Abstract
Automated cell-based high-throughput screening (HTS) is a powerful tool in drug discovery, and it is increasingly being recognized that three-dimensional (3D) models, which more closely mimic in vivo-like conditions, are desirable screening platforms. One limitation hampering the development of 3D HTS is the lack of suitable 3D culture scaffolds that can readily be incorporated into existing HTS infrastructure. We now show that β-hairpin peptide hydrogels can serve as a 3D cell culture platform that is compatible with HTS. MAX8 β-hairpin peptides can physically assemble into a hydrogel with defined porosity, permeability and mechanical stability with encapsulated cells. Most importantly, the hydrogels can then be injected under shear-flow and immediately reheal into a hydrogel with the same properties exhibited prior to injection. The post-injection hydrogels are cell culture compatible at physiological conditions. Using standard HTS equipment and medulloblastoma pediatric brain tumor cells as a model system, we show that automatic distribution of cell-peptide mixtures into 384-well assay plates results in evenly dispensed, viable MAX8-cell constructs suitable for commercially available cell viability assays. Since MAX8 peptides can be functionalized to mimic the microenvironment of cells from a variety of origins, MAX8 peptide gels should have broad applicability for 3D HTS drug discovery.
Keywords: beta-hairpin hydrogel, three-dimensional cell culture, high-throughput drug discovery, medulloblastoma
1. Introduction
High-throughput screening (HTS) remains a promising initial step in phenotypic drug discovery. However, most cell-based HTS is done using well-established two-dimensional (2D) cultures which often fail to predict in vivo efficacy of candidate compounds, thereby contributing to the high failure rate and cost of drug development. This is particularly true for cell line models of cancer [1]. While 2D cultures are more convenient and can be automated easily, 3D systems better mimic microenvironments including concentration gradients of nutrients and oxygen and mechanical cues, both of which are particularly important in tumor development [1–3]. Due to these differences, cells grown in 3D respond differently to stimuli and drug treatments, and increasing use of 3D cultures as a platform for drug discovery is expected to result in more candidate compounds with high in vivo efficacy and potential for future preclinical studies and clinical drug development.
Hydrogels are dilute polymer or supramolecular networks with a high water content, over 95% by volume, and obtain their structure through intermolecular or interfibrillar crosslinks [4–6]. Nanofibrillar hydrogels are well-suited as a 3D cell culture scaffold due to their similarity to the extracellular matrix; when properly designed, synthetic hydrogels can mimic the physical and biological properties of in vivo cell environments [7–12]. β-hairpin hydrogels consist of peptides that fold into β-hairpin conformation and then undergo hydrophobic collapse and hydrogen bonding into nanofibrils with a hydrophobic core [13]. We used MAX8 [14] (Fig. 1) as our scaffold for HTS.
Figure 1. MAX8 β-hairpin hydrogel.
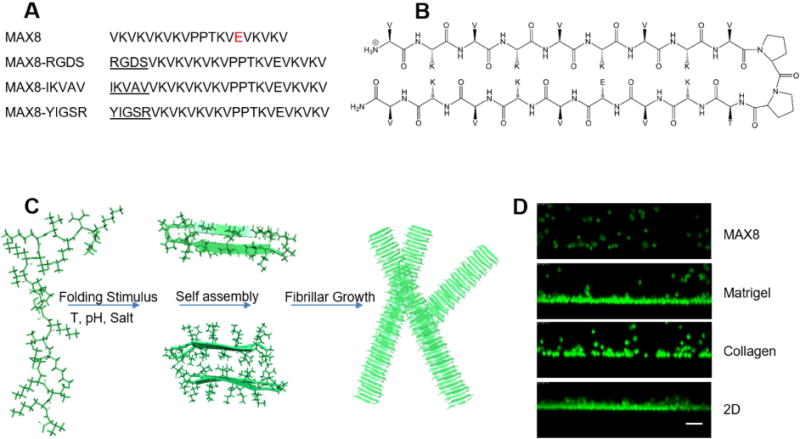
(A) Peptide sequence of MAX8. MAX8 modifications where adhesive peptide sequences were added at the N-terminal end of the peptide are shown. (B) Molecular diagram of MAX8 in folded state. (C) Peptides fold into β-hairpin conformation under physiological conditions. Top center, folded peptide viewed looking down at folded β-hairpin. After folding, hairpins undergo hydrophobic collapse and hydrogen bonding into nanofibrils with a hydrophobic core of valine side chains and diameter of ~3 nm [13, 15]. Bottom center, a view along a fibril axis with the hydrophobic valine core. The fibrils branch and entangle to form the hydrogel network [16, 17]. (D) Encapsulation of ONS-76 medulloblastoma cells in MAX8, Matrigel, collagen and 2D cultures on glass coverslips showing the even distribution of cells throughout MAX8 as cells will not settle to the bottom of the well due the fast gelation kinetics. Cells were stained with Syto 13 and XZ confocal images were acquired with a Leica TCS SP5 laser scanning confocal microscope. Bar, 100 μm.
MAX8 is a self-assembling peptide that possesses all the features of an excellent candidate for development as a 3D cell culture matrix that can be dispensed automatically using standard HTS equipment. MAX8 undergoes assembly under physiological conditions into a hydrogel with a well-defined, nanofibrillar matrix, desired porosity and stiffness and can be shear-thin injected as a solid material [18, 19]. The physical gel properties can be easily adjusted by modulating peptide sequence [14], peptide concentration or ionic strength of the culture medium to mimic the tissue environment for different cell lines [15]. With this, defined MAX8-cell constructs can be assembled at in vitro conditions to form hydrogels with known properties that can be injected and have the same properties post-injection. Due to the fast gelation kinetics, cells can be homogenously encapsulated into gel-cell constructs without settling at the bottom (Fig. 1D) [14, 18–20]. Most importantly, there is no need for additional covalent crosslinkers that may damage cells, and, unlike collagen or Matrigel that need to be handled at low temperatures, MAX8 and related β-hairpin peptides can be handled at room temperature.
