Significance
Whereas new biomolecular assays are continually being developed to achieve higher detection sensitivities, specificity is often the primary factor limiting an assay’s detection threshold—especially in single-label/signal amplification-based assays. This paper describes a sensitive and quantitative protein detection method that makes use of a dual-reporter approach with enhanced ability to distinguish true positives from false positives. This strategy gives rise to excellent sensitivity and accuracy of measurement, even for low target protein concentrations in complex biological samples.
Keywords: surface-enhanced Raman spectroscopy, SERS, protein detection, nanoparticle, ELISA
Abstract
We present a sensitive and quantitative protein detection assay that can efficiently distinguish between specific and nonspecific target binding. Our technique combines dual affinity reagents with surface-enhanced Raman spectroscopy (SERS) and chemometric analysis. We link one Raman reporter-tagged affinity reagent to gold nanoparticles and another to a gold film, such that protein-binding events create a “hot spot” with strong SERS spectra from both Raman reporter molecules. Any signal generated in this context is indicative of recognition by both affinity labels, whereas signals generated by nonspecific binding lack one or the other label, enabling us to efficiently distinguish true from false positives. We show that the number of hot spots per unit area of our substrate offers a quantitative measure of analyte concentration and demonstrate that this dual-label, SERS-linked aptasensor assay can sensitively and selectively detect human α-thrombin in 1% human serum with a limit of detection of 86 pM.
The ELISA remains an indispensable tool for detecting specific target molecules in complex biological samples (1, 2). ELISA’s reliability results from the use of two affinity reagents to capture and sandwich the target protein as a requisite for generating a reporter signal, an approach that reduces false-positive signals. This “sandwich” assay structure has inspired many groups to develop hybrid versions of ELISA that incorporate novel reporting techniques such as PCR, optical detection, and surface-enhanced Raman spectroscopy (SERS) to further improve sensitivity (3–7).
In conventional sandwich assays, the target is captured by one affinity reagent and then detected with a second affinity reagent that is linked to a reporter. The reporter signal is amplified, generating a readout that is proportional to the concentration of target as determined using a previously derived calibration function. When a reporter-linked affinity reagent binds nonspecifically to the substrate, however, it gives rise to false positives that are indistinguishable from true protein capture events (8, 9). It is obvious that the use of a single reporter-conjugated affinity reagent would generally produce a greater frequency of false positives than an assay based on two affinity reagents (both labeled with reporters), as a positive report from the former assay cannot be as confidently associated with the binding to a target protein.
In this work, we describe a sandwich-style protein detection assay that uses two Raman reporters, each conjugated with a distinct affinity reagent. This approach is less susceptible to false positives, resulting in a significantly lower limit of detection (LOD) compared with its single-reporter–based analog. To use SERS to detect binding events, we conjugate one affinity reagent to gold nanoparticles (AuNPs) and the other to a gold metal film. The AuNP-Au film sandwich structure formed in the presence of the target molecule becomes a SERS hot spot, generating a strong and unambiguous signal that indicates a true positive (10). One can analyze the SERS signals by using the traditional “analog” method to correlate the intensity of one Raman reporter to the known protein concentration, thereby developing an intensity vs. concentration calibration curve (5, 6, 10, 11). In contrast, we have opted for an improved “digital” method, in which many SERS spectra are collected in a predetermined area of the gold surface under computer control, after which true positives can be identified by using a classical least-squares (CLS) approach to measure the number of sites that report significant contributions from both reporters. Sites with no contributions from either reporter are rejected as producing no signal, whereas sites that indicate only a single reporter are rejected as false positives. In addition to being automated, the digital approach has the added benefit that one can increase the number of sites from which SERS spectra are collected, trading increased assay time for improved sensitivity and/or accuracy as required. Using this “SERS-linked sandwich assay” approach, we successfully detected human α-thrombin and show that our dual-tag approach reduces the LOD to 86 pM relative to the 248-pM LOD achieved with a single-tag approach. Additionally, we show that this approach can be used with an aptamer–antibody pair to detect tumor necrosis factor alpha (TNF-α), demonstrating the general applicability of this technique.
Results and Discussion
Assay Design and Analysis.
