Abstract
The relevance of animal models to human diseases is an area of intense scientific debate. The degree to which mouse models of lung injury recapitulate human lung injury has never been assessed. Integrating data from both human and animal expression studies allows for increased statistical power and identification of conserved differential gene expression across organisms and conditions. We sought comprehensive integration of gene expression data in experimental acute lung injury (ALI) in rodents compared with humans. We performed two separate gene expression multicohort analyses to determine differential gene expression in experimental animal and human lung injury. We used correlational and pathway analyses combined with external in vitro gene expression data to identify both potential drivers of underlying inflammation and therapeutic drug candidates. We identified 21 animal lung tissue datasets and three human lung injury bronchoalveolar lavage datasets. We show that the metasignatures of animal and human experimental ALI are significantly correlated despite these widely varying experimental conditions. The gene expression changes among mice and rats across diverse injury models (ozone, ventilator-induced lung injury, LPS) are significantly correlated with human models of lung injury (Pearson r = 0.33–0.45, P < 1E−16). Neutrophil signatures are enriched in both animal and human lung injury. Predicted therapeutic targets, peptide ligand signatures, and pathway analyses are also all highly overlapping. Gene expression changes are similar in animal and human experimental ALI, and provide several physiologic and therapeutic insights to the disease.
Keywords: acute respiratory distress syndrome, acute lung injury, gene expression, animal model, human
Clinical Relevance
Dozens of genome-wide expression studies in experimental lung injury in animals and humans have yielded important insights into its pathophysiology, but no coherent view has emerged. The degree to which murine models of lung injury recapitulate human lung injury has never been assessed. We performed integrated meta-analyses of gene expression studies in animal and human experimental lung injury to discover conserved sets of differentially expressed genes. We show that animal models recapitulate human lung injury models, including highly overlapping gene pathways, cell type enrichments, and predicted therapeutic targets and peptide ligands. These findings reinforce the relevance of animal models to human lung injury and critical illness more generally.
Acute lung injury (ALI) and the acute respiratory distress syndrome (ARDS) have a high overall mortality rate and remain difficult to treat despite clinical advances (1, 2). Animal models of ALI have shown decreased mortality with pharmacologic interventions, such as statins (3), only to have these fail in later clinical application (4–6). The experimental studies done in animal models (mostly rodents) assume that animal models of lung injury can lead to insight into the human clinical condition, but there is disagreement over the degree to which animal models recapitulate human disease in acute illness at a molecular level (7, 8). In particular, there are several possible models of experimental lung injury in animals (9), which makes interpreting results from any individual study difficult. The degrees to which animal models of lung injury are similar to one another, and whether any animal models recapitulate human lung injury, are thus open questions.
Over the past decade, dozens of studies have been published examining whole-genome expression in experimentally induced lung injury in both animals and humans. Many of these studies have been deposited in public databases, such as the National Institutes of Health (NIH) Gene Expression Omnibus (GEO) and ArrayExpress, allowing for their further study and reanalysis. Although many of these studies were very small, in aggregate, there are now hundreds of samples. In addition, the types of lung injury models studied are very diverse, which may mimic the heterogeneity seen in clinical lung injury.
Here, we sought to comprehensively examine all gene expression studies of animal and human experimental lung injury. We applied our multicohort gene expression analysis framework (10, 11) to perform separate meta-analyses of experimental ALI in animals and humans. Integrating the heterogeneity present in several different types of experimental models provides generalizable results; furthermore, cross-species analysis of animal and human data can increase statistical power (12). Thus, we here compared the resulting gene expression signatures from animal and human lung injury models and studied them for new insights into the pathophysiology of lung injury, and for potential therapeutics. We demonstrate strikingly similar gene expression changes in rodents and humans in response to lung injury, with highly overlapping gene pathways, cell type enrichment, and predicted therapeutic and inflammatory peptide ligands. These data reinforce the relevance of animal models in the study of critical illness.
Materials and Methods
We performed a systematic search for experimental lung injury studies in animals and humans in two public gene expression microarray repositories (NIH GEO, ArrayExpress) using the search terms, “ARDS,” “respiratory distress syndrome,” “ALI,” “lung injury,” “ICU,” and “mechanical ventilation.” In the rodent studies, only wild-type animals, and only control versus lung injury without any extra experimental conditions, were included in the analysis. We treated all lung injury conditions (e.g., ventilator-induced lung injury [VILI], ozone treatment, LPS treatment, etc.) as cases, and all non–lung injury conditions (nontreated, spontaneously breathing, or ventilated at optimal positive end-expiratory pressure/tidal volume) as controls. We mapped all mouse and rat genes to human homologs using biomaRt before analysis (13). We excluded probes/genes that could not be mapped to human homologs from further analysis.
In the human experimental lung injury studies, we included only bronchoalveolar lavage (BAL) studies; no experimental human lung tissue biopsy studies were found. We included only experimental conditions designed to cause lung injury (we excluded studies of only smoking, asthma, or common allergens). We also excluded all in vitro studies of human tissues or cells from the meta-analysis. We ensured that all microarray data had been log-2 transformed before analysis. For all gene analyses, we summarized probes to genes within datasets with a fixed-effect meta-analysis. For two-channel microarrays, Cy3- and Cy5-labeled samples were treated as independent.
We performed multicohort analysis of gene expression as previously described (10, 11). Briefly, we applied two meta-analysis methods: one is a DerSimonian-Laird random-effects model combining effect sizes using Hedges’ g, the other using Fisher’s sum-of-logs method combining P values. Genes set as “significant” passed a minimum false discovery rate (FDR; Benjamini-Hochberg FDR < 0.01) and a minimum effect size (1.3-fold change).
We performed cell type enrichment tests as previously described (11). Briefly, we downloaded and coquantile normalized gene expression profiles of relevant in vitro immune cell types. We then added datasets examining type I pneumocytes (14), type II pneumocytes (15, 16), and alveolar macrophages (16–18). We tested gene signatures of interest for their expression level in each in vitro cell type. We standardized the resulting scores across cell types, and calculated a P value of the resulting Z score distribution by assuming that the standardized scores are normally distributed.
We performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis with the ROntoTools R package (19). For both animal and human BAL cohorts, we analyzed the entire gene list weighted by P values. We assessed significance at 5,000 bootstraps by combining P values from perturbation accumulation and perturbation propagation analyses, followed by Benjamini-Hochberg correction.
To predict drugs that would reverse the lung injury signature, as well as extracellular signaling proteins that could be driving it, we leveraged the L1000 gene expression data generated by the NIH Library of Network-Based Cellular Signatures (LINCS) Program (NIH, Bethesda, MD). This dataset profiles the change in gene expression caused by chemical compounds and peptide ligands (cytokines, chemokines, and growth factors) across 77 cellular contexts at varying doses measured at varying time points. We used the median absolute deviation normalized data from the LINCS c3 server. We quality processed the L1000 data to obtain robust gene expression signatures from the various tested compound that could be compared with our lung injury signature. We removed signatures that had not passed LINCS internal quality-control standards and thus were not labeled as “gold”; this resulted in 12,486 chemical compounds and 311 peptide ligands.
We then removed probes not in the “bing” set of imputed probes. We collapsed the data to gene level expression by averaging the expression scores of all probes that mapped to a given gene. Each drug or peptide ligand has numerous signatures representing the induced gene expression changes in different cell lines, at different doses, and measured at different times. To create a single overall effect signature for a given drug or ligand, we took the median of gene expression for each of these specific signatures. Chemical compounds and ligands were Pearson correlated with the lung injury signatures, and P values were Benjamini-Hochberg corrected. Compounds and ligands for which the correlation FDR (q values) were < 0.05 were deemed significant.
Results
Multicohort Gene Expression Analyses
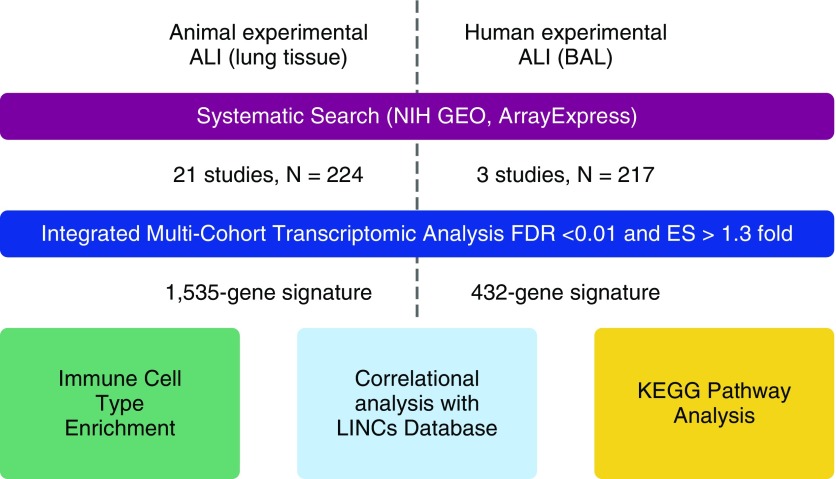
Here, we analyzed two types of studies of experimentally induced ALI: animal models that examined lung tissue, and human experiments that examined BAL fluid (Figure 1). We first performed a systematic search of NIH GEO and ArrayExpress for studies of either type. There were 21 animal lung tissue datasets (19–38) with diverse lung injury mechanisms, including ventilator-induced injury (n = 14), ozone (n = 3), intratracheal LPS (n = 3), and a hemorrhagic trauma model (n = 1) (total controls = 83, cases = 141; Table 1). No human ALI whole-lung samples were available, so we used three human experimental datasets (39–41) that studied LPS or ozone exposure via BAL (total microarrays: controls = 107, cases = 110; Table 2).
Figure 1.
Workflow schematic describing the initial gene expression meta-analyses, and the subsequent studies of significant gene sets. ALI, acute lung injury; BAL, bronchoalveolar lavage; ES, effect size; FDR, false discovery rate; GEO, Gene Expression Omnibus; KEGG, Kyoto Encyclopedia of Genes and Genomes; LINCS, Library of Network-Based Cellular Signatures; NIH, U.S. National Institutes of Health.
Table 1.
Animal Experimental Lung Injury Datasets
| First Author | Origin | Accession No. | Control | Condition | Platform | n Ctrl | n Cond |
|---|---|---|---|---|---|---|---|
| Cho | Mouse | GSE495 | Time 0 control | Hyperoxic injury (24 and 48 h) | GPL81 | 2 | 3 |
| Ma | Mouse | GSE2368 | Spontaneously breathing | VILI | GPL81 | 2 | 2 |
| Ma | Rat | GSE2368 | Spontaneously breathing | VILI | GPL85 | 2 | 2 |
| Altemeier | Mouse | GSE2411 | Spontaneuously breathing or mechanically ventilated | LPS ± MV | GPL339 | 6 | 18 |
| Dolinay | Mouse | GSE4215 | Spontaneous ventilation | VILI, LPS, or both | GPL3436 | 6 | 20 |
| Feinman | Rat | GSE6332 | Sham | Trauma/hemorrhaghic shock | GPL85 | 3 | 3 |
| Nonas | Rat | GSE7041 | Room air ventilation | VILI | GPL1355 | 6 | 6 |
| Dolinay | Mouse | GSE7742 | Spontaneously breathing | VILI | GPL5145 | 3 | 4 |
| Papaiahgari | Mouse | GSE9208 | Spontaneous ventilation | VILI | GPL8321 | 3 | 3 |
| Hong | Mouse | GSE9314 | Spontaneously breathing | VILI | GPL1261 | 4 | 4 |
| Wray | Mouse | GSE11434 | Spontaneously breathing | VILI | GPL1261 | 5 | 5 |
| Meyer | Mouse | GSE11662 | Spontaneously breathing | VILI | GPL1261 | 3 | 3 |
| Mirzapoiazova | Mouse | GSE14525 | Spontaneously breathing | VILI | GPL1261 | 3 | 3 |
| Dolinay | Mouse | GSE29920 | Time 0 control | VILI | GPL6885 | 3 | 5 |
| Park | Rat | GSE31678 | Spontaneously breathing | VILI (prone and supine) | GPL1355 | 2 | 6 |
| Oakes | Mouse | GSE38014 | Filtered air/saline | Ozone and/or Pam3Cys4 (4 and 24 h) | GPL7202 | 8 | 24 |
| Xu | Mouse | GSE48787 | Control | Intratracheal LPS | GPL13684 | 3 | 3 |
| Krebs | Rat | GSE52142 | Ventilated, optimal PEEP | VILI | GPL17896 | 6 | 6 |
| Huang | Rat | GSE57011 | Spontaneously breathing | VILI + lavage | GPL1699 | 5 | 7 |
| Spassov | Mouse | GSE58169 | Spontaneously breathing | VILI | GPL6246 | 5 | 5 |
| Verhein | Mouse | GSE58244 | Air | Ozone (6, 24, and 48 h) | GPL1261 | 3 | 9 |
Definition of abbreviations: Cond, condition; Ctrl, control; MV, mechanical ventilation; PEEP, positive end-expiratory pressure; VILI, ventilator-induced lung injury.