We chose medulloblastoma cells to establish MAX8 as a cell culture scaffold for HTS drug discovery. Medulloblastoma is the most common malignant brain tumor in children that still has high cancer-related mortality and survivors often suffer from serious therapy-related side effects [21–23]. Arising in the cerebellum, medulloblastoma is divided into four distinct subgroups: Wnt, Shh (Sonic hedgehog), and Groups 3 and 4 [22, 24]. Tumors of the Wnt group have a fairly good prognosis, but effective therapies for the other groups have yet to be identified. Human medulloblastoma cells grown in 3D neurosphere cultures express more tumor-like immature features and display increased matrix metalloproteinase (MMP) levels and invasiveness as compared to cells grown in 2D monolayers [25, 26]. Using human medulloblastoma cell lines and MAX8 we employed a liquid-handling workstation to dispense an injectable solid gel-cell mixture into 96- and 384-well plates to form reproducible hydrogel-cell constructs. We demonstrate that the RealTime-Glo MT cell viability assay is compatible with this setup and was easily optimized to provide a robust signal of viable cells. These results suggest that MAX8 provides a versatile, easy-to-use 3D cell culture scaffold that can be incorporated into standard high-throughput screening operations for drug discovery.
2. Materials and Methods
2.1. MAX8 β-hairpin peptide synthesis
The synthesis and purification of MAX8 β-hairpin peptide has been described previously in detail [14, 19]. Synthesis of MAX8 used in the current study was performed with an automated AAPPTEC peptide synthesizer, using standard Fmoc-based solid phase peptide synthesis. For functionalized peptides, the RGDS, IKVAV or YIGSR sequence was added on the N-terminus to the native MAX8 peptide VKVKVKVK-(VDPPT)-KVEVKVKV-NH2 by including the ligands in the original peptide synthesis resulting in the following peptides, RGDS-VKVKVKVK-(VDPPT)-KVEVKVKV-NH2, IKVAV-VKVKVKVK-(VDPPT)-KVEVKVKV-NH2 and, YIGSR-VKVKVKVK-(VDPPT)-KVEVKVKV-NH2 (Fig. 1A)
2.2. Oscillatory rheology
Rheology measurements were performed on an AR G2 rheometer (TA instruments) with a 20 mm stainless steel parallel plate geometry as described previously [27]. After mixing a peptide solution with buffer, samples (170 μl) were loaded immediately onto the temperature controlled (37°C) Peltier plate with a gap of 500 μm. Mineral oil was added around the circumference of the geometry to prevent dehydration of the hydrogel. Dynamic time sweep experiments (DTS) were performed to monitor the storage (G′) and loss (G″) modulus as a function of time (6 rad/s frequency, 0.2% strain) for 60 min. For shear-thinning experiments, the samples were subjected to 500 s−1 steady-state shear for 60 s, after which oscillatory measurement was performed at 6 rad/s frequency, 0.2% strain. Subsequently, recovery of the storage (G′) and loss (G″) modulus as a function of time was monitored for 30 min. Dynamic frequency (0.1–100 rad/s frequency, 1.0% strain) sweep experiments were performed to establish the frequency response of the samples. The hydrogels were well within the linear viscoelastic regime during oscillatory measurements as previously reported [28]. All measurements were done in triplicate.
2.3. Preparation of basic hydrogel-cell constructs
Human medulloblastoma DAOY cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and were cultured in Minimum Essential Medium (MEM). ONS-76 cells were kindly provided by Dr. Michael Taylor and were propagated in Dulbecco’s Minimum Essential Media (DMEM). DAOY and ONS-76 were maintained at 37°C and 5% CO2 in MEM and DMEM, respectively, supplemented with 10% fetal bovine serum and penicillin-streptomycin-glutamine, using standard 2D cell culture protocols and tissue-culture treated plastic labware. For isolation of primary cerebellar granule precursor (CGP) cells, the cerebellum was dissected from P4-6 pups of C57BL/6 mice and dissociated into single cells using the Papain Dissociation System kit (Worthington Biochemical Corp, Freehold, NJ). After filtering through a nylon mesh (70 μm pore size), the cells were briefly centrifuged and resuspended in neurobasal medium supplemented with 0.25 mM KCl and B27.
Unless otherwise indicated, MAX8-cell constructs were prepared as 0.25 wt% MAX8 hydrogels. First, the peptide was dissolved in 50 mM HEPES buffer (pH 7.4) (0.5 mg MAX8 per 100 μl of buffer) and then an equal volume of cell suspension in serum-free DMEM was added and gently mixed. Mixing the MAX8 solution with the culture medium triggers the intramolecular folding of the peptide resulting in self-assembly into a hydrogel. For additional gels with different concentrations of MAX8, the amount of peptide was adjusted but otherwise the same gel assembly protocol was followed. The same procedures were followed when ligand-functionalized MAX8 was used to prepare hydrogel-cell constructs.
2.4. Assessment of cell viability in MAX8-cell constructs
Unless noted otherwise, cell viability assays were performed in 384-well plates using the RealTime-Glo MT cell viability assay from Promega. For each experiment two stock solutions were prepared. For the first solution, lyophilized MAX8 was mixed into 50 mM HEPES (pH 7.4) buffer to create a solution of 0.5 wt% (5 mg MAX8 per ml buffer solution). The second solution was a cell mixture with 1 × 106 cells per ml (unless noted otherwise) in serum-free DMEM. The two stock solutions were thoroughly mixed 1:1 to create a gel-cell mixture (0.25 wt% MAX8 gel construct with 500 cells per μl). 4 μl was added per well to a white 384-well assay plate (Brandtech) containing 36 μl culture medium per well. The cells were allowed to equilibrate for 24 hours at 37°C and 5% CO2, after which the RealTime-Glo reagent (5x) was added in 10 μl of media to each well. The plate was incubated for 120 minutes at 37°C before measuring cell luminescence with an Envision multimode plate reader (Perkin Elmer, Waltham, MA). A series of cell growth curves were completed to determine the ideal number of cells to encapsulate. Starting with 6,400 cells per well, encapsulated in the previously described format, and diluted by half down to 100 cells per well, cell proliferation was measured every 12 hours using the RealTime-Glo MT cell viability assay. The experiment was repeated in 2D for comparison. From these growth curves, 2,000 cells per well for 3D and 1,500 for 2D cultures were chosen for further experiments in a 384-well format. For cell viability assays in 96-well plates, 64 μl of serum-containing cell culture medium was added to each well, followed by 16 μl of the 0.25 wt% MAX8 gel-cell mixture. Cell viability was determined using 20 μl of 5x RealTime-Glo reagent per well.
2.5. Treatment of MAX8-encapsulated medulloblastoma cells with chemotherapeutics
The chemotherapeutics cisplatin, vorinostat (suberanilohydroxamic acid, SAHA) and vismodegib were added at indicated concentrations into the surrounding tissue culture medium of MAX8-ONS-76 gel-cell constructs or 2D cell cultures in 384-well plates after 24 hours of cell seeding. For direct comparison, both 2D and 3D cell cultures were seeded with the BioTek microplate dispenser taking advantage of the shear thinning properties of MAX8. To limit the exposure of cultured cells to dimethyl sulfoxide (DMSO) from stock solutions of drug compound, the compound stocks were first serially diluted in DMSO and then 2.5 μl of compound was added to 1 ml of culture medium and 10 μl of this intermediate dilution was added to the well for a final DMSO concentration of 0.05%. After 48 hours of treatment the cell viability was determined using the RealTime-Glo MT cell viability assay.
