Abstract
Phycobilisome (PBS) complexes are massive light-harvesting apparati in cyanobacteria that capture and funnel light energy to the photosystem. PBS complexes are dynamically degraded during nutrient deprivation, which causes severe chlorosis, and resynthesized during nutrient repletion. PBS degradation occurs rapidly after nutrient step-down, and is specifically triggered by non-bleaching protein A (NblA), a small proteolysis adaptor that facilitates interactions between a Clp chaperone and phycobiliproteins. Little is known about the mode of action of NblA during PBS degradation. In this study, we used chemical cross-linking coupled with LC-MS/MS to investigate the interactions between NblA and phycobiliproteins. An isotopically-coded BS3 cross-linker captured a protein interaction between NblA and β-phycocyanin (PC). LC-MS/MS analysis identified the amino acid residues participating in the binding reaction, and demonstrated that K52 in NblA is cross-linked to T2 in β-PC. These results were modeled onto the existing crystal structures of NblA and PC by protein-docking simulations. Our data indicate that the C-terminus of NblA fits in an open groove of β-PC, a region located inside the central hollow cavity of a PC rod. NblA may mediate PBS degradation by disrupting the structural integrity of the PC rod from within the rod. In addition, M1-K44 and M1-K52 cross-links between the N-terminus of NblA and the C-terminus of NblA are consistent with the NblA crystal structure, confirming that the purified NblA is structurally biologically relevant. These findings provide direct evidence that NblA physically interacts with β-PC.
Keywords: cyanobacteria, phycobilisome proteolysis, nitrogen starvation, cross-linking, mass spectrometry, protein docking
Introduction
Cyanobacteria are unicellular phototrophs that naturally thrive in diverse terrestrial and marine ecosystems, where they are exposed to both scarce and abundant levels of nutrients and light levels. The versatility in cyanobacteria’s ability to acclimate to environmental changes is due to their ability to modify their metabolism rapidly, to remodel their phycobilisome (PBS) complexes dynamically (Grossman et al. 1993), and to optimize the utilization of nutrients (Schwarz and Forchhammer 2005). The light-harvesting apparati in cyanobacteria are called PBS complexes (Glazer 1982), which are massive, multi-subunit complexes found on the stromal side of the thylakoid membrane (Bogorad 1975; MacColl 1998). The PBS supramolecular membrane-bound complexes in cyanobacteria initiate photosynthesis by absorbing and transferring light energy to the reaction centers in both photosystem I (PSI) and photosystem II (PSII) (Liu et al. 2013). In nutrient-rich media, cyanobacteria appear blue-green in color owing to blue bilin pigments covalently attached to the phycobiliproteins that make up PBS complexes and green chlorophyll (Chl) pigments in the photosystems.
The soluble phycobiliproteins assemble into 3–7 megaDalton complexes (Bryant et al. 1979; Adir et al. 2006) that may account for up to 50% of the total soluble proteins in the cell, thus serving as a large cellular nitrogen reserve (Bogorad 1975). With the help of colorless linker proteins, the pigment-protein phycobiliproteins associate to form PBS complexes (Elmorjani et al. 1986; MacColl 1998). The major components of these complexes are brilliantly colored red phycoerythrin (PE), blue phycocyanin (PC), and blue-green allophycocyanin (APC) (Grossman et al. 1993; Scheer and Zhao 2008). An enormous number of phycobiliproteins make up one PBS complex. Structurally, PBS complexes may contain three hexamers of six PC rods radiating out from a bi- or tri-cylindrical APC core in a fan-shaped manner (Bogorad 1975; Williams et al. 1978; Bryant et al. 1979; Grossman et al. 1993; Ajlani and Vernotte 1998; Glazer 1982; Sidler 2004). A PC hexamer contains two PC trimers. These trimers contain three PC monomers, each of which are comprised of an α- and β-PC heterodimer (Ajlani and Vernotte 1998). There are at least 72 units of α- and β-PC in a PBS that has at least two PC hexamers in each of the six peripheral PC rods.
The plasticity of PBS restructuring is best seen during nitrogen, sulfur, or phosphorous depletion (Richaud et al. 2001), where non-diazotrophic cyanobacteria undergo bleaching (Allen and Smith 1969) owing to PC and APC transcript-synthesis repression and protein degradation (Collier and Grossman 1992). If PBS complexes do not degrade during nutrient deprivation, photosynthetic rates will remain unchanged as cellular metabolism slows down, causing excessive energy to be absorbed and harmful radical species to be produced (Adir et al. 2003). Upon exogenous addition of nutrients, cyanobacteria regain their blue-green color because phycobiliproteins are resynthesized (Schwarz and Forchhammer 2005). The processes of PBS degradation and re-synthesis play a key role for cell survival in terms of cell maintenance, growth, and development (Schwarz and Forchhammer 2005) and as a form of photoprotection (Adir et al. 2006).
The remodeling of PBS is an active, reversible, rapid, and specific process that occurs on a massive scale. A popular model of PBS degradation begins with the sequential trimming of the peripheral PC rods, starting at the most distal end, with complete degradation of the remaining PBS occurring two days after continued nutrient depletion in Synechococcus sp. PCC 7942 (hereafter referred to as Synechococcus 7942) (Collier and Grossman 1994). The degradation of these enormous light-harvesting complexes in cyanobacteria is triggered by a non-bleaching protein A (NblA) that is 7 kDa and is comprised of 59 amino acids (Collier and Grossman 1994). The Synechococcus 7942 nblA deletion strain maintains high levels of PBS after nitrogen, sulfur, or phosphorous step down (Collier and Grossman 1994). In most cyanobacteria, levels of nblA transcripts and NblA proteins are up-regulated during nitrogen, sulfur, and phosphorous stress (Collier and Grossman 1994; Baier et al. 2001), whereas PBS levels are simultaneously down-regulated (Baier et al. 2001; Li and Sherman 2002; Adir et al. 2006). For example, global transcriptome experiments in Synechococcus sp. PCC 7002 illustrate that the nblA gene is up-regulated up to 60-fold, while genes for PC are down-regulated by ~4 to 20-fold, depending on the specific nutrient starvation condition tested (i.e. carbon, nitrogen, or sulfur) (Ludwig and Bryant 2012).
NblA is an adaptor protein that triggers the degradation of PBS in cyanobacteria (Collier and Grossman 1994; Karradt et al. 2008). A single copy of nblA is present in most PBS-containing cyanobacteria and red algae, although there is low similarity between species (i.e., 30% sequence identity) (Bienert et al. 2005; Dines et al. 2008; Baier et al. 2014). Some strains have two copies of nblA; for example, Anabaena sp. PCC 7120, which only needs one nblA gene for PBS degradation (Karradt et al. 2008), and Synechocystis sp. PCC 6803, which needs both genes for PBS degradation (Baier et al. 2001). Crystal structures of NblA from Anabaena sp. PCC 7120 (Karradt et al. 2008), Thermosynechococcus vulcanus (Dines et al. 2008), and Synechococcus 7942 (Dines et al. 2008) demonstrate that the NblA monomer has a helix-loop-helix motif that undergoes homo- or hetero-dimerization to form an open four-helix bundle, which is the basic functional unit of NblA proteins (Bienert et al. 2005; Dines et al. 2008; Baier et al. 2014).