The assay begins with the conjugation of two different affinity reagents, each with its own Raman reporter, to AuNPs and gold film, respectively (Fig. 1A). In principle, this assay can be performed with any affinity reagent that can readily be linked to gold, including aptamers, antibodies, and peptides. For this initial demonstration, we chose a well-characterized thrombin aptamer pair because aptamers can be readily synthesized with an attached thiol group, allowing for easy conjugation to gold surfaces to form self-assembled monolayers, and are selected to bind to their targets with high affinities (12–14). The AuNPs were functionalized with the thrombin-binding aptamer TBA-29, labeled with the 4-nitrobenzenethiol (NBT) Raman reporter, whereas the gold film was functionalized with the thrombin-binding aptamer TBA-15 conjugated to methylene blue (MB) (15, 16). We initially incubated solutions containing various concentrations of the target protein, thrombin, and a nontarget protein (albumin) with AuNPs to enable target binding. This mixture was then transferred to the gold film; thrombin molecules that have bound to AuNPs will form a sandwich upon binding to the MB-TBA-15 aptamers on the gold surface, generating a hot spot that enhances the SERS signals of the two reporters (Fig. 1A). The sequence of incubation is important as an allosteric effect has been observed by Olmsted et al. (17), where the TBA-15 binding affinity increases when the thrombin is first bound to TBA-29. Quality-control tests were performed on random batches of AuNP probes and films to confirm reproducibility and functionality, as shown in Fig. S1.
Fig. 1.
Detection of human α-thrombin. (A) AuNP probes functionalized with TBA-29 and the Raman label NBT capture the target in solutions that contain various concentrations of thrombin and a nontarget protein, albumin. These assemblies are in turn captured by a film functionalized with MB-labeled TBA-15 to form SERS hot spots. Spectral data are collected within a designated area of the gold film by raster scanning under a Raman microscope with a 633-nm incident laser. (B) The collection of spectral data per each concentration of thrombin tested is analyzed by CLS regression to determine the contribution of each reporter to the spectra and thereby determine true protein-binding events vs. false positives. Physical locations of true- and false-positive binding events are represented in a heat map.
Fig. S1.
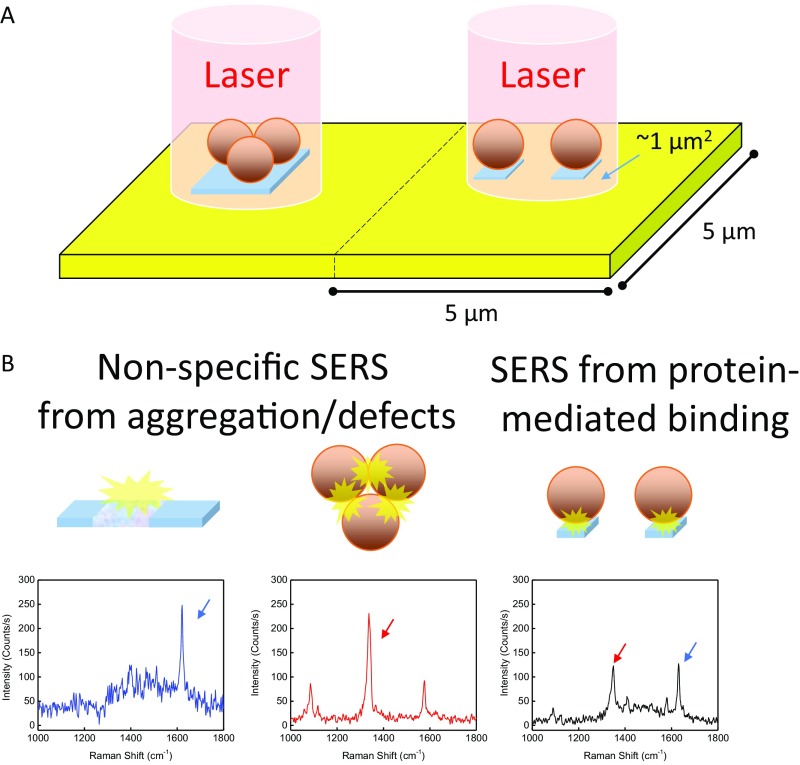
Potential sources of SERS hotspots. (A) The center of each 25-μm2 area is interrogated by a 1-μm2 focused laser beam. (B) Three types of hot spots are possible in the assay: (Left) false positives originating from defects in the film, which are dominated by the MB Raman label, with a strong peak at 1,630 cm−1; (Center) false positives from AuNP aggregates, which are dominated by the NBT Raman label with a strong peak at 1,337 cm−1; and (Right) true-positive protein-mediated binding, indicated by near-equal enhancement of MB and NBT peaks.
We collected assay data by rastering a 55-µm × 55-µm area of the gold film under a Raman microscope at 5-µm intervals, producing an 11 × 11 array of spectra totaling 121 SERS spectra per sampled area (Fig. 1A). The spectrum associated with each of the 25-μm2 areas is produced by one or more hot spots located within the 1-μm2 area irradiated by the laser (Fig. S1A). Three sets of 121 spectra were collected for each thrombin concentration. The collection of these 363 spectra, each from a different location on the substrate, took 6 min and provided sufficient precision for all samples tested over the 0.1- to 10-nM thrombin concentration range studied. Under these conditions, the entire assay requires just over 1 h in total, with 30 min required for each of the AuNP capture and gold film-binding steps, before the 6-min detection process. Finally, using CLS analysis, the spectra are categorized in terms of whether they result from one, two, or no reporters (Fig. 1B). By selectively looking at results by one or both reporters, we were able to conduct both single- and dual-tag analyses (see below) from the same set of measurements.