All studies were performed on lung tissue.
Table 2.
Experimental Human Acute Lung Injury Bronchoalveolar Lavage Datasets
| First Author | Origin | Accession No. | Control | Condition | Tissue | Platform | n Ctrl | n Cond |
|---|---|---|---|---|---|---|---|---|
| Yang | Human | GSE5272 | Saline via bronchoscope | LPS via bronchoscope | BAL fluid | GPL1708 | 82 | 84 |
| Reynier | Human | GSE40885 | Saline via bronchoscope | LPS via bronchoscope | Alveolar macrophages from BAL fluid | GPL570 | 7 | 7 |
| Leroy | Human | GSE58682 | Clean air (0 ppb) | Ozone (200 ppb) | BAL cell pellets | GPL6244 | 18 | 19 |
Definition of abbreviations: BAL, bronchoalveolar lavage; Cond, condition; Ctrl, control.
“n” refers to number of microarrays.
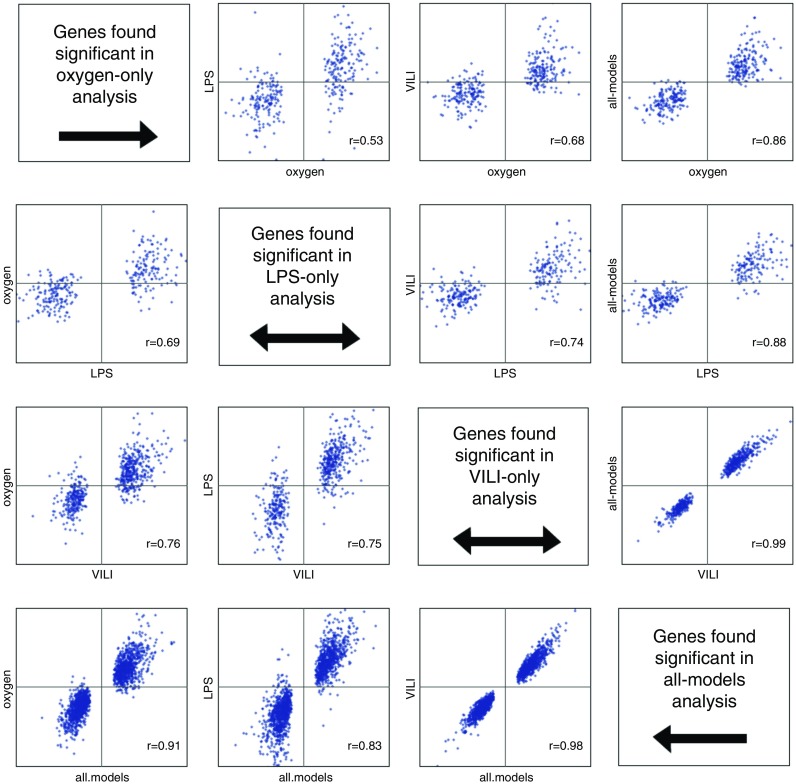
An open question is the degree of similarity in the immune response caused by different animal models of lung injury. Thus, we first performed correlational analysis of each type of experimental animal lung injury. To find genes significantly differentially expressed in experimental lung injury, we ran all cohorts from each animal model type through our integrated multicohort analysis framework, as previously described (10, 11, 42). We also performed one all-models analysis, combining comparisons of healthy lung to injured lung in all available models at once. For all analyses, we set significance thresholds of FDR less than 1% and a minimum effect size greater than 1.3-fold. We then took the genes identified as significant in each analysis, and correlated the differential effects for all model types (Figure 2). In general, intermodel correlations were high (mean r = 0.692, range = 0.53–0.76, all P < 1E−16). Furthermore, the all-models comparison was highly correlated with each individual model (mean r = 0.905, range = 0.83–0.99), indicating that the all-models meta-analysis was capturing a signal common to each model of lung injury.
Figure 2.
Correlations of meta-effect sizes between different types of experimental animal lung injury. In each graph, the x axis indicates the model type from which genes were selected as significant. VILI, ventilator-induced lung injury.
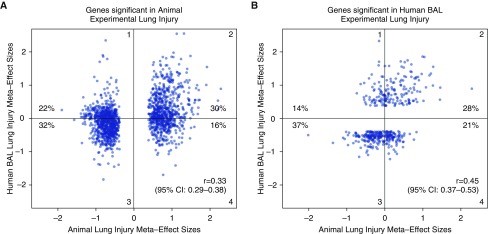
We next compared the all-models animal lung injury signature with the human BAL models of lung injury, using the same multicohort analysis framework and thresholds as described previously here. This yielded 1,535 human homologous genes in the all-models animal analysis and 430 genes in human experimental BAL analysis (see Tables E1 and E2 in the online supplement). There was a significant overlap in the number of genes significantly differentially expressed in common between the animal lung and human BAL analyses (50 genes, hypergeometric P = 0.037). We tested the correlation of the meta-effect sizes of differential gene expression between animal and human experimental lung injury, and found that including either significant genes from the animal analysis or significant genes from the human analysis resulted in significant correlation (Pearson r = 0.33 and 0.45, respectively, both P < 1E−16; Figure 3). Roughly 30–40% of the genes in each analysis had meta-effect sizes moving in opposite directions between animals and humans; this may reflect differences in model type (human intrabronchial LPS versus animal ozone, VILI, or LPS) or tissue type (human BAL versus animal lung homogenate).
