIAP2 for western blots), AbD Serotec (LPS), Invitrogen (Phospho-IKKa Ser176 Ser180 and all Alexa-conjugated anti-mouse and anti-rabbit antisera), Abcam (HOIP, β-actin, Park2 for western blots), Santa Cruz (RipK2 and HOIL-1, for western blots), Cayman chemicals (Optineurin, for western blots), BD Biosciences (p62 and Nemo, for western blots), Imgenex (IKKα, IKKβ, for western blots) and Dabco (HRP-conjugated reagents). NDP52 antisera was a kind gift from John Kendrick-Jones.
Plasmids
M5P or closely related plasmids were used to produce recombinant MLV for the expression of proteins in mammalian cells36. Open reading frames encoding human LUBAC subunits, Galectin-8, NDP52, p62, Optineurin, Nemo and IκBα were amplified by PCR. Mutations were generated by PCR and verified by sequencing. For truncations, the following domain boundaries were used: HOIP N-term (aa 1-438), HOIP C-term (aa 480-1072), HOIP PUB (aa 1-294), HOIP 3xZnF (aa 295-438), HOIP ZF+NZF1 (aa 295-379), HOIP NZF1+2 (aa 349-438), HOIP NZF1 (aa 330-379), HOIP NZF2 (aa 380-439), HOIP UBA (aa 439-636) and HOIP RBR (aa 693-1072). For imaging purposes, NDP52 lacking the SKICH domain (ΔN127) was used to prevent unwanted aggregation. For IκBα-DN, mutations S32A and S36A were introduced.
For recombinant protein production, HOIP NZF1+2 was amplified by PCR and cloned into a pETM30 vector encoding an N-terminal 6xHis-GST tag. Biotinylated HOIP NZF1+2 was cloned using primers encoding a C-terminal BirA biotinylation site into a modified pET11 vector with a ribosomal binding site followed by BirA.
Protein purification
Proteins were expressed in the E.coli strain BL21. Bacteria were grown to an OD600 of 0.7 at 37 °C before overnight induction at 16°C in the presence of 100μM IPTG (for biotinylated proteins, 1mM Biotin was added). Cells were mechanically lysed in lysis buffer (20mM Tris (pH 8.0), 150mM NaCl, 1mM DTT, protease inhibitors (Roche)) and cleared by centrifugation before incubation with equilibrated Ni-NTA resin for 2h at 4°C. Protein was then washed (20mM Tris pH 8.0, 300mM NaCl, 30mM imidazole and 1mM DTT) and eluted with elution buffer (20mM Tris pH 8.0, 150mM NaCl, 300mM imidazole and 1mM DTT) before dialysis overnight in the presence of 6xHis-TEV protease in gel filtration buffer (20mM Tris pH 8.0, 150mM NaCl and 1mM DTT). To remove the 6xHis-TEV protease and the cleaved 6xHis-GST the sample was passed through to 5ml Ni-NTA column. Flow-through was concentrated to 1ml and loaded on a Superdex 75 column equilibrated in gel filtration buffer. Protein containing fractions were pooled, concentrated and stored at -80°C.
Cell culture
Cells were grown in IMDM supplemented with 10% FCS at 37 °C in 5% CO2. All cell lines tested negative for mycoplasma. Stable cell lines were generated by retroviral transduction and selected for by antibiotic resistance. Where applicable, individual proteins carried unique resistance markers to ensure successful co-expression. Wild type and Sharpincpdm MEFs from obtained from Henning Walczack, ATG5−/− MEFs 37 from N. Mizushima, wild type, HOIP−/− and RipK2−/− HCT116s from Mads Gyrd-Hansen. Verification of genotypes is shown in Fig.S3.
Bacteria
S.Typhimurium (strain 12023), provided by D. Holden, was grown overnight in Luria broth (LB) and sub-cultured (1:33) in fresh LB for 3.5 h before infection. Such cultures were further diluted (1:5) in antibiotic-free IMDM plus 10% FCS immediately before 10μl was used to infect MEF cells in 24-well plates for 10 min at 37 °C. Following two washes with warm PBS, cells were cultured in 100 μg ml−1 gentamycin for 2 h and 20 μg ml−1 gentamycin thereafter. For p65 translocation assays, bacteria were washed 1x in warm antibiotic-free IMDM, resuspended in the same volume and 10ul used to infect cells. To enumerate intracellular bacteria, cells from triplicate wells were lysed in 1 ml cold PBS containing 0.1% Triton-X-100. Serial dilutions were plated in duplicate on TYE agar.
