Abstract
Background
Honokiol, a compound extracted from Magnolia officinalis, has antifungal activities by inducing mitochondrial dysfunction and triggering apoptosis in Candida albicans. However, the mechanism of honokiol-induced oxidative stress is poorly understood. The present investigation was designed to determine the specific mitochondrial reactive oxygen species (ROS)-generation component.
Methods/results
We found that honokiol induced mitochondrial ROS accumulation, mainly superoxide anions (O2•−) measured by fluorescent staining method. The mitochondrial respiratory chain complex I (C I) inhibitor rotenone completely blocked O2•− production and provided the protection from the killing action of honokiol. Moreover, respiratory activity and the C I enzyme activity was significantly reduced after honokiol treatment. The differential gene-expression profile also showed that genes involved in oxidoreductase activity, electron transport, and oxidative phosphorylation were upregulated.
Conclusions
The present work shows that honokiol may bind to mitochondrial respiratory chain C I, leading to mitochondrial dysfunction, accompanied by increased cellular superoxide anion and oxidative stress.
General significance
This work not only provides insights on the mechanism by which honokiol interferes with fungal cell, demonstrating previously unknown effects on mitochondrial physiology, but also raises a note of caution on the use of M. officinalis as a Chinese medicine due to the toxic for mitochondria and suggests the possibility of using honokiol as chemosensitizer.
Introduction
Currently, disseminated invasive candidiasis has an approximate estimated mortality rate of 40%, even after being treated with antifungal drugs [1,2]. Candida albicans is the major cause of candidiasis and is the fourth most commonly reported nosocomial infection [3–5]. As everyone knows, the majority of living organisms need oxygen to survive. Mitochondrial respiration, dealing with transfer of unpaired electrons to oxygen (O2), may produce reactive oxygen species (ROS) such as superoxide anion (O2•−) and subsequently dismutation of O2•− to H2O2 [6]. Within the mitochondria, the main sites of superoxide production have been localized to the electron transport chain (ETC) complexes I (C I) and Ⅲ [6–9]. The bulk of mitochondrial ROS typically arise because of electron leakage from forward electron transport onto O2 during aerobic respiration as side products. It is driven by NADH-linked substrates, C I exhibits only minimal ROS production, but the addition of a ubiquinone-site inhibitor, such as rotenone, results in a significant increase in its rate [7–9]. The other mechanism by which ETC produces large amounts of O2•− is during reverse electron transport. During reverse electron transport, driven by succinate, ROS production by C I is increased significantly, and in this case, inhibited by the addition of rotenone [7, 8]. In addition to C I, C Ⅲ is regarded as an important site of O2•− production, especially when mitochondrial respiration is suppressed by antimycin, an inhibitor of C Ⅲ[6]. O2•− is then dismutated by superoxide dismutases to H2O2 that is reduced to H2O by catalase, peroxiredoxins, and glutathione peroxidases [9].When intracellular levels of ROS are high, ROS can have deleterious effects on cellular biomolecules including protein, lipid, RNA and DNA and cause subsequent cell death [9].
Honokiol, a neolignan isolated from the oriental medicine plant Magnolia officinalis, is an interesting compound exhibiting various pharmacological activities in preclinical experimental models [10–14]. Honokiol molecule contains two phenolic groups which can exhibit antioxidant properties similar to vitamin E or polyphenols. A paradox of honokiol is that it may have both pro- and antioxidant activities [15,16]. Previous mechanism studies, including those from our own laboratory, indicated that the apoptosis induction by honokiol in C. albicans was associated with production of ROS [15]. However, we are still lacking a detailed mechanistic knowledge of the architecture of mitochondrial ROS-producing systems induced by honokiol such as of C I or C Ⅲ and detailed insights on the mechanisms controlling their activities. The present study will make an attempt to clarify specific proposed mitochondrial ROS-producing components after honokiol treatemt.
Materials and methods
Materials
Honokiol (5,5’-diallyl-2,4’-dihydroxybiphenyl) was obtained from Xi'an Yuquan Biological Technology Co., Ltd and its purity is over 98% analyzed by high-performance liquid chromatography. DCFH-DA (2′,7′-dichlorofluorescein diacetate), dihydroethidium (DHE), 5-cyano-2,3-ditolyl tetrazolium chloride (CTC), and other molecular grade chemicals were obtained from Sigma Chemicals (St. Louis, MO, U.S.A.).
Microorganisms
C. albicans wild type strain SC5314 was used in this study [17]. The strain SC5314 was cultured in YPD (yeast extract/ peptone/dextrose) broth. For agar plates, 2% (w/v) bacto agar (Difco, BD Biosciences) was added to the medium. The strain was stored as frozen stock with 15% (v/v) glycerol at –80◦C. Before each experiment, cells were freshly revived on YPD plate from the stock.
ROS determination
SC5314 cells were adjusted to 1×107 cells/mL in YPD medium and exposed to different concentration of honokiol at 37°C for 4h. Intracellular ROS concentrations were determined in liquid cultures of C. albicans after honokiol treatment. H2O2 and O2•− levels were detected by adding DCFH-DA and DHE to the culture, respectively. After staining with 10 μmol/L DCFH-DA or 5 μmol/L DHE at 37°C for 30 min, the cells were collected and washed three times with PBS. The fluorescence intensities of the resuspended cells were measured by a flow cytometer using 488nm excitation and a 515nm band-pass filter for DCF detection and a filter >560nm for DHE detection (Becton-Dickinson Immunocytometry Systems, San Jose, CA).
Determination of mitochondrial C I activity
Extraction of mitochondrial proteins was performed as previously described [18].The enzyme activity assay of NADH CoQ reductase (mitochondrial C I) was carried out according to the instruction manual of the Mitochondrial C I Assay Kit (Genmed Scientifics, Inc.). Protein quantity was estimated by BCA protein assay kit (Beyotime, China). The C I activities were all normalized by the protein content in each sample and converted to the percentage of the control group.
Respiratory activity
The tetrazolium salt CTC is frequently used as indicator of microorganisms’ respiration [19]. Reduction of CTC is an indication of respiratory enzyme activity. Respiratory activity was assessed by using CTC (5-cyano-2,3-ditolyl tetrazolium chloride), a monotetrazolium redox dye which produces a CTC-formazan (CTF) fluorescent complex (indicated by cells stained in red) when it is biologically reduced, indicating respiration (metabolic activity). Samples were stained with 2.5mM CTC for 30 min. The respiratory activity was determined by the intensity of fluorescence with flow cytometry. Images of cells were obtained using a fluorescence microscope.
Viability detection
To perform viability detection, the yeast cells were suspended at a concentration of 104 cells/mL in 5 mL YPD medium with different concentrations of compounds. The tubes were incubated at 37°C without shaking for 24 h, and then 100 μL aliquots were washed, diluted and plated for colony counts on YPD agar medium. The plates were incubated for 48 h at 37°C and colonies were counted to determine CFU per mL.
RNA isolation, cDNA library construction and sequencing
C. albicans SC5314 cells were grown overnight at 37°C. The overnight culture was used to inoculate 20 ml of YPD to an initial 1×107 cells/mL, and incubated at 37°C for an additional 4 h at 150 rpm. The cells were then harvested and RNA extraction was performed by using the hot acidic phenol method [20]. Then genomic DNA was removed from RNA sample using DNase (New England Biolabs). RNA purity was assessed using the Nanodrop-2000 (Thermo Scientific, USA). Each RNA sample had an A260:A280 ratio above 1.9 and A260:A230 ratio above 1.8. Total RNA integrity was then subsequently checked using an Agilent Technologies 2100 Bioanalyzer with an RNA Integrity Number (RIN) value greater than or equal to 8. Next, two sequencing libraries were constructed by TruSeq™RNA Sample Preparation Kit according to the product instruction (Illumina). Each library was sequenced using Illumina HiSeq2500 for 2 × 150 bp pair-end (PE) sequencing.