A NblA dimer triggers the degradation of PBS complexes by binding to ClpC and phycobiliproteins on its N-terminus (Karradt et al. 2008) and C-terminus (Bienert et al. 2005), respectively. ClpC functions in protein recognition, unfolding, and chaperoning to the membrane-bound ClpP catalytic core for degradation (Porankiewicz et al. 1999; Olinares et al. 2011). The Clp proteasome system serves as an important cellular disposal system. Clp proteins are universally found in eubacteria, including cyanobacteria, and eukaryotes (Andersson et al. 2006; Stanne et al. 2007). Cross-linking studies reveal that ClpC binds to phycobiliproteins, suggesting that the degradation of PBS is regulated by the Clp proteasome system. ClpC is an ATP-dependent, HSP100 chaperone partner that is essential and constitutively expressed in cyanobacteria and plants (Andersson et al. 2006). In Anabaena sp. PCC 7120, NblA binding activates ClpC by initiating its oligomerization into a hetero-hexamer comprised of three NblA dimers (Karradt et al. 2008).
The sequential mechanism of how NblA triggers and relies on the Clp proteasome system to disassemble PBS complexes is poorly understood. Random mutagenesis studies of NblA demonstrate that variable residues in the middle part of NblA play a crucial role in mediating PBS degradation in vivo (Dines et al. 2008). Combined with the α-helical structural similarities between NblA and phycobiliproteins, structural mimicry may be the method for NblA binding to the PBS complexes (Dines et al. 2008). Site-directed mutagenesis and pull-down assays, however, highlight several conserved amino acid residues in NblA that specifically bind to ClpC and α-PC (Bienert et al. 2005; Stanne et al. 2007; Karradt et al. 2008; Dines et al. 2008). In addition, immunoblotting shows that NblA also binds to APC subunits (Bienert et al. 2005), whereas several reports indicate that NblA does not bind to β-PC (Bienert et al. 2005; Luque et al. 2003). Interestingly, recent results show that immobilized Synechocystis sp. PCC 6803 NblA1/NblA2 can pull down β-PC from Synechocystis sp. PCC 6803 crude cell lysate (Baier et al. 2014).
In this report, we describe a combined cross-linking and mass spectrometry (MS) (Weisz et al. 2016) approach to investigate the binding patterns of NblA in the cyanobacterium Synechococcus elongatus UTEX 2973 (hereafter referred to as Synechococcus 2973) that doubles in abundance in ~ 2 hours and has only one copy of nblA that is sufficient to mediate PBS degradation (Yu et al. 2015). In vitro binding assays combined with chemical cross-linking and MS prove that the C-terminal K52 in NblA binds to the N-terminal T2 in β-PC. These experiments are coupled with protein-protein docking models (Smith et al. 2002; Gray 2006) to afford insights on how NblA interacts with the PC structure.
Results
Overexpression and purification of fusion protein
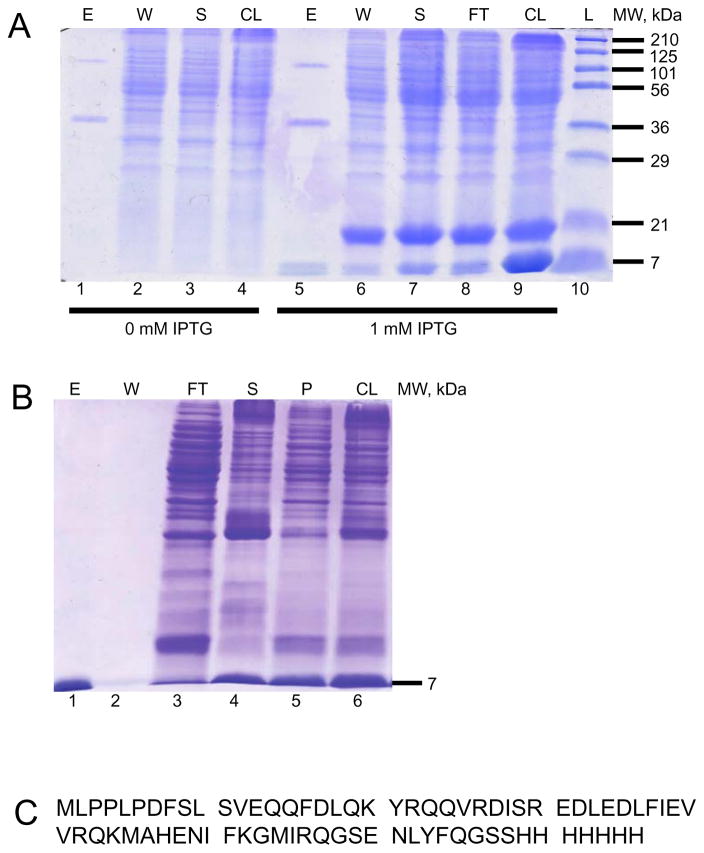
To study the binding partners of NblA, we expressed and purified NblA2973_His7 fusion protein in E. coli cells (Fig. 1a). SDS-PAGE gels confirmed that NblA2973_His7 fusion proteins were purified at the 7 kDa range (Fig. 1b). Bottom-up LC-MS/MS analysis identified 100% of the NblA2973_His7 sequence (Fig. 1c).
Fig. 1. Induction, purification, and LC-MS/MS analysis of NblA2973_His7.
a The soluble NblA2973_His7 7-kDa fusion protein was induced with 1 mM IPTG (lanes 5–9) for 96 hours at 16°C, as shown by SDS-PAGE and Coomassie Blue R250 staining. Note the two larger proteins in the elution fractions are contaminants from E. coli. b 2 M urea solubilized the NblA2973_His7 that was trapped in the pellet fraction after sonication (lane 5). NblA2973_His7 was washed and refolded extensively on the column before elution. The molecular mass and purity of NblA2973_His7 (lane 1) was verified by SDS-PAGE and Coomassie Blue R250 staining. The NblA monomer at 7 kDa and NblA dimer at 14 kDa were observed. c In-gel digestion of purified NblA2973_His7 was carried out for bottom-up LC-MS/MS. 100% coverage of the protein was detected. Abbreviations: L – Ladder, CL – Cell Lysate, FT – Flow Through, S – Supernatant, W – Wash, E – Elution, and P – Pellet.
In vitro binding assays and cross-linking
Previously, affinity columns demonstrated that purified NblA from Anabaena sp. PCC 7120 and Synechocystis sp. PCC 6803 bind to PC and APC proteins from crude cell extracts (Baier et al. 2014; Bienert et al. 2005). Our in vitro binding assays demonstrated that purified NblA2973_His7 pulled down proteins from isolated Synechococcus 2973 phycobilisomes. To capture any transient protein-protein interactions and determine the specific amino acid residues involved in the binding interactions between NblA and phycobiliproteins, we reacted the elution-fraction proteins from the in vitro assays with the isotopically labeled chemical cross-linker bis(sulfosuccinimidyl)suberate (BS3-H12/D12) (Creative Molecules, Inc., http://www.creativemolecules.com/). BS3 has a reactive group on both ends that can form covalent bonds with the primary amines in lysine residues and with the N-termini of proteins (Leitner et al. 2014). The cross-linker also reacts with the hydroxyl groups in serine, threonine, and tyrosine residues with lower yield (Sinz 2006; Rappsilber 2011). A 1:1 mixture of BS3 labeled with 12 hydrogens and 12 deuteriums facilitates the identification of true cross-links. A cross-linked peptide will be labeled 50% of the time with both light (H12) and heavy (D12) forms of BS3. Consequently, a doublet separated by 12 Daltons will be observed in the mass spectra, indicating a real cross-link. Careful analysis of the product ion (MS2) spectra permits correct identification and sequencing of the peptides that are participating in the cross-linking reaction. ZnSO4 enhanced bilin fluorescence of SDS-PAGE gels demonstrate that the binding of NblA to phycobiliproteins was captured by 1 mM BS3 cross-linker in vitro. Extensive optimizations of the ratios between proteins and cross-linker were necessary to detect cross-links. A 7:1 molar ratio of NblA:PBS in the presence of 1 mM BS3 is the best condition for capturing the binding between NblA and phycobiliproteins (Fig. 2).
