SUMMARY
The motility and signaling functions of the primary cilium require a unique protein and lipid composition that is determined by gating mechanisms localized at the base of the cilium. Several protein complexes localize to the gating zone and may regulate ciliary protein composition, however, the mechanisms of ciliary gating and the dynamics of the gating components are largely unknown. Here we used the BiFC (bimolecular fluorescence complementation) assay and report for the first time on the protein-protein interactions that occur between ciliary gating components and transiting cargoes during ciliary entry. We find that the nucleoporin Nup62 and the C-termini of the nephronophthisis (NPHP) proteins NPHP4 and NPHP5 interact with the axoneme-associated kinesin-2 motor KIF17 and thus spatially map to the inner region of the ciliary gating zone. Nup62 and NPHP4 exhibit rapid turnover at the transition zone and thus define dynamic components of the gate. We find that B9D1, AHI1 and the N-termini of NPHP4 and NPHP5 interact with the transmembrane protein SSTR3 and thus spatially map to the outer region of the ciliary gating zone. B9D1, AHI1 and NPHP5 exhibit little to no turnover at the transition zone and thus define components of a stable gating structure. These data provide the first comprehensive map of the molecular orientations of gating zone components along the inner-to-outer axis of the ciliary gating zone. These results advance our understanding of the functional roles of gating zone components in regulating ciliary protein composition.
Keywords: cilia, flagella, import, gating, nucleoporin, nephronophthisis, intraflagellar transport, kinesin, axoneme, transition zone
eTOC blurb
Cilia are organelles for cell signaling and motility. These functions require an organelle-specific protein composition that is defined by import control at the ciliary gating zone. Takao et al. use BiFC and FRAP assays to map the relative locations, gating functions, and dynamics of MKS, NPHP, and NUP components of the ciliary gating zone.
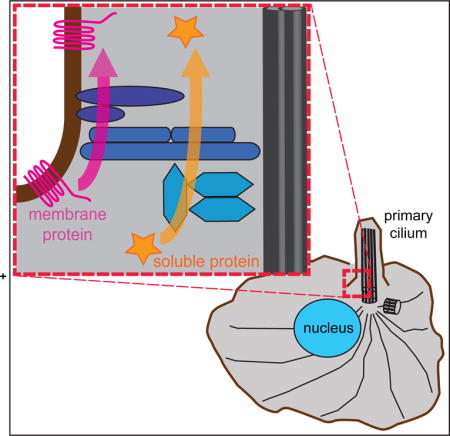
INTRODUCTION
Cilia and flagella are organelles that play critical roles in cell and organismal biology, from driving motility to sensing the extracellular environment [1]. Cilia function requires a specific protein and lipid composition that is determined by gating mechanisms located at the base of the cilium [2–4]. Defects in the generation or function of primary cilia can result in severe developmental and sometimes lethal disorders, collectively called ciliopathies [5, 6].
A large number of proteins have been localized to the base of the cilium and implicated in regulating protein entry into the compartment. First, genetic mutations associated with the ciliopathies Meckel-Gruber syndrome (MKS) and Joubert syndrome revealed a complex of proteins (the MKS complex) whose knockout or knockdown results in alteration of membrane protein composition across species [7–17]. Second, genetic mutations associated with the ciliopathy nephronophthisis (NPHP) revealed a protein complex whose knockout or knockdown results in alteration of both soluble and membrane protein composition [18–20]. Third, a number of nucleoporins (NUPs), known for their localization to and function in gating at nuclear pore complexes (NPCs) in the nuclear envelope, cause alteration of ciliary soluble protein content upon disruption of their function [21–24].
Despite the identification of MKS, NPHP and NUP proteins as components of the ciliary gating zone and their biochemical characterization into complexes, their functions in ciliary gating are largely unknown. The difficulty stems, at least in part, from an inability to separate the direct effects of loss-of-function in a particular component from indirect effects on the localization and/or function of other ciliary components. We recently developed an assay using forced crosslinking of the central nucleoporin Nup62 to rapidly and specifically block NUP complex function. We found that crosslinking Nup62 blocked active transport of cytosolic proteins into the ciliary compartment but had minimal effect on the entry of membrane proteins [24]. Similar assays have not yet been applied to determine the functions of MKS and NPHP complex proteins in ciliary gating.
It is also unclear how the structure and organization of the ciliary gating zone, including the locations of the MKS, NPHP and NUP components, relate to the paths of entry for soluble and membrane proteins. In cultured mammalian cells, MKS components and the NPHP1-4-8 module components localize to a region at the base of the cilium termed the transition zone [8–10, 13, 16, 25–27]. NPHP5-6 module components and NUPs generally localize below the transition zone [10, 21–25, 27].
To begin to address the function and organization of MKS, NPHP and NUP components in ciliary gating, we utilized the Bimolecular Fluorescence Complementation (BiFC) assay to detect protein-protein interactions between gating components and ciliary proteins transiting the gating zone. The results provide a map that for the first time integrates the molecular orientations of key gating components with their functions at the ciliary gating zone. Using fluorescence recovery after photobleaching (FRAP), we further provide the first comparison of the dynamics of MKS, NPHP, and NUP components at the ciliary gating zone. This work advances our understanding of how proteins are targeted to the ciliary compartment and has implications for understanding a large number of cellular signaling pathways and the resultant diseases of their dysfunction.
RESULTS
Using the BiFC assay to map protein-protein interactions in the ciliary gating zone
To obtain a map of the protein-protein interactions that occur during the ciliary entry process, we utilized the BiFC assay [28]. In this assay, the N-terminal or C-terminal fragments of a yellow fluorescent protein (YFP) variant (hereafter termed “YN” and “YC”, respectively) are fused to proteins of interest. The YFP fragments display little to no self-association and are non-fluorescent but when brought into close proximity via interactions of their fusion partners, the fragments assemble a YFP molecule and display fluorescence. YFP fluorescence thus provides a direct visualization of protein interactions within the cellular environment. We reasoned that when a soluble ciliary protein tagged with YN and a NUP component tagged with YC were co-expressed, the cytosolic protein would be trapped by the NUP during ciliary entry and would emit fluorescence at the cilium base (Figure 1A, left). Similarly, we reasoned that a cilium-targeted transmembrane protein tagged with YN would show an interaction with a YC-tagged transition zone protein localized at the ciliary membrane (Figure 1A, right).
Figure 1. The BiFC assay maps protein interactions at the ciliary gating zone.
