Abstract
BLM and WRN, the products of the Bloom’s and Werner’s syndrome genes, are members of the RecQ family of DNA helicases. Although both have been shown previously to unwind simple, partial duplex DNA substrates with 3′→5′ polarity, little is known about the structural features of DNA that determine the substrate specificities of these enzymes. We have compared the substrate specificities of the BLM and WRN proteins using a variety of partial duplex DNA molecules, which are based upon a common core nucleotide sequence. We show that neither BLM nor WRN is capable of unwinding duplex DNA from a blunt-ended terminus or from an internal nick. However, both enzymes efficiently unwind the same blunt-ended duplex containing a centrally located 12 nt single-stranded ‘bubble’, as well as a synthetic X-structure (a model for the Holliday junction recombination intermediate) in which each ‘arm’ of the 4-way junction is blunt-ended. Surprisingly, a 3′-tailed duplex, a standard substrate for 3′→5′ helicases, is unwound much less efficiently by BLM and WRN than are the bubble and X-structure substrates. These data show conclusively that a single-stranded 3′-tail is not a structural requirement for unwinding of standard B-form DNA by these helicases. BLM and WRN also both unwind a variety of different forms of G-quadruplex DNA, a structure that can form at guanine-rich sequences present at several genomic loci. Our data indicate that BLM and WRN are atypical helicases that are highly DNA structure specific and have similar substrate specificities. We interpret these data in the light of the genomic instability and hyper-recombination characteristics of cells from individuals with Bloom’s or Werner’s syndrome.
INTRODUCTION
DNA and RNA helicases utilise the energy derived from hydrolysis of ATP to perform several essential roles in genetic recombination, transcription, DNA repair and DNA replication (reviewed in 1). One closely related subgroup of DNA helicases, the RecQ family, includes representatives in bacteria, lower eukaryotes and mammals (reviewed in 2,3). RecQ family members share a highly conserved domain comprising approximately 450 amino acids that includes seven sequence motifs conserved in many different classes of DNA and RNA helicases. Amongst these motifs are an ATP-binding sequence (Walker A box) and a DExH box, which is a characteristic of this family. Where studied, RecQ family members have been shown to be DNA helicases that translocate in the 3′→5′ direction (4–11). To date, five RecQ family members have been identified in human cells, three of which (BLM, WRN and RECQ4) are of special interest because of their involvement in inherited disorders associated with a predisposition to cancer and/or features of premature ageing (reviewed in 3). The BLM gene is mutated in the rare disorder Bloom’s syndrome (BS) (12), which is associated with a range of phenotypes, including immunodeficiency, sub-fertility, small body size, sun-induced facial erythema and a predisposition to cancer (reviewed in 13,14). This disorder is of particular interest because affected individuals are susceptible to the full range of cancers seen in the normal population. Mutations in the WRN gene give rise to Werner’s syndrome (WS) (15), which is associated at a relative early age with many, but not all, of the features of the normal ageing process (reviewed in 16). Hence, WS individuals show loss of skin elasticity, premature greying and thinning of the hair, susceptibility to the development of cataracts and loss of subcutaneous fat. Moreover, WS individuals are also cancer prone, although to a more limited extent than is seen in BS individuals, in particular displaying an elevated incidence of sarcomas. Very recently, it has been shown that the RECQ4 gene is mutated in Rothmund–Thomson syndrome (RTS), in which affected individuals show skin and skeletal abnormalities as well as some increase in cancer incidence (17).
One feature that links these three genetic disorders at the cellular level is a propensity to display inherent genomic instability (3). In the case of BS cells, this instability is manifested as an elevated frequency of homologous recombination events, including reciprocal exchanges between sister chromatids and homologous chromosomes (13,14). WS cells do not show elevated sister chromatid exchange frequencies, but they do display increased illegitimate recombination and a high frequency of large chromosomal deletions (reviewed in 16). The genomic instability of RTS cells has not been analysed in detail, but there are suggestions of defects in responses to UV-irradiation (18).
BLM and WRN have both been purified in recombinant form and shown to possess 3′→5′ helicase activity on simple model DNA substrates (6–9). In addition, WRN possesses a 3′→5′ exonuclease activity that is absent from preparations of BLM protein (19–21). At present, however, little is known about the mechanism by which BLM and WRN recognise their DNA substrates, or whether these related helicases show major differences in substrate specificity that might provide an insight into why the clinical and cellular features of BS and WS are different. Here we have carried out a comparison of the DNA substrate specificity for the helicase activities of BLM and WRN. Both enzymes catalyse little or no unwinding of duplex DNA from blunt ends, from internal nicks or of partial duplex molecules with single-stranded 3′- or 5′-tails, but are able to initiate unwinding from ‘bubbles’ inserted internally into an otherwise blunt-ended duplex. Moreover, both enzymes efficiently unwind G-quadruplex DNA and synthetic X-junctions that model the Holliday junction recombination intermediate. These somewhat atypical substrate preferences may be relevant to the genomic instability phenotype of cells lacking one or other of these helicases.
