Abstract
Glucocorticoids exert important therapeutic effects on airway smooth muscle (ASM), yet few direct targets of glucocorticoid signaling in ASM have been definitively identified. Here, we show that the transcription factor, Krüppel-like factor 15 (KLF15), is directly induced by glucocorticoids in primary human ASM, and that KLF15 represses ASM hypertrophy. We integrated transcriptome data from KLF15 overexpression with genome-wide analysis of RNA polymerase (RNAP) II and glucocorticoid receptor (GR) occupancy to identify phospholipase C delta 1 as both a KLF15-regulated gene and a novel repressor of ASM hypertrophy. Our chromatin immunoprecipitation sequencing data also allowed us to establish numerous direct transcriptional targets of GR in ASM. Genes with inducible GR occupancy and putative antiinflammatory properties included IRS2, APPL2, RAMP1, and MFGE8. Surprisingly, we also observed GR occupancy in the absence of supplemental ligand, including robust GR binding peaks within the IL11 and LIF loci. Detection of antibody–GR complexes at these areas was abrogated by dexamethasone treatment in association with reduced RNA polymerase II occupancy, suggesting that noncanonical pathways contribute to cytokine repression by glucocorticoids in ASM. Through defining GR interactions with chromatin on a genome-wide basis in ASM, our data also provide an important resource for future studies of GR in this therapeutically relevant cell type.
Keywords: glucocorticoid receptor, airway smooth muscle, asthma, chromatin, Krüppel-like factor 15
Clinical Relevance
This study defines new mechanisms of glucocorticoid activity in airway smooth muscle (ASM) and also establishes the utility of performing chromatin immunoprecipitation sequencing in this important cell type. In addition, phospholipase C delta 1 is identified as novel repressor of ASM hypertrophy in vitro.
Airway smooth muscle (ASM) plays a central role in asthma pathophysiology (1). ASM secretes cytokines that perpetuate asthmatic airway inflammation and can also exhibit significant structural remodeling and biomechanical abnormalities that are directly linked to asthma symptoms (2). Although evidence indicates that glucocorticoids, which are central to asthma treatment, can reduce ASM contractile responses and cytokine expression (3–5), the effects of glucocorticoids on reversing structural remodeling in ASM appear to be somewhat limited and are incompletely understood (6, 7). Moreover, trials have indicated that new treatments targeting specific Th2 pathways in asthma, such as IL-5, do not fully normalize lung function in severe asthma (8). Consequently, there is an ongoing need to identify pathways and mechanisms that abrogate ASM remodeling, commonly manifested as increases in cell size (hypertrophy) and number (hyperplasia) (9).
Although it is difficult to generate data pertaining to the molecular effects of glucocorticoids on human ASM (HASM) remodeling in vivo, cultured models have emerged as a valuable tool to study ASM perturbations that are associated with severe asthma and to investigate the effects of glucocorticoids on this crucial cell type (10, 11). Work with this system has defined, in comparison to normal controls, altered gene expression in ASM from fatal asthma (12), which is also reported to exhibit a variety of persistent phenotypic abnormalities (13). Studies using cultured HASM have also established that glucocorticoids induce canonical glucocorticoid receptor (GR) signaling in this cell type, encompassing activation of antiinflammatory genes and cytokine repression (14). Furthermore, treatment with glucocorticoids can limit ASM contractile responses as well as proliferative and hypertrophic consequences of exposure to growth factors and cytokines (15–17). Although a number of mechanisms have been defined that contribute to the suppressive effects of glucocorticoids on ASM phenotype and growth, including a role for post-transcriptional control in repressing hypertrophy (17), the activity and identity of genes that are directly regulated by GR in abrogating these processes remain to be fully elucidated.
In many tissues, in addition to repressing inflammatory signals, glucocorticoids induce metabolic effects and structural remodeling, exemplified by well described effects of glucocorticoids on bone and skeletal muscle (18, 19). Although such pathways can be detrimental in some contexts, we hypothesized that glucocorticoids regulate similar pathways in ASM, possibly encompassing novel repressors of ASM proliferation and hypertrophy, with potential therapeutic benefit in the remodeled asthmatic airway. In pursuing this notion, we previously identified Krüppel-like factor 15 (KLF15), implicated in metabolic gene regulation (20), as a target of glucocorticoid signaling in ASM (14). We also showed that KLF15 represses ASM proliferation in vitro and that, in airway epithelial cells, GR and KLF15 function together to form feed-forward loops (21), an ancient regulatory system that is responsible for temporal control of stereotyped gene expression programs, such as cell differentiation and responses to environmental stimuli (22). Our work further implicated the GR-KLF15 feed-forward loop system as controlling the expression of approximately 7% of the glucocorticoid-responsive transcriptome in vivo, including skeletal muscle metabolic processes that are required for endurance exercise and therapeutic properties of glucocorticoids in treating Duchenne muscular dystrophy (23). However, whether GR and KLF15 form feed-forward loops in ASM, and potential therapeutic effects of GR-KLF15 targets on ASM remodeling, have yet to be determined. In this study, we address these areas by applying chromatin immunoprecipitation (ChIP) and reporter assays to determine whether KLF15 is directly induced by GR in ASM. To comprehensively identify sites of GR regulatory activity and novel KLF15 targets in ASM, we performed GR ChIP followed by deep sequencing (i.e., ChIP-seq) and integrated these findings with analysis of the KLF15-regulated transcriptome. We also used adenoviral transduction to test whether a newly identified KLF15 target, phospholipase C delta 1, represses ASM hypertrophy. Portions of this work have appeared previously in abstract form (24).
Materials and Methods
Cell Culture and Reagents
All experiments used primary HASM cells from a donor without asthma (HASM2) that were derived and cultured as previously described (25). Details on reagents used in this study are included in the supplemental material.
Plasmids, Transfection, and Reporter Assays
The KLF15 luciferase reporter plasmids have been described peviously (21), and existing protocols for transfections and reporter activity assays were used throughout (21, 25).
RNA Purification, Quantitative RT-PCR, and RNA-seq
For quantitative RT-PCR (qRT-PCR) experiments, HASM2 cells were grown to confluence on six-well plates, transduced with adenoviral expression constructs for KLF15 (Ad-KLF15) or green fluorescent protein (Ad-GFP) as control for approximately 17 hours, and then treated with dexamethasone (dex) or vehicle in fresh complete medium for 8 hours. RNA preparation and qRT-PCR were performed as previously described with normalization to RPL19 (21). Relative gene expression is calculated relative to the mean normalized threshold cycle (CT) value of Ad-GFP+vehicle-treated samples. Primer sequences are listed in Table E1 in the online supplement. For RNA-seq analysis, cells in biologic quadruplicate were grown to confluence on six-well plates, transduced with Ad-KLF15 or Ad-GFP for approximately 17 hours, and then incubated in fresh complete medium for 4 hours. Cells were lysed and RNA was initially purified as for qRT-PCR experiments; detailed RNA-seq methods are in the supplementary material. RNA-seq data have been deposited in Gene Expression Omnibus (GEO; no. GSE95397).