RNA-Seq data analysis and differentially expressed gene identification
Quality control of all raw reads was conducted by Fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) software. An initial filtering step was performed to exclude poor quality reads, including adaptor reads, ambiguous nucleotides and low-quality reads (reads having more than 50% bases with quality value). Then the clean reads were mapped onto the C. albicans SC5314 reference genome [21] (C_albicans_SC5314_version_A21-s02-m07-r10(http://www.candidagenome.org/download/sequence/C_albicans_SC5314/Assembly21/archive/) independently by TopHat [22] (v2.0.10). The ‘-G’ option of Tophat together with the Gene Transfer Format (GTF) file of the Ensemble gene annotation was used for read mapping. The other parameters were set to default values. Based on the Tophat alignment BAM file, HTSeq [23] (v0.6.1) was used to estimate and quantify gene expression with default parameters, yielding raw read count for each genes. Gene expression was measured in reads per kilobase of exon per million reads mapped (RPKM). Finally, edgeR [24] was used to identify the differentially expressed genes (DEGs) by pairwise comparisons. The difference was considered significant if the FDR ≤0.05.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
GO enrichment analysis provides all GO terms that are significantly enriched in DEGs, relative to the genome background, and filters the DEGs that correspond to a specific biological functions. This method firstly maps all DEGs to GO terms in the database (http://www.geneontology.org/), calculating gene numbers for every term, and then uses the hyper geometric test to find significantly enriched GO terms in the input list of DEGs. GO: TermFinder (http://smd.stanford.edu/help/GO-TermFinder/GO_TermFinder_help.shtml) was used to enrichment the differentially expressed genes. The calculated p-value goes through Bonferroni Correction, taking corrected p-value ≤0.05 as a threshold.
KEGG pathway enrichment analysis identifies significantly enriched metabolic pathways or signal transduction pathways in the whole genome background using KOBAS [25], taking the FDR≤0.05 as a threshold to identify enrichment pathway.
Quantitative real-time PCR analysis
Quantitative PCR (qPCR) was performed as described previously [20]. Primer sequences used for the amplification of specific genes are shown in S1 Table.
Statistical analysis
All data were presented as means ± standard error of the mean (S.E.M.). Graphs were generated using Microsoft Excel (Microsoft Corp., Redmond, WA). Statistical analysis was performed using SPSS 12.0 (SPSS Inc., Chicage, USA). Differences between groups were determined using analysis of variance (ANOVA).A p value < 0.05 was considered significant.
Results
Induction of oxidative stress after honokiol treatment on C. albicans
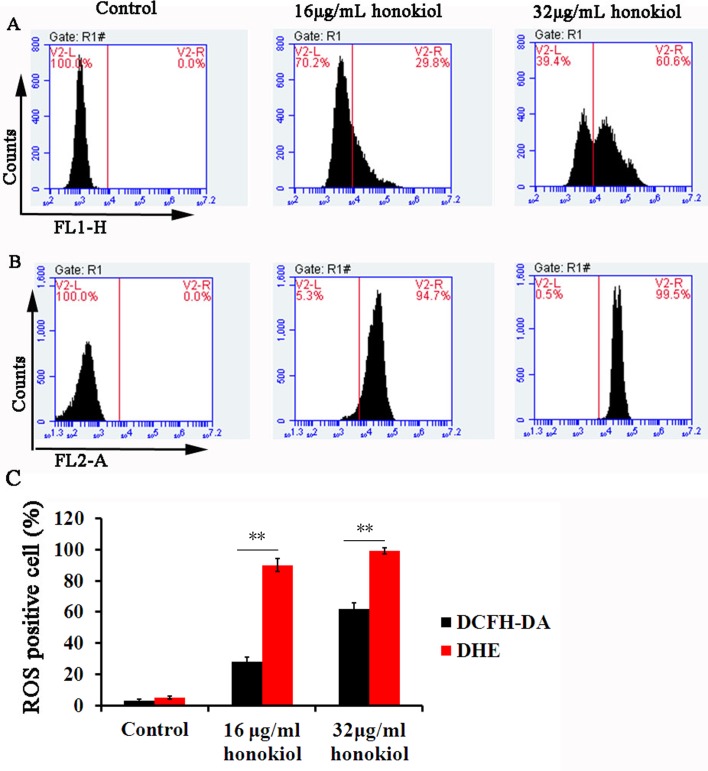
The term ROS encompasses oxygen free radicals, such as O2•− and H2O2. The levels of O2•− and H2O2 can be detected by DHE and DCFH-DA staining, respectively [26]. Mitochondria have long been established as a major source of ROS, which is generated from O2 by electron leakage or reverse electron transport originating in the mitochondrial transport chain, during respiration [6]. Fig 1A shows that control cells showed little O2•− and H2O2 accumulation, while 16μg/mL honokiol causes an increase in the fluorescence intensity of C. albicans loaded with DCFH-DA or DHE. The quantification of ROS was performed by flow cytometry. 16μg/mL honokiol induced DCF positive staining in 28.7%±3.1% cells, while most of cells (90.3±3.7%) were DHE positive staining after honokiol exposure. When addition of honokiol at 32μg/mL, the levels of ROS were further increased, 62.0%±3.6% for DCF positive cells and 99.1%±1.8% for DHE positive cells, respectively (Fig 1B). These results concluded that honokiol stimulated ROS formation, mainly O2•−.
Fig 1.
Levels of H2O2 (A) and O2•−(B) in C.albicans SC5314 after honokiol treatment. (A) and (B) ROS production was evaluated by flow cytometry. (C) The percentage of ROS-positive cells after honokiol treatment. H2O2 and O2•− were determined by fluorometric detection of the oxidation products of DCFH-DA and DHE, respectively. **p<0.01.
Effect of respiratory chain inhibitors on the oxidative burst and C. albicans killing induced by honokiol
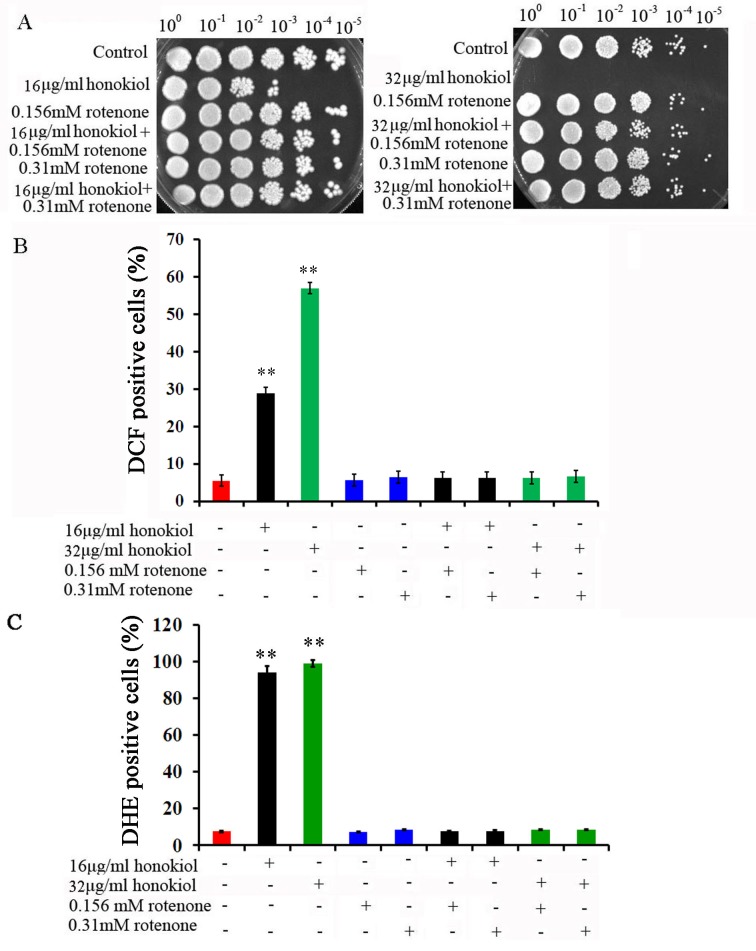
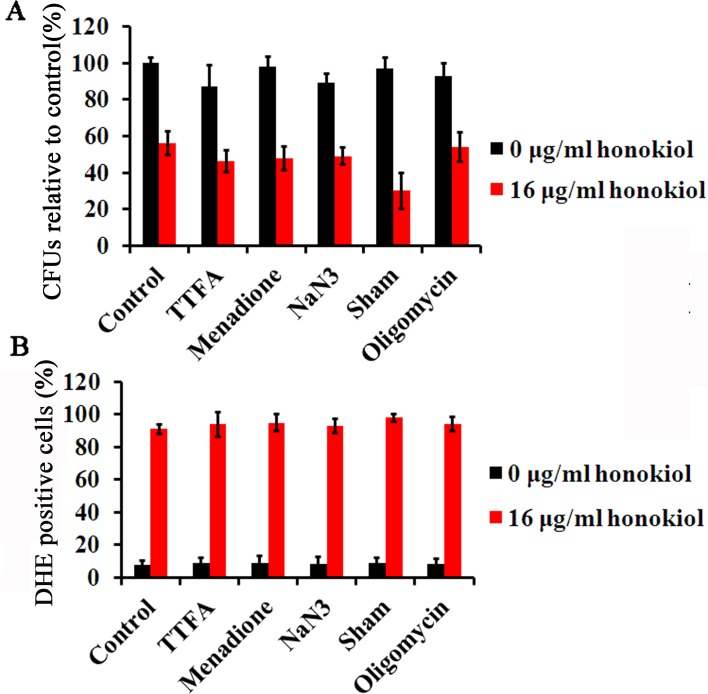