MATERIALS AND METHODS
Purification of proteins
Recombinant BLM and WRN proteins were purified as described by Karow et al. (22) and Orren et al. (23), respectively.
DNA substrates
The oligonucleotides used in this study are listed in Table 1. For each substrate a single oligonucleotide (see Table 2) was 5′-end-labelled with [γ-32P]ATP (Amersham Pharmacia Biotech) using T4 polynucleotide kinase (New England Biolabs). The labelled oligonucleotides were annealed to their unlabelled complementary strands at a 1:4 molar ratio by incubation at 100°C for 3 min followed by slow cooling to room temperature. For purification of the substrates, the annealed oligonucleotides were subjected to electrophoresis in 10% TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.0) acrylamide gels run at 200 V for 90 min at 4°C. The gels were then exposed to X-ray film and the gel slice containing the labelled substrate was excised and placed in dialysis tubing containing 0.5× TBE. The DNA sample was electroeluted in 0.5× TBE at 100 V for 2 h at 4°C. The current was reversed for 30 s before removing the labelled DNA. Samples were then dialysed against TEN (10 mM Tris–HCl, pH 8.0, 10 mM EDTA, 50 mM NaCl) for 2 h at 4°C. After dialysis, the concentration of the labelled substrate was determined using scintillation counting (Beckman LS 5000CE). G4 DNA substrates were generated as described by Sun et al. (24).
Table 1. Oligonucleotides used in this study.
The 5′-γ-32P-ATP end label is represented by and asterisk. The length of specific features of each substrate is indicated on the structure. All substrates are drawn with the 5′ end of the upper strand of the duplex on the left.
Table 2. Structures of substrates used in this study.
 |
DNA helicase assays
Standard helicase assays were performed essentially as described previously (6,23). The BLM and WRN proteins were incubated with different substrates at 37°C in helicase buffer (20 mM Tris–HCl, pH 7.5, 2 mM MgCl2, 2 mM ATP, 0.1 mg/ml BSA, 1 mM DTT). Reactions were stopped by addition of loading buffer containing 2.5% bromophenol blue and 20 mM EDTA. Samples were subjected to electrophoresis in 15% neutral gels in 1× TBE at 120 V for 2 h at 4°C. The gels were vacuum dried at 80°C for 2 h and radioactive DNA was visualised by autoradiography (Kodak BioMax MR-1). The level of unwinding of each substrate was quantified using a Storm 840 PhosphorImager (Molecular Dynamics). The rate of unwinding in each case was calculated from the gradient of the initial linear part of curves created by plotting concentration of substrate unwound against time for different concentrations of enzyme. The rate of unwinding (see Fig. 4A) was calculated from the formula
Figure 4.
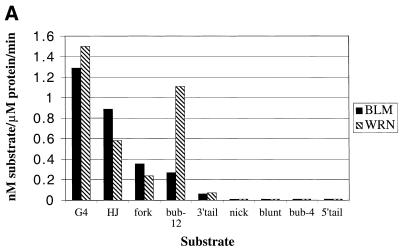
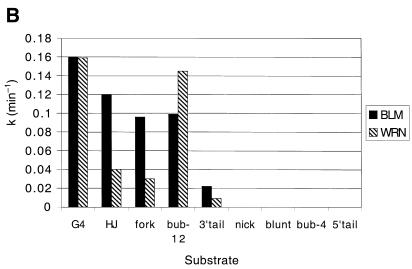
Comparative unwinding activity of the BLM and WRN helicases on different DNA substrates. (A) Rates of the unwinding reaction (nM substrate/µM protein/min) were derived as described in Materials and Methods. (B) k is the pseudo first order rate constant of DNA unwinding, which was calculated as described in Materials and Methods. Solid bars, BLM; hatched bars, WRN. The substrates are indicated along the horizontal axis: G4, OX-1T G-quadruplex; HJ, X-junction; fork, forked duplex; bub-12, duplex with 12 bp bubble; 3′-tail, 3′-tailed duplex; nick, nicked duplex; blunt, 50 bp duplex; bub-4, duplex with 4 bp bubble; 5′-tail, 5′-tailed duplex.
([ssDNA/total DNA) × nM substrate]/µM enzyme/time.
Since the helicase activities of BLM and WRN were determined under single turnover reaction conditions (excess protein over DNA), the data can be fitted to a 1-exponential equation describing pseudo first order reactions. For this analysis the fraction of DNA unwound was calculated from the formula
Fraction ssDNA = {[ssDNA/(ssDNA + dsDNA)] – [native ssDNA/(native ssDNA + native dsDNA)]} ÷ {[denatured ssDNA/(denatured ssDNA + denatured dsDNA)] – [native ssDNA/(native ssDNA + native dsDNA)]}.