ChIP–Quantitative PCR and ChIP-seq
For ChIP–quantitative PCR (qPCR) and ChIP-seq experiments, HASM2 cells were grown to confluence on three 15-cm plates per treatment group and treated for 1 hour with vehicle or dex after 17-hour incubation with Ad-GFP, Ad-KLF15, or fresh complete medium (uninfected controls). Chromatin was then prepared in biologic quintuplicate as described previously (21, 25). For ChIP-qPCR, immunoprecipitation was performed and relative factor occupancy was calculated as previously described (21). Primer sequences used for ChIP-qPCR are listed in Table E1. For ChIP-seq, library preparation, deep sequencing, and subsequent analyses were performed according to our published workflow (26). ChIP-seq data have been deposited in GEO (GSE95632).
Western Blotting
For analysis of KLF15 protein induction by dex, HASM2 cells were grown to approximately 80% confluence on 10-cm plates and treated for 24 hours with 1 μM dex or vehicle in serum-free medium. To assess effects of Ad-PLCD1 on transforming growth factor (TGF)-β–mediated induction of α-smooth muscle actin (α-SMA) protein, cells were seeded on 15-cm plates and grown to approximately 80% confluence before adenoviral transduction for approximately 17 hours and subsequent treatment with TGF-β or vehicle in fresh complete medium for 96 hours. TGF-β and vehicle treatments were replaced with fresh preparations after 48 hours. Cells were lysed and total protein was extracted, separated by SDS-PAGE, and processed by immunoblotting, as previously described (25).
Phalloidin Staining, Flow Cytometry, and Apoptosis Assay
For phalloidin staining, HASM2 cells were plated at 5,000 cells/well in 48-well plates. Cell size was additionally analyzed by flow cytometry using an LSRII flow cytometer (BD Biosciences, San Jose, CA). For apoptosis assays, cells were plated on Nunc Lab-Tek eight-well Permanox plastic chamber slides (Fisher Scientific, Denver, CO) at 25,000 cells/well. Detailed methods and experimental procedures for each of these assays are available in the supplemental material.
Results
KLF15 Is Directly Induced by GR and Regulates HASM Cell Morphology
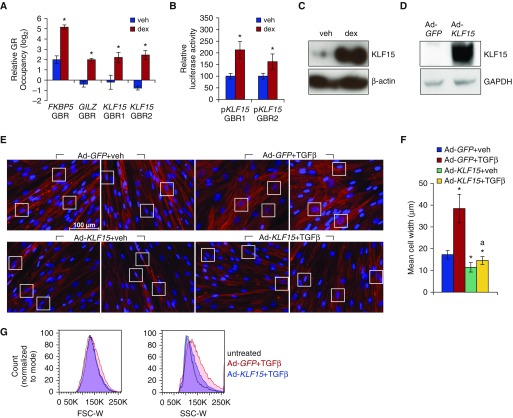
Our previous work showed that KLF15 is one of the most strongly dex-induced targets in HASM cells, and that it negatively regulates cell proliferation when overexpressed (14). We therefore sought to determine the mechanism of KLF15 induction by glucocorticoids in HASM cells and to explore whether KLF15 modulates other phenotypic characteristics of HASM cells that have been reported to be perturbed in severe asthma, such as cell size. First, based on data from other cell types (21), we used ChIP-qPCR to assess GR occupancy at two putative binding sites within the KLF15 locus in primary HASM cells treated with vehicle or dex for 1 hour. As shown in Figure 1A, dex increased GR occupancy at both of the tested KLF15 GR binding regions (GBRs) to a similar extent as the GILZ GBR, a canonical direct GR target included as a positive control for ChIP efficiency. Next, luciferase reporters containing each KLF15 GBR were transiently transfected into HASM2 cells. Both reporters exhibited significant activation in response to 8 hours of dex (Figure 1B). We then asked whether dex concordantly regulates KLF15 at the protein level by treating HASM2 cells for 24 hours with vehicle or dex and assaying KLF15 expression by Western blotting. Consistent with our previous work in other airway cell types (21), KLF15 protein expression was markedly induced by dex treatment in HASM2 cells (Figure 1C). Collectively, these findings demonstrate that KLF15 is a direct transcriptional target of GR in HASM.
Figure 1.
Krüppel-like factor 15 (KLF15) is a direct transcriptional target of glucocorticoid receptor (GR) that regulates human airway smooth muscle (HASM) cell morphology. (A) Chromatin immunoprecipitation (ChIP)–quantitative PCR (qPCR) analysis of GR occupancy in HASM2 after 1 hour of vehicle (veh) or dexamethasone (dex) treatment. Bars represent GR occupancy on a log2 scale (±SD), expressed as the mean CT value at each target region relative to the geometric mean of CT values at three negative control regions. *P ≤ 0.05 versus vehicle. (B) Luciferase activity of KLF15 reporter constructs transfected into HASM2 cells before treatment with vehicle or dex for 8 hours. Reporter activity was normalized based on a Renilla luciferase control and is expressed relative to vehicle-treated cells. Bars indicate means (±SD). *P ≤ 0.05 versus vehicle. (C) Western blot analysis of KLF15 protein expression in HASM2 cells treated for 24 hours with vehicle or 1 μM dex in serum-free medium. β-actin was used as a loading control. (D) Western blot confirming KLF15 protein induction after adenoviral KLF15 (Ad-KLF15) transduction. GAPDH served as a loading control. (E) Phalloidin staining of filamentous actin (F-actin; red) in HASM2 cells transduced with adenoviral constructs expressing green fluorescent protein (GFP) (Ad-GFP; control) or KLF15 (Ad-KLF15) approximately 17 hours before treatment with vehicle or transforming growth factor (TGF)-β for 96 hours. Nuclei were counterstained with DAPI (blue). White boxes indicate regions used to quantify cell widths in F. Scale bar: 100 μm. (F) Cell width was defined as the distance between cell borders at their widest point of separation across the nucleus. Bars show mean cell widths (±SD) from six representative measurements per treatment. *P ≤ 0.05 versus Ad-GFP+veh; aP ≤ 0.05 versus Ad-GFP+TGF-β. (G) HASM2 cell size after transduction with Ad-GFP or Ad-KLF15 approximately 17 hours before 96-hour treatment with TGF-β was characterized using flow cytometry. Cells were sorted by forward scatter (FSC-W; left) and side scatter (SSC-W; right) pulse width. GBR, GR binding region.
Glucocorticoids have previously been shown to abrogate TGF-β–induced ASM hypertrophy (17), which we corroborated (Figure E1). To determine whether KLF15 has similar effects on HASM cell morphology, we used a previously described adenoviral construct, Ad-KLF15 (27), which drives high levels of KLF15 protein expression (Figure 1D). HASM2 cells were transduced with Ad-GFP (control) or Ad-KLF15 for approximately 17 hours before treatment with vehicle or TGF-β for 96 hours. Cell morphology was subsequently visualized using a fluorescent phalloidin conjugate that stains filamentous actin. Representative fluorescent microscopy images for each treatment group are presented in Figure 1E. Transduction with Ad-KLF15 in vehicle-treated cells induced a clear visible change in their appearance, most notably, a reduction in thickness or caliber (Figure 1E, compare top left two panels to bottom left two panels). Treatment with TGF-β induced the expected hypertrophic phenotype in Ad-GFP–transduced cells, characterized by a substantial increase in cell caliber and altered organization of actin filaments (Figure 1E, top panels, right two versus left two), which was completely prevented by Ad-KLF15 transduction (Figure 1E, two top right panels versus two bottom right panels). Measurements of cell widths in each of the treatment groups, an approach commonly used to quantify atrophic effects of glucocorticoids on cultured skeletal muscle (28), were consistent with these visual observations (Figure 1F). FACS-based measurement of forward and side scatter, a standard approach to quantify ASM hypertrophy (29), also showed that transduction with Ad-KLF15 resulted in reduced cell size in comparison to GFP control (Figure 1G). Thus, KLF15 overexpression reduces TGF-β–mediated increases in cell size, a cardinal phenotypic feature of ASM remodeling.