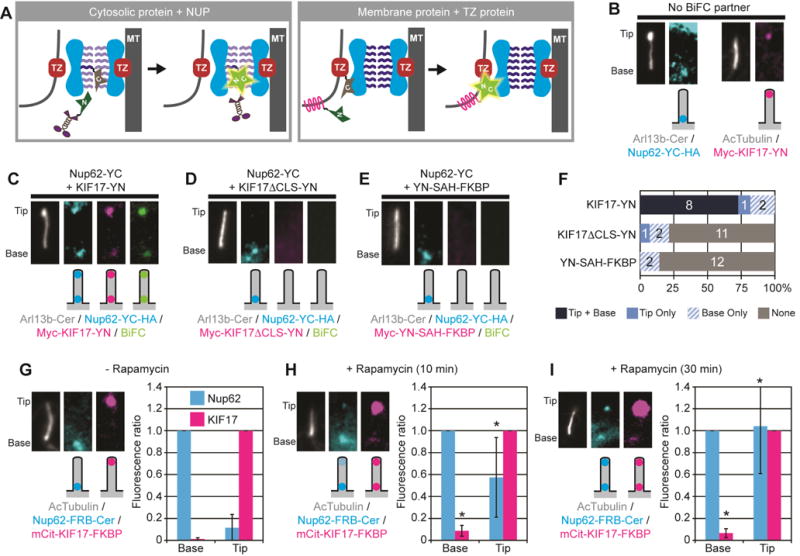
(A) Schematic of BiFC assay. The N- and C-terminal halves of a YFP (half-stars labeled N or C), when fused to proteins of interest, can reconstitute a fluorescent protein only when brought in close proximity via the interactions of their fusion partners. TZ, transition zone proteins. MT, axonemal microtubules. (B) Localization of Nup62-YC (left) and KIF17-YN (right) expressed in the absence of a BiFC partner. Arl13b-Cer or acetylated α-tubulin (AcTubulin) was used as a cilium marker. Images are of cropped regions containing the cilium, contrast-enhanced for viewing, with a schematic of the observed fluorescence localization shown below each fluorescence image. (C–E) Representative images and schematic depictions show the locations of BiFC interactions detected for Nup62-YC with (C) kinesin-2 motor KIF17-YN, (D) KIF17 with mutated CLS (KIF17ΔCLS-YN), or (E) non-ciliary protein (YN-SAH-FKBP). Nup62-YC was detected with an antibody to the HA tag (Nup62-YC-HA) whereas the KIF17 and SAH constructs were detected with an antibody to the Myc tag. See Table S1 for full description of constructs. See Figure S1 for uncropped images. (F) Quantification of the locations of BiFC interactions. The number of cells observed for each BiFC location category is indicated on the bar graph. (G–I) Time course of interaction between Nup62 and KIF17. Nup62-FRB-Cer and mCit-KIF17-FKBP were co-expressed and the cells were fixed after treatment with (G) ethanol (- Rapamycin control), (H) 10 min Rapamycin, or (I) 30 min Rapamycin. Graphs show the mean fluorescence of Nup62-FRB-Cer at the base or tip of the cilium (normalized to the fluorescence at the base) or the mean fluorescence of mCit-KIF17-FKBP at the base or tip of the cilium (normalized to the fluorescence at the tip). *p<0.01 compared to control (-rapamycin) by Student’s t-test. Error bars, S.D. n = 10–14 cells each.
Since our previous work demonstrated that the channel nucleoporin Nup62 is involved in ciliary entry of the cytosolic kinesin-2 motor KIF17 [24], we used the combination of Nup62-YC and KIF17-YN to evaluate the BiFC assay. BiFC-tagged proteins were identified by immunostaining for epitope tags and/or by fluorescent protein tags (Table S1 and Figure S1). When expressed in NIH3T3 cells without a BiFC partner, Nup62-YC localized to the base of the cilium as expected [23] whereas KIF17-YN localized to the tip of the cilium as expected [29] (Figure 1B). In cells expressing both Nup62-YC and KIF17-YN, a BiFC signal was detected in cilia with little to no fluorescence complementation in the rest of the cell (Figure 1C,F and Figure S1A). The BiFC signal at the base of the cilium (Figure 1C,F) was expected and suggests that KIF17 interacts with Nup62 during its entry into the ciliary compartment. The BiFC signal at the tip of the cilium (Figure 1C,F) was not expected and suggests that KIF17 drags Nup62 to the tip of the cilium once their association becomes irreversible due to the reconstituted YFP. To verify that the BiFC signal between Nup62-YC and KIF17-YN is due to their interaction at the ciliary base, we carried out two control experiments. First, Nup62-YC showed little to no BiFC signal when co-expressed with a KIF17 motor lacking its ciliary localization signal (KIF17ΔCLS-YN, Figure 1D,F and Figure S1B). Second, Nup62-YC showed little to no BiFC signal when co-expressed with a cytosolic protein larger than the ciliary size exclusion limit [YN-SAH-FKBP, consisting of a 30 nm long single alpha helix (SAH) and a FKBP protein] (Figure 1E,F and Figure S1C). These results support the hypothesis that the interaction between Nup62-YC and KIF17-YN occurs during ciliary entry of KIF17.
We hypothesized that KIF17-YN first interacts with Nup62-YC at the cilium base and then drags Nup62-YC to the cilium tip. To address this, we examined the time course of these interactions by replacing the BiFC assay with the rapamycin-induced heterodimerization of FKBP and FRB domains. In the absence of rapamycin, Nup62-FRB localized to the base of the cilium whereas KIF17-FKBP localized to the tip of the cilium (Figure 1G). 10 min after addition of rapamycin, a small amount of KIF17-FKBP was trapped at the cilium base and a small portion of Nup62-FRB localized to the cilium tip (Figure 1H). 30 min after addition of rapamycin, the level of KIF17-FKBP trapped at the base remained constant, presumably due to the transient nature of kinesin transport through the gating zone, whereas the amount of Nup62-FRB dragged to the cilium tip increased (Figure 1I).
Taken together, these results demonstrate that the BiFC assay is applicable for detecting interactions between gating zone components and ciliary proteins entering the compartment. In addition, as the transiting KIF17 protein is a kinesin motor that interacts directly with doublet microtubules of the axoneme, these results spatially map Nup62 to the inner region of the ciliary gating zone.
Interactions between NUPs and ciliary proteins in BiFC assay
We thus used the BiFC assay to map the spatial locations and functional interactions of gating zone components by detecting their interactions with the axoneme-associated motor KIF17, a soluble protein (Gli2, a transcription factor in the Hedgehog signaling pathway), and the transmembrane proteins SSTR3 (serotonin receptor) and GPR161 (orphan GPCR). These two transmembrane proteins were chosen because the length of their cytosolic tails (103 amino acids for SSTR3 vs 202 amino acids for GPR161) could influence their interactions with gate components. In the absence of a BiFC partner, YN-Gli2 localized to the tip of the cilium whereas SSTR3-YN and GPR161-YN localized along the ciliary membrane (Figure 2A).
Figure 2. Interactions between NUP components and transiting cilia proteins.
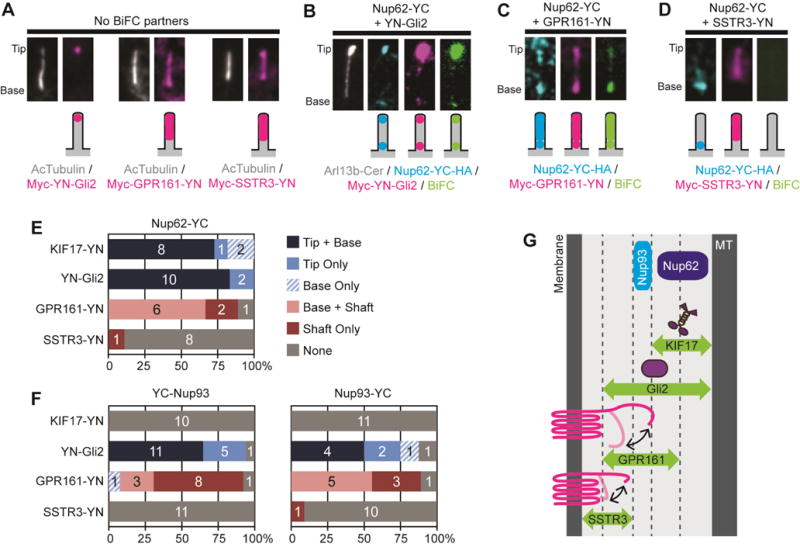
(A) Representative images showing the ciliary localizations of YN-Gli2, GPR161-YN, and SSTR3-YN expressed in the absence of a BiFC partner. (B–D) Representative images and schematic depictions show the locations of BiFC interactions detected for Nup62-YC with (B) YN-Gli2, (C) GPR161-YN, or (D) SSTR3-YN. Proteins were detected with antibodies to the epitope tags. See Table S1 for full description of constructs. See Figure S2 for uncropped images. (E,F) Quantification of the locations of BiFC interactions for (E) Nup62 or (F) Nup93 constructs. The number of cells observed for each BiFC location category is indicated on the bar graph. See Figure S3 for representative images of YC-Nup93 and Nup93-YC BiFC interactions. (G) The BiFC interactions define the locations of Nup62 and Nup93 in the inner region of the ciliary gating zone. MT, axonemal microtubules.