Fig. 2. In vitro binding and cross-linking of NblA and phycobiliproteins.
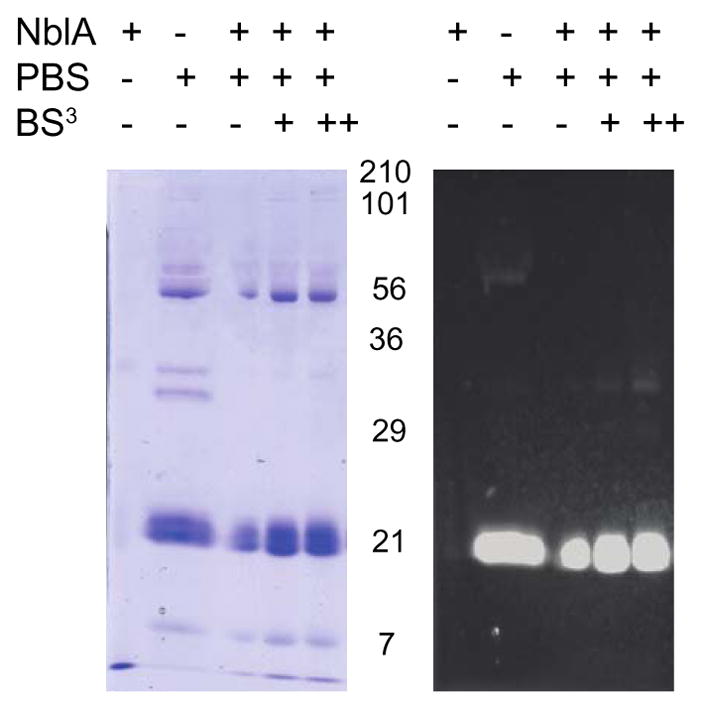
NblA2973_His7 bound NTA-Ni resin was incubated with isolated PBS from Synechococcus 2973 in the following concentrations of BS3: 0 mM (−), 0.5 mM (+), and 1.0 mM (++). Higher concentration of BS3 yielded higher molecular weight bands, suggesting that cross-links between NblA2973_His7 and PBS2973 had formed.
Mass spectrometry
It is known that the N- and C- termini of NblA bind to the ClpC chaperone and phycobiliproteins, respectively (Baier et al. 2004; Bienert et al. 2005; Karradt et al. 2008; Baier et al. 2014). Previous studies suggested that NblA preferentially binds to only α-PC (Luque et al. 2003; Bienert et al. 2005). We used a combined pull-down assay, cross-linking, and MS work-flow to provide direct evidence that NblA binds to phycobiliproteins, specifically to β-PC.
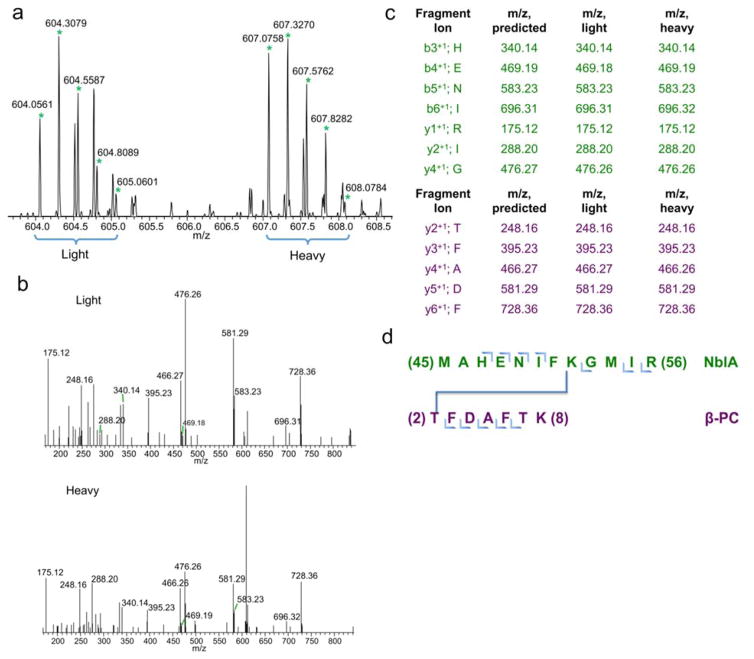
We analyzed the mass spectral doublet and the product ion (MS2) fragmentation spectra to demonstrate that K52 from NblA is cross-linked to T2 from β-PC (Fig. 3a and Fig. 3b). Manual verification of the MS2 spectra allowed confident identification of this cross-link (Fig. 3c). This cross-link suggests that the C-terminus of NblA is in close proximity to the N-terminus of β-PC (i.e., MAHENIFK52GMIR (NblA) to T2FDAFTK (β-PC)) (Fig. 3d). The “MIR” region in the C-terminus end of NblA plays a critical role in mediating PBS degradation. Sequence alignment of NblA demonstrates that the “MIR” region is conserved (Bienert et al. 2005). R56C mutagenesis in NblA inhibits PBS degradation during nutrient step-down (Dines et al. 2008). The M54 and R56 residues from NblA2973 are analogous to the L51 and K53 residues in NblA7120, and mutagenesis of L51 and K53 prohibits binding of α-PC to NblA7120 (Bienert et al. 2005; Karradt et al. 2008). Intriguingly, the T2 in the β-PC residue that is cross-linked to K52 from NblA is located inside the PC trimer, suggesting that NblA may gain access to the interior region of the PC rod via a hollow cavity inside the PC rod (Marx and Adir 2013), a region that has low but adequate solvent accessibility.
Fig. 3. C-terminus of NblA2973_His binding to the N-terminus of β-PC.
a 12 Dalton mass shift doublet in MS1 spectrum suggests a real cross-link has been detected. b MS2 fragmentations of the isotopic peaks from MS1. c In-depth analysis of all the fragments from the MS2 confirms the identity of the two proteins participating in the binding reaction. d Illustration of the T2 β-PC and M52 NblA residues are participating in the cross-linking reaction.
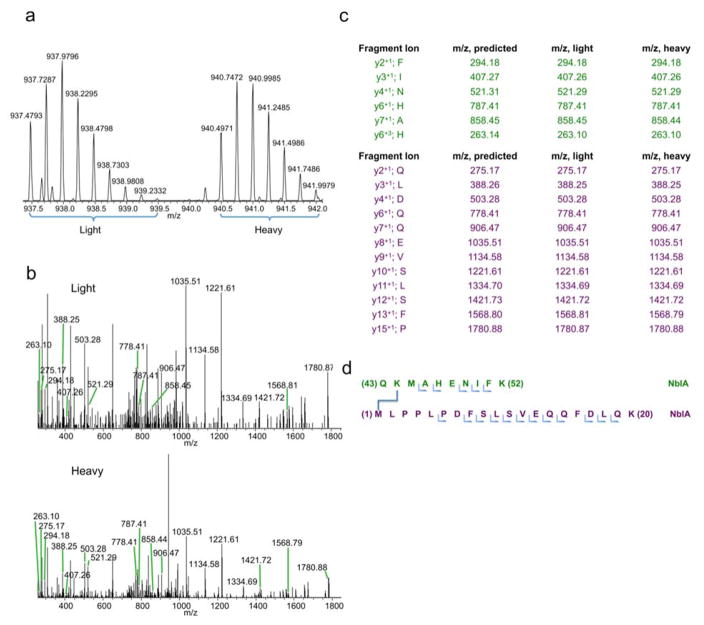
We also identified two NblA-NblA cross-links. The MS1 (Fig. 4a) and MS2 (Fig. 4b) spectra indicate that QK44MAHENIFK is cross-linked to M1LPPLPDFSLSVEQQFDLQK (Fig. 4c and 4d). A cross-link between M1LPPLPDFSLSVEQQFDLQK and MAHENIFK52GMIR also formed (not shown). These cross-links are consistent with the crystal structure of NblA, where the M1 – K44 and M1 – K52 residues are within an acceptable distance for cross-links to form. Note that these data only highlight how the N-terminus of NblA is in close proximity to the C-terminus of NblA, but do not provide insights regarding whether the cross-links formed within a NblA monomer or dimer.