Identification of the KLF15-Regulated Transcriptome in HASM Using RNA-seq
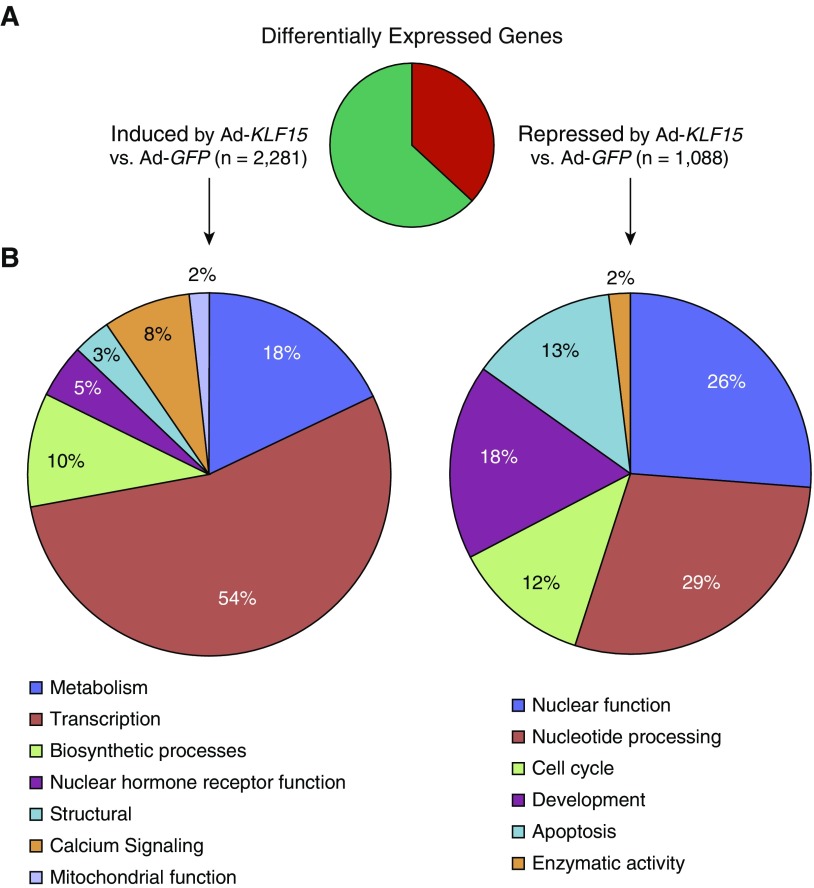
As a transcription factor, the effects of KLF15 on HASM cell morphology are likely to be mediated by downstream genes subject to transcriptional regulation by KLF15. Therefore, to identify transcriptional targets of KLF15 on a genome-wide basis in HASM, we performed deep sequencing of mRNA obtained from HASM2 cells transduced with Ad-GFP or Ad-KLF15 for approximately 17 hours. Differential expression analysis comparing quadruplicate samples for the two conditions defined approximately 3,350 genes that were regulated by KLF15 (adjusted P < 0.05) when filtered for twofold or greater expression change and 30 or greater average sequencing reads (30). As shown in Figure 2A, approximately two-thirds of this subset of KLF15-regulated genes were induced by KLF15 transduction (Table E2), whereas one-third were repressed (Table E3), consistent with the established activity of KLF15 as both a transcriptional activator and repressor (21). Gene ontology analysis of 200 of the most strongly regulated transcripts (based on adjusted P value) within gene sets significantly induced versus repressed by Ad-KLF15 revealed enrichment for a number of functional terms, including metabolic processes within the induced gene set (Figure 2B). Selected genes with particularly robust induction or repression in association with Ad-KLF15 transduction are listed in Table 1.
Figure 2.
Global analysis of KLF15-regulated transcription in HASM cells using RNA-seq. (A) Pie chart summarizing differentially expressed genes, defined as those exhibiting twofold or greater change in expression (adjusted P < 0.05) with Ad-KLF15 compared with Ad-GFP and a minimum mean read number of 30. (B) The DAVID Gene Ontology Analysis tool was applied to 200 of the top most strongly regulated transcripts (based on P value) within gene sets significantly induced (left) or repressed (right) by Ad-KLF15. We considered clusters with enrichment scores greater than 1 as significant. Pie charts illustrate distinct categories of significant functional clusters of related gene ontology terms within each gene set.
Table 1.
Selected Highly Expressed and Strongly Krüppel-Like Factor 15–Regulated Genes Identified by RNA Sequencing in Airway Smooth Muscle
| Gene Symbol | Fold Change (log2) versus Ad-GFP |
|---|---|
| CDKN1C | 8.02 |
| ABAT | 7.23 |
| LRRC15 | 6.34 |
| MKNK2 | 5.91 |
| PRELP | 5.32 |
| BMP2 | −3.58 |
| IL6 | −3.49 |
| IL33 | −3.30 |
| SERPINB2 | −3.10 |
Definition of abbreviation: Ad-GFP, adenoviral expression construct for green fluorescent protein.
ChIP-seq Analysis of GR and RNA Polymerase II Occupancy
We have previously shown that genes induced through GR-KLF15 feed-forward loops exhibit KLF15-mediated increases in GR occupancy at associated enhancers and that the set of genes induced through this system controls important metabolic pathways and physiologic effects (21, 23). We reasoned that, within the set of KLF15-induced genes in ASM, those in which GR occupancy is increased by KLF15 might include novel regulators of ASM hypertrophy and proliferation. Therefore, to identify KLF15 targets that are also regulated through GR signaling, we performed ChIP with a GR antibody followed by deep sequencing (i.e., GR ChIP-seq) using HASM2 cells treated for 1 hour with vehicle or dex after transduction with Ad-KLF15, Ad-GFP, or no virus. We also performed ChIP-seq to analyze RNA polymerase (RNAP) II occupancy under the same set of conditions. For both GR and RNAPII, two independent ChIP samples from each condition were subjected to deep sequencing. Reads were subsequently mapped to the genome, and binding peaks for both GR and RNAPII under each experimental condition were identified with model-based analysis for ChIP-seq (MACS) and visualized in the University of California, Santa Cruz (UCSC) Genome Browser using the custom track feature.