The ssDNA and dsDNA represent the amount of radioactivity in the band corresponding to the indicated DNA species. The fraction of ssDNA was plotted versus reaction time and the data were fitted to a 1-exponential equation describing pseudo first order reactions using the formula
fraction ssDNA = A × (1 – e–kt) + n.
where A is the amplitude that corresponds to the maximum fraction of ssDNA that can be generated enzymatically from the substrates, k is the pseudo first order rate constant of DNA unwinding, t is the reaction time and n is an additive constant representing the amount of ssDNA present before the reaction started. The k value for each helicase assay was calculated and plotted.
RESULTS
To analyse and compare the DNA substrate specificities for the BLM and WRN helicases, a series of oligonucleotide-based substrates was synthesised. The sequences of the various oligonucleotides employed are given in Table 1. Table 2 depicts the structures of the duplex and partial duplex DNA substrates generated following annealing of combinations of these oligonucleotides. In addition, three G-quadruplex substrates were created from single, G-rich oligonucleotides, as described previously (24). To analyse the substrate specificites of the BLM and WRN proteins, we initially determined whether either enzyme could unwind a fully blunt-ended duplex DNA molecule. This was examined for two reasons. First, RecQ, the Escherichia coli homologue of BLM and WRN, has been reported to be capable of initiating DNA unwinding from blunt ends (25). Second, if either of these helicases was capable of unwinding a blunt-ended duplex, this would preclude analysis of certain DNA structures where initiation of DNA unwinding is dependent upon entry by the helicase at an ‘internal’ site within the DNA structure, such as at a single-stranded nick or bubble. Multiple protein concentrations were tested on all the substrates. Using 25 and 50 bp blunt-ended duplexes (Table 2), we demonstrated that neither BLM nor WRN produced measurable levels of unwinding of these substrates under conditions where a substantial level of unwinding of other substrates was detectable (see Fig. 1 and data not shown). Using this observation as a starting point, we next modified the blunt-ended (50 bp) substrate in a variety of ways; by incorporation of a single-stranded nick, a 12 nt single-stranded 3′- or 5′-tail or a centrally located bubble of 4, 8 or 12 nt in length (Table 2). In addition, a fully double-stranded, synthetic X-junction (or 4-way junction), which is considered to model the Holliday junction recombination intermediate, was studied in parallel because of our earlier observation that BLM and WRN can catalyse Holliday junction branch migration (26,27). The aim of this initial part of the study was to ascertain whether BLM and/or WRN could catalyse a detectable level of unwinding of each substrate, at least under the conditions used here. We found that while neither enzyme could unwind a nicked substrate, a 5′-tailed substrate or a duplex with a 4 bp bubble, both enzymes were capable of at least some unwinding of a 3′-tailed duplex, a synthetic X-junction, a forked-duplex (with 12 bp non-complementary single-stranded tails) and a duplex with an 8 or 12 bp bubble. Figure 1 shows representative gels of the unwinding of a selection of these substrates by WRN. Very similar data were obtained with BLM (data not shown).
Figure 1.
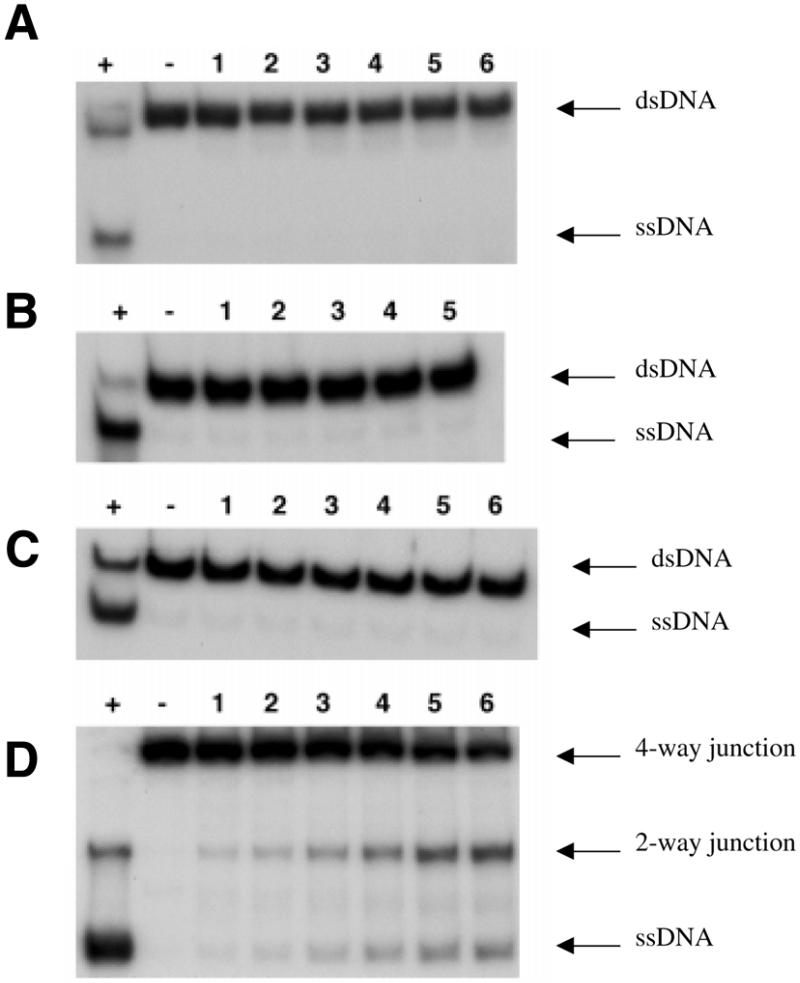
Unwinding of 32P-labelled substrates by WRN. Reactions contained 20 mM Tris–HCl, pH 7.5, 2 mM MgCl2 2 mM ATP, 0.1 mg/ml BSA, 1 mM DTT, 32P-labelled substrate and WRN. Substrates were: (A) 0.5 nM nicked duplex incubated with 10 nM WRN; (B) 1 nM 50 bp blunt-ended duplex incubated with 7.3 nM WRN; (C) 1 nM 4 bp bubble incubated with 1 nM WRN; (D) 1 nM synthetic X-junction incubated with 1.4 nM WRN. Lane +, boiled substrate; lane –, time 0; lane 1, 1 min; lane 2, 2 min; lane 3, 5 min; lane 4, 10 min; lane 5, 20 min; lane 6, 30 min incubation at 37°C. The positions of the starting substrate (dsDNA) and single-stranded DNA products (ssDNA) are indicated on the right. For the X-junction, the positions of the 4-way junction substrate and 2-way junction (splayed arm) and ssDNA products are indicated.