By design, neither the smooth gold film nor the AuNPs alone are able to produce strong SERS signals from their respective Raman reporters in the absence of target (Fig. S2 A and B). Assuming that a strong SERS signal will arise only from hot spots, we tested two analysis methods. In the analog single-tag method, the intensity of the AuNP-conjugated reporter was measured to be proportional to the concentration of the target. In the dual-tag method, strong SERS signal could indicate one of three possibilities (Fig. S1). First, it could be a false positive arising from AuNP aggregates that form either in solution before addition to the gold film or after addition onto the gold surface. Although such aggregates produce strong SERS spectra, these spectra show large contributions from the NBT reporter on the AuNP, because the hot spots are located primarily at the interstices between adjacent nanoparticles rather than between AuNPs and the gold surface. Second, false positives may arise from nanoscale defects in the film, such as very small clefts in the gold film, which are likely a result of sample handling. These primarily display the SERS spectrum of the MB reporter that was used to tag the gold film surface. Finally, there are true positives arising from protein-mediated binding of AuNPs to the gold film, which produce intense SERS spectra corresponding to both Raman tags. Examples of such SERS spectra, which have notably different contributions from those generated by the two individual tags, are shown in Fig. S1B. We used CLS to quantitatively determine the contribution of each individual reporter, making it possible to select only those true-positive signals arising from strong contributions from both reporters.
Fig. S2.
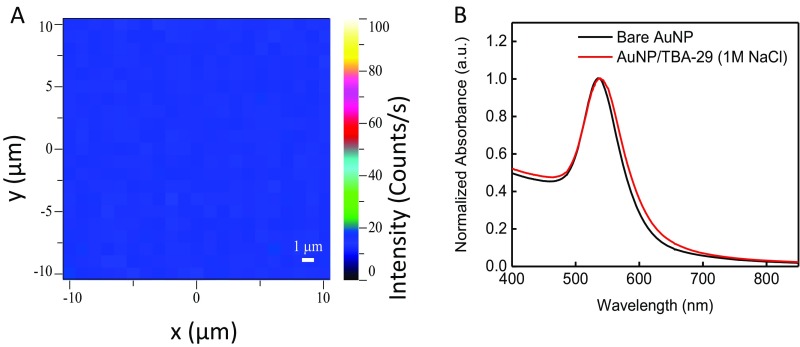
Au film and AuNPs were randomly inspected for quality control. (A) Heat map of gold film with a monolayer of MB-TBA-15. Pristine Au films have barely detectable (less than 30 counts/s) MB signal (1,630 cm−1) when excited by the 633-nm laser. The low-intensity peaks arise from both the MB molecule and the gold film in resonance with the 633-nm laser. (B) UV-visible spectroscopy of AuNPs before and after TBA-29 and NBT functionalization. When functionalization is performed properly, a slight peak shift to 542 nm is observed due to the new oligo shell on the AuNP. No significant absorbance is observed at 700 nm (indication of unstable/aggregated particles) of functionalized particles in 1 M NaCl.
Sensitivity of the SERS Assay to Changing Target Concentrations.