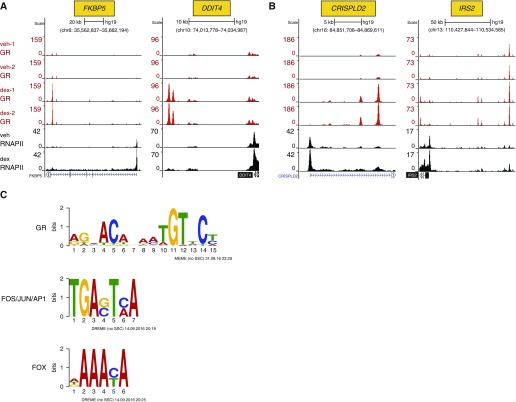
Although the primary goal of these experiments was to identify novel GR-KLF15 feed-forward targets that potentially repress ASM proliferation and hypertrophy, we began our analysis by focusing on the GR cistrome, which had not previously been reported in ASM. Using a false discovery rate (FDR) of 0.05, we identified 12,275 peaks exhibiting differential GR occupancy with dex versus vehicle treatment. To limit our analysis to sites of easily detectable dynamic interactions between GR and chromatin, we filtered for peaks with a twofold or greater change in GR occupancy with dex treatment compared with vehicle and a minimum average of 20 sequencing reads under at least one of the conditions. This subset of robust, hormone-regulated sites of GR occupancy consisted of 7,645 total peaks, of which 6,189 showed dex-inducible GR occupancy (Table E4), consistent with the well established role of glucocorticoids in causing GR nuclear localization and DNA binding (31). Visualization of selected GR binding peaks in conjunction with the RNAPII data revealed dex-induced GR occupancy and transcription within canonical GR target loci, such as FKBP5 and DDIT4 (RTP801) (Figure 3A). Sites of GR and RNAPII occupancy were also identified in association with CRISPLD2 (Figure 3B), which has been previously implicated in steroid-mediated cytokine repression in ASM (32), as well as IRS2 (Figure 3B), an inhibitor of IL4/IL13–driven allergic lung inflammation (33), among other putative antiinflammatory targets (Figure E3). Thus, these ChIP-seq data can be used to determine whether specific glucocorticoid-regulated genes are direct transcriptional targets of GR and to identify putative sites of GR-regulated enhancer activity in HASM cells.
Figure 3.
ChIP sequencing (ChIP-seq) defines genome-wide targets of dex-induced GR and RNA polymerase (RNAP) II recruitment in HASM cells. (A and B) GR (red; technical duplicates) and RNAPII (black) ChIP-seq peaks visualized in the University of California, Santa Cruz Genome Browser in vehicle- and dex-treated HASM2 samples, as indicated on the far left of A. Peak heights reflect normalized factor occupancy (reads per million reads, indicated by the vertical scale on the left of each panel) at sites within the indicated genomic loci. (A) Dex-induced GR and RNAPII recruitment in HASM2 cells at the FKBP5 and DDIT4 loci, which are canonical GR targets. (B) Examples of directly induced GR targets in HASM with reported suppressive effects in various models of allergic airway inflammation. (C) Examples of transcription factor binding site logos identified by multiple expectation maximization for motif elicitation (MEME)-ChIP as enriched within high-occupancy dex-induced GR ChIP-seq peaks. AP1, activator protein-1; DREME, discriminative regular expression motif elicitation; FOS, fos proto-oncogene; FOX, forkhead box; JUN, Jun proto-oncogene.
Analysis of GR ChIP-seq data in other cell types has revealed significant central enrichment for relatively high-affinity GR binding sites with palindromic or semipalindromic features (34, 35). Up to 60% of genomic regions associated with GR occupancy, however, are reported to lack classical GR binding sequences (34). Accordingly, GR occupancy is frequently associated with binding sites for other transcription factors in a process that is believed to be a key determinant of cell type–specific GR activity (34, 35). Therefore, to determine whether the dex-inducible GR cistrome in ASM is enriched for binding sequences for GR or other factors, we analyzed the subset of high-occupancy peaks described above using multiple expectation maximization for motif enrichment (MEME)-ChIP (36), a web-based tool kit for identifying putative binding sequences within ChIP-seq data. As expected, and supporting the quality of the data, matches for the canonical GR binding motif were centrally enriched and were identified in approximately 75% of the dex-induced GR peaks. In addition, enrichment for binding sites for other transcription factor families, including activator protein-1 (AP-1) and forkhead box (FOX), were defined (Figure 3C; see supplemental file E1 for entire MEME-ChIP analysis). Thus, similar to other cell types, GR interacts with chromatin in ASM through prototypical GR binding sites and also through engaging in combinatorial interactions with other transcription factors.
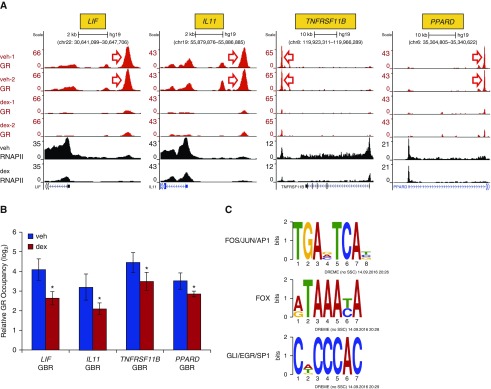
Although approximately 85% of dex-regulated GR binding peaks identified in our ChIP-seq data exhibited increased occupancy with dex, approximately 15% (1,456) of peaks had 20 or greater sequencing counts under basal culture conditions and exhibited a twofold or greater decline in GR occupancy with the addition of dex (Table E5). Visualization of such peaks within the LIF and IL11 loci, two cytokines that are strongly repressed with dex treatment, and also at the TNFRSF11B and PPARD2 loci, revealed robust GR occupancy under basal culture conditions that was curtailed upon exposure to dex (Figure 4A). Qualitatively similar results were seen in ChIP-qPCR assays interrogating GR occupancy in independent HASM2 samples (Figure 4B). Marked concomitant reduction in RNAPII recruitment to the transcriptional start site, as well as density throughout the body of each of these genes, was also observed (see Figure 4A), consistent with our published HASM microarray data in which mRNA levels of these genes was reduced after 4 hours of dex treatment (14). In contrast to the set of dex-induced sites of GR occupancy, MEME-ChIP analysis of regions with reduced GR occupancy after dex treatment did not show enrichment for the canonical palindromic GR binding site, whereas enrichment for consensus binding sites for the AP-1, FOX, and specificity protein (SP) families, among others, was observed (Figure 4C; complete MEME-ChIP analysis of these dex-reduced sites is in supplemental File E2). Further support for a functional bifurcation between sites of GR occupancy that exhibit dex-mediated increases versus decreases in GR occupancy was provided by analyzing local RNAPII occupancy. Whereas average RNAPII occupancy was increased at greater than 90% of sites in which dex treatment led to twofold or greater increase in GR occupancy, average RNAPII occupancy was decreased at greater than 70% of sites in which dex treatment reduced GR occupancy by 50% or more. In aggregate, these data suggest that GR can interact with chromatin through noncanonical binding sites in the absence of supplemental ligand. Addition of dex abrogates detection of antibody–GR complexes at these sites, frequently in association with reduced RNAPII occupancy and transcription.
Figure 4.
ChIP-seq identifies sites of GR occupancy in the absence of supplemental ligand. (A) GR (red; technical duplicates) and RNAPII (black) ChIP-seq peaks in vehicle- and dex-treated HASM2 cells, as detailed for Figure 3. Shown are representative examples of genes exhibiting robust antibody recognition of GR and RNAPII in the absence of dex that is reduced upon hormone exposure. (B) Independent validation of GR binding patterns at regions indicated by red arrows in A via ChIP-qPCR. Bars represent mean (±SD) occupancy on a log2 scale, as detailed for Figure 1. *P ≤ 0.05 versus vehicle. (C) Examples of transcription factor binding site logos identified by MEME-ChIP, as enriched within the set of GR ChIP-seq peaks with reduced occupancy after dex treatment. EGR, early growth response; GLI, GLI family zinc finger; SP1, specificity protein-1.