Having identified DNA structures that were relatively good substrates for unwinding by BLM or WRN, we then conducted a more detailed and comparative analysis of unwinding of those structures which were substrates for BLM and WRN. In order to do this, four substrates were generated based upon a common core sequence represented by oligonucleotide 1 (Table 1). Using these substrates, we aimed to eliminate, as far as possible, any variation in unwinding efficiency due to sequence differences between substrates. The substrates analysed in this part of the study were the synthetic X-junction, the 12 bp bubble-2, the fork-2 and the 3′-tailed duplex-2 (Table 2). The helicase assays using these substrates were carried out simultaneously with at least two independent preparations for each enzyme and under identical reaction conditions in order to minimise inter-experimental variation. Figure 2 shows representative gels for unwinding by BLM or WRN of the synthetic X-junction (Fig. 2A), the forked substrate (Fig. 2B), the 3′-tailed duplex (Fig. 2C) and the 12 bp bubble substrate (Fig. 2D). The level of unwinding for each substrate was quantified using a PhosphorImager (see Fig. 4). This direct comparison of the substrate specificities of BLM and WRN was repeated at least five times for each substrate and similar results were found in each case.
Figure 2.
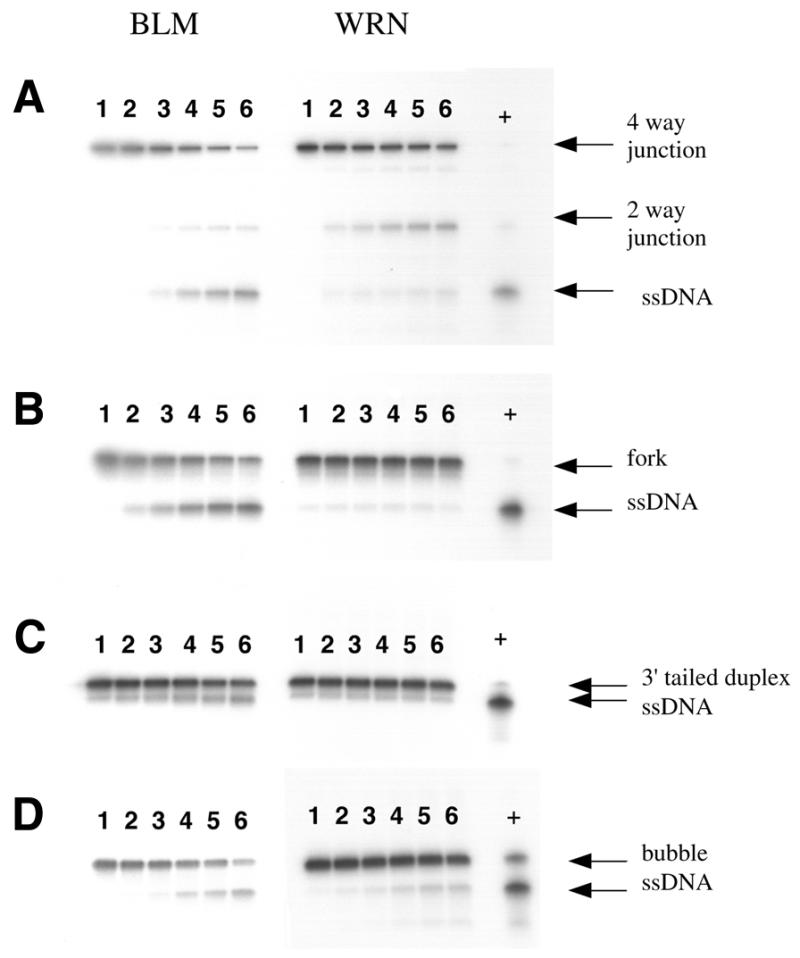
Time course of unwinding of selected DNA substrates by BLM and WRN. Reactions were performed essentially as described in the legend to Figure 1, using 1 nM substrate in each case and 20 nM BLM (set of reactions on the left, as indicated above) or 10 nM WRN (reactions on the right). WRN reactions on the bubble substrate contained 5 nM enzyme to minimise loss of substrate, which was particularly susceptible to exonucleolytic degradation. (A–D) Results for the synthetic 4-way junction, forked duplex, 3′-tailed duplex and 12 bp bubble substrate, respectively. Lanes 1–6 depict a time course of 0, 2, 4, 8, 12 and 18 min incubation at 37°C. The positions of the substrates and reaction products are indicated on the right.