2.6. Confocal microscopy
DAOY cells expressing a td-Tomato-tagged protein were encapsulated into MAX8-RGDS as described above and allowed to grow for indicated times. ONS-76 cells encapsulated in MAX8-RGDS were stained with Syto 13 (Thermo Fisher Scientific) for visualization. Z-stacks of 80–100 μm height and a step-size of 1 μm were acquired with a Leica TCS SP5 laser-scanning confocal microscope (Leica Microsystems, Mannheim, Germany) and the LSM software was used to create XZ views.
2.7. Realtime PCR
Total RNA was extracted from 2D cell cultures grown for 72 hours in 24-well plates according to standard procedures. For 3D cultures, the gel-cell constructs of three wells of a 24-well plate, all three containing 100 μl gel-cell construct, were combined after 72 hours of culture. The samples were briefly centrifuged and the supernatant was removed before adding Trizol reagent. RNA was then isolated according to manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of RNA using the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). Quantitative PCR analysis was performed with a SYBR Green PCR master mix using an ABI Prism 7900 Sequence Detection System (both from Applied Biosystems, Foster City, CA) and normalized to GAPDH. The primer sequences used were: nestin, forward 5′-GAGAACTCCCGGCTGCAAAC-3′ and reverse 5′-CTTGGGGTCCTGAAAGCTGAG-3′; gli3, forward 5′-CGAACAGATGTGAGCGAGAA-3′ and reverse 5′TTGATCAATGAGGCCCTCTC-3′; and snail1, forward 5′-GAGCCCAGGCACTATTTCA-3′ and reverse 5′-TGGGAGACACATCGGTCAGA-3′.
2.8. QC plates
Using a BioTek microplate dispenser, MAX8-ONS-76 cell constructs (2,000 cells encapsulated in 4 μl of gel) were dispensed at room temperature into 384-well plates (Brand Tech Scientific, Essex, CT) containing 36 μl of culture medium. The cells were allowed to equilibrate for 24 hours at 37°C in the presence of 5% CO2 prior to adding SAHA in two steps: intermediate dilution of 50 nl of stock solution added by pintool into 20 μl of media, followed by transfer of 10 μl of SAHA in media into QC plates. Based on the IC50 value of 0.8 μM as determined by dose response studies, a stock solution of 2 mM of SAHA was made up in DMSO, giving a final concentration of 1 μM in all 320 test wells in each 384-well QC plate (columns 3–22). DMSO was added to the high control wells (columns 1 and 23) and low control wells (columns 2 and 24; hydrogel alone, no cells) by the same two-step addition via intermediate dilution as used for SAHA. Cell viability was determined after 48 hours of incubation using the RealTime-Glo MT cell viability assay.
2.9. Statistical analysis
Data are presented as mean ± SD unless otherwise indicated. Differences between means of two groups were analyzed with a two-tailed unpaired Student’s t-test and, when applicable, P values were determined with P < 0.05 denoting statistical significance. Dose response curves were generated using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Percent inhibition for the drug compounds tested in 3D and 2D cultures were determined by non-linear regression analyses and an F-test was used to determine statistical significance between the curves.
The signal-to-noise ratio of the cell viability assay was determined as:
The Z′-factor was calculated using the following equations with μ representing the mean and σ the standard deviation.
3. Results
3.1 MAX8 β-hairpin hydrogel
MAX8 is an amphiphilic peptide with the sequence VKVKVKVK-(VDPPT)-KVEVKVKV-NH2 (Fig. 1A). Gelation can be triggered at room temperature by physiological salt concentration and pH leading to shielding of the charged lysine residues. This causes the peptide to fold into a β-hairpin and associate into bilayer fibrils that form a network through local fibril entanglement and branching (Fig. 1B, C) [14, 29]. Physical gelation triggered by physiological conditions allows for easy culture setup without requiring the addition of chemicals or organic reagents that may interfere with cell growth or phenotype. Unlike commonly used 3D matrices such as collagen and Matrigel, MAX8 gelates immediately leading to a homogenous distribution of cells throughout the gel-cell construct (Fig. 1D). One critical property of MAX8 for 3D HTS is shear thinning from the solid hydrogel state that allows for gel injection while protecting cells from shear forces and immediate recover after injection [18, 19], thus making it suitable for handling with standard automated HTS equipment.
3.2. Establishment of 3D MAX8-cell constructs
Peptide hydrogels are excellent materials to use as 3D cell culture scaffolds because of the similarities between their material properties and those of biological extracellular matrix [4]. Even in the absence of adhesive ligands, native MAX8 is compatible with cells of various origins [14, 19, 20, 30–32] including the human medulloblastoma cell lines ONS-76 (Fig. 2A) and DAOY (data not shown) as well as primary neuronal cells (Fig. 2B). Both ONS-76 cells (Fig. 2A) and primary cerebellar granule precursors (CGPs) isolated from wild-type C57BL/6 mice were viable for several days within MAX8-cell constructs as determined by the RealTime-Glo MT cell viability assay.
Figure 2. Establishment of a robust cell viability assay.