O2•− typically arises because of electron leakage or reverse electron transport from the electron transport chain onto O2 during aerobic respiration [7]. To further investigate the sites of O2•− production in mitochondria, we first used rotenone (respiratory chain C I inhibitor) to observe the effect on C. albicans killing and ROS production by honokiol. Pre-cultured with rotenone (0.156mM or 0.31mM) eliminated the antifungal activities of honokiol when the concentration was 16μg/mL (Fig 2A). We even tested the fungicide concentration 32μg/mL of honokiol when combined with two different concentrations of rotenone, yet the results were the same. In addition, ROS production (both H2O2 and O2•−) by honokiol was also completely eliminated by rotenone (Fig 2B and 2C). Rotenone can completely block both the antifungal activity of honokiol and ROS production (Fig 2). Then, we used several other respiratory chain inhibitors to observe the effect on C. albicans killing and ROS production by honokiol. Pre-cultured with thenoyltrifluoroacetone (TTFA), menadione, sodium azide (NaN3), and oligomycin showed almost no effect on honokiol-induced Candida killing and O2•− accumulation (Fig 3). Salicylhydroxamic acid (Sham), a cyanide-resistant alternative oxidase inhibitor, could slightly enhance the susceptibility to honokiol and raised the level of O2•− (Fig 3). These data indicated that the potential antifungal activity of honokiol was related to mitochondrial C I. Nevertheless, the mechanisms and consequences of such an interaction still require further investigation.
Fig 2. Effect of rotenone on C. albicans survival and ROS production after honokiol treatment.
(A) Effect of rotenone on C. albicans survival after honokiol treatment. The SC5314 cells were treated with rotenone at concentrations of 0.156mM or 0.31mM for 1h and then exposed to honokiol for 24h. Effect of rotenone on H2O2 (B) and O2•−(C) production after honokiol treatment. ROS production was evaluated by flow cytometry as described in Materials and Methods. **p<0.01.
Fig 3. Effect of other mitochondrial respiratory chain inhibitors on C. albicans survival and ROS production after honokiol treatment.
(A) Effect of respiratory chain inhibitors on C. albicans survival after honokiol treatment. The SC5314 cells were treated with respiratory chain inhibitors for 1h and then exposed to honokiol for 24h. (B) Effect of respiratory chain inhibitors on O2•− production after honokiol treatment. O2•− production was evaluated by flow cytometry as described in Materials and Methods. TTFA, thenoyltrifluoroacetone (0.125mM); menadione (0.0625mM); NaN3 (0.0025%); Sham, salicylhydroxamic acid (5mM); oligomycin (5μg/ml).
Respiratory activity in C. albicans after exposure to honokiol
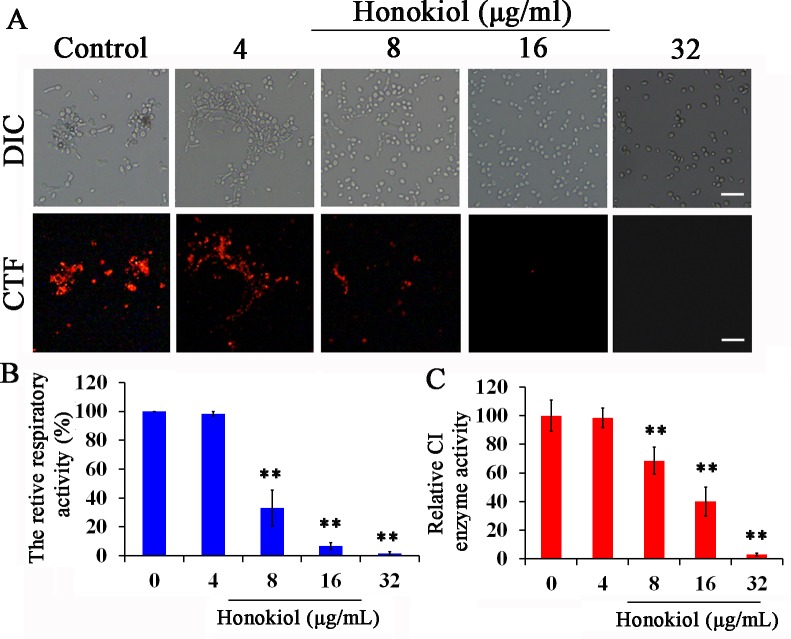
CTC staining represents an index of the respiratory activity of the cell at the time of observation [19].CTC is a soluble crystal that forms a nearly colorless non-fluorescent solution. In the electron transport system, CTC, as an artificial redox partner, was reduced to a fluorescent formazan crystal (CTF) primarily by membrane-bound NADH-dehydrogenase [19]. The accumulation of CTF red fluorescence was clearly observed in control group, but honokiol induced a concentration-dependent decrease in respiratory activity (Fig 4A). Almost no CTF accumulation was observed both in 16μg/mL and 32μg/mL of honokiol-treated cells, indicating that the respiratory activity of C. albicans was blocked by honokiol. These results were also confirmed by flow cytometry (Fig 4B). We further investigated the C I enzyme activity in C. albicans after honokiol exposure. As shown in Fig 4C, the C I enzyme activities in C. albicans after honokiol treatment at 8μg/mL and 16μg/mL were reduced to 68.5% and 39.9%, respectively. In addition, 32μg/mL honokiol could almost completely inhibit the activity of C I enzyme. There was no significant difference between 4μg/mL honokiol treatment and the control group.
Fig 4. Effect of honokiol on respiratory activity and C I enzyme activity in C. albicans.
(A) C. albicans SC5314 stained with CTC. (B) The relative fluorescence intensity was obtained by flow cytometric analysis of CTF-stained cells. (C) Effect of honokiol on C I enzyme activity. Scale bar = 10μm. **p<0.01.
The effect of honokiol on mtDNA in C. albicans
Staining with DAPI revealed that honokiol-treated cells were devoid of mtDNA, while the control group showed the punctuate staining for mtDNA when observed under fluorescence microscope (Fig 5A). In addition, we observed a significant induction of mitochondrial-encoded subunit 1 of cytochrome c oxidase (COX1) gene and subunit 6 of the F0 sector of mitochondrial F1F0 ATP synthase (ATP6) gene, compared with control (Fig 5B).
Fig 5. The effect of honokiol on mtDNA in C. albicans.