Extraction of S. Typhimurium from infected cells
A 10cm dish of mouse embryonic fibroblasts was infected with mCherry-expressing S.Typhimurium. After 3h, cells were washed twice with PBS and detached with trypsin. Cells were washed twice with PBS + 2% FCS and lysed in 1 ml lysis buffer (20mM Tris pH 7.4, 150mM NaCl, 1% Triton X-100 and 1mM Iodoacetamide) for 30 minutes at 4°C. Bacteria were recovered by centrifugation (5 min. at 13,000 rpm at 4°C) and washed twice with reaction buffer (20mM Tris pH 7.4, 150mM NaCl and 2mM DTT). DNase I (Sigma) was added to a final concentration of 100μg/ml before incubation for 30 minutes at 37°C. Bacteria were washed with reaction buffer and incubated at 37°C in the presence of buffer or Usp21 (0.2 mg/ml) for 30 minutes at 37°C. After washing with PBS + 2% FCS the bacteria were stained with primary antibodies and/or recombinant, biotinylated HOIP NZF1+2 (5μg/ml) for 30 minutes on ice.
After washing with PBS + 2% FCS bacteria were stained with fluorescently labelled secondary antibodies and/or Alexa488 labelled Streptavidin for 30 minutes on ice. Bacteria were fixed in 2% Paraformaldehyde for 30 minutes at room temperature and analysed on a LSRII flow cytometer.
RNA interference
5 × 104 cells per well were seeded in 24-well plates. The following day, cells were transfected with 6 pmol of siRNA (Invitrogen) using Lipofectamine RNAiMAX (Invitrogen) in Optimem medium (Invitrogen). Optimem was replaced with complete IMDM medium after 4 h and experiments were performed after 3 days. For ATG5−/− MEFs, 5 × 103 cells were seeded initially and siRNA transfection was repeated 2 days after first siRNA treatment.
siRNAs targeted the following sequences:
siHOIP #92 5′-CGGCAUUGACUGUCCGAAA;
siHOIP #93 5′-ACUUCACCAUUGCCCUGAA;
siHOIP #94 5′-GACCCUAACUGCAAGGUGA;
siHOIL-1 #10 5′-CUGCGAAUGUUGGAAGAUU;
siHOIL-1 #11 5′-GGGUGCAAGUAAAACCCGA;
siHOIL-1 #12 5′-ACACGUCACUCAACCCACA;
siSharpin #84 5′-GCUCAAAUACCUAAAGCAA;
siSharpin #85 5′-CCACCCACAUUGCUCCAUU;
siSharpin #86 5′-CUUUCAUCAAUGCCUCAAA;
siOptn #29 5′-GAAGCUAAAUAAUCAAGCU;
siOptn #30 5′-GCCUCGCAGUAUUCCGAUU;
siOptn #31 5′-CAAUUGAAGAACUAACCAA;
siNemo #76 5′-GGAUUGAGGAUAUGAGGAA;
siNemo #77 5′-GGAUUCGAGCAGUUAGUGA;
siNemo #78 5′-AGGCCUCUGUGAAAGCUCA;
siIKKα #61 5′-GAAAGAUCCAAAGUGUAUA;
siIKKα #62 5′-CUAUUGAUCUUACUUUGAA;
siIKKβ #73 5′-CGUUGGACAUGGAUCUUGU;
siIKKβ #75 5′-GAAGAUCGCCUGUAGCAAA;
siLRSAM1 #93 5’-GAAUAAGAAUGGAGCAGUU;
siLRSAM1 #94 5’-GAACCAGAUUAGGCUAAUA;
siPARK2 #27 5’-GGAACAACAGAGUAUUGUA;
siPARK2 #28 5’-GGAGGAUGUAUGCACAUGA;
siTRAF3 #28 5’-GGUCUACUGUCGGAAUGAA;
siTRAF3 #29 5’-GGUACAAACCAGCAGAUCA;
siTRAF6 #200 5’-GUGAGGAACUUUACUCUUA;
siTRAF6 #208 5’-UGUGGAAUGUGGAGAGGAA;
siRipK2 #49 5’-GUGUGAAGCAUGAUAUAUA;
siRipK2 #66 5’-GGAUUUAUCGCUAAACAUA;
sicIAP1 #57 5’-GAAGACUUCUCAUCAAGGA;
sicIAP1 #58 5’-GGACAAAGGAGAGCGAAGA;
sicIAP2 #54 5’-GAAGAGUGCUGACACCUUU;
sicIAP2 #55 5’-CAGCCCGUAUUAGAACAUU;
siXIAP #60 5’-GAUCGUUACUUUUGGAACA;
siXIAP #61 5’-CCAGGGUGCAAAUACCAU
The non-targeting negative control No. 1 (Invitrogen) was used as control.