Figure 3.
Unbiased correlations of meta-effect sizes between animal and human experimental lung injury. Different sets of genes are shown in each figure: (A) genes significant in animal experimental lung injury; (B) genes significant in human experimental lung injury. Genes in A may not be significant in human models, and genes significant in B may not be significant in animal models. The percent of genes in each quadrant is shown. Perfect model agreement would be nearly linear, with all genes increased in both human and animal models in quadrant 2, and all genes decreased in both human and animal models in quadrant 3 (e.g., see Figure 1 in the work by Takao and colleagues [8]). Both Pearson r values are highly significant (P < 1E−16). In each graph, the x axis indicates the model type from which genes were selected as significant. CI, confidence interval.
Cell-Type Enrichments
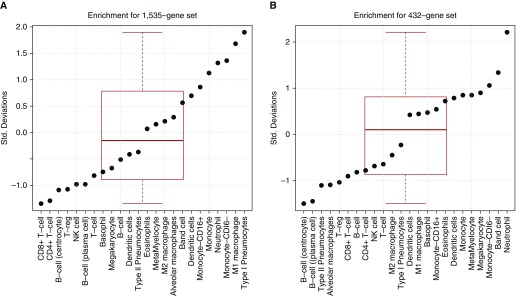
Differential gene expression signatures represent both intracellular expression changes and cell compartment shifts. Overrepresentation of a differential gene expression signature in a particular cell type can suggest that an increase in that cell type is partially responsible for the changes seen. Whereas whole lung tissue contains a complex mixture of cell types, BAL fluid contains mainly neutrophils in early lung injury (43, 44). We tested both the animal lung tissue and human BAL fluid gene expression signatures for overexpression in several in vitro cell lines, including both lung cell types (type I and type II pneumocytes, and alveolar macrophages) and immune cells, as these are the cells expected to be changing in the tissue (Figure 4). As expected, the animal lung injury signature was enriched in type I pneumocytes (P = 0.028) and M1 (IFN-γ/LPS-activated) macrophages (P = 0.046), but also trended toward enrichment in monocytes and neutrophils (P < 0.1). The human BAL fluid mainly showed enrichment for neutrophils (P = 0.01).
Figure 4.
Enrichment analysis for immune and lung cell in vitro gene expression. Shown are the immune enrichment Z scores for the experimental lung injury gene signatures in (A) animal lung tissue and (B) human BAL fluid. NK, natural killer; Std., standard; T-reg, T regulatory.
Pathway Analyses
We next studied whether the two gene expression signatures showed any overlapping pathway activations. Here, rather than use an arbitrary significance threshold for included genes, we weighted the entire differential gene expression vectors by their associated q values. This allows for accumulated pathway propagation effects to be calculated. We found that 7 of the top 10 most significantly activated pathways overlap between animal and human lung injury (Table 3). In particular, the mitogen-activate protein kinase (MAPK) signaling pathway is the top-ranked pathway in human ALI and the second-ranked pathway for animal ALI. Notably, the NF-κB pathway and the tuberculosis pathway (which is primarily driven by IFN response) were both in the top 10 lists of both human and rodent lung injury gene expression signatures.
Table 3.
KEGG Pathways Enriched in Animal and Human Experimental Lung Injury
| Animal Lung Pathways | Combined FDR | Human BAL Pathways | Combined FDR |
|---|---|---|---|
| hsa04060: Cytokine–cytokine receptor interaction | 0.0004 | hsa04010: MAPK signaling | 1.72E−05 |
| hsa04010: MAPK signaling pathway | 0.0012 | hsa04062: Chemokine signaling | 1.72E−05 |
| hsa05146: Amoebiasis | 0.0013 | hsa04740: Olfactory transduction | 1.72E−05 |
| hsa05152: Tuberculosis | 0.0013 | hsa04932: Nonalcoholic fatty liver disease | 1.72E−05 |
| hsa04062: Chemokine signaling pathway | 0.0013 | hsa05146: Amoebiasis | 1.72E−05 |
| hsa04064: NF-κB signaling pathway | 0.0013 | hsa05152: Tuberculosis | 1.72E−05 |
| hsa05166: HTLV-I infection | 0.0029 | hsa05169: Epstein-Barr virus infection | 1.72E−05 |
| hsa04110: Cell cycle | 0.0029 | hsa04060: Cytokine–cytokine receptor interaction | 2.11E−05 |
| hsa04932: Nonalcoholic fatty liver disease | 0.0036 | hsa04064: NF-κB signaling | 2.11E−05 |
| hsa04115: p53 signaling pathway | 0.0041 | hsa05161: Hepatitis B | 2.11E−05 |
Definition of abbreviations: BAL, bronchoalveolar lavage; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; HTLV-1, human T-cell lymphotropic virus type 1; MAPK, mitogen-activate protein kinase.
The top 10 significant pathways are shown for each condition. Common pathways are in boldface type.
Therapeutic Targets Predicted by Lung Injury Gene Signatures
We next studied whether the lung injury signatures could be used to predict therapeutic drug actions or find drug targets. For both the animal lung and BAL analyses, the effect sizes from all significant genes were compared with gene expression measurements from cell lines treated with various drugs and ligands in the LINCS database (http://lincscloud.org/). Here, a drug that induces a gene expression pattern that is negatively correlated with the meta-effect sizes found in disease is reasoned to be potentially protective, as it may reverse the underlying gene expression signature of the given condition (45). Similarly, a ligand with a gene expression pattern that is positively correlated with a disease signature could be a potential causative signaling agent (and eventual drug target) of the given condition. The LINCS database lists 12,486 drugs and small molecules that have been tested for gene expression changes in multiple cell lines and that passed our quality control. We tested the expression vectors of all of these agents for correlation with the animal lung and human BAL signatures.