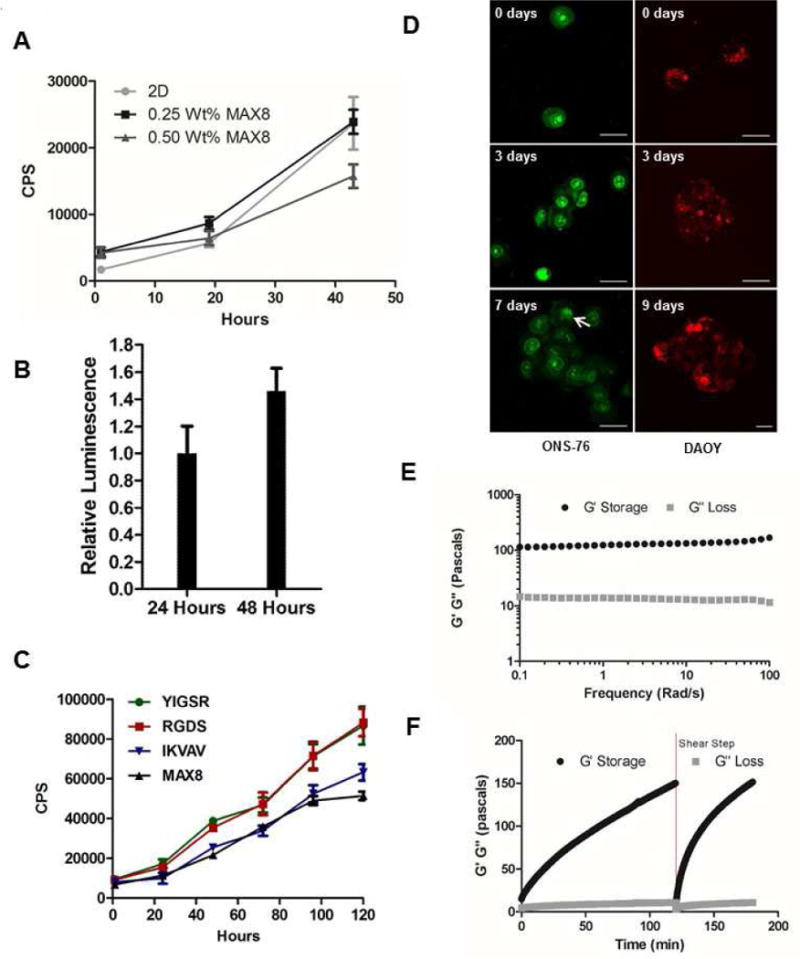
(A) ONS-76 cells (10,000 cells/well in a 96-well plate) were encapsulated in 0.25 wt % or 0.5 wt % MAX8, and cell viability was determined using the RealTime-Glo MT cell viability assay at indicated time points. Bars represent standard deviation (SD) of the mean, n = 3. CPS, counts per second. (B) Cell viability of primary mouse cerebellar granule precursor cells from C57BL/6 mice (CGP; 50,000 cells/well seeded in a 96-well plate) encapsulated in 0.5 wt % MAX8. Signal measured 48 hours after cell encapsulation was normalized relative to the baseline signal obtained after cells were allowed to equilibrate for 24 hours. Data represent mean and SD of five determinations. (C) Cell viability of ONS-76 cells encapsulated in 0.5 wt % native MAX8 or MAX8 functionalized with RGDS, IKVAV or YIGSR sequences. Cell viability was determined at indicated time points using the RealTime-Glo MT cell viability assay. Bars represent SD of the mean, n = 3. CPS, counts per second. (D) Confocal microscopy images of ONS-76 cells and DAOY cells cultured in MAX8-RGDS for indicated time points. The arrow indicates a mitotic cell. Bar, 25 μm. (E, F) Oscillatory rheology of MAX8-RGDS showing clear hydrogel behavior with the time sweep in F showing gelation kinetics before and after shear thinning (red dotted line) with immediate rehealing of MAX8-RGDS after network disruption. Black lines, G′ (storage modulus); grey lines, G″ (loss modulus).
Encapsulation of ONS-76 cells in MAX8 revealed that cell proliferation decreased with increasing MAX8 concentrations (Fig. 2A). This could be due to reduced diffusion of growth factors from the culture medium into the gel at higher peptide concentrations [33, 34] or increased stiffness of the hydrogel matrix itself. Addition of the RGDS ligand, a peptide sequence found in fibronectin that interacts with integrins and supports cell adhesion [35], enhanced the proliferation of ONS-76 cells encapsulated in MAX8-RGDS-cell constructs 1.7-fold over native MAX8 (Fig. 2C). Even after five days in culture, cells encapsulated in MAX8-RGDS showed robust proliferation while cells grown in native MAX8 appeared to plateau. MAX8-RGDS was not only compatible with ONS-76 cells but also with DAOY cells which grew in small cell spheres (Fig. 2D). Tagging of MAX8 with IKVAV or YIGSR, both ligands that are normally observed in laminin protein and support neuronal differentiation [13, 36], increased the proliferation of ONS-76 cells when compared to unmodified MAX8 (Fig. 2C). This increase in cell proliferation was mostly due to the presence of the adhesive sequences since the addition of the peptide sequence did not substantially change material properties: MAX8-RGDS showed a storage modulus/stiffness in frequency sweep measurements (Fig. 2E) as well as shear thinning properties (Fig. 2F) that were similar to that observed in unmodified MAX8 [14]. Based on these results we chose MAX8 tagged with RGDS sequence at 0.25 wt % peptide concentration to establish a 3D HTS screening platform.
3.3. Comparison of MAX8-cell constructs with 2D cultures
It is well established for cells of various origins [1–3, 37–39], including medulloblastoma [25, 26], that cells cultured in 2D differ from those in 3D cultures. We compared the expression profiles of three differentiation and stem cell markers in ONS-76 cells grown in 2D monolayers and in 3D hydrogels with MAX8 and RGDS-tagged MAX8 cell constructs (Fig. 3A). These markers have higher expression levels in tumors from Smo/Smo mice, a transgenic medulloblastoma mouse model, when compared to normal cerebellum isolated from wild-type C57BL/6 mice (Fig. 3B). Interestingly, while some variations were observed between hydrogel constructs with native MAX8 or RGDS-tagged MAX8, all 3D cultures had higher mRNA levels of nestin, snail and gli3 compared to ONS-76 monolayers, suggesting that culturing medulloblastoma cells in 3D supports a cancer cell phenotype more closely to the one observed in tumor tissue.
Figure 3. Comparison of 2D and 3D cultures.