When co-expressed with Nup62-YC, YN-Gli2 was trapped by Nup62-YC at the cilium base and it also dragged Nup62-YC to the cilium tip (Figure 2B,E), with little interaction in the rest of the cell (Figure S2A). SSTR3-YN showed no detectable BiFC interactions with Nup62-YC (Figure 2D,E and Figure S2C), however, GPR161-YN was trapped by Nup62-YC at the cilium base and it also dragged Nup62-YC to the cilium shaft (Figure 2C,E and Figure S2B). It may be that the longer cytosolic tail of GPR161 allows it to come in proximity of Nup62-YC during ciliary entry (Figure 2G) even though Nup62 does not play a critical role in regulating its entry [24]. Quantification of the ciliary content of Nup62-YC, KIF17-YN, and SSTR3-YN demonstrates that the ability to interact and reconstitute the YFP (positive BiFC) is not dependent on the levels of protein expression (Figure S2D). These results suggest that Nup62 interacts with KIF17, Gli2 and GPR161 but not SSTR3 during the entry of these proteins into the ciliary compartment (Figure 2G).
To further examine the interactions between NUPs and ciliary proteins at the ciliary gating zone, we applied the BiFC assay to nucleoporin Nup93 that forms part of a scaffold complex of the NPC [30] and localizes to the cilium base (Figure S3A and [23]). We generated YC-tagged versions of Nup93 and tested the interactions with transiting ciliary proteins. Neither YC-Nup93 nor Nup93-YC displayed a BiFC interaction with the axoneme-associated motor KIF17-YN (Figure 2F and Figure S3 B,F,J), or with the transmembrane protein SSTR3-YN (Figure 2F and Figure S2 E,I). However, both Nup93 constructs displayed BiFC interactions at the base and tip of the cilium with the cytosolic protein YN-Gli2 (Figure 2F and Figure S3 C,G,J) and BiFC interactions at the base and/or shaft of the cilium with GPR161-YN (Figure 2F and Figure S3 D,H). The ability to form a BiFC interaction was independent of protein expression level (Figure S3J). The fact that Nup93 is unable to interact with the axoneme-associated motor KIF17 or the transmembrane protein SSTR3 (Figure 2F) suggests that Nup93 occupies a central position in the inner-outer axis of the ciliary gating zone (Figure 2G). These results also provide strong support for the model that NUP components play a functional role in ciliary gating [3,4].
MKS components interact with membrane proteins and IFT cargoes during ciliary gating
We next applied the BiFC assay to examine the spatial locations and functional interactions of members of the MKS complex. We focused on B9D1 and AHI1 due to the proper localization of tagged versions to the base of the cilium (Figure 3A) [8, 31]. For YC-B9D1, a BiFC interaction could not be reliably detected with the cytosolic protein KIF17-YN (Figure 3E and Figure S4A,E) whereas a BiFC interaction was observed with YN-Gli2 at the base and/or tip of most cilia (Figure 3E and Figure S4B). When YC-B9D1 was co-expressed with the transmembrane proteins SSTR3-YN or GRP161-YN, strong BiFC interactions were observed and YC-B9D1 was dragged into the shaft of the cilium due to these interactions (Figure 3B,E and Figure S4C,D). The ability to form a BiFC interaction was independent of protein expression level (Figure S4E). We hypothesized that the interactions between YC-B9D1 and the transmembrane proteins occurs during transit of SSTR3 and GPR161 through the ciliary gating zone. In support of this, BiFC interactions were not detected for YC-B9D1 with two transmembrane proteins excluded from the ciliary compartment, caveolin and transferrin receptor (Figure S5).
Figure 3. Interactions between MKS components and transiting cilia proteins.
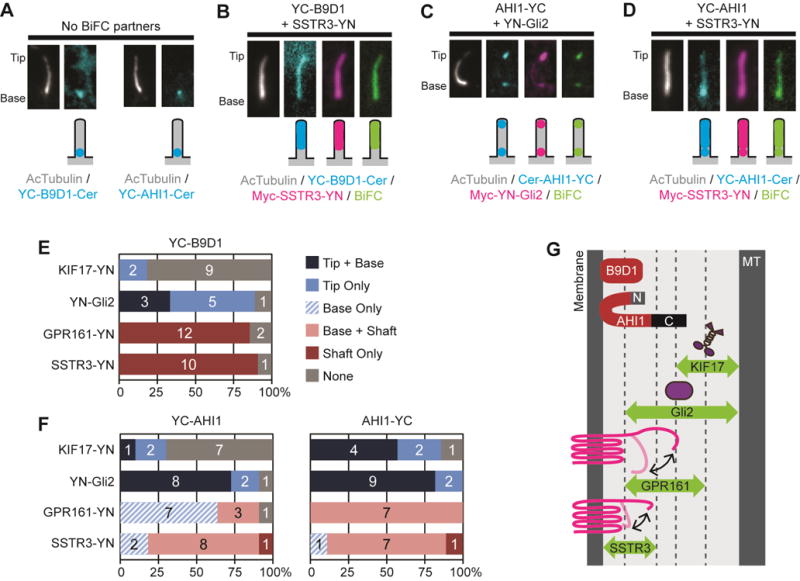
(A) Representative images showing the ciliary localizations of the MKS components YC-B9D1 and YC-AHI1 expressed in the absence of a BiFC partner. (B–D) Representative images and schematic depictions of locations of BiFC interactions for select B9D1 and AHI1 combinations. Proteins were detected using fluorescent tags or antibodies to the epitope tags. See Table S1 for full description of constructs. See Figure S4 for images of YC-B9D1 and YC-AHI1 BiFC interactions. (E,F) Quantification of the locations of the BiFC interactions for (E) B9D1 or (F) AHI1 constructs. The number of cells observed for each BiFC location category is indicated on the bar graph. (G) The BiFC assay defines the positions and orientations of B9D1 and AHI1 at the outer region of the ciliary gating zone.
For AHI1, the ability to interact with ciliary proteins and reconstitute YFP depended on where the split-YFP fragment was located. When the YC fragment was placed at the N-terminus of AHI1 (YC-AHI1), BiFC interactions were not reliably observed for KIF17-YN whereas ciliary BiFC signal was detected for YN-Gli2, SSTR3-YN and GPR161-YN (Figure 3D,F and Figure S4F-I). When the YC fragment was placed at the C-terminus of AHI1 (AHI1-YC), ciliary BiFC signals were observed with both cytosolic proteins (KIF17-YN and YN-Gli2, Figure 3C,F and Figure S4J,K) and with both transmembrane proteins (SSTR3-YN and GPR161-YN, Figure 3F and Figure S4L,M). These results suggest that the AHI1 is oriented within the gating zone such that its N-terminus is localized close to the ciliary membrane whereas its C-terminus is localized in the central region of the gating zone and along the entry path of both soluble and transmembrane proteins (Figure 3G).