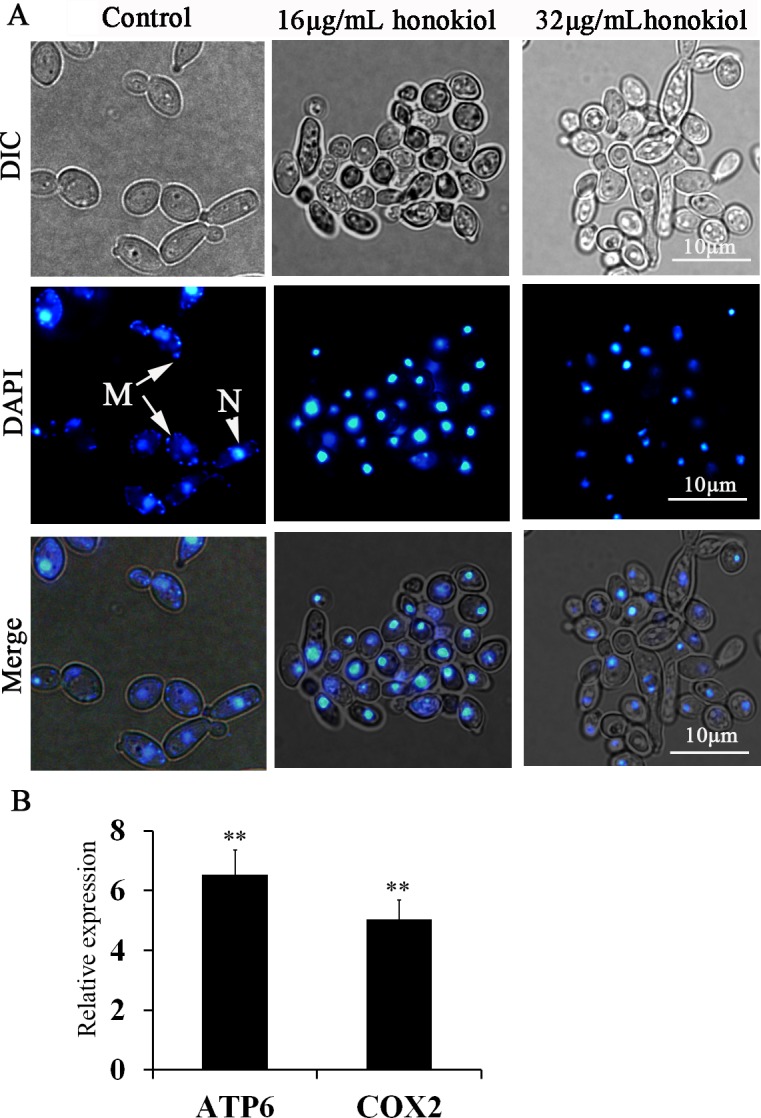
(A) Nuclear and mtDNA was stained with DAPI. Nuclear DNA stained brightly, while the mitochondrial nucleoids displayed punctuate cytoplasmic staining. N, nucleus; M, mtDNA. (B) qRT-PCR analyzed the expression of mtDNA genes. Expression is relative to control group and the data are presented as the average of three biological replicates each normalized to ACT1. **p<0.01.
Honokiol treatment increases the sensitivity to cell wall-perturbing agents
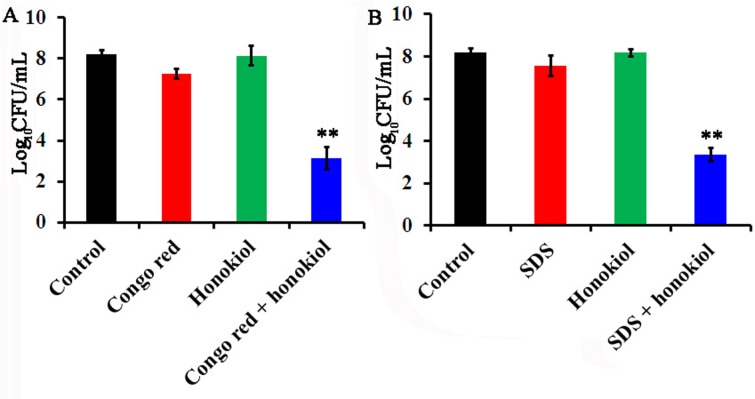
Mitochondrial dysfunction in C. albicans is reported to be linked with loss of cell wall integrity [27–29]. Therefore, we compared the sensitivities of C. albicans to the cell-wall perturbing agents Congo red (4μg/mL) or SDS (0.01%) alone or in combination with honokiol (4μg/mL). As shown in Fig 6, honokiol significantly increased the sensitivity of the cell wall-perturbing agents against C. albicans.
Fig 6. Honokiol treatment increases the sensitivity to cell wall-perturbing agents against C. albicans.
(A) Honokiol treatment increases the sensitivity to Congo red. (B) Honokiol treatment increases the sensitivity to SDS. Cells were treated with honokiol (4μg/mL), cell-wall perturbing agents (Congo red, 4μg/mL; SDS, 0.01%) or their combination for 24 h. Viability detections were described in Materials and Methods. **p<0.01.
Transcriptional analysis of honokiol-treated C. albicans
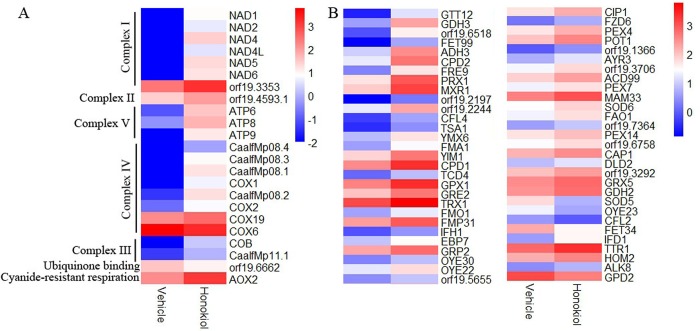
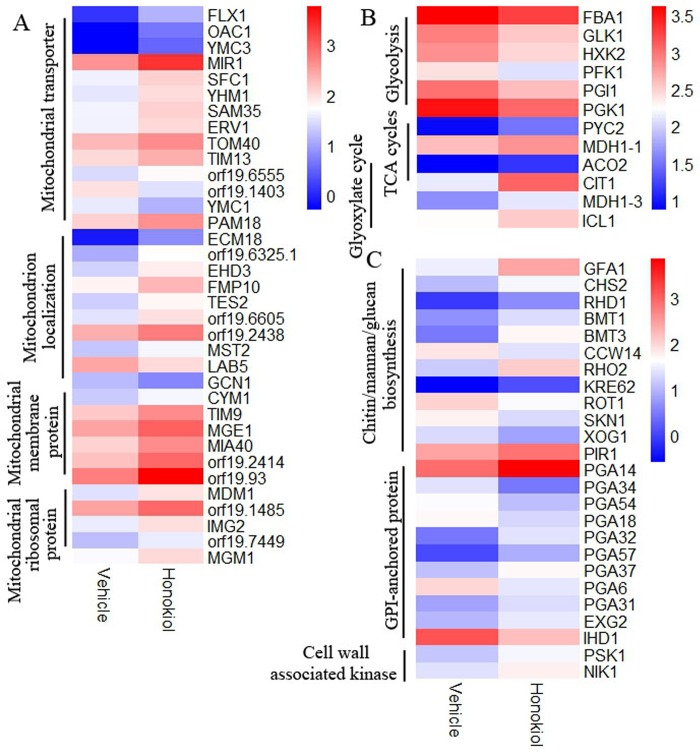
In order to find pathway and processed involved in generating superoxide of C. albicans treatment by honokiol, we performed a genome-wide transcriptome analysis of C. albicans SC5314 after honokiol treatment, 1152 genes were differentially expressed in vehicle-treated group vs. honokiol-treated group, of which 837 were significantly upregulated and 315 genes were significantly downregulated. RNA sequencing reveals 116 genes involved in mitochondrial and oxidation reduction were differentially expressed in honokiol-treated group (Fig 7). Because honokiol decreased mitochondrial respiratory chain C I enzyme activity and induced mitochondrial dysfunction, we focused on genes related to mitochondrial function. The genes encoding mitochondrial ETC proteins of CⅠ(NAD1, NAD2, NAD4, NAD4L, NAD5, NAD6, and orf19.3353), CⅡ (orf19.4593.1), CⅢ (COB and CaalfMp11.1), CⅣ (COX1, CaalfMp08.1, CaalfMp08.2, CaalfMp08.4, COX2, and COX19), CV (ATP9, ATP6, and ATP8), and alternative oxidase (AOX2, involved in a cyanide-resistant respiratory pathway) were upregulated after honokiol exposure (Fig 7A). Honokiol, having pro- and antioxidant activities, strongly induced the expression of genes involved in the oxidation–reduction process (Fig 7B). Besides the genes involved in mitochondrial electron transport and oxidation–reduction process, the mitochondrial transporter genes (TOM40, OAC1, FLX1, YMC3, SFC1, YHM1, SAM35, ERV1, TIM13, MIR1, PAM18, and orf19.6555), mitochondrial membrane (CYM1, TIM9, MGE1, MIA40, orf19.2414, and orf19.93) and ribosomal protein (MDM1, orf19.1485, IMG2, orf19.7449,and MGM1), and genes involved in mitochondrion localization (ECM18, orf19.6325.1, EHD3, FMP10, TES2, orf19.6605, orf19.2438, MST2, LAB5, and GCN1) are significantly upregulated (Fig 8A). In addition, the expression of genes involved in carbon metabolism, such as glycolysis, three carboxylic acid (TAC) cycles, and glyoxylate cycle were changed. Honokiol treatment downregulated the transcriptional levels of glycolysis (FBA1, GLK, HXK2, PFK1, PGI1, and PGK1) while upregulated three carboxylic acid cycles (ACO2, CIT1, and MDH1-1), and glyoxylate cycle genes (ACO2, CIT1, MDH1-3, and ICL1) (Fig 8B). We note that the transcription in several categories of cell wall protein (RBR2, SCW4, DSE1), cell wall chitin/hexosamine biosynthesis (GFA1, CHS2), phosphopeptidomannan, mannosyltransferase (RHD1, BMT1, BMT3), glucan synthase (RHO2, KRE62, PIR1), and GPI-anchored cell wall proteins (EXG2, PGA14, PGA31, PGA32, PGA57, and PGA37) were all upregulated expression (Fig 8C).