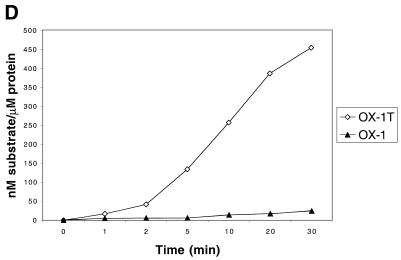
It has been reported previously that the WRN protein unwinds G-quadruplex DNA, but that it has a very limited substrate range, being specific for a structure derived from the fragile X chromosome region (28). Given that BLM has the ability to disrupt several different G-quadruplex sequences (24), we further examined this apparent difference in the substrate specificities of the BLM and WRN enzymes. We tested the specificity of WRN for unwinding three different synthetic G4 substrates (Table 2). The G4-TP substrate represents a consensus repeat from the murine immunoglobulin Sγ2b switch region, while G4-OX-1T and G4-OX-1 contain the Oxytricha telomeric repeat sequence. Both G4-TP and G4-OX-1T contain a single-stranded 3′-DNA tail of 7 nt in length, whereas G4-OX-1 lacks this 3′-tail. In our DNA unwinding assays WRN protein was able to efficiently unwind the 3′-tailed G4-TP and G4-OX-1T substrates to generate single-stranded DNA products (Fig. 3). However, the G4-OX-1 substrate was not unwound to any appreciable extent by WRN (Fig. 3).
Figure 3.
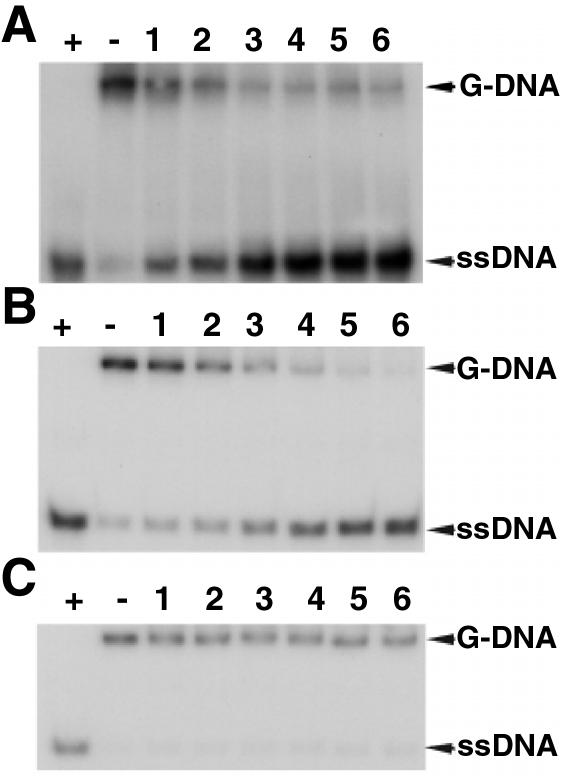
WRN can unwind a variety of G4 DNA substrates. (A) Time course of unwinding of 1 nM G4-TP substrate by 5 nM WRN. (B) Time course of unwinding of 1 nM G4-OX-IT substrate by 1 nM WRN. (C) Time course of unwinding of 1 nM G4-OX-1 substrate by 1 nM WRN. Lane +, boiled substrate; lane –, no enzyme; lanes 1–6, times of incubation at 37°C of 1, 2, 5, 8, 16 and 24 min, respectively. The positions of the G-DNA substrate and the ssDNA products are indicated on the right. (D) Quantification of the data from (B) and (C), comparing the rates of unwinding of the G4-OX-1T substrate containing a 7 nt 3′-tail and the equivalent G4-OX-1 substrate lacking the tail.