NPHP components interact with cytosolic and membrane proteins during ciliary gating
To gain an understanding of the spatial locations and functional roles of NPHP module proteins in ciliary entry, we focused on NPHP4 and NPHP5 (Figure 4A). The BiFC interactions of NPHP4 depended on the location of the YC fragment fusion. When the YC fragment was fused to the N-terminus of NPHP4 (YC-NPHP4), a BiFC interaction was reliably detected only with SSTR3-YN (Figure 4B,E and Figure S6A–D,I). These results suggest that the N-terminus of NPHP4 resides close to the ciliary membrane where it can engage with the shorter tail of SSTR3 but not the longer tail of GPR161 (Figure 4G). When the YC fragment was fused to the C-terminus of NPHP4 (NPHP4-YC), strong BiFC interactions were observed at the base and tip of cilia with both of the cytosolic proteins, KIF17-YN and YN-Gli2 (Figure 4C,E and Figure S6E,F), whereas no BIFC interactions were detected with either of the transmembrane proteins, SSTR3-YN and GPR161-YN (Figure 4E and Figure S6G,H). The ability to form a BiFC interaction was independent of protein expression level (Figure S6I). These results suggest that the C-terminus of NPHP4 is positioned away from the ciliary membrane and may lie close to the axonemal microtubules (Figure 4G).
Figure 4. Interactions between NPHP components and transiting cilia proteins.
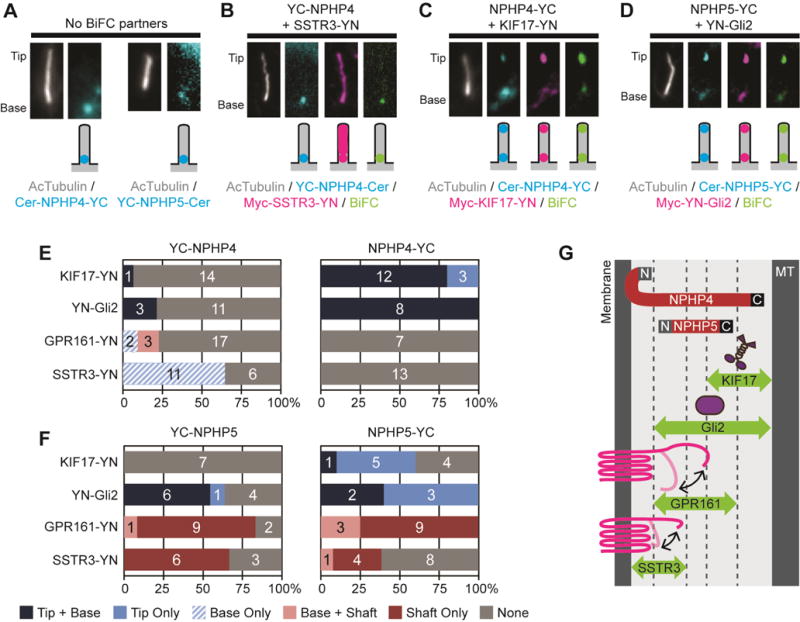
(A) Representative images showing the ciliary localizations of NPHP4-YC and YC-NPHP5 expressed without a BiFC partner. (B–D) Representative images and schematic depictions of BiFC interactions for select NPHP4 and NPHP5 combinations. Proteins were detected with fluorescent tags or with antibodies to the epitope tags. See Table S1 for full description of the constructs. See Figure S6 for representative images. (E,F) Quantification of the locations of the BiFC interactions for (E) NPHP4 or (F) NPHP5 constructs. The number of cells observed for each BiFC location category is indicated on the bar graph. (G) The BiFC assay defines the positions and orientations of NPHP4 and NPHP5 within the ciliary gating zone.
In contrast to the restricted interactions observed for NPHP4 (Figure 4E), numerous BiFC interactions were observed for NPHP5 (Figure 4D,F and Figure S6J-Q). These results suggest that NPHP5 is centrally located within the ciliary gating zone such that it can interact with both soluble and transmembrane proteins as they transit the gating zone (Figure 4G). The absence of a BiFC interaction between YC-NPHP5 and KIF17-YN suggests that the N-terminus of NPHP5 is oriented toward the ciliary membrane whereas the minimal BiFC interactions between NPHP5-YC and SSTR3-YN suggests that the C-terminus of NPHP5 is oriented toward the axonemal microtubules (Figure 4G).
Dynamics of ciliary gating zone components
One of the most surprising findings in the BiFC assay was that the ciliary gating components could be dragged from their resident location to that of the interacting partner. For example, Nup62-YC was dragged to the tip of the cilium by KIF17-YN (Figure 1C,F) and YC-B9D1 was dragged to the ciliary shaft by SSTR3-YN (Figure 3B,E). These results suggest that components of the ciliary gating zone are not static within the macromolecular complex but rather undergo dynamic associations with the complex.
To directly measure the dynamic nature of ciliary gating zone components, we applied a fluorescence recovery after photobleaching (FRAP) assay. For the NUP component Nup62-mCit, ~70% fluorescence recovery was obtained within 5 min after photobleaching (Figure 5A,F), suggesting that the majority of Nup62 protein at the base of the cilium is highly mobile. In contrast, the MKS components B9D1 (Figure 5B) and AHI1 (Figure 5C) displayed a large immobile fraction, suggesting that these are stable components of the gating zone. For the NPHP components, NPHP4 appeared to be a relatively dynamic component, with ~60% recovery within 5 min after photobleaching (Figure 5D,F), whereas NPHP5 appeared to be a stable component, with little to no fluorescence recovery (Figure 5E). These results suggest that NPHP components can vary greatly in their dynamics and that NPHP subcomplexes are likely to play distinct structural (stable) or functional (dynamic) roles at the ciliary gating zone.
Figure 5. Dynamics of ciliary gating zone proteins.
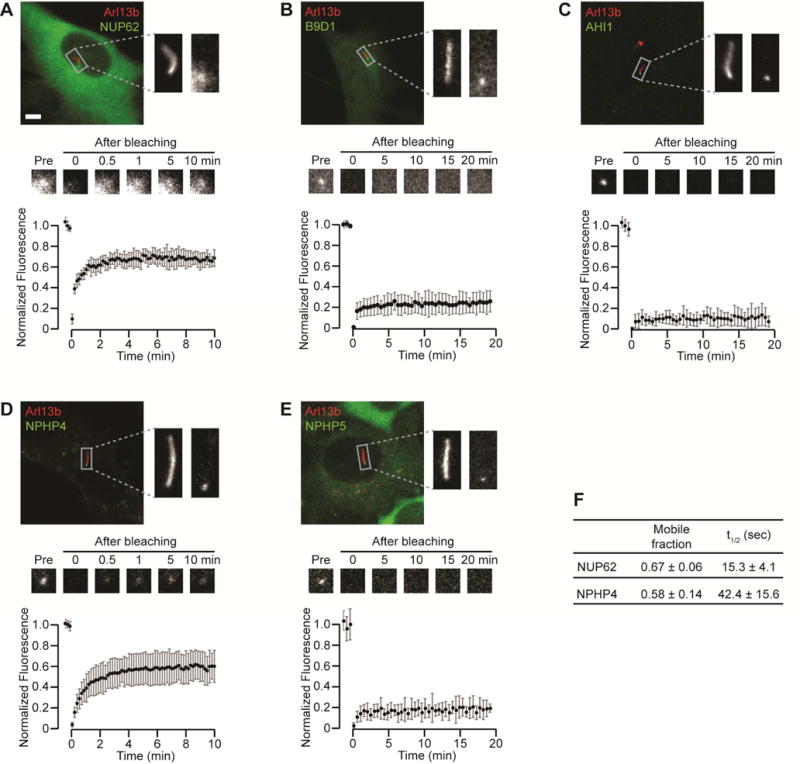
(A–E) FRAP analysis of the gating components (A) Nup62-mCit, (B) B9D1-NeonGreen, (C) EGFP-AHI1, (D) mCherry-NPHP4 (pseudocolored green), and (E) EGFP-NPHP5. Arl13b-mCherry or Arl13b-mCit was used as a cilium marker (red in all images). The top panels show representative fluorescence images with magnified images of the boxed region containing the primary cilium. The middle panels show representative time-lapse images of fluorescence recovery at the cilium base. The bottom panels show fluorescence recovery curves. n = 7–14 cells each. Graphs show the mean +/− S.D. fluorescence intensity (A.U.=Arbitrary Units) over time. Scale bar, 5 μm. (F) The mobile fraction and half recovery time (t1/2) were calculated for the dynamic components Nup62 and NPHP4 by fitting an exponential curve to the fluorescence recovery data.
