INTRODUCTION
Students often struggle when they are expected to apply the facts they have learned to their conceptual understanding of the cellular consequences of biological processes (1). I describe a hands-on classroom activity employed in a junior-level genetics course that engages students in the practical implications of the central dogma of molecular biology as well as extending central dogma understanding by exploring the consequences of changing the genetic material. This activity was performed during the “Regulation of Eukaryotic Gene Expression” section of the course, after students had learned about transcription, translation, and mutation.
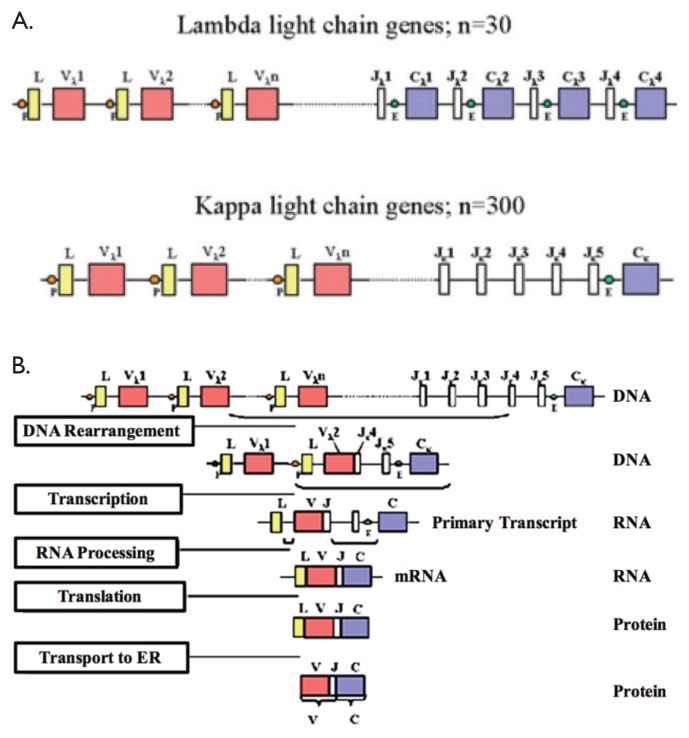
Students model the central dogma of molecular biology in this activity. They form a simulated VDJ coding region, using recombination, transcription, and translation to polypeptide. The process of VDJ recombination involves random recombination of a chromosome (Fig. 1A) to remove large segments of the genome (Fig. 1B “DNA Rearrangement”) to generate a chromosome capable of being transcribed to pre-mRNA (Fig. 1B “Transcription”). This pre-mRNA will be spliced to a final product (“RNA Processing”) that will be translated (“Translation”) to produce polypeptides to incorporate into antibodies unique to each B cell lineage. After students “recombine” their paper chromosome to resemble Figure 1B, they transcribe the mRNA and then use a codon table to deduce the final polypeptide sequence.
FIGURE 1.
B Cell Receptor Structure and Recombination. A) Chromosomal gene structure of B Cell Receptor (BCR) light chain genes before recombination. Lines denote introns and rectangles denote potential coding regions. B) Process of BCR kappa light chain rearrangement. Recombination produces a gene structure in which one linker, one variable, one joining, and the constant region are translated after transcription and splicing. Used with permission from Mayer (2). P = promoter; L = leader; V = variable region; J = joining region; C = constant region; E = enhancer.