The combined data for unwinding by BLM and WRN of all of the substrates analysed are shown in Figure 4. It should be noted that these data were based on the lowest amount of enzyme required for unwinding after multiple BLM and WRN protein concentrations were tested on each substrate. The data were analysed in two ways. Firstly, the rate of the unwinding was calculated as nM substrate unwound/µM enzyme/min (Fig. 4A). Secondly, since the helicase activities of BLM and WRN were determined under single turnover reaction conditions (excess protein over DNA), the data were fitted to a 1-exponential equation describing pseudo first order rate constants (k) for DNA unwinding (Fig. 4B). These two methods of data analysis generated qualitatively similar results, with the ranking order for the efficiency of unwinding of each substrate being the same in each case. BLM had a preference for G4 DNA and the synthetic X-junction, but could also unwind the forked duplex and the duplex with a 12 bp bubble efficiently. However, BLM showed only very limited unwinding of the 3′-tailed duplex (at a rate up to 10-fold lower than for the aforementioned substrates). WRN unwound the G4 and 12 bp bubble substrates more efficiently that any of the other substrates, but like BLM was also active on the synthetic X-junction and the forked duplex. Again, it was notable that the 3′-tailed duplex was very poorly unwound by WRN. Moreover, neither BLM nor WRN showed appreciable unwinding of the nicked duplex, the blunt-ended duplex, the duplex with a 4 bp bubble or the 5′-tailed duplex (Fig. 4A and B).
DISCUSSION
We have shown that the BLM and WRN proteins are DNA structure-specific helicases with similar substrate specificities. This specificity is not only unusual amongst previously characterised helicases, but also significantly different from that of other RecQ family helicases characterised from lower organisms (reviewed in 1–3).
Our previous analyses have shown that BLM cannot form a stable complex with a blunt-ended duplex (26). Consistent with this, we have shown here that the level of BLM and WRN unwinding of blunt-ended, fully double-stranded DNA substrates is below the level of detection under the assay conditions employed. In contrast, E.coli RecQ protein, like several other helicases such as RecBCD, can initiate duplex DNA unwinding from a blunt-ended terminus (25; reviewed in 1,29). The addition of a short single-stranded tail (a 3′-tail for 3′→5′ helicases or a 5′-tail for a 5′→3′ helicases) to a duplex molecule is sufficient to create a good substrate for many helicases, including those incapable of unwinding from blunt ends. Indeed, this has been shown to be the case with Sgs1p, the Saccharomyces cerevisiae homologue of BLM and WRN. In their studies Bennett et al. (30) concluded that Sgs1p had a requirement for a 3′-tail of at least 3 nt and used the junction of single-stranded and double-stranded DNA as a key element in the substrate recognition process. In our studies, as expected, a 5′-tailed duplex was not a substrate for BLM or WRN. Much more surprising was the finding that 3′-tailed DNA was a very poor substrate for BLM and WRN compared to many of the other substrates tested, including those with fully blunt-ended termini such as the bubble substrate. These data indicate that BLM and WRN utilise different features of DNA structure to effect substrate recognition than do RecQ, Sgs1p and many other 3′→5′ helicases. Our data suggest that the following features of DNA substrate structure are not required for the action of BLM or WRN: (i) BLM and WRN are unlikely to use the single-stranded/double-stranded junction as a primary substrate recognition feature, unlike Sgs1p (30); (ii) BLM and WRN are apparently not loading onto a single-stranded DNA tail or terminus and then tracking along the tail until the duplex portion of the substrate is encountered; (iii) BLM and WRN appear not to have a requirement for a DNA terminus at all, since the bubble substrate with blunt-ended termini was an efficient substrate. Although we cannot eliminate the possibility that, as part of the substrate recognition/unwinding process, BLM and/or WRN require the bubble structure to be in close proximity to a DNA end, we consider this unlikely because DNA molecules with a 12 bp bubble located at a greater distance from the duplex ends also serve as substrates for BLM and WRN (unpublished data). Moreover, it is clear that the bubble itself is a critical feature of the recognition process given that a duplex molecule of identical sequence (except the bubble region) is not bound or unwound by BLM. Taken together, these data show conclusively that the BLM and WRN helicases have no absolute requirement for a single-stranded 3′-tail to effect duplex DNA unwinding. While this holds true for certain substrates, notably the bubble and X-junction DNAs, it is not a universal feature of unwinding catalysed by BLM and WRN, since the presence of a 3′-tail dramatically enhanced the efficiency with which the telomeric G-quadruplex DNA was disrupted.
Through utilisation of the same core sequence for the oligonucleotide-based substrates we were able, as far as is possible, to eliminate variations caused by the influence of nucleotide sequence. Although we are cautious about interpreting the combined data that establish a ‘ranking order’ of substrates for BLM and WRN (Fig. 4), there are some general conclusions that can be drawn. First, several DNA structures were apparently not substrates for unwinding by BLM or WRN. This does not rule out the possibility that under different conditions or at far higher protein concentrations, BLM or WRN might show detectable unwinding of some of these substrates. Second, as a substrate for unwinding by either BLM or WRN, G-quadruplex DNA is as good, if not better, than the best of the conventional B-form DNA substrates analysed (see also 24). Third, forked DNA molecules, be they classical fork structures or bubble structures, that comprise at least one junction between duplex DNA and two single strands are better substrates for BLM and WRN than is 3′-tailed DNA, which comprises duplex DNA and only one single strand. Fourth, BLM and WRN are efficient at unwinding X-junction and 12 nt bubble substrates with otherwise blunt-ended duplex termini, suggesting a key requirement for utilising ‘internal’ structural features of DNA to initiate unwinding. Fifth, both BLM and WRN require that the bubble structure be >4 nt in order to produce any measurable DNA unwinding. The 8 nt bubble structure was a substrate for BLM and WRN, but to only a limited extent (data not shown). It may be relevant to the functions of BLM and/or WRN that DNA repair reactions, such as those catalysed by the nucleotide excision repair (NER) machinery to remove bulky covalent adducts from DNA, take place within a ‘bubble’ created by helicases (XPB and XPD for NER). Whether the efficient recognition of bubble structures by BLM and WRN is important for some aspect of DNA repair in human cells will require further investigation.