The relative stability of B9D1-mNeonGreen in the FRAP assay (Figure 5B) appeared to be at odds with the ability of YC-B9D1 to be dragged to the ciliary tip by YN-Gli2 and to the ciliary shaft by SSTR3-YN and GPR161-YN in the BiFC assay (Figure 3E and Figure S4B–D). We reasoned that the time course of the BiFC assay (typically 16h) is likely to be longer than the turnover rates of even stable gating components such as B9D1. To further examine the dynamics of B9D1 and the time course of its interactions, we utilized rapamycin to drive interactions between FRB-B9D1 and SSTR3-FKBP. In the absence of rapamycin, FRB-B9D1 localized to the base of the cilium and SSTR3-FKBP localized to the shaft of the cilium (Figure 6A). After 30 min of rapamycin treatment, the transfer of FRB-B9D1 into the cilium could be observed in only a few cells (Figure 6B,D). Longer rapamycin treatment resulted in an increase in the number of cells with co-localization of FRB-B9D1 and SSTR3-FKBP in the cilium (Figure 6C,D). These results suggest that the lifetime of B9D1 at the base of the cilium is on the order of several hours and that this turnover rate allows B9D1 to be dragged into the cilium when stably associated with transiting proteins in the BiFC assay.
Figure 6. Dynamics of the MKS component B9D1 at the base of the cilium.
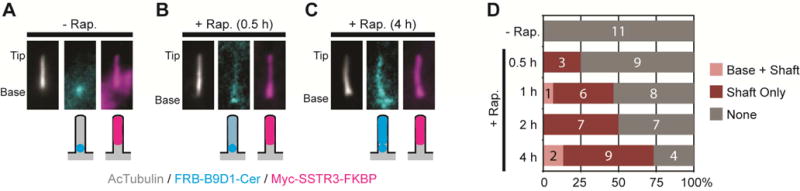
(A) Representative images and schematic depictions of locations of FRB-B9D1 and SSTR3-FKBP. Cells were treated with ethanol (- Rapamycin control) or rapamycin for the indicated times. (B) Quantification of the locations of the FRB-B9D1-Cer and Myc-SSTR3-FKBP interactions in cilia. The number of cells observed for each location category is indicated on the bar graphs.
DISCUSSION
To gain insight into the localizations, interactions, and functions of ciliary gating components, we used the BiFC assay and report for the first time on the protein-protein interactions that occur between ciliary gating components and transiting cargoes during ciliary entry. We also provide the first comparison of the dynamics of ciliary gating components in mammalian cells. Together these data provide a map of the spatial and temporal organization of the ciliary gating zone (Figure 7).
Figure 7. Model of the spatial organization and dynamics of ciliary gating components.
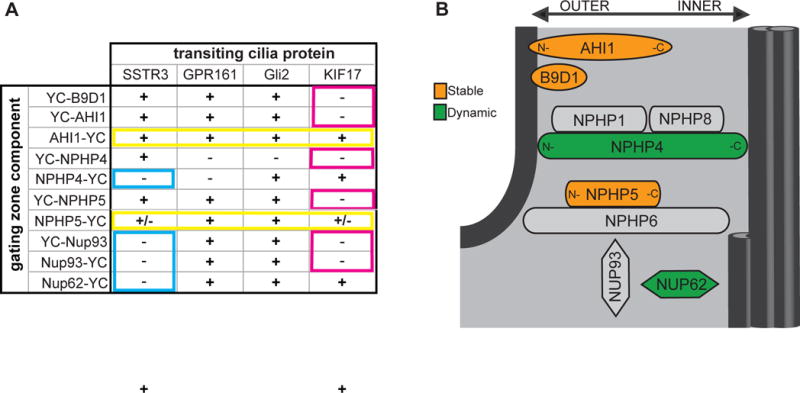
(A) Summary of BiFC interactions. (+) = BiFC signal detected in cilium, (−) = little to no BIFC signal detected. Blue boxes: Nup62, Nup93 and the C-terminus of NPHP4 are unable to interact with the short cytoplasmic tail of transmembrane protein SSTR3 and thus localize to the inner region of the ciliary gating zone. Pink boxes: B9D1, Nup93, and the N-termini of AHI1, NPHP4, and NPHP5 are unable to interact with the KIF17 motor protein and are thus oriented to the outer region of the gating zone. Yellow boxes: the C-termini of AHI1 and NPHP5 interact with all transiting proteins and are thus widely accessible throughout the ciliary gating zone. (b) Schematic of the localizations and dynamics of ciliary gating components. The relative positioning of ciliary gating zone components along the inner-to-outer axis is shown. The relative dynamics of the components is indicated by the color shading where orange = stable components, green = dynamic components, gray = unknown.
Nup62 is likely to be one of the inner-most components of the ciliary gating zone as it displays BiFC interactions with both cytosolic proteins (KIF17 and Gli2) whereas Nup93 likely resides peripheral to Nup62 as it displays BiFC interactions only with Gli2 (Figure 7A,B). Both NUPs appear to reside below the transition zone (Figure 7B), perhaps at the transition fibers or basal appendages of the centriole. Interestingly, both NUP components displayed interactions with the long cytoplasmic tail of the transmembrane protein GPR161. Thus, although Nup62 does not play a functional role in regulating the transit of membrane proteins through the ciliary gating zone [24], the tail segments of some transmembrane proteins may reach to the inner region of the gating zone where Nup62 and Nup93 reside. This is similar to nuclear gating where the long cytoplasmic tails of some transmembrane proteins can extend into the NPC [32]. Alternatively, the ability of GPR161 but not SSTR3 to interact with NUP components may be due to other differences such as their interaction partners or time of transit through the gating zone.
We found that Nup62 is a highly dynamic component of the ciliary gating zone (Figure 7B) as it shows rapid exchange between ciliary and cytoplasmic pools in a FRAP assay (Figure 5A) and co-localization with the transiting KIF17 motor within 10 min of forced interaction (Figure 1H). The short residence time of Nup62 within the ciliary gating zone is similar to its dynamics in the NPC [33–35]. The rapid mobility of nucleoporins such as Nup62 may enable structural and compositional flexibility of both nuclear and ciliary barriers. Consistent with this, forced crosslinking of Nup62 resulted in attenuated entry of cytosolic proteins into both the nuclear and the ciliary compartments [24].