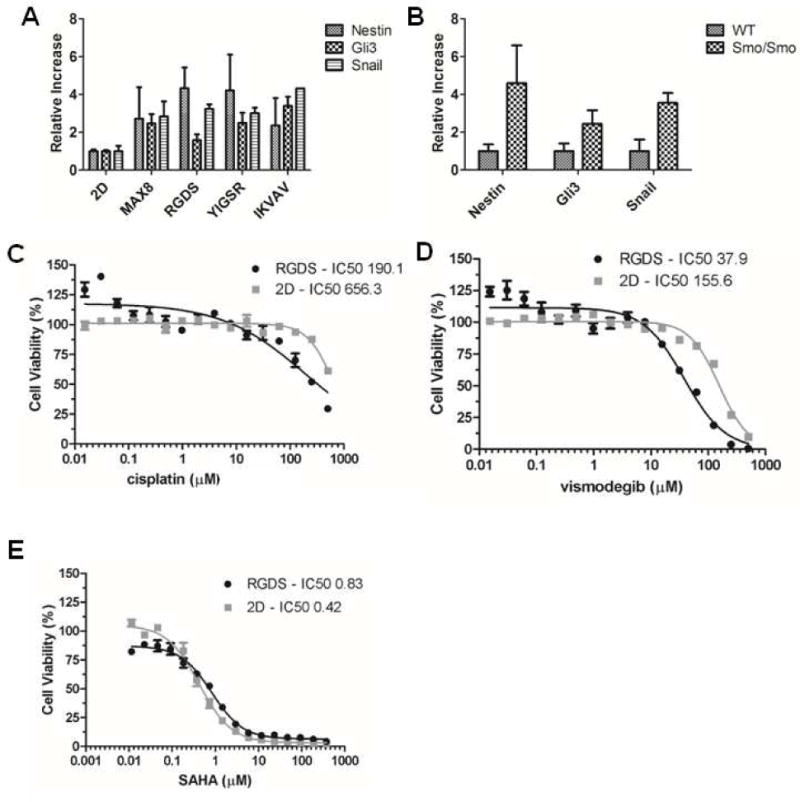
(A, B) Relative quantity of nestin, snail and gli3 mRNA in ONS-76 cells cultured in monolayers and in 3D hydrogel constructs, native or tagged with indicated adhesive peptides, as measured by qRT-PCR (A) and in Smo/Smo medullo-blastoma tumors and cerebella of wild-type mice (B). Bars represent SD of the mean, n = 3. (C–E) Dose response curves of cisplatin, vismodegib and vorinostat (SAHA) in ONS-76 cells cultured in 3D MAX8-RGDS constructs and in 2D monolayers. Cell viability was determined using the RealTime-Glo MT cell viability assay. Bars represent SD of the mean, n = 3. The 2D and 3D dose response curves for each test compound were compared in whole using the F-test. The test determined a significant difference in each case (p < 0.001). The t-test was used to compare the sets of IC50 values with the differences in all three sets being statistical significant (p < 0.001). The IC50 values of test compounds in 2D and 3D cultures are indicated in μM concentrations.
We also tested the sensitivity of ONS-76 cells in MAX-RGDS cell constructs and in 2D monolayers to commonly used chemotherapeutics such as vismodegib and cisplatin as well as the histone deacetylase (HDAC) inhibitor vorinostat (SAHA), which is in clinical trials for medulloblastoma. Interestingly, we found that cells cultured in 3D hydrogels were more sensitive to cisplatin and vismodegib as compared to 2D monolayers whereas the opposite was found for vorinosat (Fig. 3C–E). Thus, when medulloblastoma cells are grown in 3D we did not observe a general shift in drug sensitivity, but the cells showed a distinct drug response profile when compared to 2D monolayers.
3.4. Feasibility of MAX8 constructs as a 3D scaffold for automated HTS
The fast gelation kinetics at room temperature and under physiological conditions is a critical property that makes MAX8 suitable for automated handling by standard HTS equipment and allowed us to dispense MAX8-cell mixtures into microplates using pipets and reagent dispensers. We tested the compatibility of MAX8-RGDS cell constructs with the commercially available RealTime-Glo MT cell viability assay. This is a nonlytic bioluminescent method to measure cell viability in real time and determines the number of viable cells by measuring the reduction potential and thus metabolism of cells. We chose this assay due to its capability for longitudinal tracking of cell growth in a single sample. This assay showed a strong correlation between signal and number of viable cells (Fig. 4A) making it well suited for cytotoxicity studies.
Figure 4. Validation of 3D-HTS assay.
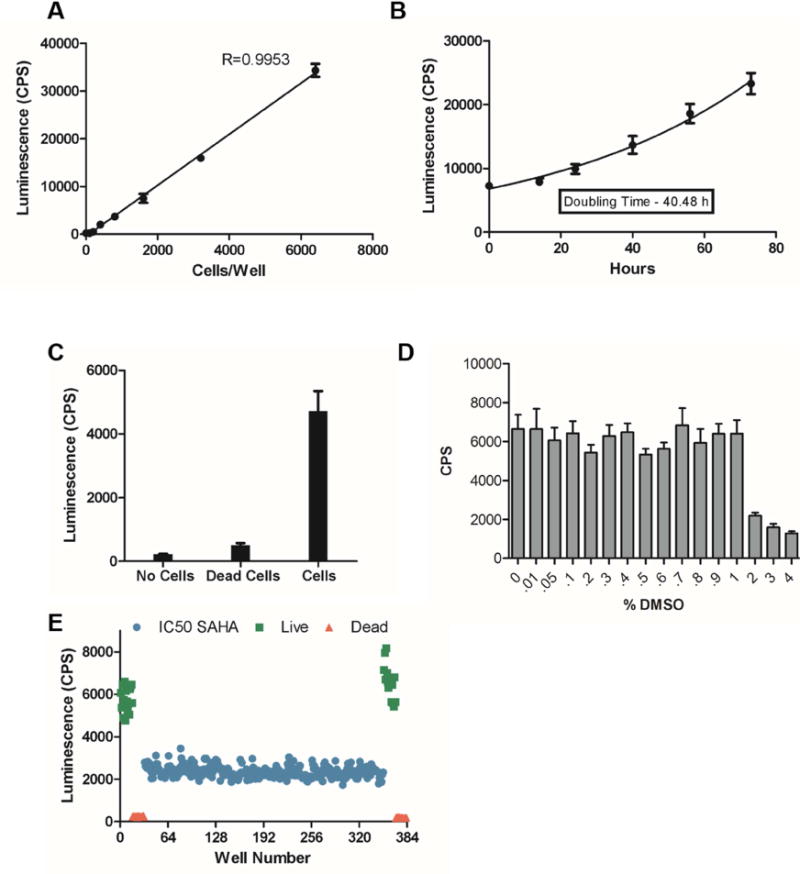
(A) Cell viability signal measured from an increasing number of cells encapsulated in 0.25 wt% MAX8 using the RealTime-Glo MT cell viability assay. N = 5; CPS, counts per second. (B) Cell growth of ONS-76 cells encapsulated in 0.25 wt% MAX8 tagged with the RGDS sequence. (C) The screen statistics calculated using untreated control cells, cells treated with ethanol to induce cell death (dead cells), and wells without cells (no cells). Z′-factor, 0.58; Signal to noise, 9.5. (D) DMSO tolerance of ONS-76 cell in MAX8 after 48 h of treatment using a 384-well plate setup. (E) Quality Control (QC) plate to determine the statistical robustness of the assay. A 384-well plate was set up with each well containing 2,000 cells encapsulated in 4 μL of 0.25 wt% MAX8-RGDS in 36 μL media. Test cells were treated with 1 μM vorinostat (blue) for 48 hours and compared to positive DMSO treated (green) and negative controls (red). Each symbol represents a single well. Z′-factor, 0.58.