KLF15 Overexpression Alters GR Occupancy in ASM
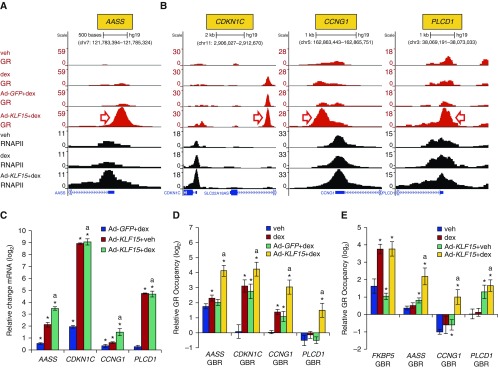
Next, we turned our attention to changes in GR occupancy associated with KLF15 overexpression, which was the underlying motivation for the ChIP-seq experiments. Differential binding analysis revealed 2,811 sites with 20 or greater sequencing reads, where GR occupancy in the presence of dex was increased twofold or greater in association with Ad-KLF15 transduction in comparison to control Ad-GFP (FDR < 0.05). Differential binding analysis also identified 2,246 such sites that exhibited twofold or greater decreased GR occupancy (FDR < 0.05) with Ad-KLF15 transduction. As KLF15-induced genes have been implicated in controlling catabolic processes in other studies (20), we focused primarily on genes with occupancy patterns suggesting cooperative induction by GR and KLF15. This pattern is exemplified by the AASS locus where increased recruitment of GR and RNAPII with combined dex treatment and Ad-KLF15 transduction in comparison to Ad-GFP control was evident (Figure 5A), consistent with our findings in airway epithelial cells (21). Examples of additional loci in which GR occupancy is increased by KLF15 (Figure 5B) include CDKN1C and CCNG1, which both repress proliferation in other cell types (37, 38), and PLCD1, which encodes the Δ-1 member of the phospholipase C family (39) and disrupts cytoskeletal architecture in cancer cells. Expression of each of these genes was also increased by Ad-KLF15 transduction in our RNA-seq analysis, which we confirmed by qRT-PCR (Figure 5C). We also used ChIP-qPCR to validate the GR ChIP-seq results at each of these loci, as shown in Figure 5D. ChIP-qPCR using a GR antibody raised against a different epitope (see supplemental material) further indicated that KLF15 was able to recruit GR to some sites under basal culture conditions (Figure 5E), suggesting that GR occupancy in the absence of supplemental ligand can be modified by heterologous transcription factors. Taken together, these data show that GR occupancy is altered by KLF15 at thousands of sites across the genome, including within regulatory elements for genes that potentially repress ASM proliferation and hypertrophy.
Figure 5.
Ad-KLF15 enhances dex-induced GR and RNAPII recruitment to genes that are putative regulators of HASM phenotype. (A and B) GR (red) and RNAPII (black) ChIP-seq peaks in control (uninfected) and adenovirus-transduced HASM2 cells treated with vehicle or dex, as indicated on the far left of A. (A) GR and RNAPII occupancy patterns at AASS, which is an established GR-KLF15 feed-forward loop target. (B) Examples of KLF15-induced genes (identified by RNA-seq) exhibiting binding patterns consistent with GR-KLF15 feed-forward loop-based regulation. (C) Independent validation of transcriptional induction of genes in A and B by Ad-KLF15 and/or 8 hours of dex using qPCR. Bars depict mean (±SD) CT values on a log2 scale relative to Ad-GFP+veh-treated cells. *P ≤ 0.05 versus Ad-GFP+veh; aP ≤ 0.05 versus Ad-GFP+dex. (D) ChIP-qPCR validation of GR binding patterns at regions indicated by red arrows in A and B. Bars indicate mean (±SD) GR occupancy on a log2 scale. *P ≤ 0.05 versus vehicle; aP ≤ 0.05 versus Ad-GFP+dex. (E) GR occupancy in uninfected or Ad-KLF15–transduced HASM2 cells treated with vehicle or dex, as assessed by ChIP-qPCR using a different GR antibody than in A, B, and D. *P ≤ 0.05 versus vehicle; aP ≤ 0.05 versus dex. PLCD1, phospholipase C delta 1.
The KLF15 Target, PLCD1, Represses TGF-β–Induced ASM Hypertrophy
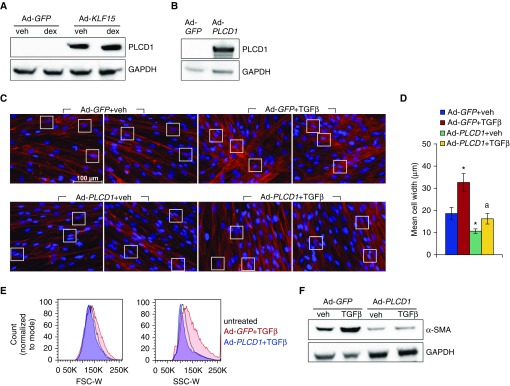
PLCD1 is known to be a target of KLF15 in the heart in vivo and has also been shown to reduce actin fiber caliber in cancer cells (21, 39). We were therefore interested in whether PLCD1 could influence ASM hypertrophy. First, we used Western analysis to verify that PLCD1 protein expression was increased by Ad-KLF15 (Figure 6A). Next, we generated an adenoviral expression vector for PLCD1 (Ad-PLCD1), and determined that transduction of ASM with Ad-PLCD1 led to increased PLCD1 protein levels (Figure 6B). We then tested whether PLCD1 overexpression alters ASM hypertrophy by transducing cells with Ad-PLCD1 or Ad-GFP control approximately 17 hours before treatment with TGF-β or vehicle for 96 hours. Phalloidin staining was used to visualize the actin cytoskeleton, which revealed an obvious reduction in cell caliber associated with Ad-PLCD1 transduction (Figure 6C). Quantification of mean cell widths confirmed the visually identified differences in cellular morphology (Figure 6D), which did not appear to be a consequence of apoptosis (Figure E2). These findings were further corroborated using FACS-based measurement of forward and side scatter (Figure 6E). Moreover, analysis of α-SMA protein showed that Ad-PLCD1 transduction abrogated the inductive effect of TGF-β on α-SMA expression observed in Ad-GFP–transduced cells (Figure 6F). Taken together, these data define PLCD1 as a novel repressor of TGF-β–mediated ASM hypertrophy.
Figure 6.
PLCD1 is a KLF15 target in HASM that prevents TGF-β–mediated hypertrophy. (A) Western blot analysis of PLCD1 protein expression in HASM2 cells transduced with Ad-GFP or Ad-KLF15 approximately 17 hours before 24 hours of vehicle or dex exposure. (B) Verification of PLCD1 induction by Western blotting after approximately 17-hour incubation with Ad-PLCD1 followed by approximately 72-hour recovery in fresh complete medium. (C) Phalloidin staining of F-actin (red) in HASM2 cells transduced with Ad-GFP or Ad-PLCD1 constructs approximately 17 hours before treatment with vehicle or TGF-β for 96 hours. White boxes highlight regions used for quantification of cell widths in D. Scale bar: 100 μm. (D) Mean cell widths (±SD), calculated as described for Figure 1. *P ≤ 0.05 versus Ad-GFP+veh; aP ≤ 0.05 versus Ad-GFP+TGF-β. (E) HASM2 cell size after transduction with Ad-GFP or Ad-PLCD1 approximately 17 hours before 96-hour treatment with TGF-β was characterized using flow cytometry. Cells were sorted according to FSC-W (left) and SSC-W (right) pulse width. (F) Western blot analysis of α-smooth muscle actin (α-SMA) protein expression in cells treated as described for C. GAPDH was used as a loading control.