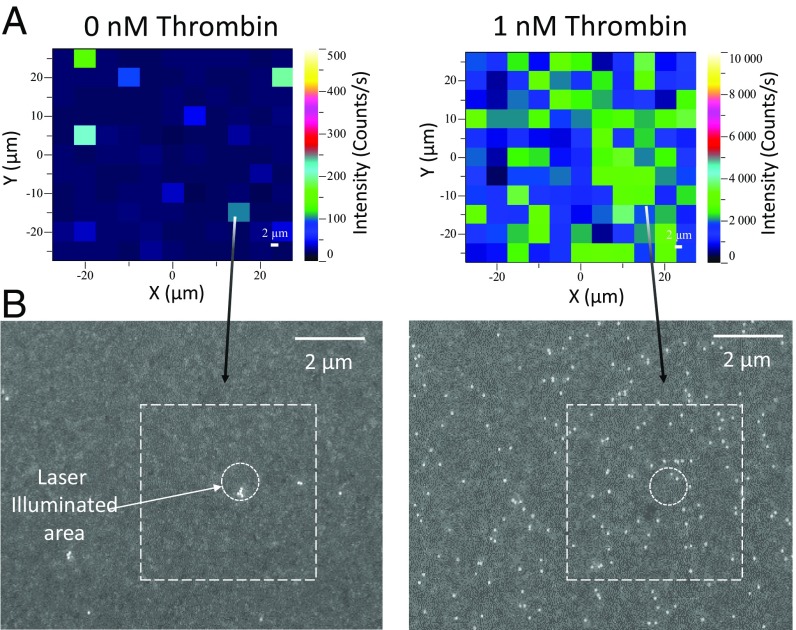
We examined how SERS signal intensity changes as a function of thrombin concentration, which also helped us to establish the baseline resulting from nonspecific binding. We performed assays with known thrombin concentrations (0 nM, 0.1 nM, 0.2 nM, 0.5 nM, 1 nM, 5 nM, and 10 nM) in a buffered salt solution containing 5 µM BSA, equivalent to the albumin concentration found in 1% human serum. Fig. 2A illustrates the relative increase of the 1,337-cm−1 SERS peak (the NO2 stretching vibration) produced by NBT-conjugated AuNPs in the absence (Fig. 2A, Left) or presence (Fig. 2A, Right) of thrombin. Scanning electron micrographs (SEM) of the same areas (Fig. 2B and Fig. S3) confirm the increased number of AuNPs bound to the film in the presence of thrombin. A major advantage of the SERS assay is that nonspecific binding of particles is very rare in our system due to the electrostatic repulsion by the dense layer of oligonucleotides on the Au surface. The assay proved to be sensitive over a wide range of target molecule concentrations, and measurable SERS signals were observed even from smaller numbers of AuNPs on the film, confirming that the assay is sensitive at low target concentrations (Fig. S3).
Fig. 2.
(A) False-color heat map of the intensity of the 1,337-cm−1 band, indicating SERS hot spots measured for (Left) 0 nM and (Right) 1 nM thrombin solution. Maximum intensity for each map is (Left) 500 counts/s and (Right) 10,000 counts/s. The different scale ranges are indicative of the assay’s sensitivity to thrombin. (B) Scanning electron micrographs of the same samples.
Fig. S3.
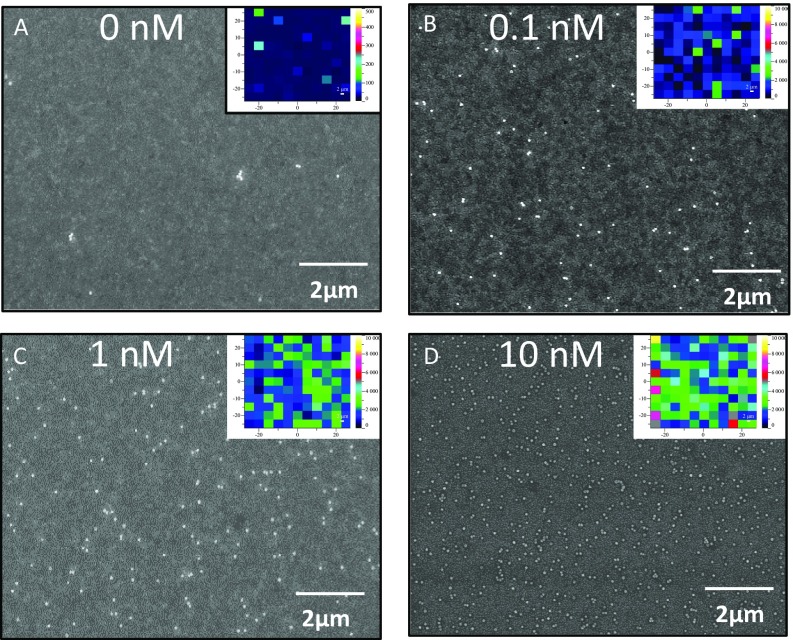
SEM and heat map (Insets) of thrombin assays. (A–D) Maps displaying assay results with 0 nM, 0.1 nM, 1 nM, and 10 nM thrombin in buffer. Maximum intensity scale for A is 500 counts, whereas for B–D maximum intensity is 10,000 counts, indicating strong SERS with increased protein binding.
In the absence of thrombin, very few AuNPs bound to the Au surface compared with samples containing thrombin, indicating very low levels of nonspecific binding. Those few particles primarily consisted of AuNP aggregates, whereas the vast majority of AuNPs remained dispersed in samples with thrombin (Fig. 2B). This important feature can help distinguish true from false positives in the CLS analytical process. Although heat map and micrograph analyses are helpful as a rough visual assessment of target binding at moderate or high concentrations of thrombin, the modest background NBT SERS signal observed in the absence of thrombin (Fig. 2A) limits detection capabilities at low thrombin concentrations, highlighting the primary drawback of single-reporter assays. This limitation is greatly ameliorated by quantitatively determining each of the two reporters’ relative contribution to each spectrum, using CLS regression. This presents a systematic breakdown of the signal composition, revealing the underlying configuration of the hot spot that produces such a spectrum. This allows us to locate signals produced by aggregate outliers and reveals that intense spectra in samples without thrombin primarily arise from a single reporter.