Fig. 4. C-terminus of NblA2973_His binding to the N-terminus of NblA.
a 12 Dalton mass shift doublet in MS1 chromatogram suggests a real cross-link has been detected. b MS2 fragmentations of the isotopic peaks from MS1. c In-depth analysis of all the fragments from the MS2 confirms the identity of the two proteins participating in the binding reaction. d Illustration of the M1 NblA and K44 NblA residues are participating in the cross-linking reaction.
Modeling
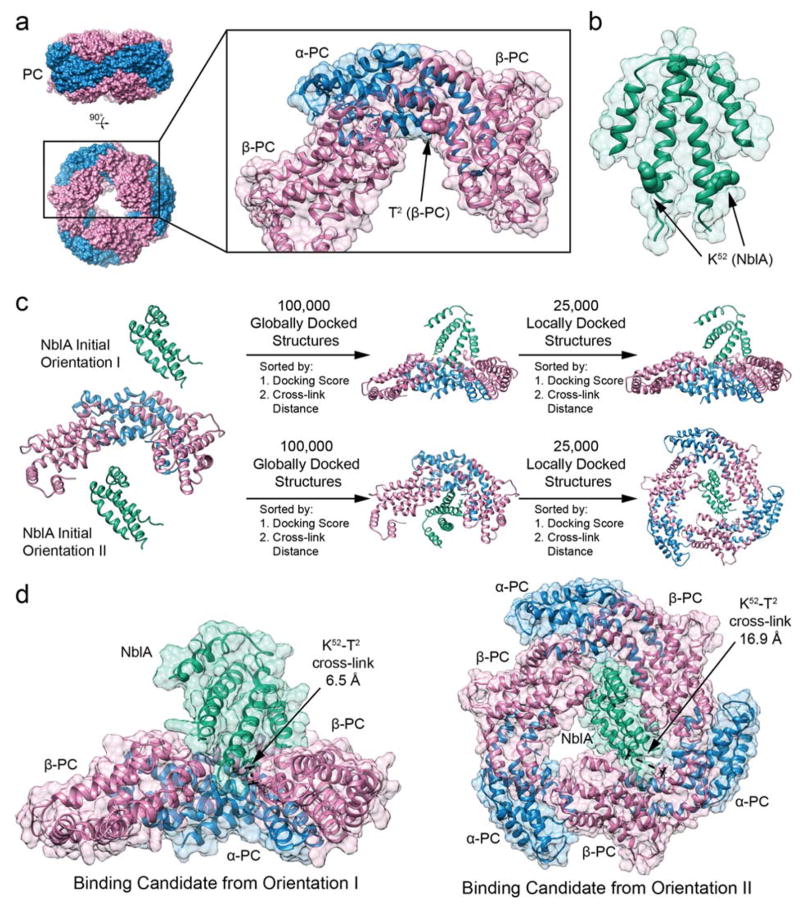
Structural analysis of the cross-links revealed that the N-terminus of β-PC is located in the interior region of the PC trimer, which is potentially accessible from above or inside the rod structure (Fig. 5a). For the C-terminus of NblA to bind to the N-terminus of β-PC, NblA may need to enter the central hollow cavity of the PC rod (Fig. 5a) or interact with a disassembled PBS complex. We speculate that the capping linker protein, CpcD, needs to dissociate from the distal end of the PC rod first for NblA to gain access to the hollow cavity inside the PC rod. It is likely that nutrient starvation triggers CpcD to dissociate from the PC rod before NblA binds to β-PC. Separation of CpcD from PC rods may speed up the degradation of the peripheral PC rods by making PC more accessible (de Lorimier et al. 1990, BBA).
Fig. 5. Molecular Docking of NblA dimer to PC.
a X-ray structure of Phycocyanin (PC) from Synechococcus 7942 (PDB ID: 4H0M), showing top and side view of hexameric structure. Monomeric sub-unit highlighted including one α-PC (blue) and one β-PC (pink), with an additional β-PC (pink) to define the NblA binding pocket. The N-terminus of β-PC is located in the interior hollow cavity of the PC rod. b X-ray structure of NblA dimer from Synechococcus 7942 (PDB ID: 3CS5), where residue K52 is highlighted due to cross-link with β-PC. c Protein-protein docking protocol using RosettaDock. Two initial orientations are chosen, simulating an NblA dimer (green) docking to PC from above (orientation I) and from inside the rod (orientation II). First, 100,000 globally docked candidates structures are generated, sorted by docking score and subsequently sorted by closest cross-link between T2 (β-PC) and K52 (NblA). Global docking details are presented in Fig. S2. Second, the best globally docked candidate is the initial structure for generating 25,000 locally docked candidate structures, sorted again by docking score and subsequently sorted by closest cross-link between T2 (β-PC) and K52 (NblA). For orientation II, the trimeric PC structure (three sub-units each of α-PC and β-PC) is used to accurately define the NblA binding pocket inside the PC rod. Local docking details are presented in Fig. S3. d Binding candidates for NblA dimers docked to PC from the two initial orientations. Residues T2 from β-PC and K52 from NblA are highlighted due to cross-link.
Interestingly, the interior region of the PC rod contains a groove near the T2 residue in β-PC that cross-links to NblA. This spatial vacancy is of similar shape to that of the α-helices near the C-terminus of an NblA dimer. Docking calculations (Fig. 5b) reveal this geometrical arrangement to be conducive to NblA – PC binding and to bringing the K52 residue on NblA in close proximity to T2 on β-PC (Fig. 5c), suggesting an initial mode of NblA – PC binding (detailed information on the modeling approach is contained in the Experimental Procedures section).
Discussion
Combining isotope-encoded chemical cross-linking and MS affords a powerful structural proteomics tool to investigate protein-protein interactions and establish low-resolution structures and their conformational changes (Sinz 2014; Bricker et al. 2015). But despite the power of this method, there are many challenges to analyzing cross-links correctly. For example, the tryptic digest of cross-linked proteins is an extremely complex mixture for LC-MS/MS analysis. In addition, cross-links tend to be in low-abundance compared to the un-linked peptides. Lastly, unambiguous identification can be difficult, especially with a large candidate protein database. To obviate these problems, we used a mixture of isotopically labeled BS3 and Protein Prospector, taking advantage of the characteristic doublet in the mass chromatograms to achieve high throughput.
In this study, we were particularly interested in discovering the interactions between NblA and phycobiliproteins to understand how NblA mediates PBS degradation. We induced a C-terminal His-tagged NblA protein from Synechococcus 2973 with IPTG (Fig. 1a) and purified NblA2973_His7 from E. coli with urea to increase the solubility and yield (Fig. 1b). Optimizations of the molar ratios between NblA2973_His7, isolated PBS complexes from Synechococcus 2973, and BS3 cross-linker demonstrated that a key to cross-linking is the cross-linker concentration. In the absence of the isotopically labeled BS3 H12/D12 cross-linker, we observed a faint band with an approximate MW of 34 kDa. In the presence of 1 mM BS3, we found a denser band, suggesting that BS3 captured the transient protein-protein interactions in the cross-linking reactions. We tentatively assigned a band at 34 kDa as a dimer of two phycobiliproteins. In addition, the large band that migrated to the area corresponding to 21 kDa potentially could be a combination of α- and β-PC and α- and β-APC (Fig. 2).