The extent of overlap between potential targets in animal and human lung injury models is highly significant (hypergeometric P < 1E−13). A total of 181 drugs are significantly negatively correlated with the lung injury signatures from both humans and animals (q < 0.05, Pearson r range = −0.5 to −0.08 for animals and −0.16 to −0.42 in humans; Table 4 and Table E3A). The list of common targets includes several previously implicated in lung injury, including multiple MAPK/extracellular signal regulated kinase (ERK) inhibitors (including fostamatinib, PD98059, and ERK inhibitor 11E) (46–48) and multiple statins (including rosuvastatin, simvastatin, and atorvastin). In addition, many of the implicated drugs have never been tested in ALI, including, for example, AS605240, a PI3K inhibitor that has been previously associated with a reduction in bleomycin-induced lung fibrosis (49), but has never been studied in ALI. The full ranked drug lists are supplied in Table E3B.
Table 4.
Top 20 Drugs that Are Anticorrelated with Lung Injury Gene Signature in Both Human and Animal Models
| Drug | Effect Human | q Value Human | Animal Effect size | Animal q Value |
|---|---|---|---|---|
| PD98059 | −0.418 | 1.1E−09 | −0.467 | 2.7E−48 |
| AZ628 | −0.395 | 1.5E−08 | −0.463 | 2.3E−47 |
| Fostamatinib | −0.392 | 2.1E−08 | −0.491 | 4.1E−54 |
| AS605240 | −0.374 | 1.2E−07 | −0.434 | 4.5E−41 |
| ERK inhibitor 11E | −0.367 | 2.4E−07 | −0.478 | 5.2E−51 |
| AS703026 | −0.357 | 5.8E−07 | −0.345 | 5.2E−25 |
| PD0325901 | −0.355 | 6.5E−07 | −0.321 | 1.7E−21 |
| PD184352 | −0.353 | 7.6E−07 | −0.362 | 9.7E−28 |
| Atorvastatin | −0.352 | 8.7E−07 | −0.468 | 1.2E−48 |
| Orteronel | −0.349 | 1.1E−06 | −0.212 | 2.2E−09 |
| BRDK70153099 | −0.340 | 2.4E−06 | −0.088 | 2.5E−02 |
| BRDK27064849 | −0.339 | 2.6E−06 | −0.109 | 4.4E−03 |
| BRDK29622226 | −0.321 | 1.1E−05 | −0.331 | 7.2E−23 |
| Selumetinib | −0.320 | 1.2E−05 | −0.383 | 3.2E−31 |
| BRDK04986062 | −0.313 | 2.0E−05 | −0.182 | 4.7E−07 |
| Lapatinib | −0.305 | 3.7E−05 | −0.387 | 6.6E−32 |
| BRDK87010407 | −0.299 | 6.0E−05 | −0.101 | 9.0E−03 |
| Vemurafenib | −0.299 | 6.1E−05 | −0.231 | 4.8E−11 |
| OM137 | −0.296 | 7.2E−05 | −0.233 | 3.2E−11 |
| Trametinib | −0.295 | 7.7E−05 | −0.305 | 2.4E−19 |
Definition of abbreviation: ERK, extracellular signal regulated kinase.
We also explored correlations between the animal and human gene signatures and the gene expression profiles of the 311 peptide ligands (cytokines, chemokines, and growth factors) present in the LINCS database. We hypothesized that ligands that induce an expression profile highly correlated to one identified in a disease may be potentially driving the underlying response. We found that the top 15 positively correlated ligands for both animal lung and human BAL gene expression signatures share 9 overlapping ligands, including TNF and IL-1/IL-1A (hypergeometric P < 1E−9; the complete ranked peptide ligand lists are supplied in Table E4). Given the well established links between IL-1, TNF, and ARDS, this lends credibility to the LINCS methodology. Other ligands include several MAPK activators, such as betacellulin, heparin-binding epidermal growth factor-like growth factor, oncostatin M, and epiregulin.
Discussion
ARDS is a complex syndrome with a high mortality rate, and for which there are no proven pharmacological or molecular therapeutics. Several groups have turned to gene expression profiling as a way to better understand the underlying pathophysiology of lung injury. Human lung tissue biopsies are rarely obtained at the time of ARDS because of the high potential morbidity in a critically ill patient. Thus, mouse models and human BAL profiling are used as surrogates. The question about how relevant these are to both each other and also, thus, the injured human lung is critical. Here, we have performed comprehensive multicohort analyses of gene expression in both animal and human experimental lung injury. We showed not only that gene expression changes across disparate animal injury models are consistent, but also that they reflect changes in human BAL fluid in response to injury. These similarities are reflected in the correlation of meta-effect sizes of individual genes, pathway analysis, cell type enrichment, and predicted therapeutic targets.
The degree to which animal models of disease mimic clinical disease in the gene expression space has been an area of active debate, particularly in critical care (7, 8). In experimental lung injury, we found a significant overlap and effect size correlation between the gene expression signatures of animal lung tissue and human BAL fluid. Notably, the correlation seen in meta-effect sizes between animal models and human models (Pearson r = 0.33, 0.45) were much higher than were previously reported for single-study comparisons of acute injuries between mouse injury models and human clinical disease (burns, trauma) and models of illness (endotoxemia) (Pearson r = 0.00–0.09) (7). Overall, this difference may be evidence that the extremely low correlation identified by Seok and colleagues (7) was caused by both difference in circumstance (comparison of models of disease with clinical illness) and differences in organism (mouse versus human).
The higher correlation between animal and human lung injury models seen here may be due to higher similarity in model conditions, the use of a meta-effect size rather than single-dataset comparisons, or both. Furthermore, despite the fact that some 35–38% of significant genes changed in the opposite direction in different species, the animal and human experimental models shared similar KEGG pathway activations, predicted therapeutics, and predicted driving ligands. This suggests that advanced network analyses may be a more robust way to compare models than simple correlations. Our findings overall suggest that animal lung injury models may generally recapitulate human experimental lung injury models at a molecular level.