A recent paper by Shen and Loeb (31) showed that bubble structures are a good substrate for the exonuclease activity of WRN. Consistent with this, we noted that the bubble substrate was readily destroyed by WRN exonuclease, necessitating a reduction in the concentration of WRN used in unwinding reactions using this substrate. This suggests the possibility that the helicase and exonuclease functions of WRN on bubble structures are linked, possibly as a result of the helicase action creating a substrate for the exonuclease activity. This would be consistent with the requirement for ATP for the exonuclease activity of WRN on this substrate (31).
Our previous analyses have shown that BLM is an oligomeric ring protein, probably comprising six subunits (22). If, as it would appear, BLM can initiate unwinding through accessing duplex DNA from an ‘internal’ single-stranded entry site, such as a bubble, this would seem to eliminate the possibility that rings of BLM protein thread onto single-stranded termini prior to translocation along the DNA molecule. The current study does not address how DNA binding is effected by BLM, but recent work on another hexameric ring helicase, T7 helicase-primase, has indicated a ring opening/closing mechanism for binding to single-stranded DNA, as opposed to a ‘threading’ model or de novo assembly of rings on the DNA (32). Further work will be required to determine whether BLM utilises a DNA binding mechanism similar to that of the T7 enzyme.
We have shown previously that BLM unwinds a variety of different G-quadruplex structures in DNA (24). In contrast, a previous study by Fry and Loeb (28) concluded that while WRN could unwind G-quadruplex DNA, this activity of WRN was restricted to the particular structure formed using the sequence derived from the fragile X genomic locus. This pointed to an exquisite sequence or structural specificity for WRN that was not a feature of the action of BLM, and possibly to an important functional distinction between these two related helicases. However, our data indicate that WRN is active on (at least) two additional G-quadruplex forming sequences and is, therefore, a general G-quadruplex disrupting enzyme, like BLM. We infer from the inability of WRN to unwind the G4-OX-1 substrate but ability to unwind G4-OX-1T that WRN requires a single-stranded 3′-tail to initiate unwinding of G-quadruplex DNA. The lack of a 3′-tail on the non-fragile X substrates used by Fry and Loeb (28) probably accounts for the lack of unwinding found in their study.
We have shown previously that BLM and WRN promote branch migration of a Holliday junction recombination intermediate (26,27). Perhaps not surprisingly, therefore, both enzymes could unwind the synthetic 4-way junction (X-junction) used here as a model for a Holliday junction. The product of branch migration on a 4-way junction is a 2-way junction (effectively a forked structure). We infer from the lack of evidence of 3-way junction products (comprising 3 of the 4 original strands) in Figure 2 that BLM and WRN disrupt the 4-way junction first by promoting branch migration and then by subsequently unwinding the resulting forked structure to produce single-stranded oligonucleotide products. Since under the conditions under which we performed the assays (presence of Mg2+) the synthetic 4-way junction forms a so-called stacked X-structure (33), which does not contain any single-stranded DNA at its core, it is difficult to envisage BLM/WRN recognising any single-stranded features of this structure. Indeed, we have shown previously that BLM binds to X-junctions with enhanced specificity compared to single-stranded DNA or partial duplex molecules (26). A question still unresolved from this and previous studies, therefore, is whether any single feature of DNA structure is utilised for substrate recognition by BLM and WRN. Given that such diverse structures as the bubble, the X-junction and G-quadruplex DNA are all unwound much more efficiently by BLM and WRN than is a simple partial duplex molecule, it would seem unlikely that this will prove to be the case.
In conclusion, we have defined a number of the structural features of DNA molecules that permit them to be efficient substrates for unwinding by the BLM and WRN helicases. Our data indicate that the substrate specificities of the BLM and WRN proteins are similar, but nevertheless atypical amongst helicases. It seems unlikely with what we know at present that differences in substrate specificity will be a fundamental distinguishing feature of the BLM and WRN helicases responsible for defining their specific roles in cellular DNA metabolism.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the Hickson and Bohr laboratories for useful discussions, Dr C. Norbury for critical reading of the manuscript and Mrs J. Pepper for preparation of the manuscript. This work was supported by the Imperial Cancer Research Fund (P.M., J.K.K. and I.D.H.). J.K.K. was a Boehringer Ingelheim Fonds Fellow.