Microscopy
MEF cells were grown on glass cover slips before infection. After infection, cells were washed twice with warm PBS and fixed in 4% paraformaldehyde or ice-cold methanol (phospho-IKKα stained samples only) for 20 min. Cells were washed twice in PBS and then simultaneously permeabilised and blocked in PBSB (PBS, 0.01% saponin, 2% BSA). Coverslips were incubated with primary followed by secondary antibodies for 1 h in PBSB. Samples were mounted in mounting medium with DAPI (Vector Laboratories) for confocal or Prolong Antifade mounting medium (Invitrogen) for super resolution microscopy. Marker positive bacteria were counted by eye amongst at least 100 bacteria per coverslip using a widefield microscope. Confocal images were taken with a ×63, 1.4 numerical aperture objective on either a Zeiss 710 or a Zeiss 780 microscope. Live imaging was performed on a Nikon Eclipse Ti equipped with an Andor Revolution XD system and a Yokogawa CSU-X1 spinning disk unit. Super resolution images were acquired using an Elyra S1 structured illumination microscope (Carl Zeiss Microscopy Ltd,Cambridge,UK). The system has four laser excitation sources (405nm,488nm,561nm and 640nm) with fluorescence emission filter sets matched to these wavelengths. SIM Images were obtained using a 63X 1.4 NA oil immersion lens with grating projections at 3 rotations and 5 phases in accordance with the manufacturers instructions. The number of Z planes varied with sample thickness. Super resolution images were calculated from the raw data using Zeiss ZEN software. Pearsons’s correlation coefficients were calculated from n>5 images using Imaris software.
Western blot
Cells were lysed in Lysis buffer (20mM Tris pH7.4, 150mM NaCl, 0.1% Triton X-100, 10% glycerol plus 1mM phenylmethylsulfonyl fluoride (PMSF), 1mM benzamidine, 2μg/ml aprotinin, 5ug/ml leupeptin, 1mM dithiothreitol (DTT)) before clearing by centrifugation and addition of SDS loading buffer. Samples were then separated on 4–12% denaturing gels (Invitrogen) and visualized by immuno-blotting using ECL detection reagents (Amersham Bioscience).
Reverse Transcriptase PCR
Total RNA from siRNA-treated MEF cells was extracted using the RNeasy Plus Mini Kit (Qiagen) followed by conversion into cDNA using the SuperScript III reverse transcriptase kit (Invitrogen) according to the manufacturer’s protocol. Gene expression was quantified using specific primer pairs with a Power SYBR qPCR green kit (Applied Biosystems) by following the manufacturer’s protocol. Relative amounts of cDNA were calculated using the ddCt method and normalized to β-actin cDNA levels in each sample. The following primers were used:
IKBKG (Nemo)
5’- CAGGTCCCATAAGGCTACAAG
5’- TTCGTTCAGGCATACACAGG
Sharpin
5’- AGAAGGAGTATTTGCAGGAGC
5’-TGGAGCAGGGAGTAAAGGAG
ACTB (β-actin)
5’- TGACAGCATTGCTTCTGTGTAAATT
5’-ATTGGTCTCAAGTCAGTGTACAGGC
Traf3
5’- GCACCTGTAGTTTTAAGCGC
5’- TGAAGGATCTTGGACTCGTTG
Traf6
5’- GAACTGAGACATCTCGAGGATC
5’- AGAGGACAGCTTTGATCATGG
XIAP
5’- CTGAAAAAACACCACCGCTAAC
5’- CTAAATCCCATTCGTATAGCTTCTTG
LRSAM1
5’- CTCGAGAATGAGGTCCTTGG
5’- GCTGACAGCAGCAGACGTG
RipK2
5’- CTCGTGTTCCTTGGCTGTAA
5’- CAATGGCTTCCCTCTTACTCTG
Biacore
SPR experiments were performed on a Biacore 2000 instrument (GE Healthcare) as described previously38. CM5 chips (GE Healthcare) were functionalized with a 1:1 mix of EDC/NHS, and one flow channel per chip was immediately quenched with 1M ethanolamine pH 8.0 to serve as a reference. For the other channels, the differently linked diUb were diluted to 100 ng/μl in 20 mM sodium acetate pH 5.0 and injected until a response of ~2000 RU was reached. Protein samples at different concentrations were prepared in SPR buffer (20 mM Tris pH 7.4, 150 mM NaCl, 1 mM DTT) and injected for 60 s followed by 150 s of dissociation in SPR buffer at 20 °C. The Kd was determined from fitting triplicate experiments using the reference-corrected equilibrium response in Prism 6 (GraphPad Software).