We analyzed by MS the cross-linked samples, and found not only that NblA interacts with β-PC but also the specific amino acid residues participating in the binding sites. To start, the K52 –T2 (Fig. 3d) cross-link between NblA-β-PC provides data on the specific amino acid residues involved in the binding reaction. The C-terminal end of β-PC is positioned inside in the central hollow cavity of a PC rod. Protein docking studies suggest that NblA fits in the open groove near the N-terminus of β-PC, via one or a combination of structural mimicry (Dines et al. 2008) including binding to conserved residues (Karradt et al. 2008) by electrostatic- and/or van der Waals’ forces. The open groove is also in close proximity to the N-termini of both α and β-PC. Interestingly, studies in Anabaena sp. PCC 7120 show that NblA binds to amino acid residues 16–39 of α-PC (Bienert et al. 2005). The binding interaction between NblA-β-PC has implications about PBS rearrangement and degradation. In addition, the M1 – K44 (Fig. 4d) and M1 – K52 cross-links between NblA-NblA are consistent with proper NblA refolding on the column after solubilization by urea.
Confocal imaging shows that NblA:GFP preferentially co-localizes to photosynthetic membranes where PBS complexes are present, as opposed to interacting with dissociated phycobiliproteins dispersed throughout the cell (Sendersky et al. 2014). Given that NblA only functions as a proteolysis adaptor, the key factors that actually degrade phycobiliproteins belong to the Clp proteasome system. The chaperone, ClpC, has the ability to unfold and shuttle proteins to the ClpP catalytic protease core, which is usually located in the membrane (Olinares et al. 2011). ClpC oligomerizes into hetero-hexamers with NblA to form NblA/ClpC structures that are several hundred kDa in size (Kirstein et al. 2006; Karradt et al. 2008) and can bind to PBS complexes, according to cross-linking studies (Stanne et al. 2007). These findings imply that NblA facilitates the interactions between ClpC and phycobiliproteins (Karradt et al. 2008) in PBS complexes that are still attached to the thylakoid membrane.
It is unlikely that these large NblA/ClpC complexes could fit inside the inner hollow cavity of the PC rod if PBS complexes are rigid structures when all colorless linker proteins and colored phycobiliproteins are present. NblA may mediate the degradation of PBS after other factors promote the disassembly of PBS complexes. Given that the PC rods are degraded sequentially from the distal end towards the core-proximal end during sulfur starvation (Collier et al 1994), perhaps the extension or dissociation of the capping linker protein, CpcD, may provide NblA the space to gain access and recruit ClpC to the N-terminal end of β-PC. The binding of NblA to β-PC may occur while the PC hexamers are attached to the PBS complex or after PC hexamers are dissociated from the PBS complexes. Once NblA binds to and mediates the degradation of β-PC with the help of ClpC, the structural integrity of PBS may become compromised as the structure becomes more exposed, accelerating the degradation of the PC rods by making them more accessible for degradation by the Clp proteins. Although compelling, experimental verification is needed to validate several aspects of this model.
In summary, we successfully discovered that NblA binds to the N-terminus of β-PC, which is a region located in the interior region of a PC trimer (Fig. 5), by using a combined cross-linking and MS method to determine the phycobiliproteins that NblA could interact with. In a fully assembled PC rod, the interior region of a PC trimer forms an internal hollow cavity. This finding opens new directions of research to elucidate the sequential mechanism of how NblA mediates PBS degradation and to determine how many copies of NblA bind simultaneously to the PC rods. Densitometric analyses of the LiDS gels of His6-NblA1-phycobiliprotein complexes from Tolypothrix PCC 7601 indicate that the molar ratio of NblA1/PC(α+β) is 0.65, after correcting for the difference in molecular mass between these subunits (Luque et al. 2003). This suggests that the number of NblA needed to trigger PBS degradation is less than the amount of PC available. Although binding between NblA and APC proteins was not detected by gels in Tolypothrix sp. PCC 7601 (Luque et al. 2003), recent imaging studies show that NblA can actually interact with the APC proteins (Sendersky et al. 2015). Further analysis of NblA binding to the APC core will provide more insight on how NblA triggers degradation of the PBS complexes. Lastly, the mechanistic details of how NblA/ClpC hetero-oligomer denatures and shuttles phycobiliproteins to the proteolytic catalytic center remain unclear. Further explorations of how the Clp proteasome system regulates the levels of phycobiliproteins should offer more insights into the roles of Clp proteins in cyanobacteria, other eubacteria, and plants.
Experimental Procedures
Bacterial Strains and Growth Conditions
All E. coli strains were grown under standard conditions (Sambrook and Russell 2001), and supplemented with 100 μg/mL Ampicillin (Amp). E. coli cultures induced with 1 mM IPTG were kept at 16 °C for 96 h. Details about strains, plasmids, and oligonucleotides are listed in Table S1.
Overexpression, purification of NblA2973_His7 Fusion Protein in E. coli
Overexpression of nblA2973_His from Synechococcus 2973 was driven by the T7 promoter in pET21a expression vector. We placed a TEV cleavage site between nblA and the His tag. The resultant plasmid, pSL2432, was transformed into the E. coli expression host BL21DE3 to make pSL2436 (Fig. S1). The E. coli cells transformed with pSL2436 were grown in LB at 37 °C to O.D.600 0.5. WT NblA2973 with ENLYFQGSSHHHHHHH appended on the C-terminus end was induced with 1 mM IPTG (Sigma) in SL2436 E. coli cells at 16 °C for 96 h (Baier et al 2014) (Fig. 1a). Cell pellets were dissolved in lysis buffer A (50 mM Tris pH 8, 20 mM Imidazole, 500 mM NaCl, 10% glycerol, 1% Tween 20) in the presence of a protease inhibitor cocktail, DNase, and lysozyme and sonicated to lyse cells. The cell lysate was centrifuged for 1 h at 20,200 x g and 4 °C. The resultant supernatant was saved. A large fraction of NblA proteins were still trapped in the pellet fraction, despite prolonged IPTG induction of the proteins at low temperature. Consequently, 2 M urea was used to increase the solubility of NblA2973_His7 fusion proteins and not denature the NblA dimers (Dines et al. 2007). Further processing of this pellet fraction started with a wash step containing Triton X-100 to remove cell membrane and wall debris by re-suspending with wash buffer B (50 mM Tris pH 8, 20 mM imidazole, 500 mM NaCl, 10% glycerol, 0.5% Triton X-100, and 2 M urea). Five 20 min centrifugations at 20,200 x g and 4 °C were carried out with buffer A. Then the pellet was re-suspended in wash buffer C (50 mM Tris pH 8, 20 mM imidazole, 500 mM NaCl, 10% glycerol, and 2 M urea) to remove Triton X-100, and submitted to two rounds of centrifugation for 20 min at 20,198.7 x g and 4 °C to recover the pellet. Another round of sonication and centrifugation was carried out to release the NblA2973_His7 fusion proteins. All supernatant fractions containing NblA2973_His7 fusion proteins were incubated with Ni-NTA resin. Before elution, proteins were refolded on the column with an extensive wash step in the absence of urea. To do so, wash buffer B was combined with wash buffer D (50 mM Tris pH 8, 20 mM imidazole, 500 mM NaCl, and 10% glycerol) to create a linear gradient from 2 M to 0 M urea (with [NaCl] constant in the buffer) at 1 mL/min. A step gradients of 100, 200, 300, 400, and 500 mM imidazole were used to elute the bound protein with an elution buffer E (50 mM Tris pH 8, 100–500 mM Imidazole, 500 mM NaCl, 10% glycerol).