This activity integrates students’ understanding of DNA structure and function, recombination, transcription, and translation into one compact exercise that reinforces the enduring understanding of gene mutation and gene product formation. Students are further asked to reflect on the similarities and differences in their variable regions to reinforce the concepts of DNA mutation leading to polypeptide changes, as students must repeat, apply, extend, and reflect to truly internalize a concept (3). This activity differs from others already published as it combines chromosomal structure, function, and recombination with central dogma. This exercise could be an excellent companion to many existing central dogma classroom activities, such as performing transcription and translation with paper ribosomes, tRNA, and amino acids (4–6) or beads representing amino acids (7). The exercise described in Norflus and Allen (8), a depiction of VDJ recombination using models and animations that focuses on the recombination process, would be a terrific addition to the described activity. These two exercises differ, as the present exercise spotlights the changes to the protein product as chromosome structural changes are made, while Norflus and Allen’s activity covers the recombination process itself. Alternatively, students could model VDJ recombination with pipe cleaners (9) and then perform this activity to deduce the polypeptide consequences of the recombination. This described activity concentrates on the gene product, as contrasted with the techniques described in (8,9) that are constructed around the process (8) and structural outcome (9) of VDJ recombination. The novelty of this activity arises from its integration of the kinesthetic activity of chromosome assembly and folding with its fusion of deliberate iteration to illustrate the central dogma of molecular biology.
At the end of this activity students should be able to:
Describe how recombination leads to enhanced genetic variability.
Apply the central dogma of molecular biology by relating DNA to mRNA to polypeptide sequence.
Analyze how changes in DNA lead to changes in gene products.
Instructor procedures
The instructor describes VDJ recombination to students, similar to Figure 1, and can extend activities as in (8,9).
Students are given the handout (Fig. 2A and Supplementary Material). It is a paper model with six linker/variable regions, five joining regions, and one constant region, simulating the chromosomal region of the B cell kappa light chain.
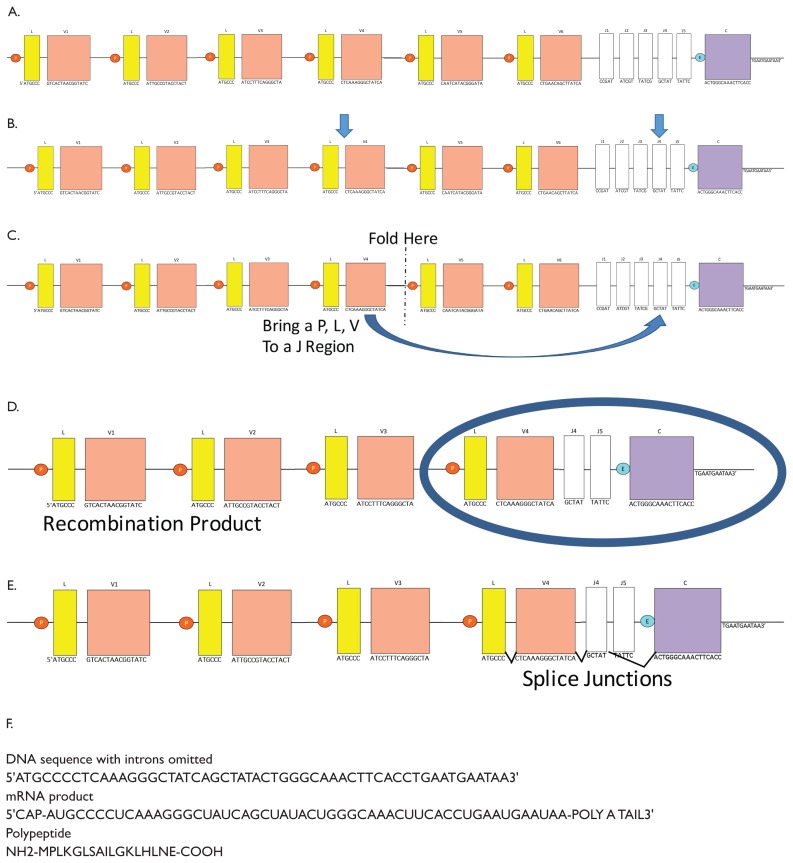
The instructor demonstrates how to fold the model (Fig. 2A–C).
Students are directed to use the paper model to simulate DNA recombination to explore the concepts of the central dogma of molecular biology (Figs. 2 and 3). The paper should be folded such that a linking and variable region (Fig. 1A, L and V) is brought close to a joining region (J) to model the process outlined in Figure 1. Students’ products should differ (examples in Fig. 3).
FIGURE 2.