A total of 181 drugs are inversely correlated with lung injury gene expression signatures in both animal and human experimental models. Several are drugs that converge on the ERK signaling pathway; the importance of the ERK/MAPK pathway in mouse models of ventilator-induced lung injury has been previously established (37, 46, 50, 51). This drugs list also includes many statins. Although large clinical trials of new statins in established ARDS have been largely negative (5, 6), these agents are effective at ameliorating experimental lung injury (3). Thus, the appearance of a statin in the top 10 lists of predicted drugs confirms that drugs, the actions of which are inversely correlated with gene expression signatures, may indeed reverse LPS-mediated lung injury (45). The large number of drugs and peptide ligands that could potentially inhibit both the human and animal lung injury gene signatures further supports the high relevance of animal models to human ARDS drug discovery. Furthermore, our analytic strategy of including diverse mechanisms of lung injury (VILI, LPS, ozone, hyperoxia) increases the likelihood that this pathway has broad clinical relevance.
Gene expression profiles from complex tissues represent an overall signature from a mixture of cell types. Changes in gene expression signatures may thus represent changes within cells, or may represent changes in cell type proportions. Here, the gene expression signatures were reassuringly overrepresented in the known abundant cell types (type 1 pneumocytes in lung, and neutrophils in early lung injury BAL fluid). Furthermore, several potential cytokine drivers of the lung injury signature were identified by the LINCS analysis; some are well known (IL-1, TNF), others less so (β-cellulin, heparin-binding EGF-like growth factor, oncostatin M). Combinations of these cell types and cytokines should be a focus of further mechanistic inquiry.
Our study has several important limitations. First, though we find that human and animal models of lung injury are substantially overlapping, no clinical samples of ARDS were used, so the relevance to clinical disease is unclear. Our analysis is limited to using models of lung injury, because patients rarely undergo lung biopsy for ARDS, particularly early in the course of disease, and thus no data from lung biopsies or BALs from clinical ARDS were available for study. Thus, we focused on the most pertinent available lung samples, accepting that lung injury models (LPS, ozone, etc.) are not a perfect mimic of clinical ARDS.
Another important limitation of the study is that the available human and animal lung injury models are still substantially different: although whole-lung samples are feasible in a mouse, in human volunteers the injury was certainly less severe and the collected sample was BAL, not biopsy/whole lung. The cell type enrichment shows important similarities between human and animal lung injury, but, again, is limited by the sample available. Type 2 pneumocytes, for example, changed the most in animal whole-lung models; these cells would presumably be reflected very poorly in BAL from LPS-injured human cells. These disparate injury models and samples should bias toward the null, and our conclusions likely underestimate the similarity between human and animal response to injury.
In summary, the present study has several important findings. By combining 21 animal and 3 human datasets, we are able to detect, much more powerfully, important gene expression changes in response to lung injury. We found that the gene expression response to lung injury is highly similar between injury models as disparate as VILI, LPS, and ozone. Most importantly, we provide substantial evidence, using pathway analysis, cell enrichment, and the highly overlapping predicted inhibitory drug and peptide list, that animal models of lung injury do, in fact, recapitulate human experimental lung injury.
Acknowledgments
Acknowledgments
The authors thank the many authors who contributed the gene expression data reanalyzed here, without whom this study would not have been possible.
Footnotes
This work was supported by a Stanford Child Health Research Institute Young Investigator Award through the Institute for Immunity, Transplantation and Infection (T.E.S.), a Society for University Surgeons Resident Research Award (T.E.S.), National Heart, Lung, and Blood Institute grant K23 HL125663 (A.J.R.), and by National Institute of Allergy and Infectious Diseases grants 1U19AI109662, U19AI057229, U54I117925, and U01AI089859 (P.K.).
Author Contributions: T.E.S., P.K., and A.J.R. conceived the study; T.E.S. performed the experiments; S.L. and P.K. produced the LINCS tools; T.E.S., P.K., and A.J.R. analyzed and interpreted data; all authors revised the manuscript and approved its final version.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0395OC on March 21, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Briel M, Meade M, Mercat A, Brower RG, Talmor D, Walter SD, Slutsky AS, Pullenayegum E, Zhou Q, Cook D, et al. Higher vs lower positive end-expiratory pressure in patients with acute lung injury and acute respiratory distress syndrome: systematic review and meta-analysis. JAMA. 2010;303:865–873. doi: 10.1001/jama.2010.218. [DOI] [PubMed] [Google Scholar]
- 2.Guérin C, Reignier J, Richard JC. Prone positioning in the acute respiratory distress syndrome. N Engl J Med. 2013;369:980–981. doi: 10.1056/NEJMc1308895. [DOI] [PubMed] [Google Scholar]
- 3.Singla S, Jacobson JR. Statins as a novel therapeutic strategy in acute lung injury. Pulm Circ. 2012;2:397–406. doi: 10.4103/2045-8932.105028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosma KJ, Taneja R, Lewis JF. Pharmacotherapy for prevention and treatment of acute respiratory distress syndrome: current and experimental approaches. Drugs. 2010;70:1255–1282. doi: 10.2165/10898570-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McAuley DF, Laffey JG, O’Kane CM, Perkins GD, Mullan B, Trinder TJ, Johnston P, Hopkins PA, Johnston AJ, McDowell C, et al. HARP-2 Investigators; Irish Critical Care Trials Group. Simvastatin in the acute respiratory distress syndrome. N Engl J Med. 2014;371:1695–1703. doi: 10.1056/NEJMoa1403285. [DOI] [PubMed] [Google Scholar]