Isolation of intact PBS
The NblA-mediated PBS degradation mechanism was studied in cyanobacterium Synechococcus 2973. Preparation of PBS from crude Synechococcus 2973 was as described by Ajlani et al 1995. A sucrose gradient containing 0.8 M potassium phosphate pH 7.0 buffer was used in combination with ultracentrifugation (SW41Ti rotor at 209,490 x g and 20 °C) to isolate intact PBS from Synechococcus 2973. Sucrose gradients consisted of 3 mL 1.0 M sucrose, 2.5 mL 0.75 M sucrose, 2.5 mL 0.5 M sucrose, and 2.0 mL 0.25 M sucrose.
In vitro binding assays
Ni-NTA (Qiagen, Germantown, MD) resins bound with NblA2973_His7 fusion proteins (0.5 mL) were mixed with PBS in 1 M sucrose and 0.8 M potassium phosphate conditions via affinity chromatography. In brief, NblA2973_His7 was first incubated with Ni-NTA resin for 16 h at 9 °C with gentle agitation. Unbound NblA2973_His7 proteins were removed by five-column volumes of wash buffer F (10 mM HEPES pH 7.8, 50 mM NaCl, and 5 mM imidazole). The NblA-loaded resin was incubated with ~10 mL of isolated intact PBS in 1.0 M sucrose and 0.8 M potassium phosphate overnight at 9 °C with gentle agitation, and then incubated at room temperature for 1 h with gentle agitation. The molar ratios of NblA:PBS were optimized to a 7:1 ratio. Prior to the elution step, unbound proteins were removed by two-column volumes of wash buffer G (10 mM HEPES pH 7.8, 50 mM NaCl, and 5 mM imidazole). Bound proteins were eluted with 100 μL buffer H (10 mM HEPES pH 7.8, 50 mM NaCl, and 200 mM imidazole) in three fractions. The elution fractions were quickly submitted to the cross-linking step.
Chemical Cross-linking
To capture any transient protein interactions of NblA and PBS, the elution fractions from the in vitro pull-down assays were incubated for 16 h at 9 °C the isotopically labeled bis(sulfosuccinimidyl)suberate chemical cross-linker (BS3-H12/D12) (Creative Molecules, Inc., http://www.creativemolecules.com/) that had been dissolved in water to a final concentration of 1 mM. Since the cross-linker was added directly to the elution fractions, the buffer for the cross-linking step included 10 mM HEPES pH 7.8, 50 mM NaCl, and 200 mM imidazole. This protocol should allow reactive groups on both ends of BS3 to bind covalently to the primary amines of lysine and at the N-terminus (Leitner et al. 2014) that are within a 11.4 Å distance. Unbound proteins were washed with HEPES buffer. The BS3 cross-linking reactions were quenched during an incubation with 50 mM Tris-HCl to BS3 for 20 min. Samples were heated at 70 °C for 10 min before analysis with SDS-PAGE, Coomassie Brilliant Blue R250, and UV-enhanced illumination of bilin pigments (Fig. 2).
Sample Digestion, LC-MS/MS, and Data Processing
For MS analysis, cross-linked proteins were precipitated with acetone by using a 2D-cleanup Kit (GE Healthcare, Chicago, IL) and digested in-solution with LysC (Wako Chemicals, Richmond, VA) and Trypsin (Promega, Madison, WI) by following a previously published method (Leitner et al. 2014). In brief, protein pellets were re-suspended in 20 μL 8 M urea (Sigma-Aldrich, St. Louis, MO), then incubated with 2.5 mM tris(2-carboxyethyl)phosphine (Sigma-Aldrich, St. Louis, MO) at 37 °C for 30 min and 5 mM iodoacetamide (Sigma-Aldrich, St. Louis, MO) for 30 min at room temperature (21 °C). LysC (Wako Chemicals, Richmond, VA) stock solution (2 μL of 0.5 μg/μL) was added to each cross-linked sample at 37 °C for 2 h. The protein solution was diluted into 100 mM Tris (Sigma-Aldrich, St. Louis, MO) and incubated with trypsin overnight at 37 °C. Formic acid (0.1%, Sigma-Aldrich, St. Louis, MO) was added to the samples to quench the digestion.
LC-MS/MS with an Ultimate 3000 Nano LC system (Thermo Scientific Dionex, Sunnyvale, CA) attached on-line to a Q Exactive Plus mass spectrometer (Thermo Fisher, Waltham, MA) was used to analyze the cross-linked sample digests. A 5 μL aliquot of each sample was loaded onto a guard column (Acclaim PepMap100, 100 μm × 2 cm, C18, 5 μm, 100 Å; Thermo Scientific Dionex) in Solvent A (water with 0.1% formic acid), and the peptides in the sample were separated on a custom-packed C18 reversed-phase column (Magic, 0.075 mm ×150 mm, 5 μm, 120 Å, Michrom Bioresources, Inc., Auburn, CA) at a flow rate of 4.5 μL/min. A linear 90-min gradient from 5–95% solvent B (80% acetonitrile, 20% water, 0.1% formic acid), followed by a 10-min hold at 95% solvent B, was used for peptide elution. As the peptides eluted, they were admitted into the mass spectrometer via a PicoView Nanospray Source (PV550, New Objective, Inc., Woburn, MA) at a spray voltage of 1.8 kV. The mass spectrometer was operated in the positive-ion mode with standard data-dependent acquisition settings as described previously (Liu et al. 2016) with the following modifications: for each precursor-ion scan, the top 15 ions with minimal intensity of 4 × 104 counts were selected for fragmentation with an isolation width of 3.0 m/z and normalized collision energy of 30% of maximum. Data processing and analysis was by Protein Prospector, see references cited in Liu et al (2016).
Protein Modeling
To gain insight into the NblA–PC binding modes, a two-step protein-protein docking methodology was used to elucidate likely structural motifs. First, the NblA protein dimer was globally docked from two potential starting configurations onto the PC monomeric sub-unit, consisting of an α-PC and a β-PC protein monomer, with an additional β-PC protein necessary to define the NblA binding pocket. The two starting conformations for docking the NblA protein dimer onto the PC sub-unit were from above the distal end of the PC rod (Orientation I) and from inside the PC rod (Orientation II) (Fig. 5b and Fig. S2). Second, the most likely structural binding mode from each starting configuration was retained and used as an initial structure for a local protein docking routine (Fig. 5b and Fig. S3). The best scoring structures from this local search were retained as the most likely structural motifs for NblA – PC binding (Fig. 5c).
Global Protein Docking
Initial models for the NblA protein dimer and the monomeric sub-unit of PC were obtained from their respective X-ray crystal structures; NblA was obtained from Synechococcus 7942 (PDB ID: 3CS5) (Dines et al. 2008) and PC was obtained from Synechococcus 7942 (PDB ID: 4H0M) (Marx and Adir 2013). For each of the two starting configurations 100,000 globally docked models of the NblA–PC binding motif were generated, using the protein-protein docking protocol (Gray et al. 2003; Wang et al. 2007) implemented in the Rosetta 3.4 software package (Leaver-Fay et al. 2011) (Fig. 5b and Fig. S2).