Fig 7.
Transcriptional profiling of mitochondrial electron transport (A) and oxidation-reduction process (B) in C. albicans SC5314 after honokiol treatment. The expression level of genes which are represented by log2 (RPKM) values are indicated from blue to red.
Fig 8. Transcriptional profiling of genes in response to honokiol treatment.
Functional annotation for genes are shown: (A) genes of mitochondrial transporter, mitochondrion localization, mitochondrial membrane protein, and mitochondrial ribosomal protein. (B) genes involved in carbon metabolism, such as glycolysis, TAC cycles, and glyoxylate cycle. (C) genes involved in cell wall. The expression level of genes which are represented by log2 (RPKM) values are indicated from blue to red.
GO assessment
Gene annotation and classification with GO terms for C. albicans was performed in Candida Database. The DEGs were mapped to GO ontologies of cellular components, molecular functions, and biological processes for GO enrichment analysis (Table 1). GO analysis revealed DEGs in honokiol treatment were enriched for 9 GO terms. The largest number of DEGs were enriched in oxidoreductase activity for GO molecular function (GO:0016491). Changes in biological processes included ribosome biogenesis (GO:0042254), electron transport and oxidative phosphorylation (GO:0042775, GO:0006119, GO:0042773, GO:0022904). In addition, the term enrichment for cellular component were respiratory chain (GO:0070469 and GO:0005746) and proteasome complex (GO:0000502).
Table 1. GO enrichment analysis.
| Term | TermName | DEGs number | Gene name | Padja | |||
|---|---|---|---|---|---|---|---|
| Ontology: Biological process | |||||||
| GO:0042254 | ribosome biogenesis | 31 | orf19.5991,RRS1,NOC4,orf19.6234,UTP21,DIM1,SPB4,YTM1,NOG1,KRR1,orf19.6828,IMP4,DBP8,RPS8A,HCA4,UBI3,orf19.1646,PES1,MRT4,NSA1,YST1,orf19.3393,NEP1,RCL1,TSR1,NOP5,RPL8B,RPL82,orf19.3778,GAR1,orf19.1388 | 0.00578 | |||
| GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 14 | CaalfMp11.1,COX1,CaalfMp08.4,CaalfMp08.1,COB,NAD4L,NAD1,NAD4,CaalfMp08.2,COX2,NAD5,NAD2,NAD6,CaalfMp08.3 | 0.00604 | |||
| GO:0006119 | oxidative phosphorylation | 14 | NAD6,CaalfMp08.3,COX2,CaalfMp08.2,NAD5,NAD2,NAD4L,NAD1,NAD4,CaalfMp11.1,COX1,CaalfMp08.1,CaalfMp08.4,COB | 0.01208 | |||
| GO:0042773 | ATP synthesis coupled electron transport | 14 | NAD5,NAD2,CaalfMp08.2,COX2,CaalfMp08.3,NAD6,CaalfMp08.4,CaalfMp08.1,COB,CaalfMp11.1,COX1,NAD4,NAD1,NAD4L | 0.01208 | |||
| GO:0022904 | respiratory electron transport chain | 14 | CaalfMp08.3,NAD6,NAD5,NAD2,CaalfMp08.2,COX2,NAD4,NAD1,NAD4L,CaalfMp08.1,COB,CaalfMp08.4,CaalfMp11.1,COX1 | 0.04127 | |||
| Ontology: Molecular function | |||||||
| GO:0016491 | oxidoreductase activity | 111 | XYL2,NAD4L,OYE22,orf19.3706,ADH4,FET99,DLD2,orf19.6518,orf19.7288,CaalfMp11.1,CaalfMp08.1,ADH3,PXP2,orf19.7306,EBP7,orf19.1366,SSP96,orf19.5565,orf19.2175,MXR1,IFM3,orf19.1639,NAD4,OYE23,orf19.2782,orf19.1438,orf19.5728,orf19.1340,orf19.2312,AOX2,YIM1,POX1,NAD2,GDH3,GPD2,SOD6,orf19.6066,IFD3,orf19.4567,spliced|Uncharacterized,orf19.2177,orf19.1844,orf19.86,FRE7,POX18,PRX1,NAD1,RNR1,orf19.1461,ERO1,RNR22,ERV1,TRX1,orf19.6341,NAD6,CFL2,orf19.1802,CFL4,GRP2,orf19.7204,FMO1,GRE2,FOX2,IFD6,ARG5,6,FET34,ADH2,orf19.4476,COB,HOM2,SOD5,orf19.5978,orf19.320,MIA40,orf19.7364,CaalfMp08.2,FRE9,orf19.6838,NAD5,TTR1,COX6,orf19.355,FRE30,orf19.5879,orf19.3537,orf19.1709,COX2,PUT2,ALK2,IFK2,orf19.6869,orf19.6306,COX1,CaalfMp08.4,CaalfMp08.3,orf19.3711,orf19.2394,MDH1-3,orf19.6398,GDH2,YMX6,orf19.2197,orf19.5655,TSA1,orf19.7495,orf19.2244,orf19.22.1,orf19.6143,orf19.6758,orf19.3292,ALK8 | 0.00949 | |||
| Ontology: Cellular component | |||||||
| GO:0005746 | mitochondrial respiratory chain | 14 | CaalfMp08.3,NAD6,NAD5,NAD2,CaalfMp08.2,COX2,NAD4,NAD1,NAD4L,CaalfMp08.4,CaalfMp08.1,COB,CaalfMp11.1,COX1 | 0.00149 | |||
| GO:0000502 | proteasome complex | 18 | orf19.6973,orf19.213,RPT2,PUP2,orf19.1993,PRE8,RPN2,RPN3,PUP3,ECM29,RPN1,RPN10,PRE9,PRE10,orf19.2755,orf19.5961,CIC1,orf19.2301 | 0.00854 | |||
| GO:0070469 | respiratory chain | 15 | NAD1,NAD4L,NAD4,COX1,CaalfMp11.1,CaalfMp08.1,AOX2,CaalfMp08.4,COB,NAD6,CaalfMp08.3,CaalfMp08.2,COX2,NAD2,NAD5 | 0.0169 | |||
aSignificantly enriched representative GO terms (adjusted p-value<0.05) are listed in table.