Supplementary Material
Acknowledgements
This work was supported by the MRC (U105170648 to FR, U105192732 to DK) and the Wellcome Trust (WT104752MA) and Boehringer Ingelheim Fonds Ph.D. fellowships (to CP and MAM). We thank Noboru Mizushima for ATG5-/- MEFs, Henning Walczack for Sharpincpdm MEFs, Mads Gyrd-Hansen for RipK2-/- and HOIP-/- HCT116s, and Emma Werner for technical help.
Footnotes
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heath RJ, et al. RNF166 Determines Recruitment of Adaptor Proteins during Antibacterial Autophagy. CellReports. 2016;17:2183–2194. doi: 10.1016/j.celrep.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manzanillo PS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franco LH, et al. The Ubiquitin Ligase Smurf1 Functions in Selective Autophagy of Mycobacterium tuberculosis and Anti-tuberculous Host Defense. Cell Host and Microbe. 2017;21:59–72. doi: 10.1016/j.chom.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrin A, Jiang X, Birmingham C, So N, Brumell J. Recognition of Bacteria in the Cytosol of Mammalian Cells by the Ubiquitin System. Current Biology. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Thurston TLM, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 9.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyle KB, Randow F. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Current Opinion in Microbiology. 2013;16:1–10. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7:22–26. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirisako T, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokunaga F, et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 15.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 16.Fiskin E, Bionda T, Dikic I, Behrends C. Global Analysis of Host and Bacterial Ubiquitinome in Response to Salmonella Typhimurium Infection. Molecular Cell. 2016;62:967–981. doi: 10.1016/j.molcel.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 17.Collins CA, et al. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5:e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita N, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. The Journal of Cell Biology. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita H, et al. Mechanism Underlying I B Kinase Activation Mediated by the Linear Ubiquitin Chain Assembly Complex. Mol Cell Biol. 2014;34:1322–1335. doi: 10.1128/MCB.01538-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stieglitz B, et al. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature. 2013;503:422–426. doi: 10.1038/nature12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas TL, et al. Recruitment of the Linear Ubiquitin Chain Assembly Complex Stabilizes the TNF-R1 Signaling Complex and Is Required for TNF-Mediated Gene Induction. Molecular Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Sato Y, et al. Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc Natl Acad Sci USA. 2011;108:20520–20525. doi: 10.1073/pnas.1109088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Bloor S, et al. Signal processing by its coil zipper domain activates IKK gamma. Proc Natl Acad Sci USA. 2008;105:1279–1284. doi: 10.1073/pnas.0706552105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kageyama S, et al. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell. 2011;22:2290–2300. doi: 10.1091/mbc.E10-11-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Muhlinen N, et al. LC3C, Bound Selectively by a Noncanonical LIR Motif in NDP52, Is Required for Antibacterial Autophagy. Molecular Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, et al. Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy. Science signaling. 2013;6 doi: 10.1126/scisignal.2003730. ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichimura Y, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 30.de Jong MF, Liu Z, Chen D, Alto NM. Shigella flexneri suppresses NF-κB activation by inhibiting linear ubiquitin chain ligation. Nat Microbiol. 2016;1:16084. doi: 10.1038/nmicrobiol.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caruso R, Warner N, Inohara N, Núñez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huett A, et al. The LRR and RING Domain Protein LRSAM1 Is an E3 Ligase Crucial for Ubiquitin-Dependent Autophagy of Intracellular Salmonella Typhimurium. Cell Host and Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Wijk SJL, et al. Fluorescence-Based Sensors to Monitor Localization and Functions of Linear and K63-Linked Ubiquitin Chains in Cells. Molecular Cell. 2012 doi: 10.1016/j.molcel.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randow F, Youle RJ. Self and Nonself: How Autophagy Targets Mitochondria and Bacteria. Cell Host and Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 36.Randow F, Sale JE. Retroviral transduction of DT40. Subcell Biochem. 2006;40:383–386. doi: 10.1007/978-1-4020-4896-8_30. [DOI] [PubMed] [Google Scholar]
- 37.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 38.Michel MA, et al. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Molecular Cell. 2015;58:95–109. doi: 10.1016/j.molcel.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.