- 6.Truwit JD, Bernard GR, Steingrub J, Matthay MA, Liu KD, Albertson TE, Brower RG, Shanholtz C, Rock P, Douglas IS, et al. National Heart, Lung, and Blood Institute ARDS Clinical Trials Network. Rosuvastatin for sepsis-associated acute respiratory distress syndrome. N Engl J Med. 2014;370:2191–2200. doi: 10.1056/NEJMoa1401520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. Inflammation and Host Response to Injury, Large Scale Collaborative Research Program. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matute-Bello G, Frevert CW, Martin TR. Animal models of acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2008;295:L379–L399. doi: 10.1152/ajplung.00010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khatri P, Roedder S, Kimura N, De Vusser K, Morgan AA, Gong Y, Fischbein MP, Robbins RC, Naesens M, Butte AJ, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweeney TE, Shidham A, Wong HR, Khatri P. A comprehensive time-course–based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med. 2015;7:287ra71. doi: 10.1126/scitranslmed.aaa5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 13.Durinck S, Spellman P, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SM, Chan RW, Gardy JL, Lo CK, Sihoe AD, Kang SS, Cheung TK, Guan YI, Chan MC, Hancock RE, et al. Systems-level comparison of host responses induced by pandemic and seasonal influenza A H1N1 viruses in primary human type I–like alveolar epithelial cells in vitro. Respir Res. 2010;11:147. doi: 10.1186/1465-9921-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballard PL, Lee JW, Fang X, Chapin C, Allen L, Segal MR, Fischer H, Illek B, Gonzales LW, Kolla V, et al. Regulated gene expression in cultured type II cells of adult human lung. Am J Physiol Lung Cell Mol Physiol. 2010;299:L36–L50. doi: 10.1152/ajplung.00427.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Nikrad MP, Phang T, Gao B, Alford T, Ito Y, Edeen K, Travanty EA, Kosmider B, Hartshorn K, et al. Innate immune response to influenza A virus in differentiated human alveolar type II cells. Am J Respir Cell Mol Biol. 2011;45:582–591. doi: 10.1165/rcmb.2010-0108OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodruff PG, Koth LL, Yang YH, Rodriguez MW, Favoreto S, Dolganov GM, Paquet AC, Erle DJ. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med. 2005;172:1383–1392. doi: 10.1164/rccm.200505-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazeros A, Harvey BG, Carolan BJ, Vanni H, Krause A, Crystal RG. Overexpression of apoptotic cell removal receptor MERTK in alveolar macrophages of cigarette smokers. Am J Respir Cell Mol Biol. 2008;39:747–757. doi: 10.1165/rcmb.2007-0306OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho HY, Reddy SP, Debiase A, Yamamoto M, Kleeberger SR. Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radic Biol Med. 2005;38:325–343. doi: 10.1016/j.freeradbiomed.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Ma SF, Grigoryev DN, Taylor AD, Nonas S, Sammani S, Ye SQ, Garcia JG. Bioinformatic identification of novel early stress response genes in rodent models of lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;289:L468–L477. doi: 10.1152/ajplung.00109.2005. [DOI] [PubMed] [Google Scholar]
- 22.Altemeier WA, Matute-Bello G, Gharib SA, Glenny RW, Martin TR, Liles WC. Modulation of lipopolysaccharide-induced gene transcription and promotion of lung injury by mechanical ventilation. J Immunol. 2005;175:3369–3376. doi: 10.4049/jimmunol.175.5.3369. [DOI] [PubMed] [Google Scholar]
- 23.Feinman R, Deitch EA, Aris V, Chu HB, Abungu B, Caputo FJ, Galante A, Xu D, Lu Q, Colorado I, et al. Molecular signatures of trauma–hemorrhagic shock–induced lung injury: hemorrhage- and injury-associated genes. Shock. 2007;28:360–368. doi: 10.1097/shk.0b013e318048565b. [DOI] [PubMed] [Google Scholar]
- 24.Acharya SD, Brooks MM, Evans RW, Linkov F, Burke LE. Weight loss is more important than the diet type in improving adiponectin levels among overweight/obese adults. J Am Coll Nutr. 2013;32:264–271. doi: 10.1080/07315724.2013.816607. [DOI] [PubMed] [Google Scholar]
- 25.Papaiahgari S, Yerrapureddy A, Reddy SR, Reddy NM, Dodd-O JM, Crow MT, Grigoryev DN, Barnes K, Tuder RM, Yamamoto M, et al. Genetic and pharmacologic evidence links oxidative stress to ventilator-induced lung injury in mice. Am J Respir Crit Care Med. 2007;176:1222–1235. doi: 10.1164/rccm.200701-060OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong SB, Huang Y, Moreno-Vinasco L, Sammani S, Moitra J, Barnard JW, Ma SF, Mirzapoiazova T, Evenoski C, Reeves RR, et al. Essential role of pre–B-cell colony enhancing factor in ventilator-induced lung injury. Am J Respir Crit Care Med. 2008;178:605–617. doi: 10.1164/rccm.200712-1822OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wray C, Mao Y, Pan J, Chandrasena A, Piasta F, Frank JA. Claudin-4 augments alveolar epithelial barrier function and is induced in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2009;297:L219–L227. doi: 10.1152/ajplung.00043.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer NJ, Huang Y, Singleton PA, Sammani S, Moitra J, Evenoski CL, Husain AN, Mitra S, Moreno-Vinasco L, Jacobson JR, et al. GADD45a is a novel candidate gene in inflammatory lung injury via influences on Akt signaling. FASEB J. 2009;23:1325–1337. doi: 10.1096/fj.08-119073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirzapoiazova T, Moitra J, Moreno-Vinasco L, Sammani S, Turner JR, Chiang ET, Evenoski C, Wang T, Singleton PA, Huang Y, et al. Non-muscle myosin light chain kinase isoform is a viable molecular target in acute inflammatory lung injury. Am J Respir Cell Mol Biol. 2011;44:40–52. doi: 10.1165/rcmb.2009-0197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolinay T, Kim YS, Howrylak J, Hunninghake GM, An CH, Fredenburgh L, Massaro AF, Rogers A, Gazourian L, Nakahira K, et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med. 2012;185:1225–1234. doi: 10.1164/rccm.201201-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park MS, He Q, Edwards MG, Sergew