To prepare for the docking simulation, initial structures were first relaxed with all heavy atoms constrained using the Rosetta’s relax protocol. This initial step minimized steric clashes due to irregularities in the X-ray crystal structures. Next, the docking partners were “pre-packed” by using Rosetta’s docking prepack protocol, which moved the docking partners out of contact, optimized the side chains, and then brought the docking partners back to their initial positions. Subsequently, global protein-protein docking was performed by generating 100,000 decoys of the NblA – PC binding motif. For global docking, the NblA dimer was moved out of contact with PC, randomly oriented, and brought back into contact with PC. During this step, PC was held in its initial orientation. Next, the rigid-body orientation of NblA was energy minimized, and positions of NblA and PC side-chains were optimized before scoring the docked conformation (Leaver-Fay et al. 2013).
For each starting conformation, the top 10 scoring structures were retained, and the distance between residues K52 on NblA and T2 on β-PC was calculated to accommodate a cross-link between these residues. The ProDy package (Bakan et al. 2011) was used to extract atomic level structure from residues K52 on NblA and T2 on β-PC, and to estimate the cross-linking distance from the centroids of the two residues. The cross-linking distances were ranked from lowest to highest, and the binding motif with the lowest cross-linking distance was retained for subsequent local protein docking simulations (Fig. S2).
Local Protein Docking
By using the best globally docked NblA–PC structure from both starting conformations, a second local protein docking search was performed (Fig. S3). This refining step was similar to global docking except that the NblA dimer was not randomly oriented prior to rotamer sampling and side-chain optimization. Instead, the initial structure was randomly perturbed by using a Gaussian function for the translational and rotational components, where starting structures were within a 3 Å translation and an 8° rotation of the initial structure. Local protein docking was performed for 25,000 decoys of the NblA – PC binding motif at each starting conformation. In this local docking step, for Orientation I (above distal end of PC rod), the PC monomeric sub-unit was retained since steric hindrance with the full trimeric PC structure is not problematic. On the other hand, for Orientation II (inside the PC rod), the full trimeric PC structure was utilized for local docking refinement. For each starting conformation, the top 10 structures were sorted according to their total docking scores, and the estimated cross-linking distance from K52 on NblA and T2 on β-PC was calculated from the centroids of the two residues. The binding motifs with the lowest cross-linking distance as candidate structures were retained.
Supplementary Material
Acknowledgments
We thank Drs. Aparna Nagarajan, Michelle Liberton, Haijun Liu, Mark Bathe, and all the members of the Pakrasi and Bathe laboratories for collegial discussions and advice. Funding for this study was provided by the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under award number DE-SC 0001035. AYN has been supported by an NSF Graduate Research Fellowship grant number DGE-1143954. Funding for WPB has been provided by Army Research Office MURI award W911NF1210420. WPB also acknowledges the Office of Naval Research DURIP awards N00014-13-1-0664 and N00014-15-1-2830, which funded the high-performance computational cluster in the Bathe laboratory used for protein modeling. Mass spectrometry was supported in part by the NIH, NIGMS Grant No. P41GM103422 to MLG. We also thank Drs. Antje Baier, Anne Karradt, and Wolfgang Lockau for the generous gifts of NblA plasmids.
Footnotes
Author Contributions – AYN and HBP conceived the project; AYN, and HZ performed the experiments; AYN, DAW, and HZ analyzed the mass spectrometry data; WPB designed and implemented the docking models, and analyzed the docking modeling data; MLG and HBP contributed to the development of the measurement capabilities used; AYN, WPB, DAW, MLG, and HBP wrote the manuscript.
References
- Adir N, Dines M, Klartag M, McGregor A, Melamed-Frank M. Complex Intracellular Structures in Prokaryotes. Springer Berlin Heidelberg; Berlin, Heidelberg: 2006. Assembly and disassembly of phycobilisomes; pp. 47–77. [DOI] [Google Scholar]
- Adir N, Zer H, Shochat S, Ohad I. Photoinhibition - a historical perspective. Photosynth Res. 2003;76:343–370. doi: 10.1023/A:1024969518145. [DOI] [PubMed] [Google Scholar]
- Ajlani G, Vernotte C. Construction and characterization of a phycobiliprotein-less mutant of Synechocystis sp. PCC 6803. Plant Mol Biol. 1998;37:577–580. doi: 10.1023/A:1005924730298. [DOI] [PubMed] [Google Scholar]
- Ajlani G, Vernotte C, DiMagno L, Haselkorn R. Phycobilisome core mutants of Synechocystis sp. PCC 6803. Biochim et Biophys Acta. 1995;1231:189–196. doi: 10.1016/0005-2728(95)00086-X. [DOI] [Google Scholar]
- Allen MM, Smith AJ. Nitrogen chlorosis in blue-green algae. Arch Mikrobiol. 1969;69:114–120. doi: 10.1007/BF00409755. [DOI] [PubMed] [Google Scholar]
- Andersson FI, Blakytny R, Kirstein J, Turgay K, Bukau B, Mogk A, Clarke AK. Cyanobacterial ClpC/HSP100 protein displays intrinsic chaperone activity. J Biol Chem. 2006;281:5468–5475. doi: 10.1074/jbc.M509661200. [DOI] [PubMed] [Google Scholar]
- Baier A, Winkler W, Korte T, Lockau W, Karradt A. Degradation of phycobilisomes in Synechocystis sp. PCC 6803: evidence for essential formation of an NblA1/NblA2 heterodimer and its codegradation by a Clp protease complex. J Biol Chem. 2014;289:11755–11766. doi: 10.1074/jbc.M113.520601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier K, Lehmann H, Stephan DP, Lockau W. NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology. 2004;150:2739–2749. doi: 10.1099/mic.0.27153-0. [DOI] [PubMed] [Google Scholar]
- Baier K, Nicklisch S, Grundner C, Reinecke J, Lockau W. Expression of two nblA-homologous genes is required for phycobilisome degradation in nitrogen-starved Synechocystis sp. PCC 6803. FEMS Microbiol Lett. 2001;195:35–39. doi: 10.1016/S0378-1097(00)00541-3. [DOI] [PubMed] [Google Scholar]
- Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert R, Baier K, Volkmer R, Lockau W, Heinemann U. Crystal structure of NblA from Anabaena sp. PCC 7120, a small protein playing a key role in phycobilisome degradation. J Biol Chem. 2005;281:5216–5223. doi: 10.1074/jbc.M507243200. [DOI] [PubMed] [Google Scholar]
- Bogorad L. Phycobiliproteins and complementary chromatic adaptation. Ann Rev Plant Physiol. 1975;26:369–401. doi: 10.1146/annurev.pp.26.060175.002101. [DOI] [Google Scholar]
- Bricker TM, Mummadisetti MP, Frankel LK. Recent advances in the use of mass spectrometry to examine structure/function relationships in photosystem II. J Photochem Photobiol B. 2015;15:227–246. doi: 10.1016/j.jphotobiol.2015.08.031. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Guglielmi G, de Marsac NT, Castets A-M, Cohen-Bazire G. The structure of cyanobacterial phycobilisomes: a model. Arch Microbiol. 1979;123:113–127. [Google Scholar]