KEGG analysis
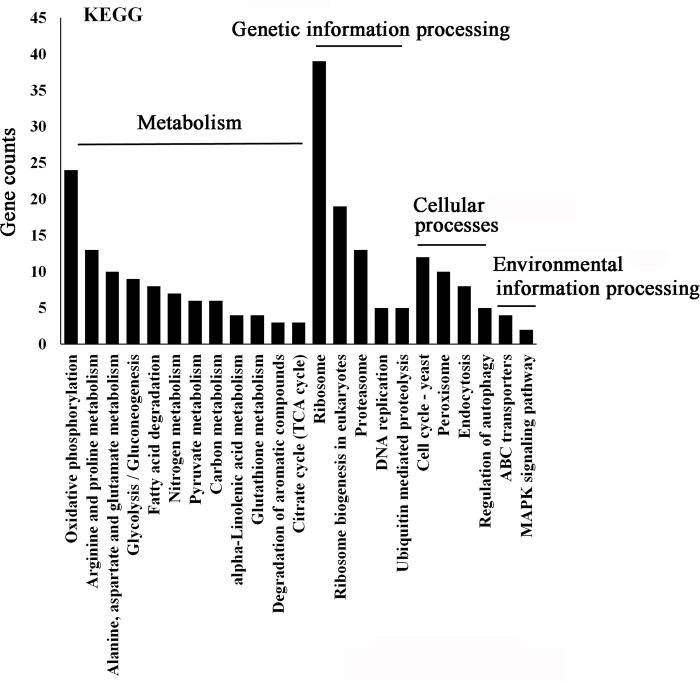
We also employed the KEGG pathway database to determine if the DEGs belong to particular signaling pathways. KEGG pathway mapping is the process to map molecular datasets, especially large-scale datasets, to the KEGG pathway maps for biological interpretation of higher-level systemic functions. The signaling pathway hypothesized to be affected by honokiol treatment mainly included signaling pathways related to metabolism (oxidative phosphorylation, arginine and proline metabolism, alanine, aspartate and glutamate metabolism,glycolysis/gluconeogenesis, fatty acid degradation, nitrogen metabolism, pyruvate metabolism, carbon metabolism, alpha-linolenic acid metabolism, glutathione metabolism, degradation of aromatic compounds, and citrate cycle), genetic information processing (ribosome, ribosome biogenesis in eukaryotes, proteasome, DNA replication, and ubiquitin mediated proteolysis), cellular processes (cell cycle, peroxisome, endocytosis, and regulation of autophagy), and environmental information processing (ABC transporters and MAPK signaling pathway) (Fig 9).
Fig 9. KEGG pathway annotation in response to honokiol treatment.
The bottom x-axis indicates he main category. The y-axis indicates the number of genes in a specific category. The results are summarized in the following four main categories: (1) metabolism;(2) genetic information processing; (3) cellular precesses; (4) environmental information processing.
Discussion
Candida species are a group of opportunistic fungal pathogens in humans, particularly among immunocompromised and hospitalized population [1–3]. Only few classes of antifungals such as azoles, flucytosine, polyenes, allylamines, and echinocandins are available for the treatment of Candida infections [30]. However, the rising drug resistance is an inevitable problem [31]. Therefore, it is necessary to discover new antifungal agents or other safer alternatives to improve the effectiveness of treatment of Candida infections [32,33]. Honokiol is an interesting small-molecule exhibiting pro- and antioxidant activities, similar with vitamin C and E [14]. Past studies have also demonstrated that honokiol has antifungal activities (MIC rang, 8–32 μg/mL) and when used in combination with fluconazole, can exhibit synergistic activity against clinical isolates of fluconazole-resistant C. albicans [20,34,35].To better understand how honokiol works on a cellular level, we sought to explore its antifungal mechanism and found honokiol caused an increase of intracellular ROS, lipid peroxidation, and protein oxidation in a dose-dependent manner in C. albicans [15]. This study will aim to clarify specific mitochondrial ROS-generation components that are involved in honokiol treatemt. Mitochondria have always been considered as a primary source of ROS like the superoxide anion radicals, which are converted from O2 by electron leakage from the mitochondrial transport chain during cellular respiration [6,7].
In the current study, we demonstrated that superoxide radicals was mainly generated and accumulated in mitochondria after honokiol exposure by using fluorescent ROS probes DHE and DCFH-DA (Fig 1). The mitochondrial respiratory chain is the major source of intracellular ROS generation. About 1–2% of the molecular oxygen consumed during normal physiological respiration is converted into superoxide radicals who serve as the precursor of most ROS [6]. There is growing evidence that most of the superoxide radicals generated by mitochondria is produced by C I and C Ⅲ[7,8]. To obtain an insight into intra-mitochondrial site of honokiol-induced overproduction of superoxide radicals, we used specific respiratory chain inhibitors. Only rotenone can completely block superoxide radicals generation and deprived of fungicidal activity of honokiol (Figs 2 and 3). Since rotenone is a C I inhibitor, we hypothesis that honokiol may compete with rotenone at or close to the quinone binding sites of C I or the binding site of honokiol is upstream from the rotenone binding site. In addition, we also found mitochondrial NADH dehydrogenase activity was decreased after honokiol, indicating the C I activities were inhibited (Fig 4). Consistent with the above results, respiratory activity was decreased, which may be associated with the inhibition of C I (Fig 4). In view of the fact that O2•− production is completely inhibited by the addition of a specific C I ubiquinone–site inhibitor rotenone that prevents C I binding to CoQ, we concluded the mechanism by which honokiol produces large amounts of O2•− is specifically through respiratory C I reverse electron transport [36,37].
One important target of ROS is the mitochondrial DNA due to the close proximity to the electron transport chain, the major locus for free radical production, and the lack of protective histones [38]. DAPI staining revealed that honokiol treatment caused loss of mtDNA, especially after 32μg/mL honokiol treatment, the cells were completely devoid of mtDNA. However, we observed increased transcription of mitochondrial genes (ATP6 and COX2) after honokiol exposure (Fig 5). It is possible that cells after honokiol exposure attempts to overcome the ROS-induced mtDNA damage by increased mtDNA genes expression, which reflects a compensatory mechanism [39,40].
We identified a number of DEGs from honokiol treatment. After honokiol treatment, we observed increased expression of genes involved in mitochondrial respiratory electron transport chain, oxidoreductase activity, and ribosome biogenesis (Fig 7 and Table 1). C I is the first step in the respiratory chain and is located in the inner mitochondrial membrane [7]. The molecular mass of CI is around 800kDa and is composed of at least 39 subunit proteins [29]. To date, the CI structure in C. albicans remains somewhat unclear. C. albicans has 14 core CI genes of bacterial progenitor that are proton-pumping NADH: ubiquinone oxidoreductases, seven of which are encoded by mitochondrial DNA (NAD1, NAD2, NAD3, NAD4, NAD4L, NAD5, and NAD6), the other subunit genes are nuclear-encoded. Since methods to selectively mutate the mitochondrial genome in C. albicans are unavailable, the core CI genes encoded by mitochondrial DNA in C. albicans are not studied yet [29]. Our further work is required to clarify the interaction of honokiol with mitochondrial C I.
There are many lines of evidence to suggest that oxidative stress is associated with the action of honokiol [14,15]. Honokiol could trigger a multifaceted response (Fig 10). It activates stress response pathway to tolerate ROS toxicity, including increased expression of genes involved in the oxidative stress response (CAP1, GTT12, PRX1, MXR1, GPX1, TRX1, IFH1, and SOD6), proteolysis and protein degradation (UBC4, UBC15, UBC8, and UBP6), and autophagy (ATG8, ATG9, ATG27, orf19.3998, APG13, and PRB1) (Fig 8 and S2 Table). CAP1, an AP-1 bZIP transcription factor, was significantly upregulated after honokiol treatment. Cap1 regulates a variety of oxidative stress response including the cellular antioxidant defense system, carbohydrate metabolism and energy metabolism, protein degradation, ATP-dependent RNA helicase, and resistance pathways [41]. Redox homeostasis in cells is important for maintaining proper cellular functions [42]. Honokiol targets on mitochondrial respiratory chain C I, inducing ROS accumulation, disruption of intracellular redox homeostasis, irreversible oxidative modifications of lipid, protein or DNA, and activation of autophagy or apoptosis signaling pathway.