Identification of False Positives and Creation of a Calibration Curve from True-Positive Results.
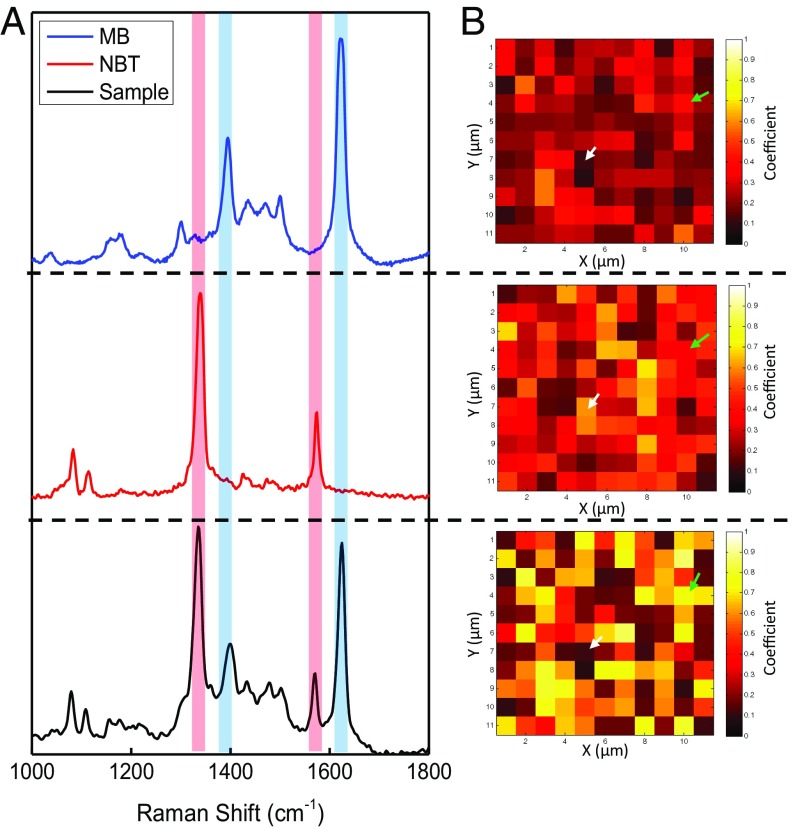
For CLS analysis, we fitted a linear combination of the individual SERS spectra of the two reporter molecules (Fig. 3A, Top and Middle) plus an error residual to correct for spectra showing minimal SERS features obtained from the raster scans (Fig. 3A, Bottom). Details are specified in Data Processing. Fig. 3A, Bottom shows a representative spectrum from a single rastered spot, 1 of 363 total spectra measured for the 0.5-nM thrombin sample. Careful examination of the peaks in the combined spectrum reveals a slight difference in the ratio of major to minor peak heights shown in the reference spectra (Fig. 3 A and B). Whereas both tags have major peaks with distinct Raman shifts, some of the minor peaks are common to both reporters. This means that when both molecules are enhanced together, the intensity of these overlapping peaks is summed and may therefore differ relative to the minor peak intensities observed in the SERS spectra from the individual reporters. The magnitudes of the two coefficients returned by the CLS analysis for each rastered spot are assumed to be proportional to the relative contribution of each tag at that location. In cases where the spectra showed very weak or no SERS signals, CLS returned low contributions for the two reporters and high residual values due to poor fit. These data were categorized as blanks. In the coefficient heat map shown in Fig. 3B, those locations are given the darkest color. The signals detected were divided into three categories depending on the reporter contributions present in the recorded spectra. AuNP aggregates nonspecifically bound to the surface of the gold film in the absence of target produce a SERS signal dominated by NBT. Likewise, surface defects in the gold film produce signals that are dominated by the SERS spectrum of MB. In contrast, SERS hot spots created by protein binding tend to be more evenly spaced across the surface of the film and produce spectra with relatively equal contributions from both MB and NBT (Fig. S3). Spectra containing both Raman reporters with a signal-to-noise ratio equal to or exceeding 3:1 were counted as true positives. We constructed a calibration curve by plotting the percentage of true positives of a scanned region vs. the known thrombin concentration (Fig. 4A). The calibration curve is monotonic relative to thrombin concentration; although the curve is linear at low concentrations, this linearity is lost at higher concentrations (>1 nM). This is because at low thrombin concentrations, most of the 1-μm2 illuminated areas reporting true positives contain a single AuNP, whereas at higher concentrations a single laser spot is likely to illuminate several AuNPs at once, such that the assay is no longer digital. The remaining spectra containing either one of the two tags can either be ignored or be further analyzed to determine the source of the nonspecifically produced signals. We emphasize that no background subtraction was required, due to the inherently ultralow nonspecific binding of our system, which represents a major advantage of our methodology.