Discussion
ASM abnormalities, including excessive cytokine production and pathologic remodeling, are implicated in contributing to severe and steroid-refractory asthma. However, few primary targets of GR signaling and genomic sites of GR-directed regulatory activity in ASM had been unequivocally defined in past studies. Here, we used a combination of ChIP and reporter assays to prove that KLF15 is directly induced by GR in ASM. Through integrating GR ChIP-seq data with the effects of KLF15 overexpression on both GR occupancy and the ASM transcriptome, we identified the KLF15 target, PLCD1, as a novel repressor of ASM hypertrophy. Our GR ChIP-seq data also comprehensively delineate sites of GR regulatory activity in ASM, thus defining novel antiinflammatory mechanisms and pathways.
Despite the importance of ASM in asthma pathogenesis and as a target of glucocorticoid-based therapies, interactions between GR and chromatin had not previously been defined on a genome-wide basis in ASM. Thus, our ChIP-seq data provide important new insights into mechanisms that underpin GR activity in this therapeutically relevant cell type. For example, our data defined inducible GR binding peaks associated with IRS2 (Figure 3B), which was recently implicated as an inhibitor of allergic lung inflammation (33). Robustly inducible GR occupancy was also observed within the CRISPLD2, MFGE8, RAMP1, and APPL2 loci (Figure 3B and Figure E3), each of which has antiinflammatory properties in other biologic contexts (40–42). It will be of interest to determine whether GR occupancy or expression of these genes is altered in ASM cells derived from patients that have succumbed to fatal asthma. Although technically challenging, determining whether GR occupies similar binding sites in material obtained directly from patients would aid in generalizing our data to the clinical effects of glucocorticoids. In that regard, the SYNPO2 and FAM129A loci, which have previously been identified as responsive to glucocorticoids in patient samples (43), exhibit numerous sites of GR occupancy in our ChIP-seq data, including several inducible GBRs (Table E4). To aid in future studies of GR signaling in ASM, the ChIP-seq data we generated have been deposited in the GEO, and custom tracks within the UCSC Genome Browser will be made available upon request.
Our data showed evident GR occupancy at sites throughout the genome in the absence of supplemental ligand. In a number of cases, significant associations between GR and chromatin, visualized as peaks within the UCSC Genome Browser (e.g., see Figure 4), were found in presumptive regulatory regions for cytokines. These peaks were lost after 1 hour of dex treatment, along with concomitant declines in RNAPII occupancy. We have previously reported significant GR occupancy in the absence of supplemental hormone in different cell types (26), and nuclear GR in ASM under basal culture conditions has also been reported (44). Moreover, a recent study reported that a subset of regulatory regions for inflammatory genes, frequently in association with signal transducer and activator of transcription (STAT) family binding sequences, exhibited monomeric GR occupancy in murine liver that was reduced after systemic administration of dex (45). We found no enrichment for canonical dimeric GR binding sites within the sequences in which GR occupancy was reduced by dex in ASM, suggesting that these peaks also represent occupancy by monomeric GR (46). Whereas recruitment of corepressors by tethered monomeric GR has long been implicated in repressive effects of glucocorticoids (47), the data from our work and others suggests a distinct mechanism in which monomeric GR functions as an activator at specific loci in concert with other transcription factors, potentially including the STAT family in murine liver and the AP-1 and FOX families in ASM. In both cellular contexts, transcriptional enhancement mediated by monomeric GR at selected genomic loci appears to be abrogated by GR interacting with supplemental glucocorticoids. This could occur though a ligand-induced conformational change in GR, which is the accepted mechanistic basis for glucocorticoid-induced nuclear translocation of cytoplasmic GR (34). In that regard, although our data are most consistent with ligand-induced loss of GR occupancy at certain sites, the ChIP assay does not directly measure associations between a specific transcription factor and chromatin, but instead identifies and quantifies antibody interactions with protein–DNA complexes. It is thus formally possible that the GR antibody we employed recognizes a non-GR protein that responds dynamically to dex treatment. Alternatively, upon the addition of dex, GR may not evacuate occupancy sites, but, instead, dex may induce GR to adopt a conformation that is less recognized by the antibody within the context of loci (e.g., LIF and IL11) that exhibit declines in GR peak size with dex treatment. Additional studies are clearly needed to fully elucidate the significance of presumptive GR occupancy in the absence of supplemental hormone and the contribution of these GR–chromatin interactions to gene regulation in ASM.
In addition to uncovering genomic sites of GR action in ASM with and without dex treatment, we performed GR ChIP-seq analysis in the setting of KLF15 overexpression to determine whether KLF15 can modulate GR binding patterns in ASM, as we have observed in other cell types. Our data show that KLF15 overexpression dramatically altered GR occupancy on a genome-wide basis, with a roughly equal number of sites exhibiting significant increases compared with decreases in GR occupancy associated with KLF15 expression. This is concordant with our published work using Klf15 knockout mice in which KLF15 influenced the expression of approximately 7% of GR-regulated genes through enhancing or repressing GR activity, depending on the genomic context. Whereas we have shown previously that GR and KLF15 physically associate (48), the molecular basis for KLF15 selectively enhancing or reducing GR occupancy at specific loci, and the role of GR interactions with ligand in this process, remain to be determined. Future studies are also needed to characterize alterations in GR occupancy in ASM that occur in the context of physiologic changes in KLF15 expression in ASM.
Although there are limitations in applying overexpression to define physiologically germane pathways, through integrating ChIP-seq and RNA-seq data generated after transduction with Ad-KLF15, we identified PLCD1 as a KLF15-induced gene in ASM, the promoter of which exhibited increases in GR occupancy with KLF15 overexpression. We chose to further investigate the effect of PLCD1 on ASM hypertrophy, as public expression data from Klf15 knockout mice suggest that KLF15 regulates PLCD1 in vivo, and PLCD1 mRNA was increased in ASM after 24 hours of dex treatment in our published microarray data; moreover, it had been previously shown in other cell types that PLCD1 decreases the cofilin:phosphocofilin ratio (39), which can influence actin polymerization (49). Similar to effects observed with transduction of KLF15, overexpression of PLCD1 altered ASM cytoskeletal architecture, as manifested by a decrease in cell size. This was associated with a pronounced reduction in TGF-β–induced α-SMA expression, a marker of hypertrophy. Manipulating the actin cytoskeleton has been previously proposed as a therapeutic strategy for treating ASM abnormalities in asthma (50), and our data implicate augmentation of PLCD1 activity, either through or independently of the GR–KLF15 pathway, as a potential tactic in this approach. In that regard, knockdown of PLCD1 did not appear to prevent the repressive effect of KLF15 on α-SMA expression (Figure E4), suggesting redundancy, compensation, or that PLCD1 is not directly responsible for glucocorticoid- or KLF15-mediated repression of ASM hypertrophy. Additional experiments are needed to definitively determine the mechanistic basis for the effects of PLCD1 on ASM hypertrophy, the role of PLCD1 in mediating downstream effects of GR and KLF15, and whether PLCD1 can repress ASM hypertrophy in vivo.