The encapsulation of medulloblastoma cells into MAX8-RGDS hydrogel allowed for detection of a robust measure of proliferating cells (Fig. 4B) with a calculated Z′-factor of 0.58 (Fig. 4C). In preparation for HTS screening, we tested the DMSO tolerance of ONS-76 cells in gel-cell constructs and determined the overall quality of our 3D HTS setup. ONS-76 cells were viable at DMSO concentrations of up to 1% (Fig. 4D), therefore, 0.05% DMSO introduced by 50 nL pintool delivery of test compounds in an HTS screen should not be a concern. Furthermore, dispensation of MAX8-RGDS/ONS-76 hydrogel-cell mixtures into 384-well cells using a Janus microplate dispenser proved to be reproducible and reliable (Fig. 4E). The QC plate had a Z′-factor of 0.58, positive control CV of 12%, and LD50 control CV of 11%. The controls were within acceptable ranges showing the assay provides statistically robust results.
4. Discussion
3D HTS is a rapidly expanding section of the drug discovery process that is predicated on the idea that using a disease model that is a more accurate recapitulation of the in vivo environment will provide more clinically actionable results. However, until recently, most 3D culture technologies had limited automation possibilities, scalability and reproducibility. Here we have shown that the MAX8 β-hairpin hydrogel with its well-defined material characteristics, unique solution assembly, and flow shear properties can overcome these limitations and provide a suitable 3D cell culture scaffold for HTS. Medulloblastoma cells incorporated into MAX8 and automatically dispensed into 384-well plates showed robust cell proliferation with the proliferation rate depending on peptide concentration. The addition of ligand peptides found in extracellular matrix such as RGDS, IKVAV and YIGSR increased medulloblastoma cell proliferation within 3D hydrogel-cell constructs. Differences in cell phenotype of cells cultured in hydrogels and in monolayers were confirmed by dose response studies to commonly used chemotherapeutics and mRNA levels of cancer stem cell markers in cells grown in MAX8 were more similar to tumor tissue from a transgenic mouse medulloblastoma model. Using the RealTime-Glo MT cell viability assay we standardized a sensitive HTS-compatible cell viability assay with a robust Z-score. As MAX8 has been shown previously to be compatible with a variety of cell lines and primary cells [14, 18–20] and can be incorporated into standard HTS equipment, we expect MAX8 to have broad applicability as a versatile cell culture scaffold for 3D HTS.
Thus far, there are several different approaches to creating a 3D HTS method, but each comes with its own set of challenges and difficulties [1, 37–39]. The best studied methods employ spheroids or hydrogels. Spheroids, which are a group of cells growing in a sphere designed to represent a tumor, may be created in several ways. The hanging drop method utilizes gravity to induce spheroid formation in specifically designed cell culture plates. Low attachment surfaces, nanoparticle magnets, and cell stirring have also been used to create spheroids. Spheroids have been shown to produce mass transport similar to that found in vivo, and are able to maintain cell-cell and some cell-matrix interactions if the cells deposit their own extracellular matrix proteins. Challenges for their use in drug discovery include difficulty forming and maintaining uniform spheroids and making tissue-like spheroids that may contain multiple cell types for a more in vivo like tumor environment. Overall, these complexities in spheroid generation and the complexity of assay methods has slowed their adoption by HTS labs.
Cultures in 3D matrices employing hydrogels that mimic the environment of tumors provide an appealing alternative to the use of spheroids. Among 3D culture matrices, natural or appropriately designed hydrogels most closely resemble the material and biological properties of extracellular matrix [4, 6]. Hydrogels of natural origin that are commonly used are collagen and Matrigel. They are usually cytocompatible but their chemical and mechanical properties cannot readily be modified to mimic different tissue environments. Moreover, they comprise complex and ill-defined scaffolds that can vary mechanically, morphologically, and biochemically between preparations, leading to inconsistent screening results. The need to handle these matrices at cold temperatures complicates automated liquid handling. Moreover, the slow gelation kinetics makes for non-uniform distribution of encapsulated cells, often leading to an undesirable additional monolayer on the bottom of the well (Fig. 1D).
In contrast to the limitations of hydrogels derived from natural sources, synthetically produced hydrogels function as well-defined, reproducible scaffolds that can easily be tuned by controlling properties such as porosity, permeability and mechanical stability to provide a standardized, reproducible and biologically active cell environment [1–4]. Here we have shown that the MAX8 β-hairpin hydrogel with its well-defined material characteristics, unique solution assembly, and shear thinning injectable solid properties provides a suitable 3D cell culture scaffold for HTS. MAX8’s shear thinning properties that, due to the lack of intermolecular covalent bonds between individual hairpins, allow for the hydrogel to fracture into domains under shear and percolate back together once the shear force seizes while at the same time displaying a plug flow velocity profile with consistent velocities through the majority of the tube’s cross section make it particularly well suited for dispensing with automated liquid handling equipment. MAX8 is compatible with a commercially available cell viability assay and can be incorporated into a standard HTS setup. Medulloblastoma cells encapsulated into MAX8 and dispensed into 384-well plates using liquid handling automation showed robust cell proliferation dependent on peptide concentration. As expected, primary cultures of mouse cerebellar granule cells proliferated at a slower rate, but were nevertheless compatible with native MAX8. The addition of adhesion peptides found in extracellular matrix such as RGDS, IKVAV, and YIGSR increased ONS-76 medulloblastoma cell proliferation within 3D hydrogel-cell constructs (Fig. 2C). When compared to standard 2D cultures, ONS-76 cells in 3D MAX8 constructs were more sensitive to cisplatin and vismodegib, while having a similar sensitivity toward vorinostat (SAHA), suggesting a 3D-specific cell phenotype. Differences in the phenotype of cells cultured in hydrogels and in monolayers were further confirmed by determining the mRNA levels of the stem cell markers nestin and snail1. Snail1 and gli3 mRNA levels further served as markers for activation of the Shh signaling cascade. Expression of nestin, and Shh activation levels changed with the inclusion of adhesion peptides, suggesting that MAX8 can be tuned to mimic important environmental factors altering cell phenotype. Since cancer stem cells are main players for chemoresistance against a variety of drugs including cisplatin, temozolomide, etoposide and doxorubicin and in various cancers [36], designing stem cell-promoting hydrogels could be important for cancer drug discovery. Using the RealTime-Glo MT cell viability assay, we standardized a sensitive HTS-compatible cell viability assay with a robust Z′-factor. As MAX8 has been shown previously to be compatible with a variety of other cell lines and primary cells [14, 18–20] and can be incorporated into standard HTS equipment, we expect it to have broad applicability as a versatile cell culture scaffold for 3D HTS.