A, Riches DW, Albert RK, Douglas IS. Mitogen-activated protein kinase phosphatase-1 modulates regional effects of injurious mechanical ventilation in rodent lungs. Am J Respir Crit Care Med. 2012;186:72–81. doi: 10.1164/rccm.201109-1593OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oakes JL, O’Connor BP, Warg LA, Burton R, Hock A, Loader J, Laflamme D, Jing J, Hui L, Schwartz DA, et al. Ozone enhances pulmonary innate immune response to a Toll-like receptor-2 agonist. Am J Respir Cell Mol Biol. 2013;48:27–34. doi: 10.1165/rcmb.2012-0187OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu W, Zhu Y, Ning Y, Dong Y, Huang H, Zhang W, Sun Q, Li Q. Nogo-B protects mice against lipopolysaccharide-induced acute lung injury. Sci Rep. 2015;5:12061. doi: 10.1038/srep12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krebs J, Tsagogiorgas C, Pelosi P, Rocco PR, Hottenrott M, Sticht C, Yard B, Luecke T. Open lung approach with low tidal volume mechanical ventilation attenuates lung injury in rats with massive brain damage. Crit Care. 2014;18:R59. doi: 10.1186/cc13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spassov S, Pfeifer D, Strosing K, Ryter S, Hummel M, Faller S, Hoetzel A. Genetic targets of hydrogen sulfide in ventilator-induced lung injury—a microarray study. PLoS One. 2014;9:e102401. doi: 10.1371/journal.pone.0102401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhein KC, McCaw Z, Gladwell W, Trivedi S, Bushel PR, Kleeberger SR. Novel roles for Notch3 and Notch4 receptors in gene expression and susceptibility to ozone-induced lung inflammation in mice. Environ Health Perspect. 2015;123:799–805. doi: 10.1289/ehp.1408852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolinay T, Kaminski N, Felgendreher M, Kim HP, Reynolds P, Watkins SC, Karp D, Uhlig S, Choi AM. Gene expression profiling of target genes in ventilator-induced lung injury. Physiol Genomics. 2006;26:68–75. doi: 10.1152/physiolgenomics.00110.2005. [DOI] [PubMed] [Google Scholar]
- 38.Dolinay T, Wu W, Kaminski N, Ifedigbo E, Kaynar AM, Szilasi M, Watkins SC, Ryter SW, Hoetzel A, Choi AM. Mitogen-activated protein kinases regulate susceptibility to ventilator-induced lung injury. PLoS One. 2008;3:e1601. doi: 10.1371/journal.pone.0001601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang C, Xiao X, Chintagari NR, Breshears M, Wang Y, Liu L. MicroRNA and mRNA expression profiling in rat acute respiratory distress syndrome. BMC Med Genomics. 2014;7:46. doi: 10.1186/1755-8794-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang IV, Tomfohr J, Singh J, Foss CM, Marshall HE, Que LG, McElvania-Tekippe E, Florence S, Sundy JS, Schwartz DA. The clinical and environmental determinants of airway transcriptional profiles in allergic asthma. Am J Respir Crit Care Med. 2012;185:620–627. doi: 10.1164/rccm.201108-1503OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynier F, de Vos AF, Hoogerwerf JJ, Bresser P, van der Zee JS, Paye M, Pachot A, Mougin B, van der Poll T. Gene expression profiles in alveolar macrophages induced by lipopolysaccharide in humans. Mol Med. 2012;18:1303–1311. doi: 10.2119/molmed.2012.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leroy P, Tham A, Wong H, Tenney R, Chen C, Stiner R, Balmes JR, Paquet AC, Arjomandi M. Inflammatory and repair pathways induced in human bronchoalveolar lavage cells with ozone inhalation. PLoS One. 2015;10:e0127283. doi: 10.1371/journal.pone.0127283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andres-Terre M, McGuire H, Pouliot Y, Sweeney T, Tato C, Khatri P. Transcriptional signatures of viral infection across multiple respiratory viruses derived from integrated, multi-cohort analysis. Immunity. 2015;43:1199–1211. doi: 10.1016/j.immuni.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiland JE, Davis WB, Holter JF, Mohammed JR, Dorinsky PM, Gadek JE. Lung neutrophils in the adult respiratory distress syndrome: clinical and pathophysiologic significance. Am Rev Respir Dis. 1986;133:218–225. doi: 10.1164/arrd.1986.133.2.218. [DOI] [PubMed] [Google Scholar]
- 45.Matute-Bello G, Downey G, Moore BB, Groshong SD, Matthay MA, Slutsky AS, Kuebler WM Acute Lung Injury in Animals Study Group. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol. 2011;44:725–738. doi: 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Paola R, Crisafulli C, Mazzon E, Genovese T, Paterniti I, Bramanti P, Cuzzocrea S. Effect of PD98059, a selective MAPK3/MAPK1 inhibitor, on acute lung injury in mice. Int J Immunopathol Pharmacol. 2009;22:937–950. doi: 10.1177/039463200902200409. [DOI] [PubMed] [Google Scholar]
- 48.Di Paola R, Galuppo M, Mazzon E, Paterniti I, Bramanti P, Cuzzocrea S. PD98059, a specific MAP kinase inhibitor, attenuates multiple organ dysfunction syndrome/failure (MODS) induced by zymosan in mice. Pharmacol Res. 2010;61:175–187. doi: 10.1016/j.phrs.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Pamuk ON, Can G, Ayvaz S, Karaca T, Pamuk GE, Demirtas S, Tsokos GC. Spleen tyrosine kinase (Syk) inhibitor fostamatinib limits tissue damage and fibrosis in a bleomycin-induced scleroderma mouse model. Clin Exp Rheumatol. 2015;33(4) suppl 91:S15–S22. [PubMed] [Google Scholar]
- 50.Wei X, Han J, Chen ZZ, Qi BW, Wang GC, Ma YH, Zheng H, Luo YF, Wei YQ, Chen LJ. A phosphoinositide 3-kinase-gamma inhibitor, AS605240 prevents bleomycin-induced pulmonary fibrosis in rats. Biochem Biophys Res Commun. 2010;397:311–317. doi: 10.1016/j.bbrc.2010.05.109. [DOI] [PubMed] [Google Scholar]
- 51.Damarla M, Hasan E, Boueiz A, Le A, Pae HH, Montouchet C, Kolb T, Simms T, Myers A, Kayyali US, et al. Mitogen activated protein kinase activated protein kinase 2 regulates actin polymerization and vascular leak in ventilator associated lung injury. PLoS One. 2009;4:e4600. doi: 10.1371/journal.pone.0004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sio SW, Ang SF, Lu J, Moochhala S, Bhatia M. Substance P upregulates cyclooxygenase-2 and prostaglandin E metabolite by activating ERK1/2 and NF-κB in a mouse model of burn-induced remote acute lung injury. J Immunol. 2010;185:6265–6276. doi: 10.4049/jimmunol.1001739. [DOI] [PubMed] [Google Scholar]