Fig. 3.
Graphical summary of the results of the CLS analysis used to determine the relative contributions of the two Raman tags for a representative assay with 0.5 M thrombin. (A) SERS spectra in the range from 1,000 cm−1 to 1,800 cm−1, showing reference SERS spectra of (Top) MB, (Middle) NBT, and (Bottom) a single representative spectrum from the raster scans after an assay has been conducted. The red and blue shaded bars highlight prominent peaks associated with NBT and MB, respectively. (B) Heat maps based on the coefficients returned by CLS analysis for (Top) MB, (Middle) NBT, and (Bottom) the sum of the NBT and MB coefficients for sites with above-threshold values. Brighter color indicates a greater contribution by that tag. Green arrows indicate an example of a true-positive sector, where both reporter coefficients are above threshold. White arrows show an example of a false-positive sector, where one of the two reporters is below threshold.
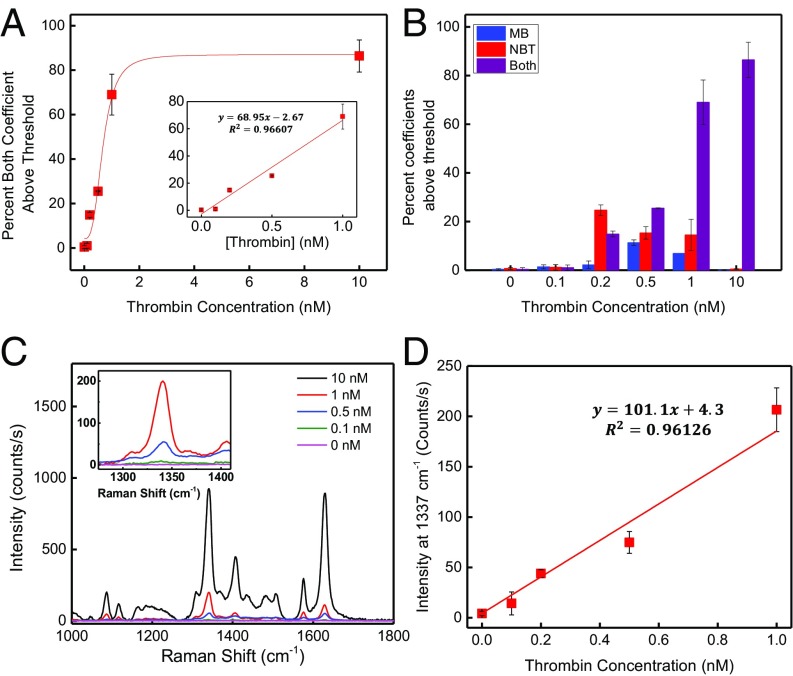
Fig. 4.
Summary of results from single- and dual-reporter SERS assays. (A) Standard thrombin-binding curve in 5 µM albumin for the dual-reporter assay. Inset shows the low-concentration portion of the graph, expanded for clarity. (B) Summary of results from CLS analysis expressed as percentage of contribution from each tag in the dual-reporter assay. Blue bars (MB only) indicate signal derived from film defects, whereas red bars (NBT only) indicate spectra derived from AuNP aggregates. Purple bars indicate true protein-binding events. (C) Average intensity of all 121 spectra as a function of thrombin concentration for the single-reporter SERS assay. Inset shows the 1,337-cm−1 region expanded for clarity. (D) Intensity of the 1,337-cm−1 peak as a function of thrombin concentration for the single-reporter assay.
For samples with no thrombin, we found that an average of 0.6% of spectra contained strong NBT contributions. This represents the false-positive rate derived solely from nonspecific binding of AuNP aggregates, as shown in Fig. 2A. An additional 0.4% of the spectra exhibited high values for the MB coefficient with subthreshold contribution from NBT, representing hot spots produced by defects in the gold film. Thus, a total of 1% of the rastered area produced false-positive signals derived from nonspecific sources. Importantly, only 0.3% of the spectra showed above-threshold contributions from both NBT and MB tags.