- Collier JL, Grossman AR. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J Bacteriol. 1992;174:4718–4726. doi: 10.1128/jb.174.14.4718-4726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier JL, Grossman AR. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO Journal. 1994;13:1039–1047. doi: 10.1002/j.1460-2075.1994.tb06352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dines M, Sendersky E, David L, Schwarz R, Adir N. Structural, functional, and mutational analysis of the NblA protein provides insight into possible modes of interaction with the phycobilisome. J Biol Chem. 2008;283:30330–30340. doi: 10.1074/jbc.M804241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dines M, Sendersky E, Schwarz R, Adir N. Crystallization of sparingly soluble stress-related proteins from cyanobacteria by controlled urea solublization. J Struct Biol. 2007;158:116–121. doi: 10.1016/j.jsb.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Elmorjani K, Thomas JC, Sebban P. Phycobilisomes of wild type and pigment mutants of the cyanobacterium Synechocystis PCC 6803. Arch Mikrobiol. 1986;146:186–191. doi: 10.1007/BF00402349. [DOI] [Google Scholar]
- Glazer AN. Phycobilisomes: structure and dynamics. Annu Rev Microbiol. 1982;36:173–198. doi: 10.1146/annurev.mi.36.100182.001133. [DOI] [PubMed] [Google Scholar]
- Gorl M, Sauer J, Baier T, Forchhammer K. Nitrogen-starvation-induced chlorosis in Synechococcus PCC 7942: adaptation to long-term survival. Microbiology. 1998;144:2449–2458. doi: 10.1099/00221287-144-9-2449. [DOI] [PubMed] [Google Scholar]
- Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/S0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- Gray JJ. High-resolution protein-protein docking. Curr Opin Struct Biol. 2006;16:183–193. doi: 10.1016/j.sbi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Grossman AR, Schaefer MR, Chiang GG, Collier JL. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol Rev. 1993;57:725–749. doi: 10.1128/mr.57.3.725-749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karradt A, Sobanski J, Mattow J, Lockau W, Baier K. NblA, a key protein of phycobilisome degradation, interacts with ClpC, a HSP100 chaperone partner of a cyanobacterial Clp protease. J Biol Chem. 2008;283:32394–32403. doi: 10.1074/jbc.M805823200. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Schlothauer T, Dougan DA, Lilie H, Tischendorf G, Mogk A, Bukau B, Turgay K. Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 2006;25:1481–1491. doi: 10.1038/sj.emboj.7601042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Jacak R, Song Y, Kellogg EH, Thompson J, Davis IW, Pache RA, Lyskov S, Gray JJ, Kortemme T, Richardson JS, Havranek JJ, Snoeyink J, Baker D, Kuhlman B. Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 2013;523:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc. 2014;9:120–137. doi: 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- Li H, Sherman LA. Characterization of Synechocystis sp. strain PCC 6803 and Δnbl mutants under nitrogen-deficient conditions. Arch Microbiol. 2002;178:256–266. doi: 10.1007/s00203-002-0446-y. doi:10:1007/s00203-002-0446-y. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang H, Niedzwiedzki DM, Prado M, He G, Gross ML, Blankenship RE. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science. 2013;342:1104–1107. doi: 10.1126/science.1242321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Orf GS, Lu Y, Jiang J, King JD, Wolf NR, Gross ML, Blankenship RE. Dramatic domain rearrangements of the cyanobacterial orange carotenoid protein upon photoactivation. Biochemistry. 2016;55:1003–1009. doi: 10.1021/acs.biochem.6b00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorimier R, Bryant DA, Stevens SE. Genetic analysis of a 9-kDa phycocyanin-associated linker polypeptide. Biochim Biophys Acta. 1990;1019:29–41. doi: 10.1016/0005-2728(90)90121-J. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Bryant DA. Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. Strain PCC 7002 to nutrient limitations and different nitrogen sources. Front Microbiol. 2012;3:145. doi: 10.3389/fmicb.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque I, Ochoa de Alda JA, Richaud C, Zabulon G, Thomas JC, Houmard J. The NblAI protein from the filamentous cyanobacterium Tolypothrix PCC 7601: regulation of its expression and interactions with phycobilisome components. Mol Microbiol. 2003;50:1043–1054. doi: 10.1046/j.1365-2958.2003.03768.x. [DOI] [PubMed] [Google Scholar]
- MacColl R. Cyanobacterial phycobilisomes. J Struct Biol. 1998;124:311–334. doi: 10.1006/jsbi.1998.4062. [DOI] [PubMed] [Google Scholar]
- Marx A, Adir N. Allophycocyanin and phycocyanin crystal structures reveal facets of phycobilisome assembly. Biochim Biophys Acta. 2013;1827:311–318. doi: 10.1016/j.bbabio.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Olinares PD, Kim J, van Wijk KJ. The Clp protease system; a central component of the chloroplast protease network. Biochim Biophys Acta. 2011;1807:999–1011. doi: 10.1016/j.bbabio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Porankiewicz J, Wang J, Clarke AK. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol Microbiol. 1999;32:449–458. doi: 10.1046/j.1365-2958.1999.01357.x. [DOI] [PubMed] [Google Scholar]
- Rappsilber J. The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richaud C, Zabulon G, Joder A, Thomas JC. Nitrogen or sulfur starvation differentially affects phycobilisome degradation and expression of the nblA gene in Synechocystis strain PCC 6803. J Bacteriol. 2001;183:2989–2994. doi: 10.1128/JB.183.10.2989-2994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- Scheer H, Zhao KH. Biliprotein maturation: the chromophore attachment. Mol Microbiol. 2008;68:263–276. doi: 10.1111/j.1365-2958.2008.06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Forchhammer K. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology. 2005;151:2503–2514. doi: 10.1099/mic.0.27883-0. [DOI] [PubMed] [Google Scholar]
- Sendersky E, Kozer N, Levi M, Garini Y, Shav-Tal Y, Schwarz R. The proteolysis adaptor, NblA, initiates protein pigment degradation by interacting with the cyanobacterial light-harvesting complexes. Plant J. 2014;79:118–126. doi: 10.1111/tpj.12543. [DOI] [PubMed] [Google Scholar]
- Sendersky E, Kozer N, Levi M, Moizik M, Garini Y, Shav-Tal Y, Schwarz R. The proteolysis adaptor, NblA, is essential for degradation of the core pigment of the cyanobacterial light-harvesting complex. Plant J. 2015;83:845–852. doi: 10.1111/tpj.12931. [DOI] [PubMed] [Google Scholar]
- Sidler W. Phycobilisome and phycobiliprotein structures. In: Bryant DA, editor. The Molecular Biology of Cyanobacteria. Kluwer Academic Publishers; Dordrecht: 2004. pp. 139–216. [Google Scholar]
- Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrom Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- Sinz A. The advancement of chemical cross-linking and mass spectrometry for structural proteomics: from single proteins to protein interaction networks. Expert Rev Proteomics. 2014;11:733–743. doi: 10.1586/14789450.2014.960852. [DOI] [PubMed] [Google Scholar]
- Smith GR, Sternberg MJE. Prediction of protein-protein interactions by docking methods. Curr Opin Struct Biol. 2002;12:28–35. doi: 10.1016/S0959-440X(02)00285-3. [DOI] [PubMed] [Google Scholar]
- Stanne TM, Pojidaeva E, Andersson FI, Clarke AK. Distinctive types of ATP-dependent Clp proteases in cyanobacteria. J Biol Chem. 2007;282:14394–14402. doi: 10.1074/jbc/M700275200. [DOI] [PubMed] [Google Scholar]
- Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Weisz DA, Gross ML, Pakrasi HB. The use of advanced mass spectrometry to dissect the life-cycle of Photosystem II. Front Plant Sci. 2016;7:1–25. doi: 10.3389/fpls.2016.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams VP, Glazer AN. Structural studies on phycobiliproteins. I. Bilin-containing peptides of C-phycocyanin. J Biol Chem. 1978;253:202–211. [PubMed] [Google Scholar]
- Yu J, Liberton M, Cliften PF, Head RD, Jacobs JM, Smith RD, Koppenaal DW, Brand JJ, Pakrasi HB. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep. 2015;5:1–10. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.