The MKS complex proteins B9D1 and AHI1 contain lipid-binding domains and both proteins showed BiFC interactions with transiting transmembrane proteins (Figure 7A), indicating that these MKS proteins map to the outer region of the ciliary gating zone (Figure 7B) and are thus positioned to regulate the entry of membrane proteins into the ciliary compartment. This is consistent with recent super-resolution imaging in mammalian cells that localized the MKS module components TMEM67, TMEM237, and AHI1 to a ring whose diameter corresponds to that of the ciliary membrane [12, 27, 31]. As B9D1 and AHI1 are peripheral membrane proteins, we were surprised to find that they are stable components of the ciliary gating zone (Figure 7B). Both proteins show a large immobile fraction in the FRAP assay (Figure 5), consistent with recent analysis of MKS module proteins in C. elegans [12]. In addition, the transiting SSTR3 transmembrane protein required several hours of forced interaction to drag B9D1 out of the transition zone (Figure 6). We therefore propose that that the MKS complex plays both a structural role in the transition zone and a functional role in regulating entry of membrane proteins.
In contrast to the NUP and MKS components which localize largely to the inner and outer regions of the ciliary gating zone, respectively, the NPHP components appear to be more widely distributed. For NPHP5, the BiFC interactions indicate that this protein is accessible throughout the inner-outer axis of the ciliary gating zone (Figure 7A). NPHP5 forms a complex with CEP290/NPHP6, and the localization of these proteins along the longitudinal axis of the ciliary gating zone ranges from basal body to transition zone [19, 25, 27, 36–38]. CEP290 is a dynamic component of the transition zone in Chlamydomonous [19] and we were thus surprised to find that NPHP5 is largely immobile at the cilium base in mammalian cells (Figure 7B). Being a stable component of the ciliary gating zone, NPHP5 may play a structural role and NPHP6 may regulate its turnover and/or function during ciliary gating.
For NPHP4, its N-terminal region likely occupies the most outer position within the ciliary gating zone as it interacts only with the transmembrane protein SSTR3 (Figure 7A). However, the C-terminal region of NPHP4 maps to the innermost region of the ciliary gating zone, close to the doublet microtubules of the axoneme, based on its ability to interact only with the cytosolic proteins KIF17 and Gli2 (Figure 7A). This is consistent with reports that NPHP4’s partner protein NPHP8 (RPGRIP1L) localizes close to the doublet microtubules in mammalian cells [12, 27], NPHP4 shows genetic interactions with OSM-3 (KIF17 homologue) in C. elegans [39], and NPHP4 can co-immunoprecipitate α-tubulin [40]. The ability of NPHP4 to physically span the inner-to-outer axis of the ciliary gating zone suggests that NPHP4, together with its partner proteins NPHP1 and NPHP8 [14, 25], may play a structural role and thereby impact the gated entry of both membrane and cytosolic proteins into the ciliary compartment [18, 20]. However, NPHP4 is a dynamic component of the cilium base in mammalian cells (Figure 7B) despite its apparent stability at the base of Chlamydomonas flagella [18]. These results may reflect differences in the structural organization of the ciliary gating zone among species and indicate that further work is clearly needed to uncover the localization and function of NPHP subcomplexes.
A major advantage to the BiFC assay is that it enables direct visualization of protein interactions in living cells with limited cell perturbation and without specialized equipment or data processing [41, 42]. BiFC can report on interactions that are weak and/or transient, direct or indirect, and occur among a subpopulation of proteins. In our hands, the BiFC interactions were independent of protein expression level, however, like other protein-protein interaction assays, the BiFC assay is subject to both false positive interactions (e.g. nonspecific self-assembly of the fluorophore) and false negative interactions (e.g. interacting proteins are unable to reconstitute YFP). Techniques such as superresolution and electron microscopy are needed to confirm the spatial localizations of gate components suggested by our interaction analysis.
The finding that components of the ciliary gate are dynamic seems, at first glance, antithetical to the function of ciliary gating. However, there are many examples in biology where a macromolecular complex may itself be a stable entity and yet the components of the complex undergo rapid turnover, including intermediate filaments, adherens junctions, and nuclear pore complexes [34, 43–45]. In addition, the ciliary gate itself has been shown to be dynamic in terms of migrating along the axoneme during Drosophila spermatogenesis [7], and turnover of gate components may facilitate this migration event. Finally, given then ability of cilia to shed ectosomes from their tips [46–50], gating/sorting mechanisms likely exist at both the base and the tip of cilia that together regulate the protein and lipid content of the organelle.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kristen J. Verhey (kjverhey@umich.edu). The use of mNeonGreen [51] is covered by an MTA with Allele Biotechnology.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Male NIH 3T3 cells, purchased from ATCC Cat#CRL-1658 RRID:CVCL_0594, were grown in DMEM (Gibco) supplemented with 10% fetal clone III (Hyclone) and 1% GlutaMAX (Gibco) at 37°C and 5% CO2.
METHOD DETAILS
Antibodies and plasmids
Commercial antibodies for acetylated α-tubulin (1:10,000, Sigma, 6-11B-1) and the epitope tags Myc (1:500, Sigma, C3956) and HA (1:1,000, Covance, 16B12) were used. Secondary antibodies for immunofluorescence were from Jackson ImmunoResearch and Invitrogen (1:500). All plasmids were constructed by subcloning via convenient restrictions sites or PCR into a pEGFP-N1 or pEGFP-C1 backbone. All constructs utilize full-length proteins with cDNAs derived from the following plasmids: pN1-Arl13b-mCherry [23], pN1-KIF17-mCitrine [52], pCMV-Nup62-EGFP3 (purchased from EUROSCARF [34]), pCDNA3-6xMYC-Gli2 [53], pCMV-SSTR3-EGFP (gift of Dr. Brad Yoder [54]), pDONR-GRP161 (purchased from DNASU), pENTR-NPHP4 and pENTR-NPHP5 (gifts of Dr. Friedhelm Hildebrandt [55]), pGLAP5-B9D1 (gift of Dr. Jeremy Reiter [9]), and pC3-EGFP-AHI1 (Addgene #30494, gift of Dr. Joseph Gleeson [56]). The 30 nm SAH domain from Trichomonas vaginalis Kelch-motif family protein has been described [57, 58].
BiFC constructs
We began our studies using fragments of the YFP variant Venus [N-terminal aa1–172, (Addgene plasmid #22010) and C-terminal aa155–238/A206K (Addgene plasmid #22011), gifts from Dr. Chang-Deng Hu] because of its high efficiency in BiFC [59]. However, even the low levels of Venus self-assembly resulted in a background cytoplasmic fluorescence that obscured the positive BiFC signal. Previous work indicated that the optimal choice of YFP variants depends on the experimental conditions [60], and we found that the combination of an N-terminal fragment (aa1–172) of the YFP variant mCitrine with the C-terminal fragment (aa155–238) of Venus resulted in low self-assembly (and low background fluorescence) but high BiFC fluorescence. Thus, for the work reported in this paper, the YFP N-terminal (YN) fragment comprises the N-terminal half of mCitrine whereas the YFP C-terminal (YC) fragment comprises the C-terminal half of Venus. Protein expression and localization was assessed by immunostaining for epitope tags and/or by fluorescent protein tags, however, for simplicity only the YN or YC tag is indicated throughout the main text. See Table S1 for detailed information about all of the BiFC constructs.
Serum-starvation and transfection
NIH 3T3 cells were switched to serum-free media (to arrest cells in G1 and increase the number of ciliated cells in the population), transfected 8 h later using 1.0–1.5 μg of DNA plus 3–4.5 μl of Trans-IT (Mirus) per 35 mm dish, and then fixed or imaged after an additional 16 h. Generally, 20–30% of cells expressed both fusion proteins. The entire slide was scanned for a visual assessment of protein expression levels and only cells expressing the fusion proteins at the lowest levels were imaged and analyzed. Each experiment was repeated ≥3 times and the data were pooled. For FRAP analysis, cell lines stably expressing B9D1-NeonGreen, EGFP-AHI1, mCherry-NPHP4, or EGFP-NPHP5 were generated using FRT plasmids and integration into NIH 3T3 Flp-In cells (Life Technologies RRID:CVCL_U422 ) by selection in 150 μg/ml hygromycin B as described [24].