References
- 1.Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- 2.Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- 3.Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- 4.Seki M., Miyazawa,H., Tada,S., Yanagisawa,J., Yamaoka,T., Hoshino,S., Ozawa,K., Eki,T., Nogami,M., Okumura,K. et al. (1994) Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res., 22, 4566–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puranam K.L. and Blackshear,P.J. (1994) Cloning and characterization of RECQL, a potential human homologue of the Escherichiacoli DNA helicase RecQ. J. Biol. Chem., 269, 29838–29845. [PubMed] [Google Scholar]
- 6.Karow J.K., Chakraverty,R.K. and Hickson,I.D. (1997) The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J. Biol. Chem., 272, 30611–30614. [DOI] [PubMed] [Google Scholar]
- 7.Gray M.D., Shen,J.C., Kamath-Loeb,A.S., Blank,A., Sopher,B.L., Martin,G.M., Oshima,J. and Loeb,L.A. (1997) The Werner syndrome protein is a DNA helicase. Nat. Genet., 17, 100–103. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki N., Shimamoto,A., Imamura,O., Kuromitsu,J., Kitao,S., Goto,M. and Furuichi,Y. (1997) DNA helicase activity in Werner’s syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res., 25, 2973–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J.C., Gray,M.D., Oshima,J. and Loeb,L.A. (1998) Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res., 26, 2879–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umezu K. and Nakayama,H. (1993) RecQ DNA helicase of Escherichia coli: characterisation of the helix-unwinding activity with emphasis on the effect of single-stranded DNA-binding protein. J. Mol. Biol., 230, 1145–1150. [DOI] [PubMed] [Google Scholar]
- 11.Bennett R.J., Sharp,J.A. and Wang,J.C. (1998) Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem., 273, 9644–9650. [DOI] [PubMed] [Google Scholar]
- 12.Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s Syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- 13.German J. (1993) Bloom Syndrome: a Mendelian prototype of somatic mutational disease. Medicine, 72, 393–406. [PubMed] [Google Scholar]
- 14.German J. (1995) Bloom’s syndrome. Dermatol. Clin., 13, 7–18. [PubMed] [Google Scholar]
- 15.Yu C., Oshima,J., Fu,Y., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Najura,J., Miki,T., Ouais,S. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 16.Shen J.C. and Loeb,L.A. (2000) The Werner syndrome gene: the molecular basis of RecQ helicase-deficiency diseases. Trends Genet., 16, 213–220. [DOI] [PubMed] [Google Scholar]
- 17.Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nat. Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- 18.Vasseur F., Delaporte,E., Zabot,M.T., Sturque,M.N., Barrut,D., Savary,J.B., Thomas,L. and Thomas,P. (1999) Excision repair defect in Rothmund Thomson Syndrome. Acta Dermatol. Venereol. (Stockh.), 79, 150–152. [DOI] [PubMed] [Google Scholar]
- 19.Huang S., Li,B., Gray,M.D., Oshima,J., Mian,I.S. and Campisi,J. (1998) The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nat. Genet., 20, 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen J.C., Gray,M.D., Oshima,J., Kamath-Loeb,A.S., Fry,M. and Loeb,L.A. (1998) Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J. Biol. Chem., 273, 34139–34144. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki N., Shiratori,M., Goto,M. and Furuichi,Y. (1999) Werner syndrome helicase contains a 5′→3′ exonuclease activity that digests DNA and RNA strands in DNA/DNA and RNA/DNA duplexes dependent on unwinding. Nucleic Acids Res., 27, 2361–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow J.K., Newman,R.H., Freemont,P.S. and Hickson,I.D. (1999) Oligomeric ring structure of the Bloom’s syndrome helicase. Curr. Biol., 9, 597–600. [DOI] [PubMed] [Google Scholar]
- 23.Orren D.K., Brosh,R., Nehlin,J.O., Machwe,A., Gray,M.D. and Bohr,V.A. (1999) Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res., 27, 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H., Karow,J.K., Hickson,I.D. and Maizels,N. (1998) The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem., 273, 27587–27592. [DOI] [PubMed] [Google Scholar]
- 25.Harmon F.G. and Kowalczykowski,S.C. (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev., 12, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karow J.K., Constantinou,A., Li,J.L., West,S.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Constantinou A., Tarsounas,M., Karow,J.K., Brosh,R.M., Bohr,V.A., Hickson,I.D. and West,S.C. (2000) Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep., 1, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fry M. and Loeb,L.A. (1999) Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- 29.West S.C. (1996) DNA helicases: new breeds of translocating motors and molecular pumps. Cell, 86, 177–180. [DOI] [PubMed] [Google Scholar]
- 30.Bennett R.J., Keck,J.L. and Wang,J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol., 289, 235–248. [DOI] [PubMed] [Google Scholar]
- 31.Shen J.C. and Loeb,L.A. (2000) Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res., 28, 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahnert P., Picha,K.M. and Patel,S.S. (2000) A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J., 19, 3418–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lilley D.M.J. (1999) Structures and interactions of helical junctions in nucleic acids. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, New York, NY, pp. 471–498.