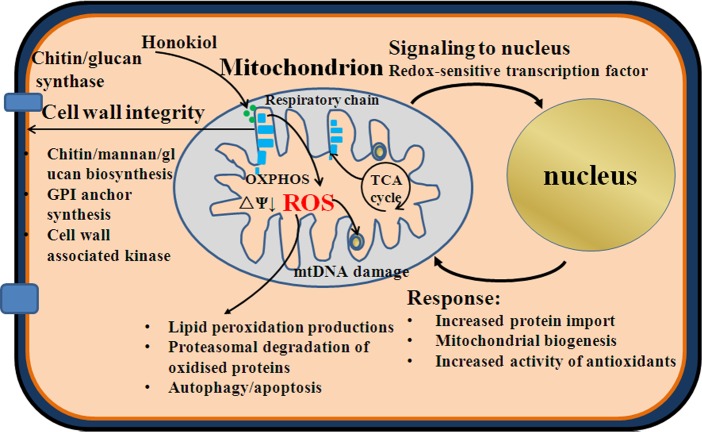
Fig 10. A model of the cellular response to honokiol treatment.
Honokiol targets on the mitochondrial respiratory electron transport chain C I inducing ROS production (mainly O2·−) and mitochondria dysfunction. ROS induces mtDNA damage, mtΔψ decrease, oxidation of proteins and lipids. The transcriptome indicates that the cellular stress response increases including increased protein import to mitochondria, upregulate the expression of antioxidant enzyme genes, remove oxidized proteins by the ubiquitin-dependent proteasome system. Mitochondrial dysfunction in C. albicans is associated with loss of cell wall integrity. (1) affect the activity of wall synthesis enzymes in plasma membrane; (2) impinge on GPI anchor synthesis; (3)activation of cell wall integrity pathway upon cell wall stress. mtΔψ: mitochondrial membrane potential.
There are many lines of evidence to suggest that mitochondrial dysfunction in C. albicans is known to be linked with loss of cell wall integrity [27–29]. The fungal cell wall has vital roles in growth, survival, morphogenesis, and pathogenicity. Critical for the coordination of these activities is the dynamic nature of the cell wall, its ability to respond to external and internal stimuli [43]. Altered sensitivities to cell wall perturbing agents indicate modified cell wall properties [43,44]. In our study, honokiol treatment resulted in an increased sensitivity to Congo red and SDS (Fig 6). In addition, the genes for cell wall synthesis have three major changes in transcriptional levels (Fig 8): 1). chitin/mannan/glucan biosynthesis, such as upregulated CHS2 which encodes chitin synthase and BMT3 which encodes β-mannosyltransferase is required for β-mannan elongation either in phosphopeptidomannan or in phospholipomannan [44]. Reduced glucanase gene expression, for example, SKN1 encoding a N-glycosylated type II membrane protein with a predicted role in β-1,6-glucan synthesis was downregulated after honokiol exposure; 2). GPI anchor protein, such as PGA31, EXG2, PGA32, PGA57, and PGA37 were upregulated. GPI-anchored proteins are important cell surface proteins in C.albicans, and they have important impact on the adhesion,morphogenesis and cell wall synthesis [44]; 3). cell wall associated kinase. such as PSK1(PAS kinase), a putative serine/threonine protein kinase, was upregulated after honokiol exposure. It has been reported that inhibition of cell wall biogenesis by caspofungin also causes an increase in PSK1 expression [44]. Honokiol treatment caused mitochondrial dysfunction and then damaged the cell wall integrity, thus, increased sensitivity to cell wall perturbing agents.
The present work shows that honokiol may target mitochondrial respiratory chain C I, leading to mitochondrial dysfunction, accompanied by increased cellular superoxide anion and oxidative stress (Fig 10). Use of antifungal agents that are mitochondrial respiratory chain C I inhibitors can also be toxic to mammalian cells associated with aging and age-related diseases such as Parkinson’s disease [45]. However, potential side effects of mitochondrial respiratory chain inhibitors as antifungal agents can be reduced if effective dosage can be diminished. Such a reduction can be achieved by use of honokiol as a chemosensitizer to azoles agents or echinocandins. This work not only provides insights on the mechanism by which honokiol interferes with fungal cell, demonstrating previously unknown effects on mitochondrial physiology, but also raises a note of caution on the use of M. officinalis as a Chinese medicine due to the toxic for mitochondria and suggests the possibility of using honokiol as chemosensitizer.
Supporting information
(DOC)
(XLSX)
Data Availability
The sequences reported in this paper have been deposited as raw reads in the GenBank SRA database (accession no. SRX2977968 and SRX2977967).
Funding Statement
This study was funded by the National Natural Science Foundation of China grants (81302814 - Dr. Lingmei Sun) and (81703574 - Kai Liao); Jiangsu Province Natural Science Foundation (20140624 - Kai Liao); Fundamental Research Funds for the Central Universities (2242017K40096 - Dr. Lingmei Sun); and Fundamental Research Funds for the Central Universities (2242017K40093 - Kai Liao). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alexander BD, Perfect JR. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs.1997; 54: 657–678. [DOI] [PubMed] [Google Scholar]
- 2.Dos SAP, McArthur CP, Africa CW. Multi-drug resistant oral Candida species isolated from HIV-positive patients in South Africa and Cameroon. Diagn Microbiol Infect Dis. 2014; 79: 222–227. doi: 10.1016/j.diagmicrobio.2013.09.016 [DOI] [PubMed] [Google Scholar]
- 3.Wisplinghoff H, Seifert H, Wenzel RP, Edmond MB. Current trends in the epidemiology of nosocomial bloodstream infections in patients with hematological malignancies and solid neoplasms in hospitals in the United States. Clin Infect Dis. 2003; 36: 1103–1110. doi: 10.1086/374339 [DOI] [PubMed] [Google Scholar]
- 4.Wisplinghoff H, Seifert H, Tallent SM, Bischoff T, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in pediatric patients in United States hospitals: epidemiology, clinical features and susceptibilities. Pediatr Infect Dis. 2003; J 22: 686–691. [DOI] [PubMed] [Google Scholar]
- 5.Beck-Sague C, Jarvis WR. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. National Nosocomial Infections Surveillance System. J Infect Dis. 1993; 167: 1247–1251. [DOI] [PubMed] [Google Scholar]
- 6.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009; 417: 1–13. doi: 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirst J, King MS, Pryde KR. The production of reactive oxygen species by complex I. Biochem Soc Trans. 2008; 36: 976–980. doi: 10.1042/BST0360976 [DOI] [PubMed] [Google Scholar]
- 8.Orr AL, Ashok D, Sarantos MR, Shi T, Hughes RE, Brand MD. Inhibitors of ROS production by the ubiquinone-binding site of mitochondrial complex I identified by chemical screening. Free Radic Biol Med. 2013; 65: 1047–1059. doi: 10.1016/j.freeradbiomed.2013.08.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014; 94: 909–950. doi: 10.1152/physrev.00026.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Fu X, Liu Y, Wang Y, Huo B, Guo Y, et al. Hypoglycemic effect and mechanism of honokiol on type 2 diabetic mice. Drug Des Devel Ther. 2015; 9: 6327–6342. doi: 10.2147/DDDT.S92777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jangra A, Dwivedi S, Sriram CS, Gurjar SS, Kwatra M, Sulakhiya K, et al. Honokiol abrogates chronic restraint stress-induced cognitive impairment and depressive-like behaviour by blocking endoplasmic reticulum stress in the hippocampus of mice. Eur J Pharmacol. 2016; 770: 25–32. doi: 10.1016/j.ejphar.2015.11.047 [DOI] [PubMed] [Google Scholar]
- 12.Pan J, Lee Y, Wang Y, You M. Honokiol targets mitochondria to halt cancer progression and metastasis. Mol Nutr Food Res. 2016; 60: 1383–1395. doi: 10.1002/mnfr.201501007 [DOI] [PubMed] [Google Scholar]
- 13.Wang XD, Wang YL, Gao WF. Honokiol possesses potential anti-inflammatory effects on rheumatoid arthritis and GM-CSF can be a target for its treatment. Int J Clin Exp Pathol. 2015; 8: 7929–7936. [PMC free article] [PubMed] [Google Scholar]