Microscopy
For BiFC interaction analysis, the cilium itself and the expressed proteins were detected by fluorescent protein tags or with antibodies to epitope tags (see Table S1). The cells were fixed with 3.7% paraformaldehyde for 10 min, quenched with 50 mM NH4Cl in PBS for 5 min, permeabilized with 0.2% TX-100 in PBS for 5 min, and blocked in 0.2% fish skin gelatin in PBS for 20 min. The cells were stained with primary and secondary antibodies for 1h each at room temperature. Fluorescence images were obtained using an inverted epifluorescence microscope (Nikon TE2000-E) with 60x oil immersion objective (N.A. 1.4) and a Photometrics CoolSnap HQ camera. Cells were selected for imaging based on the expression of both BiFC partner proteins (Cer fluorescence or staining for the Myc or HA tags) and then the BiFC image (YFP filter) was obtained without prior observation. The same imaging parameters (camera, time of acquisition, filter sets, etc) were used for each fluorescence channel across experiments and thus the arbitrary units of fluorescence are comparable across figures and panels.
For FKBP-FRB interaction analysis, 20 ng/ml rapamycin (EMD Milipore) was added to the culture medium for the indicated times before fixation or the same volume of ethanol was added as a control. The cells were fixed, stained, and imaged as described above for the BiFC assay.
For analysis of protein dynamics by FRAP, the cells were imaged live and analyzed as described [61]. A Nikon A1 confocal system with 60x water immersion objective (N.A. 1.20) equipped with a stage-top incubator was used. Fluorescence intensities were measured using Image J software (NIH). After background subtraction, the fluorescence intensities of the ROI were normalized against the averaged intensities of the three pre-photobleaching images.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all figures, n represents the number of cells imaged across 3 or more experiments. For quantification of BiFC localizations, the fluorescence intensities at the base, tip, and shaft of the cilium was measured using Image J for each channel (each interacting protein and BiFC) and the background fluorescence of the cytoplasmic region next to the cilium was subtracted. A positive BiFC interaction was noted if the fluorescence intensity in the YFP channel was higher than that of the negative control (no BiFC interaction). The base versus tip of the cilium was defined based on the acetylated tubulin or Arl13b staining. For example, for acetylated tubulin staining, the base of the cilium is brighter and thicker and cytoplasmic microtubules can often be seen emanating from the base. In addition, gating zone components can localize adjacent to the cilium marker, whereas the localization of cilium tip proteins (e.g. KIF17 and Gli2) generally overlaps with the cilium marker.
For Nup62-FRB and KIF17-FKBP interaction analysis (Figure 1 G–I), the fluorescence intensities at the base or tip of cilia were quantified as follows. First, small ROIs at the tip and base of a cilium were selected and the mean fluorescence of each ROI was obtained in both the Nup62 and KIF17 channels. After background subtraction, the Nup62 fluorescence at both the base and the tip was divided by the Nup62 base fluorescence whereas the KIF17 fluorescence at both the base and the tip was divided by the KIF17 tip fluorescence. Thus, the graphs in Figure 1G–I indicate the base (base/base) versus tip (tip/base) fluorescence for Nup62 or the base (base/tip) versus tip (tip/tip) fluorescence for KIF17 and thus graphically depict how the amount of Nup62-FRB at the tip increased relative to the base whereas the amount of KIF17-FKBP at the base increased relative to the tip during the time course. The data in Figure 1G–I and Figure 5 are presented as the mean +/− standard deviation. A two-tailed Student’s t test (Excel) was used to examine the significance of fluorescence intensity differences. No methods were used to determine whether the data met assumptions of the statistical approach.
Supplementary Material
HIGHLIGHTS.
-
-
regulated entry at the ciliary gating zone defines the composition of the organelle
-
-
NUP and NPHP module proteins interact with an IFT motor transiting the gate
-
-
MKS and NPHP module proteins interact with membrane proteins transiting the gate
-
-
NUP, NPHP, and MKS components differ in their turnover at the gating zone
Acknowledgments
This work was funded by a grant from NIH to K.J.V (R01GM116204) and a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science to D.T. We gratefully acknowledge S. Lentz and the Morphology and Image Analysis Core of the Michigan Diabetes Research and Training Center funded by NIH 5P60DK20572 from the National Institute of Diabetes & Digestive & Kidney Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
D.T. and K.J.V designed the experiments; D.T., L.W. and A.B. performed the experiments and analyzed the data. D.T. and K.J.V wrote the manuscript. All authors proofread the manuscript.
References
- 1.Fry AM, Leaper MJ, Bayliss R. The primary cilium: guardian of organ development and homeostasis. Organogenesis. 2014;10:62–68. doi: 10.4161/org.28910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madhivanan K, Aguilar RC. Ciliopathies: the trafficking connection. Traffic. 2014;15:1031–1056. doi: 10.1111/tra.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takao D, Verhey KJ. Gated entry into the ciliary compartment. Cell Mol Life Sci. 2016;73:119–127. doi: 10.1007/s00018-015-2058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhey KJ, Yang W. Permeability barriers for generating a unique ciliary protein and lipid composition. Curr Opin Cell Biol. 2016;41:109–116. doi: 10.1016/j.ceb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis EE, Katsanis N. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr Opin Genet Dev. 2012;22:290–303. doi: 10.1016/j.gde.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basiri ML, Ha A, Chadha A, Clark NM, Polyanovsky A, Cook B, Avidor-Reiss T. A migrating ciliary gate compartmentalizes the site of axoneme assembly in Drosophila spermatids. Curr Biol. 2014;24:2622–2631. doi: 10.1016/j.cub.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2012;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- 9.Dowdle WE, Robinson JF, Kneist A, Sirerol-Piquer MS, Frints SG, Corbit KC, Zaghloul NA, van Lijnschoten G, Mulders L, Verver DE, et al. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet. 2011;89:94–110. doi: 10.1016/j.ajhg.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen VL, Li C, Bowie RV, Clarke L, Mohan S, Blacque OE, Leroux MR. Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J. 2015;34:2537–2556. doi: 10.15252/embj.201488044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambacher NJ, Bruel AL, van Dam TJ, Szymanska K, Slaats GG, Kuhns S, McManus GJ, Kennedy JE, Gaff K, Wu KM, et al. TMEM107 recruits ciliopathy proteins to subdomains of the ciliary transition zone and causes Joubert syndrome. Nat Cell Biol. 2016;18:122–131. doi: 10.1038/ncb3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberson EC, Dowdle WE, Ozanturk A, Garcia-Gonzalo FR, Li C, Halbritter J, Elkhartoufi N, Porath JD, Cope H, Ashley-Koch A, et al. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J Cell Biol. 2015;209:129–142. doi: 10.1083/jcb.201411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, Bialas NJ, Stupay RM, Chen N, Blacque OE, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams CL, Winkelbauer ME, Schafer JC, Michaud EJ, Yoder BK. Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditis elegans ciliogenesis. Mol Biol Cell. 2008;19:2154–2168. doi: 10.1091/mbc.E07-10-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yee LE, Garcia-Gonzalo FR, Bowie RV, Li C, Kennedy JK, Ashrafi K, Blacque OE, Leroux MR, Reiter JF. Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling. PLoS Genet. 