Using this approach, we were able to generate a reliable binding curve as a function of thrombin concentration in solution (Fig. 4A), which yielded an LOD of 86 pM (18). The LOD was obtained by taking the signal from a blank sample and adding it to the SD obtained from the signal recorded from the lowest tested sample concentration (100 pM). As shown in Fig. 4B, spectra containing only one tag occur with approximately double the frequency of spectra containing two tags, especially at low target concentrations. The frequency of false positives is further validated by the results obtained from analyzing the data from a one-reporter approach. We produced a calibration curve by plotting the average intensity of the 1,337-cm−1 band corresponding to the NBT-conjugated AuNPs for the 121 collected spectra and triplicates against the known thrombin concentration (Fig. 4 C and D). With one-reporter assays, any detectable signal is considered to be produced by a protein capture; from the data shown in Fig. 2A, we know this to be a false assumption. In the range of 0–1.0 nM, the SERS intensity increased linearly with thrombin concentration in our one-reporter assay. However, the LOD was calculated to be 248 pM in this scenario, a 2.9-fold decrease in sensitivity relative to the two-reporter assay. This decrease in assay sensitivity can be attributed to the inability of the one-reporter assay to reject false-positive signals and demonstrates the extent to which nonspecific binding in the one-reporter system can confound assay sensitivity.
To demonstrate how the sensitivity can be altered by simply changing parameters, we challenged the assay with 50 pM thrombin, which is below the detection limit of this assay (86 pM). As shown in Fig. S4, in a collection of 300 spectra, the total count for a 50-pM sample is indistinguishable from that of a 100-pM sample due to high levels of error. However, upon increasing the number of spectra collected to 1,000 spectra (and thereby increasing the collection time from 6 min to ∼17 min), the error decreases, and this makes it possible to confidently distinguish the difference between a 50-pM sample and a 100-pM sample. With 2,000 spectra (collection time of ∼33 min), the difference is drastic. An increase in binding is also noted when the binding time for AuNP-target complex to film is increased. By further increasing the number of sites from which spectra are collected or increasing target incubation time during the assay, one can further improve the sensitivity and resolution of the assay at the expense of greater assay time (19).
Fig. S4.
Improving sensitivity by increasing interrogation area or assay time. (A) The error or deviation can be tuned in our assay by simply increasing the number of sites from which spectra are collected. As the number of collected spectra increases, the error decreases and the resolution of the assay improves, resulting in the ability to distinguish 50 pM and 100 pM, at the expense of greater data collection time. (B) If the assay is given greater time to proceed, more particles in solution are bound to the film at lower concentrations. This is demonstrated by performing the assay with 0.1 nM thrombin. By extending the time to allow the AuNP–thrombin complex to bind onto the Au film for 60 min rather than 30 min, we observe nearly double the total binding.
Detection in Diluted Serum.
Finally, we investigated how the assay performs in a biologically relevant medium, by testing its capacity to detect thrombin in 1% human serum. Serum is collected by allowing the blood plasma to clot until factors are depleted, after which the clot is removed by centrifugation. Although serum theoretically should be free of clotting factors, trace amounts remain after extraction. To determine how much endogenous thrombin is in the sample, we obtained a reference value using a commercial dual-antibody ELISA kit, with which we determined the concentration to be 183 ± 4 pM. When we analyzed the same sample using the one-reporter approach, we observed a low-intensity NBT signal, but this was indistinguishable from noise. This observation is unsurprising, given that the measured concentration is below the assay’s LOD of 248 pM. Using the two-reporter approach, we determined the concentration to be 111 ± 24 pM after testing seven individually prepared samples, which is within the range reported by the ELISA.
Detection of TNF-α.