One prerequisite for successful 3D drug discovery using hydrogels is to create reproducible 3D matrix-cell constructs that will maintain a constant volume, do not disintegrate during extended culture times, and minimally interact with structurally diverse compounds present in screening libraries. We have shown previously that MAX8 β-hairpin hydrogels do not swell or dissolve in aqueous solutions; instead, they remain as intact solid, porous hydrogels over several weeks [33]. Here, our rheology studies showed that the addition of adhesion moieties to the MAX8 peptide resulted in a hydrogel with similar properties as MAX8 in its native form (Fig. 2D, E) suggesting that a hydrogel can easily be tuned without affecting its physical properties. Moreover, our previous drug encapsulation studies demonstrated that encapsulating structurally diverse compounds in the millimolar range did not significantly change the physical properties of MAX8 [27, 30, 40]. Therefore, MAX8 provides a versatile and tunable cell culture matrix with predictable physical properties suitable for 3D drug discovery.
5. Conclusions
The majority of HTS drug discovery is being carried out in cells cultured in 2D, but compelling evidence suggests that cells grown in non-physiological 2D conditions differ from cells grown in the more in vivo like 3D systems. For example, human medulloblastoma cells grown in 3D cultures express increasingly immature features and vary in drug response when compared to cells grown on tissue culture plates. It is increasingly being recognized that 3D models are of critical importance to better predict the toxicity and efficacy of potential drugs. This is of particular importance in oncology drug discovery where there is a fine line between therapeutic benefit and toxic side effects. Commonly used natural 3D matrices such as collagen or Matrigel provide an in vivo like environment, but the capabilities to modify chemical and mechanical properties are limited. This can be overcome by synthetic matrices that can be optimized to mimic the native extracellular matrix by porosity, permeability and mechanical stability and that can provide a biologically active environment for cells to proliferate and differentiate. MAX8 β-hairpin peptide hydrogels meet all these requirements and thus provide a versatile, HTS-compatible 3D high-throughput matrix for drug repurposing and to discover new candidate compounds with high in vivo efficacy for future drug development.
Acknowledgments
This work was supported by funds from ACS RSG-09-021-01-CNE, the NIH IDeA program, with grants from the National Institute of General Medical Sciences NIGMS (P20-GM103464, P30-GM114736, and U54-GM104941), the DO Believe Foundation and the Nemours Foundation. We would like to thank Dr. Jobayer Hossain for providing statistical expertise. We acknowledge the support of the National Institute of Standards and Technology, U.S. Department of Commerce, in providing the neutron research facilities used in this work. This work utilized facilities supported in part by the National Science Foundation under Agreement No. DMR-0944772. This manuscript was prepared under cooperative agreement 70NANB12H239 from NIST, U.S. Department of Commerce. The statements, findings, conclusions, and recommendations are those of the authors and do not necessarily reflect the view of NIST or the U.S. Department of Commerce.
Abbreviations
- 2D
two-dimensional
- 3D
three-dimensional
- CGP
cerebellar granule precursor cell
- CPS
counts per second
- CV
coefficient of variation
- DMEM
Dulbecco’s Minimum Essential Medium
- DMSO
dimethyl sulfoxide
- HTS
high-throughput screening
- SAHA
suberanilohydroxamic acid
- Shh
sonic hedgehog
- Smo
smoothened
- QC
quality control
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential conflicts of interest: The authors declare no conflict of interest.
References
- 1.Montanez-Sauri SI, Beebe DJ, Sung KE. Microscale screening systems for 3D cellular microenvironments: platforms, advances, and challenges. Cell Mol Life Sci. 2015;72:237–249. doi: 10.1007/s00018-014-1738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimlin LC, Casagrande G, Virador VM. In vitro three-dimensional (3D) models in cancer research: an update. Mol Carcinog. 2013;52:167–182. doi: 10.1002/mc.21844. [DOI] [PubMed] [Google Scholar]
- 3.Ravi M, Paramesh V, Kaviya SR, Anuradha E, Solomon FD. 3D cell culture systems: advantages and applications. J Cell Physiol. 2015;230:16–26. doi: 10.1002/jcp.24683. [DOI] [PubMed] [Google Scholar]
- 4.Worthington P, Pochan DJ, Langhans SA. Peptide Hydrogels - Versatile Matrices for 3D Cell Culture in Cancer Medicine. Front Oncol. 2015;5:92. doi: 10.3389/fonc.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caliari SR, Burdick JA. A practical guide to hydrogels for cell culture. Nat Methods. 2016;13:405–414. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai L, Dewi RE, Heilshorn SC. Injectable Hydrogels with In Situ Double Network Formation Enhance Retention of Transplanted Stem Cells. Adv Funct Mater. 2015;25:1344–1351. doi: 10.1002/adfm.201403631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan D, Du X, Shi J, Zhou N, Zhou J, Xu B. Mixing biomimetic heterodimers of nucleopeptides to generate biocompatible and biostable supramolecular hydrogels. Angew Chem Int Ed Engl. 2015;54:5705–5708. doi: 10.1002/anie.201412448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar VA, Shi S, Wang BK, Li IC, Jalan AA, Sarkar B, Wickremasinghe NC, Hartgerink JD. Drug-triggered and cross-linked self-assembling nanofibrous hydrogels. J Am Chem Soc. 2015;137:4823–4830. doi: 10.1021/jacs.5b01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draper ER, Eden EG, McDonald TO, Adams DJ. Spatially resolved multicomponent gels. Nat Chem. 2015;7:848–852. doi: 10.1038/nchem.2347. [DOI] [PubMed] [Google Scholar]
- 11.Wan S, Borland S, Richardson SM, Merry CL, Saiani A, Gough JE. Self-assembling peptide hydrogel for intervertebral disc tissue engineering. Acta Biomater. 2016;46:29–40. doi: 10.1016/j.actbio.2016.09.033. [DOI] [PubMed] [Google Scholar]
- 12.Haigh JN, Chuang Y-m, Farrugia B, Hoogenboom R, Dalton PD, Dargaville TR. Hierarchically Structured Porous Poly(2-oxazoline) Hydrogels. Macromolecular Rapid Communications. 2016;37:93–99. doi: 10.1002/marc.201500495. [DOI] [PubMed] [Google Scholar]