2015;11:e1005627. doi: 10.1371/journal.pgen.1005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C, Malicki J. Nephrocystins and MKS proteins interact with IFT particle and facilitate transport of selected ciliary cargos. EMBO J. 2011;30:2532–2544. doi: 10.1038/emboj.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awata J, Takada S, Standley C, Lechtreck KF, Bellve KD, Pazour GJ, Fogarty KE, Witman GB. Nephrocystin-4 controls ciliary trafficking of membrane and large soluble proteins at the transition zone. J Cell Sci. 2014;127:4714–4727. doi: 10.1242/jcs.155275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jauregui AR, Nguyen KC, Hall DH, Barr MM. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J Cell Biol. 2008;180:973–988. doi: 10.1083/jcb.200707090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Viso F, Huang F, Myers J, Chalfant M, Zhang Y, Reza N, Bewersdorf J, Lusk CP, Khokha MK. Congenital Heart Disease Genetics Uncovers Context-Dependent Organization and Function of Nucleoporins at Cilia. Dev Cell. 2016;38:478–492. doi: 10.1016/j.devcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endicott SJ, Basu B, Khokha M, Brueckner M. The NIMA-like kinase Nek2 is a key switch balancing cilia biogenesis and resorption in the development of left-right asymmetry. Development. 2015;142:4068–4079. doi: 10.1242/dev.126953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takao D, Dishinger JF, Kee HL, Pinskey JM, Allen BL, Verhey KJ. An assay for clogging the ciliary pore complex distinguishes mechanisms of cytosolic and membrane protein entry. Curr Biol. 2014;24:2288–2294. doi: 10.1016/j.cub.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang L, Szymanska K, Jensen VL, Janecke AR, Innes AM, Davis EE, Frosk P, Li C, Willer JR, Chodirker BN, et al. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am J Hum Genet. 2011;89:713–730. doi: 10.1016/j.ajhg.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang TT, Su J, Wang WJ, Craige B, Witman GB, Tsou MF, Liao JC. Superresolution Pattern Recognition Reveals the Architectural Map of the Ciliary Transition Zone. Sci Rep. 2015;5:14096. doi: 10.1038/srep14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoelz A, Glavy JS, Beck M. Toward the atomic structure of the nuclear pore complex: when top down meets bottom up. Nat Struct Mol Biol. 2016;23:624–630. doi: 10.1038/nsmb.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YL, Sante J, Comerci CJ, Cyge B, Menezes LF, Li FQ, Germino GG, Moerner WE, Takemaru K, Stearns T. Cby1 promotes Ahi1 recruitment to a ring-shaped domain at the centriole-cilium interface and facilitates proper cilium formation and function. Mol Biol Cell. 2014;25:2919–2933. doi: 10.1091/mbc.E14-02-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk CP, Poolman B, Veenhoff LM. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333:90–93. doi: 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- 33.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 35.Sakiyama Y, Mazur A, Kapinos LE, Lim RY. Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat Nanotechnol. 2016;11:719–723. doi: 10.1038/nnano.2016.62. [DOI] [PubMed] [Google Scholar]
- 36.Barbelanne M, Song J, Ahmadzai M, Tsang WY. Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Hum Mol Genet. 2013;22:2482–2494. doi: 10.1093/hmg/ddt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole JF, Helou J, Attanasio M, et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 38.Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masyukova SV, Landis DE, Henke SJ, Williams CL, Pieczynski JN, Roszczynialski KN, Covington JE, Malarkey EB, Yoder BK. A Screen for Modifiers of Cilia Phenotypes Reveals Novel MKS Alleles and Uncovers a Specific Genetic Interaction between osm-3 and nphp-4. PLoS Genet. 2016;12:e1005841. doi: 10.1371/journal.pgen.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mollet G, Silbermann F, Delous M, Salomon R, Antignac C, Saunier S. Characterization of the nephrocystin/nephrocystin-4 complex and subcellular localization of nephrocystin-4 to primary cilia and centrosomes. Hum Mol Genet. 2005;14:645–656. doi: 10.1093/hmg/ddi061. [DOI] [PubMed] [Google Scholar]
- 41.Kerppola TK. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol. 2008;85:431–470. doi: 10.1016/S0091-679X(08)85019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kudla J, Bock R. Lighting the Way to Protein-Protein Interactions: Recommendations on Best Practices for Bimolecular Fluorescence Complementation Analyses. Plant Cell. 2016;28:1002–1008. doi: 10.1105/tpc.16.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon KH, Yoon M, Moir RD, Khuon S, Flitney FW, Goldman RD. Insights into the dynamic properties of keratin intermediate filaments in living epithelial cells. J Cell Biol. 2001;153:503–516. doi: 10.1083/jcb.153.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Beco S, Gueudry C, Amblard F, Coscoy S. Endocytosis is required for E-cadherin redistribution at mature adherens junctions. Proc Natl Acad Sci U S A. 2009;106:7010–7015. doi: 10.1073/pnas.0811253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Silva M, Haas LA, Morsci NS, Nguyen KC, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. Elife. 2015;4:e05242. doi: 10.7554/eLife.05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nager AR, Goldstein JS, Herranz-Perez V, Portran D, Ye F, Garcia-Verdugo JM, Nachury MV. An Actin Network Dispatches Ciliary GPCRs into Extracellular Vesicles to Modulate Signaling. Cell. 2017;168:252–263 e214. doi: 10.1016/j.cell.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, Pusapati GV, Setou M, Rohatgi R, Reiter JF, Ikegami K, et al. Dynamic Remodeling of Membrane Composition Drives Cell Cycle through Primary Cilia Excision. Cell. 2017;168:264–279. doi: 10.1016/j.cell.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carpenter BS, Barry RL, Verhey KJ, Allen BL. The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. J Cell Sci. 2015;128:1034–1050. doi: 10.1242/jcs.162552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Connor AK, Malarkey EB, Berbari NF, Croyle MJ, Haycraft CJ, Bell PD, Hohenstein P, Kesterson RA, Yoder BK. An inducible CiliaGFP mouse model for in vivo visualization and analysis of cilia in live tissue. Cilia. 2013;2:8. doi: 10.1186/2046-2530-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schafer T, Putz M, Lienkamp S, Ganner A, Bergbreiter A, Ramachandran H, Gieloff V, Gerner M, Mattonet C, Czarnecki PG, et al. Genetic and physical interaction between the NPHP5 and NPHP6 gene products. Hum Mol Genet. 2008;17:3655–3662. doi: 10.1093/hmg/ddn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lancaster MA, Louie CM, Silhavy JL, Sintasath L, Decambre M, Nigam SK, Willert K, Gleeson JG. Impaired Wnt-beta-catenin signaling disrupts adult renal homeostasis and leads to cystic kidney ciliopathy. Nat Med. 2009;15:1046–1054. doi: 10.1038/nm.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Norris SR, Soppina V, Dizaji AS, Schimert KI, Sept D, Cai D, Sivaramakrishnan S, Verhey KJ. A method for multiprotein assembly in cells reveals independent action of kinesins in complex. J Cell Biol. 2014;207:393–406. doi: 10.1083/jcb.201407086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sivaramakrishnan S, Sung J, Ali M, Doniach S, Flyvbjerg H, Spudich JA. Combining single-molecule optical trapping and small-angle x-ray scattering measurements to compute the persistence length of a protein ER/K alpha-helix. Biophys J. 2009;97:2993–2999. doi: 10.1016/j.bpj.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 60.Saka Y, Hagemann AI, Piepenburg O, Smith JC. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development. 2007;134:4209–4218. doi: 10.1242/dev.010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takao D, Verhey KJ. Methods for Studying Ciliary Import Mechanisms. Methods Mol Biol. 2016;1454:1–14. doi: 10.1007/978-1-4939-3789-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