Despite decades of efforts to improve corticosteroid-based therapies through combatting resistance or developing selective ligands (47), clinical successes have been largely limited to improved pharmacokinetic profiles, localized delivery systems, and enhanced potency. It is thus noteworthy that most of the accepted mechanisms believed to underpin both efficacy of glucocorticoids and resistance in airway disease were developed without the benefit of deep sequencing–based genomics methodology, which has proven capable of defining new therapeutic targets and mechanisms in many other disease contexts. In this work, we used ChIP-seq and transcriptome profiling to uncover a potential role for PLCD1 in repressing ASM hypertrophy. Our data also highlight numerous new inductive pathways and targets through which glucocorticoids may exert antiinflammatory effects. In addition, our data suggest that nuclear localization of GR in the absence of supplemental ligand has an important biologic role in this cell type. Additional validation of these ASM pathways and their mechanistic underpinnings may aid in defining a new set of pharmacologic targets for steroid-resistant asthma.
Acknowledgments
Acknowledgments
Chromatin immunoprecipitation sequencing was performed at the University of Colorado Cancer Center (Denver, CO), which is supported by P30-CA046934 National Cancer Institute. The authors thank Katrina Diener (University of Colorado Denver, Denver, CO); Kendra Walton, Sonia Leach, and Courtney Frasch (National Jewish Health, Denver, CO) for assistance with FACS protocols and analysis.
Footnotes
This work was supported in part through National Institutes of Health grant R01HL109557 (A.N.G.).
Author Contributions: S.K.S. and V.K. designed, performed, and interpreted experiments; T.D. and T.L.P. analyzed data; V.K., T.D., and R.A.P. edited the manuscript; R.A.P. participated in study design; S.K.S. and A.N.G. co-wrote the manuscript; A.N.G. conceived the project and interpreted data.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0357OC on April 4, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Doeing DC, Solway J. Airway smooth muscle in the pathophysiology and treatment of asthma. J Appl Physiol (1985) 2013;114:834–843. doi: 10.1152/japplphysiol.00950.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black JL, Panettieri RA, Jr, Banerjee A, Berger P. Airway smooth muscle in asthma: just a target for bronchodilation? Clin Chest Med. 2012;33:543–558. doi: 10.1016/j.ccm.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pera T, Penn RB. Bronchoprotection and bronchorelaxation in asthma: new targets, and new ways to target the old ones. Pharmacol Ther. 2016;164:82–96. doi: 10.1016/j.pharmthera.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tliba O, Amrani Y, Panettieri RA., Jr Is airway smooth muscle the “missing link” modulating airway inflammation in asthma? Chest. 2008;133:236–242. doi: 10.1378/chest.07-0262. [DOI] [PubMed] [Google Scholar]
- 5.Prakash YS. Airway smooth muscle in airway reactivity and remodeling: what have we learned? Am J Physiol Lung Cell Mol Physiol. 2013;305:L912–L933. doi: 10.1152/ajplung.00259.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durrani SR, Viswanathan RK, Busse WW. What effect does asthma treatment have on airway remodeling? Current perspectives. J Allergy Clin Immunol. 2011;128:439–448. [Quiz, 449–450]. doi: 10.1016/j.jaci.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Bergeron C, Hauber HP, Gotfried M, Newman K, Dhanda R, Servi RJ, Ludwig MS, Hamid Q. Evidence of remodeling in peripheral airways of patients with mild to moderate asthma: effect of hydrofluoroalkane-flunisolide. J Allergy Clin Immunol. 2005;116:983–989. doi: 10.1016/j.jaci.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 8.Bel EH, Ortega HG, Pavord ID. Glucocorticoids and mepolizumab in eosinophilic asthma. N Engl J Med. 2014;371:2434. doi: 10.1056/NEJMc1412892. [DOI] [PubMed] [Google Scholar]
- 9.James AL, Elliot JG, Jones RL, Carroll ML, Mauad T, Bai TR, Abramson MJ, McKay KO, Green FH. Airway smooth muscle hypertrophy and hyperplasia in asthma. Am J Respir Crit Care Med. 2012;185:1058–1064. doi: 10.1164/rccm.201110-1849OC. [DOI] [PubMed] [Google Scholar]
- 10.Panettieri RA., Jr Isolation and culture of human airway smooth muscle cells. Methods Mol Med. 2001;56:155–160. doi: 10.1385/1-59259-151-5:155. [DOI] [PubMed] [Google Scholar]
- 11.Halayko AJ, Tran T, Ji SY, Yamasaki A, Gosens R. Airway smooth muscle phenotype and function: interactions with current asthma therapies. Curr Drug Targets. 2006;7:525–540. doi: 10.2174/138945006776818728. [DOI] [PubMed] [Google Scholar]
- 12.Himes BE, Koziol-White C, Johnson M, Nikolos C, Jester W, Klanderman B, Litonjua AA, Tantisira KG, Truskowski K, MacDonald K, et al. Vitamin D modulates expression of the airway smooth muscle transcriptome in fatal asthma. PLoS One. 2015;10:e0134057. doi: 10.1371/journal.pone.0134057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright DB, Trian T, Siddiqui S, Pascoe CD, Ojo OO, Johnson JR, Dekkers BG, Dakshinamurti S, Bagchi R, Burgess JK, et al. Functional phenotype of airway myocytes from asthmatic airways. Pulm Pharmacol Ther. 2013;26:95–104. doi: 10.1016/j.pupt.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Masuno K, Haldar SM, Jeyaraj D, Mailloux CM, Huang X, Panettieri RA, Jr, Jain MK, Gerber AN. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am J Respir Cell Mol Biol. 2011;45:642–649. doi: 10.1165/rcmb.2010-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart AG, Fernandes D, Tomlinson PR. The effect of glucocorticoids on proliferation of human cultured airway smooth muscle. Br J Pharmacol. 1995;116:3219–3226. doi: 10.1111/j.1476-5381.1995.tb15127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore PE, Laporte JD, Gonzalez S, Moller W, Heyder J, Panettieri RA, Jr, Shore SA. Glucocorticoids ablate IL-1β–induced β-adrenergic hyporesponsiveness in human airway smooth muscle cells. Am J Physiol. 1999;277:L932–L942. doi: 10.1152/ajplung.1999.277.5.L932. [DOI] [PubMed] [Google Scholar]
- 17.Goldsmith AM, Hershenson MB, Wolbert MP, Bentley JK. Regulation of airway smooth muscle α-actin expression by glucocorticoids. Am J Physiol Lung Cell Mol Physiol. 2007;292:L99–L106. doi: 10.1152/ajplung.00269.2006. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann K, Koenen M, Schauer S, Wittig-Blaich S, Ahmad M, Baschant U, Tuckermann JP. Molecular actions of glucocorticoids in cartilage and bone during health, disease, and steroid therapy. Physiol Rev. 2016;96:409–447. doi: 10.1152/physrev.00011.2015. [DOI] [PubMed] [Google Scholar]
- 19.Bodine SC, Furlow JD. Glucocorticoids and skeletal muscle. Adv Exp Med Biol. 2015;872:145–176. doi: 10.1007/978-1-4939-2895-8_7. [DOI] [PubMed] [Google Scholar]