- 13.Nagy-Smith K, Moore E, Schneider J, Tycko R. Molecular structure of monomorphic peptide fibrils within a kinetically trapped hydrogel network. Proc Natl Acad Sci U S A. 2015;112:9816–9821. doi: 10.1073/pnas.1509313112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, Lamm MS, Pochan DJ, Schneider JP. Controlling hydrogelation kinetics by peptide design for three-dimensional encapsulation and injectable delivery of cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozbas B, Kretsinger J, Rajagopal K, Schneider JP, Pochan DJ. Salt-triggered peptide folding and consequent self-assembly into hydrogels with tunable modulus. Macromolecules. 2004;37:7331–7337. [Google Scholar]
- 16.Yucel T, Micklitsch CM, Schneider JP, Pochan DJ. Direct observation of early-time hydrogelation in beta-hairpin peptide self-assembly. Macromolecules. 2008;41:5763–5772. doi: 10.1021/ma702840q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sathaye S, Zhang H, Sonmez C, Schneider JP, MacDermaid CM, Von Bargen CD, Saven JG, Pochan DJ. Engineering complementary hydrophobic interactions to control beta-hairpin peptide self-assembly, network branching, and hydrogel properties. Biomacromolecules. 2014;15:3891–3900. doi: 10.1021/bm500874t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan C, Altunbas A, Yucel T, Nagarkar RP, Schneider JP, Pochan DJ. Injectable solid hydrogel: mechanism of shear-thinning and immediate recovery of injectable [small beta] -hairpin peptide hydrogels. Soft Matter. 2010;6:5143–5156. doi: 10.1039/C0SM00642D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan C, Mackay ME, Czymmek K, Nagarkar RP, Schneider JP, Pochan DJ. Injectable solid peptide hydrogel as a cell carrier: effects of shear flow on hydrogels and cell payload. Langmuir. 2012;28:6076–6087. doi: 10.1021/la2041746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled beta-hairpin peptide hydrogel surfaces. Biomaterials. 2005;26:5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 21.Fossati P, Ricardi U, Orecchia R. Pediatric medulloblastoma: Toxicity of current treatment and potential role of protontherapy. Cancer Treatment Reviews. 2009;35:79–96. doi: 10.1016/j.ctrv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Northcott PA, Dubuc AM, Pfister S, Taylor MD. Molecular subgroups of medulloblastoma. Expert Rev Neurother. 2012;12:871–884. doi: 10.1586/ern.12.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, Pfister SM. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van Meeteren A, van Vuurden D, Clifford SC, Pietsch T, von Bueren AO, Rutkowski S, McCabe M, Collins VP, Backlund ML, Haberler C, Bourdeaut F, Delattre O, Doz F, Ellison DW, Gilbertson RJ, Pomeroy SL, Taylor MD, Lichter P, Pfister SM. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Annabi B, Rojas-Sutterlin S, Laflamme C, Lachambre MP, Rolland Y, Sartelet H, Beliveau R. Tumor environment dictates medulloblastoma cancer stem cell expression and invasive phenotype. Mol Cancer Res. 2008;6:907–916. doi: 10.1158/1541-7786.MCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Diaz PC, Burton TL, Burns SC, Hung JY, Penalva LO. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer. 2008;8:280. doi: 10.1186/1471-2407-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altunbas A, Lee SJ, Rajasekaran SA, Schneider JP, Pochan DJ. Encapsulation of curcumin in self-assembling peptide hydrogels as injectable drug delivery vehicles. Biomaterials. 2011 doi: 10.1016/j.biomaterials.2011.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Branco MC, Nettesheim F, Pochan DJ, Schneider JP, Wagner NJ. Fast dynamics of semiflexible chain networks of self-assembled peptides. Biomacromolecules. 2009;10:1374–1380. doi: 10.1021/bm801396e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hule RA, Nagarkar RP, Altunbas A, Ramay HR, Branco MC, Schneider JP, Pochan DJ. Correlations between structure, material properties and bioproperties in self-assembled beta-hairpin peptide hydrogels. Faraday Discussions. 2008;139:251–264. doi: 10.1039/b717616c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsey S, Piatt JH, Worthington P, Sonmez C, Satheye S, Schneider JP, Pochan DJ, Langhans SA. Beta Hairpin Peptide Hydrogels as an Injectable Solid Vehicle for Neurotrophic Growth Factor Delivery. Biomacromolecules. 2015;16:2672–2683. doi: 10.1021/acs.biomac.5b00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haines-Butterick LA, Salick DA, Pochan DJ, Schneider JP. In vitro assessment of the pro-inflammatory potential of beta-hairpin peptide hydrogels. Biomaterials. 2008;29:4164–4169. doi: 10.1016/j.biomaterials.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinthuvanich C, Haines-Butterick LA, Nagy KJ, Schneider JP. Iterative design of peptide-based hydrogels and the effect of network electrostatics on primary chondrocyte behavior. Biomaterials. 2012;33:7478–7488. doi: 10.1016/j.biomaterials.2012.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branco MC, Pochan DJ, Wagner NJ, Schneider JP. Macromolecular diffusion and release from self-assembled beta-hairpin peptide hydrogels. Biomaterials. 2009;30:1339–1347. doi: 10.1016/j.biomaterials.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branco MC, Pochan DJ, Wagner NJ, Schneider JP. The effect of protein structure on their controlled release from an injectable peptide hydrogel. Biomaterials. 2010;31:9527–9534. doi: 10.1016/j.biomaterials.2010.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 36.Zhao J. Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacol Ther. 2016;160:145–158. doi: 10.1016/j.pharmthera.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan SL, Baird AM, Vaz G, Urquhart AJ, Senge M, Richard DJ, O’Byrne KJ, Davies AM. Drug Discovery Approaches Utilizing Three-Dimensional Cell Culture. Assay Drug Dev Technol. 2016;14:19–28. doi: 10.1089/adt.2015.670. [DOI] [PubMed] [Google Scholar]
- 38.Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadler M, Walter S, Walzl A, Kramer N, Unger C, Scherzer M, Unterleuthner D, Hengstschlager M, Krupitza G, Dolznig H. Increased complexity in carcinomas: Analyzing and modeling the interaction of human cancer cells with their microenvironment. Semin Cancer Biol. 2015;35:107–124. doi: 10.1016/j.semcancer.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Sun JE, Stewart B, Litan A, Lee SJ, Schneider JP, Langhans SA, Pochan DJ. Sustained release of active chemotherapeutics from injectable-solid beta-hairpin peptide hydrogel. Biomater Sci. 2016;4:839–848. doi: 10.1039/c5bm00538h. [DOI] [PMC free article] [PubMed] [Google Scholar]