Instructions on how to fold the “chromosome.” A) Full length chromosome. B) P (promoter), L (linker), V (variable), and J (joining) regions chosen. C) Fold the left side of the paper toward the right side, so that a PVL region unit is brought close to a J region. D) Example recombination product with coding region circled. E) Chromosome after recombination with splice junctions shown; the closest promoter to the enhancer region (E) is used. F) example DNA, mRNA, and polypeptide results.
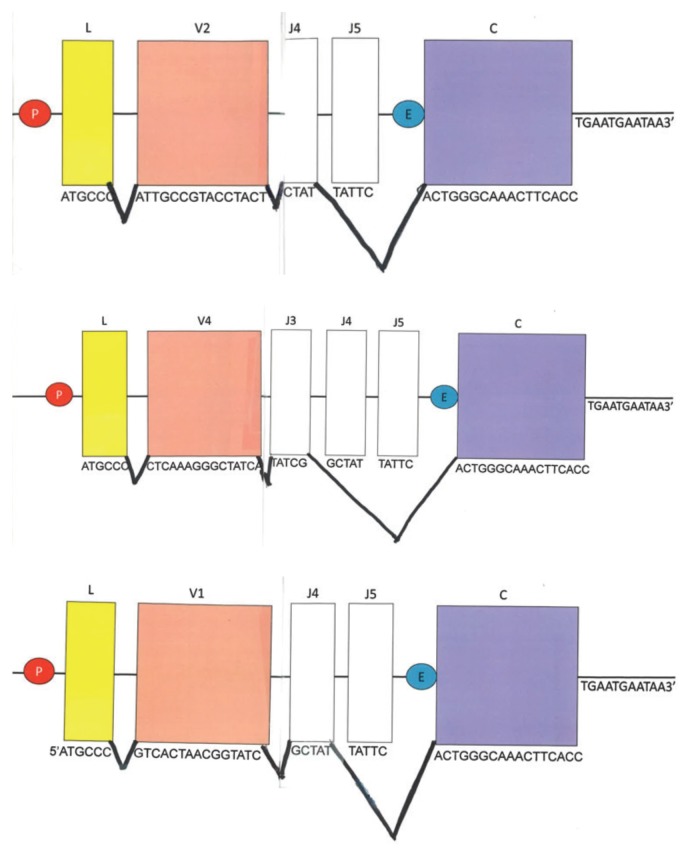
FIGURE 3.
Example assignment folds. One P (promoter) – L (linker) – V (variable) region is recombined next to a J (joining region). This DNA model is then transcribed and translated to produce the mock kappa chain. Note the introns are not shown in the student handout. This is to focus the student on the application of the central dogma. Splicing is represented under the DNA strand.
Student procedures
With the associated handout (resembling Fig. 2A, found in the Supplemental Material), you are to generate 4 (four) kappa light chains, using Figures 1 and 2 as a guide.
Tape the pages of the handout together to form a structure that looks like Figure 2A (A to B to C to D, left to right).
Note the structure of the chromosome. It has lines to indicate introns, and blocks to indicate potential coding region exons. L = leader, V = variable, J = joining, and C = constant. E indicates an enhancer region, and P is a promoter region.
Choose a P/L/V combination and a J for your chromosome. In our example in 2B, we have chosen P/L/V4 and J4, denoted with arrows.
Now fold your paper chromosome (Fig. 2C) to bring your chosen L/V next to the chosen J region, as in 2D.
The part of the chromosome that has the P region closest to your E region becomes the coding region, circled in 2D.
Notice that your new recombination product has both introns and exons. Insert splice junctions between the L and V you chose, the J you chose, and the C. Notice that you may be skipping one or more J regions, as in 2E. Each product should have only one J region.
Now generate your sequences. Figure 2F shows examples generated from Figure 2E.
Once you have your sequences, write a paragraph to compare and contrast the four kappa light chains you have produced. What are the similarities and differences? Focus on the DNA, mRNA, and polypeptides produced. This section should have at least one sentence per comparison, for a total of six. You’ll need an introductory sentence or two and a concluding sentence as well. The concluding sentence needs to be a meta-analysis of what you have learned by doing this activity.