- 20.Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, Cline GW, Kim JK, Peroni OD, Kahn BB, et al. Regulation of gluconeogenesis by Krüppel-like factor 15. Cell Metab. 2007;5:305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasse SK, Mailloux CM, Barczak AJ, Wang Q, Altonsy MO, Jain MK, Haldar SM, Gerber AN. The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol Cell Biol. 2013;33:2104–2115. doi: 10.1128/MCB.01474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasse SK, Gerber AN. Feed-forward transcriptional programming by nuclear receptors: regulatory principles and therapeutic implications. Pharmacol Ther. 2015;145:85–91. doi: 10.1016/j.pharmthera.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison-Nozik A, Anand P, Zhu H, Duan Q, Sabeh M, Prosdocimo DA, Lemieux ME, Nordsborg N, Russell AP, MacRae CA, et al. Glucocorticoids enhance muscle endurance and ameliorate Duchenne muscular dystrophy through a defined metabolic program. Proc Natl Acad Sci USA. 2015;112:E6780–E6789. doi: 10.1073/pnas.1512968112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerber A, Kadiyala V, Panettieri R, Sasse S. Novel mechanisms of steroid function in airway smooth muscle defined through chip-seq [abstract] New York: American Thoracic Society; 2016. p. A4344. [Google Scholar]
- 25.Sasse SK, Altonsy MO, Kadiyala V, Cao G, Panettieri RA, Jr, Gerber AN. Glucocorticoid and TNF signaling converge at A20 (TNFAIP3) to repress airway smooth muscle cytokine expression. Am J Physiol Lung Cell Mol Physiol. 2016;311:L421–L432. doi: 10.1152/ajplung.00179.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadiyala V, Sasse SK, Altonsy MO, Berman R, Chu HW, Phang TL, Gerber AN. Cistrome-based cooperation between airway epithelial glucocorticoid receptor and NF-κB orchestrates anti-inflammatory effects. J Biol Chem. 2016;291:12673–12687. doi: 10.1074/jbc.M116.721217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, et al. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci USA. 2007;104:7074–7079. doi: 10.1073/pnas.0701981104. [Published erratum appears in Proc Natl Acad Sci USA 104:13851.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. FOXO transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng H, Dokshin GA, Lei J, Goldsmith AM, Bitar KN, Fingar DC, Hershenson MB, Bentley JK. Inhibition of glycogen synthase kinase-3β is sufficient for airway smooth muscle hypertrophy. J Biol Chem. 2008;283:10198–10207. doi: 10.1074/jbc.M800624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arango-Lievano M, Lambert WM, Jeanneteau F. Molecular biology of glucocorticoid signaling. Adv Exp Med Biol. 2015;872:33–57. doi: 10.1007/978-1-4939-2895-8_2. [DOI] [PubMed] [Google Scholar]
- 32.Himes BE, Jiang X, Wagner P, Hu R, Wang Q, Klanderman B, Whitaker RM, Duan Q, Lasky-Su J, Nikolos C, et al. RNA-seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One. 2014;9:e99625. doi: 10.1371/journal.pone.0099625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dasgupta P, Dorsey NJ, Li J, Qi X, Smith EP, Yamaji-Kegan K, Keegan AD. The adaptor protein insulin receptor substrate 2 inhibits alternative macrophage activation and allergic lung inflammation. Sci Signal. 2016;9:ra63. doi: 10.1126/scisignal.aad6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijsing SH. Mechanisms of glucocorticoid-regulated gene transcription. Adv Exp Med Biol. 2015;872:59–81. doi: 10.1007/978-1-4939-2895-8_3. [DOI] [PubMed] [Google Scholar]
- 35.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eggermann T, Binder G, Brioude F, Maher ER, Lapunzina P, Cubellis MV, Bergadá I, Prawitt D, Begemann M. CDKN1C mutations: two sides of the same coin. Trends Mol Med. 2014;20:614–622. doi: 10.1016/j.molmed.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Zhao L, Samuels T, Winckler S, Korgaonkar C, Tompkins V, Horne MC, Quelle DE. Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways. Mol Cancer Res. 2003;1:195–206. [PubMed] [Google Scholar]
- 39.Mu H, Wang N, Zhao L, Li S, Li Q, Chen L, Luo X, Qiu Z, Li L, Ren G, et al. Methylation of PLCD1 and adenovirus-mediated PLCD1 overexpression elicits a gene therapy effect on human breast cancer. Exp Cell Res. 2015;332:179–189. doi: 10.1016/j.yexcr.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 40.Kudo M, Khalifeh Soltani SM, Sakuma SA, McKleroy W, Lee TH, Woodruff PG, Lee JW, Huang K, Chen C, Arjomandi M, et al. Mfge8 suppresses airway hyperresponsiveness in asthma by regulating smooth muscle contraction. Proc Natl Acad Sci USA. 2013;110:660–665. doi: 10.1073/pnas.1216673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsujikawa K, Yayama K, Hayashi T, Matsushita H, Yamaguchi T, Shigeno T, Ogitani Y, Hirayama M, Kato T, Fukada S, et al. Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1–deficient mice. Proc Natl Acad Sci USA. 2007;104:16702–16707. doi: 10.1073/pnas.0705974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeo JC, Wall AA, Luo L, Condon ND, Stow JL. Distinct roles for appl1 and appl2 in regulating Toll-like receptor 4 signaling in macrophages. Traffic. 2016;17:1014–1026. doi: 10.1111/tra.12415. [DOI] [PubMed] [Google Scholar]
- 43.Yick CY, Zwinderman AH, Kunst PW, Grünberg K, Mauad T, Fluiter K, Bel EH, Lutter R, Baas F, Sterk PJ. Glucocorticoid-induced changes in gene expression of airway smooth muscle in patients with asthma. Am J Respir Crit Care Med. 2013;187:1076–1084. doi: 10.1164/rccm.201210-1886OC. [DOI] [PubMed] [Google Scholar]
- 44.Hu A, Josephson MB, Diener BL, Nino G, Xu S, Paranjape C, Orange JS, Grunstein MM. Pro-asthmatic cytokines regulate unliganded and ligand-dependent glucocorticoid receptor signaling in airway smooth muscle. PLoS One. 2013;8:e60452. doi: 10.1371/journal.pone.0060452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim HW, Uhlenhaut NH, Rauch A, Weiner J, Hübner S, Hübner N, Won KJ, Lazar MA, Tuckermann J, Steger DJ. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 2015;25:836–844. doi: 10.1101/gr.188581.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiller BJ, Chodankar R, Watson LC, Stallcup MR, Yamamoto KR. Glucocorticoid receptor binds half sites as a monomer and regulates specific target genes. Genome Biol. 2014;15:418. doi: 10.1186/s13059-014-0418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Bosscher K, Haegeman G, Elewaut D. Targeting inflammation using selective glucocorticoid receptor modulators. Curr Opin Pharmacol. 2010;10:497–504. doi: 10.1016/j.coph.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Sasse SK, Zuo Z, Kadiyala V, Zhang L, Pufall MA, Jain MK, Phang TL, Stormo GD, Gerber AN. Response element composition governs correlations between binding site affinity and transcription in glucocorticoid receptor feed-forward loops. J Biol Chem. 2015;290:19756–19769. doi: 10.1074/jbc.M115.668558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuno K. Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell Signal. 2013;25:457–469. doi: 10.1016/j.cellsig.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Lavoie TL, Dowell ML, Lakser OJ, Gerthoffer WT, Fredberg JJ, Seow CY, Mitchell RW, Solway J. Disrupting actin–myosin–actin connectivity in airway smooth muscle as a treatment for asthma? Proc Am Thorac Soc. 2009;6:295–300. doi: 10.1513/pats.200808-078RM. [DOI] [PMC free article] [PubMed] [Google Scholar]