- 14.Hahm ER, Sakao K, Singh SV. Honokiol activates reactive oxygen species-mediated cytoprotective autophagy in human prostate cancer cells. Prostate. 2014; 74: 1209–1221. doi: 10.1002/pros.22837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Liao K, Hang C, Wang D. Honokiol induces reactive oxygen species-mediated apoptosis in Candida albicans through mitochondrial dysfunction. PLoS One. 2017; 12: e172228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dikalov S, Losik T, Arbiser JL. Honokiol is a potent scavenger of superoxide and peroxyl radicals. Biochem Pharmacol. 2008; 76: 589–596. doi: 10.1016/j.bcp.2008.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993; 134: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao J, Cao H, Liang D, Liu Y, Zhang H, Zhao H, et al. Taxol, a microtubule stabilizer, prevents ischemic ventricular arrhythmias in rats. J Cell Mol Med. 2011; 15: 1166–1176. doi: 10.1111/j.1582-4934.2010.01106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi T, Mito T, Watanabe N, Suzuki T, Shiraishi A, Ohashi Y. Use of 5-cyano-2,3-ditolyl-tetrazolium chloride staining as an indicator of biocidal activity in a rapid assay for anti-Acanthamoeba agents. J Clin Microbiol. 2012; 50: 1606–1612. doi: 10.1128/JCM.06461-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun LM, Liao K, Liang S, Yu PH, Wang DY. Synergistic activity of magnolol with azoles and its possible antifungal mechanism against Candida albicans. J Appl Microbiol. 2015; 118: 826–838. doi: 10.1111/jam.12737 [DOI] [PubMed] [Google Scholar]
- 21.Skrzypek MS, Arnaud MB, Costanzo MC, Inglis DO, Shah P, Binkley G. et al. New tools at the Candida Genome Database: biochemical pathways and full-text literature search. Nucleic Acids Res. 2010; 38: D428–D432. doi: 10.1093/nar/gkp836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25: 1105–1111. doi: 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31: 166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26: 139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006; 34: W720–W724. doi: 10.1093/nar/gkl167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomes A, Fernandes E, Lima JL. Fluorescence probes used for detection of reactive oxygen species. J Biochem Biophys Methods. 2005; 65: 45–80. doi: 10.1016/j.jbbm.2005.10.003 [DOI] [PubMed] [Google Scholar]
- 27.She X, Khamooshi K, Gao Y, Shen Y, Lv Y, Calderone R, et al. Fungal-specific subunits of the Candida albicans mitochondrial complex I drive diverse cell functions including cell wall synthesis. Cell Microbiol. 2015; 17: 1350–1364. doi: 10.1111/cmi.12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.She X, Zhang L, Chen H, Calderone R, Li D. Cell surface changes in the Candida albicans mitochondrial mutant goa1Delta are associated with reduced recognition by innate immune cells. Cell Microbiol. 2013; 15: 1572–1584. doi: 10.1111/cmi.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, She X, Calderone R. Functional diversity of complex I subunits in Candida albicans mitochondria. Curr Genet. 2016; 62: 87–95. doi: 10.1007/s00294-015-0518-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondaryk M, Kurzatkowski W, Staniszewska M. Antifungal agents commonly used in the superficial and mucosal candidiasis treatment: mode of action and resistance development. Postepy Dermatol Alergol. 2013; 30: 293–301. doi: 10.5114/pdia.2013.38358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang C, Dong D, Yu B, Cai G, Wang X, Ji Y, et al. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J Antimicrob Chemother. 2013; 68: 778–785. doi: 10.1093/jac/dks481 [DOI] [PubMed] [Google Scholar]
- 32.Lehan PH, Furcolow ML, Brasher CA, Larsh HW. Therapeutic trials with the newer antifungal agents. Antibiot Annu. 1956; 467–469. [PubMed] [Google Scholar]
- 33.Johnson MD, Perfect JR. Use of antifungal combination therapy: agents, order, and timing. Curr Fungal Infect Rep. 2010; 4: 87–95. doi: 10.1007/s12281-010-0018-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin J, Guo N, Zhang J, Ding Y, Tang X, Liang J, et al. The synergy of honokiol and fluconazole against clinical isolates of azole-resistant Candida albicans. Lett Appl Microbiol. 2010; 51: 351–357. doi: 10.1111/j.1472-765X.2010.02900.x [DOI] [PubMed] [Google Scholar]
- 35.Teodoro GR, Ellepola K, Seneviratne CJ, Koga-Ito CY. Potential use of phenolic acids as anti-Candida agents: A Review. Front Microbiol. 2015; 6: 1420 doi: 10.3389/fmicb.2015.01420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Sanchez R, Hernandez-Esquivel L, Rivero-Segura NA, Marin-Hernandez A, Neuzil J, Ralph SJ, et al. Reactive oxygen species are generated by the respiratory complex II-evidence for lack of contribution of the reverse electron flow in complex I. FEBS J. 2013; 280: 927–938. doi: 10.1111/febs.12086 [DOI] [PubMed] [Google Scholar]
- 37.Scialo F, Sriram A, Fernandez-Ayala D, Gubina N, Lõhmus M, Nelson G, et al. Mitochondrial ROS produced via reverse electron transport extend animal lifespan. Cell Metab. 2016; 23: 725–734. doi: 10.1016/j.cmet.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunnon K, Keohane A, Pidsley R, Newhouse S, Riddoch-Contreras J, Thubron EB, et al. Mitochondrial genes are altered in blood early in Alzheimer's disease. Neurobiol Aging. 2017; 53: 36–47. doi: 10.1016/j.neurobiolaging.2016.12.029 [DOI] [PubMed] [Google Scholar]
- 39.Evdokimovsky EV, Ushakova TE, Kudriavtcev AA, Gaziev AI. Alteration of mtDNA copy number, mitochondrial gene expression and extracellular DNA content in mice after irradiation at lethal dose. Radiat Environ Biophys. 2011; 50: 181–188. doi: 10.1007/s00411-010-0329-6 [DOI] [PubMed] [Google Scholar]
- 40.Pereira GC, Branco AF, Matos JA, Pereira SL, Parke D, Perkins EL, et al. Mitochondrially targeted effects of berberine [Natural Yellow 18, 5,6-dihydro-9,10-dimethoxybenzo(g)-1,3-benzodioxolo(5,6-a) quinolizinium] on K1735-M2 mouse melanoma cells: comparison with direct effects on isolated mitochondrial fractions. J Pharmacol Exp Ther. 2007; 323: 636–649. doi: 10.1124/jpet.107.128017 [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, De Micheli M, Coleman ST, Sanglard D, Moye-Rowley WS. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol Microbiol. 2000; 36: 618–629. [DOI] [PubMed] [Google Scholar]
- 42.Ayer A, Gourlay CW, Dawes IW. Cellular redox homeostasis, reactive oxygen species and replicative ageing in Saccharomyces cerevisiae. FEMS Yeast Res. 2014; 14: 60–72. doi: 10.1111/1567-1364.12114 [DOI] [PubMed] [Google Scholar]
- 43.Tada R, Latge JP, Aimanianda V. Undressing the fungal cell wall/cell membrane—the antifungal drug targets. Curr Pharm Des. 2013; 19: 3738–3747. [DOI] [PubMed] [Google Scholar]
- 44.Plaine A, Walker L, Da CG, Mora-Montes HM, McKinnon A, Gow NA, et al. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet Biol. 2008; 45: 1404–1414. doi: 10.1016/j.fgb.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Distelmaier F, Koopman WJ, van den Heuvel LP, Rodenburg RJ, Mayatepek E, Willems PH, et al. Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain. 2009; 132: 833–842. doi: 10.1093/brain/awp058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(XLSX)
Data Availability Statement
The sequences reported in this paper have been deposited as raw reads in the GenBank SRA database (accession no. SRX2977968 and SRX2977967).