Rubric
Students earn full credit if all directions are followed: four recombination products are turned in, each product has an associated mRNA and polypeptide sequence labeled correctly, and the paragraph is complete and has introductory and concluding sentence(s). Rubrics for both the exercise and the essay can be found in Appendix 3.
CONCLUSION
Implementation
This activity focuses on using VDJ recombination as a tool to generate different genes and guides students through the process of developing gene products. I have encountered one difficulty with the activity: students were sometimes confused as to how to assemble the paper and then “recombine” the molecule. A short instructor-led demonstration showing how the linker and variable region should be recombined next to a joining region enhances students’ understanding of the way the model should be assembled (Fig. 2 and Fig. 3). Instructor concerns may include the volume of grading, since each student is generating four unique recombination products and the associated mRNA and polypeptides. There are many websites that will transcribe and translate input DNA that instructors can use to grade the assignment. Alternatively, to further enhance the students’ hands-on time with the activity, students can swap their recombinants and grade each other’s for accuracy.
Educational implications
Students are shown the codon table and are expected to be able to utilize it to translate a polypeptide in class levels that range from mid-grades to high school to first year of college. However, students struggle with the practical application of the codon table to a DNA sequence. Students are required to develop, transcribe, and translate several polypeptides to reinforce the concept that changes in DNA lead to changes in mRNA that in turn lead to changes in the polypeptide sequence. Furthermore, having the students write about the relationship between the gene and the gene product helps to activate additional areas of the brain to promote long-term retention and conceptual understanding (10).
SUPPLEMENTAL MATERIALS
ACKNOWLEDGMENTS
The author declares that there are no conflicts of interest.
Footnotes
Supplemental materials available at http://asmscience.org/jmbe
REFERENCES
- 1.Tanner K, Allen D. Approaches to biology teaching and learning: understanding the wrong answers—teaching toward conceptual change. CBE Life Sci Educ. 2005;4:112–117. doi: 10.1187/cbe.05-02-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer G. Genetics of immunoglobulins. In: Hunt RC, editor. Microbiology and immunology on-line. University of South Carolina School of Medicine; Columbia, SC: 2003. http://www.microbiologybook.org/mayer/IgGenetics2000.htm. [Google Scholar]
- 3.Felder RM, Brent R. Teaching and learning STEM: a practical guide. Jossey Bass; San Francisco, CA: 2016. [Google Scholar]
- 4.McLean JL, Schuman E. Using magnets and classroom flipping to promote student engagement and learning about protein translation in a large microbiology class. J Microbiol Biol Educ. 2016;17(2):288–289. doi: 10.1128/jmbe.v17i2.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yung S, Primm T. Nucleotide manipulatives to illustrate the central dogma. J Microbiol Biol Educ. 2015;16(2):274–277. doi: 10.1128/jmbe.v16i2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall K, Dunitz J, Shields Build-a-polypeptide: a hands-on worksheet to enhance student learning in an introductory biology course. J Microbiol Biol Educ. 2014;15:307–309. doi: 10.1128/jmbe.v15i2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBruyn JM. Teaching the central dogma of molecular biology using jewelry. J Microbiol Biol Educ. 2012;13(1):62–64. doi: 10.1128/jmbe.v13i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norflus F, Allen NC. Use of computer models and animations to teach about B Cell (antibody) and T Cell recombination (TCR) J Microbiol Biol Educ. 2016;17:292–293. doi: 10.1128/jmbe.v17i2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breer M, Christensen B, Taylor J. Models in movies: teaching abstract concepts in concrete models. J Microbiol Biol Educ. 2012;13(1):80–82. doi: 10.1128/jmbe.v13i1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunel M, Hand B, Prain V. Writing for learning in science: a secondary analysis of six studies. Int J Sci Math Educ. 2007;5:615–637. doi: 10.1007/s10763-007-9082-y. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.