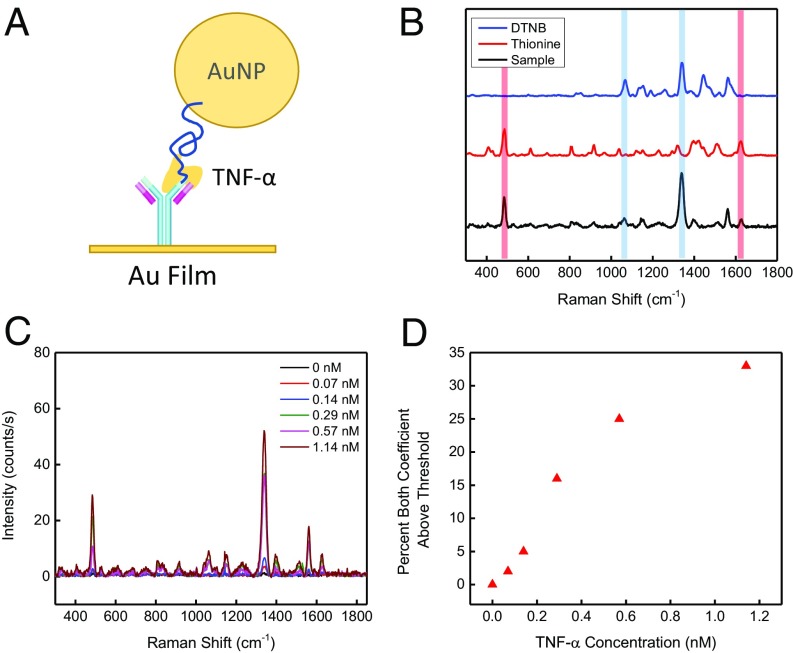
We chose to use aptamers in this initial demonstration of the SERS assay for their robustness, ease of chemical modification, and cost-effectiveness, but our assay design can also be used with other affinity reagents. We performed a proof of concept assay with a commercially available aptamer–antibody pair for TNF-α (Fig. 5A). We used the same procedure described above to prepare AuNPs functionalized with a TNF-α aptamer and thionine as Raman label. For the gold film, we used 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), a Raman label with a carboxylic acid functional group, to label the anti–TNF-α antibodies (Fig. 5B). As with the thrombin assay, the SERS signal intensity changes as a function of TNF-α concentration with equally sparse nonspecific binding (Fig. 5C). We performed assays with known TNF-α concentrations (0 nM, 0.07 nM, 0.14 nM, 0.29 nM, 0.57 nM, and 1.14 nM) in a buffered salt solution containing 5 µM BSA. As before, we obtained a total of three replicates of 121 spectra to produce a standard curve of true TNF-α binding events. We observed that binding increases monotonically relative to TNF-α concentrations (Fig. 5D) and thereby demonstrated the ease with which our assay could be applied to other targets.
Fig. 5.
Detection of TNF-α using an antibody–aptamer reagent pair. (A) Schematic of the TNF-α detection system, in which labeled antibodies are immobilized on the AuNP film and TNF-α aptamers (blue line) are immobilized on AuNPs. (B) The AuNPs and gold films were respectively labeled with the Raman tags thionine and DTNB. (C) As the concentration of TNF-α protein increases in the solution, an increase in Raman intensity of both labels is observed. (D) Standard TNF-α binding curve in a background of 5 µM albumin.
Data Processing
SERS signals were collected at 5-µm intervals over a 50-µm × 50-µm area on the gold film, yielding a total of 121 spectra. Three samples were measured for each thrombin concentration tested. The following thrombin concentrations were tested in a mixture with 5 µM BSA: 0 nM, 0.1 nM, 0.2 nM, 0.5 nM, 1 nM, 5 nM, and 10 nM. The resulting data were subjected to three types of analysis.
Single-Reporter Analysis.
We created a calibration curve that plots the mean intensity of one or more selected Raman bands against the known concentration of the analyte. In our experiments, the 121 individual spectra were averaged, and the intensity of the mean 1,337-cm−1 band corresponding to the NBT label on the AuNPs was plotted against the known thrombin concentration.
Dual-Reporter Analysis.
The obtained reference spectra and replicates of the 121 SERS spectra from each thrombin sample were normalized over the 1,000-cm−1 to 1,800-cm−1 spectral range, which includes most of the prominent features associated with MB and NBT (Materials and Methods). CLS analysis determined the values of the two coefficients for each of the two components (MB and NBT), each ranging in value between 0 and 1, which best fitted the experimentally measured SERS spectra over the 1,000-cm−1 to 1,800-cm−1 range, plus an additional baseline value. We established threshold values for the two coefficients of 0.14 for NBT and 0.2 for MB, reflecting the relative contributions of these two reporters to the overall spectrum, by assessing the minimum values that give rise to a peak signal-to-noise ratio of 3:1. MB has an absorption maximum near 660 nm (23), and upon irradiating the substrate with the 633-nm laser, we observed strong MB fluorescence in the spectra even in the absence of target. To avoid counting this natural fluorescence as true MB reporting, we set the MB threshold higher to 0.2, at which point we no longer observed false-positive contributions from MB.
The data were then separated into three categories: (i) neither reporter above threshold (zero), (ii) only one reporter above threshold (false positive), and (iii) both reporters above threshold (true positive). The total number of spectra in each category was determined, and the fraction of spectra above threshold was correlated with the known thrombin concentration.
Reporter Coefficient Heat Map.
To visualize the separation of the coefficients in the scanned area, as in Fig. 3B, we replotted the coefficients determined from the two-reporter analysis according to their location coordinates. Fig. 3B, Top represents the coefficient of NBT for the 121 scanned sectors, whereas Fig. 3B, Middle represents the coefficient of MB for the same areas. We used three steps to enhance those pixels in Fig. 3B, Bottom. First, sectors below threshold for both reporters had their coefficients automatically set to 0 to represent no signal. Second, pixels or spectra above threshold for both reporters had their coefficients summed. Finally, pixels with one or the other tag below threshold were represented by the corresponding coefficient that was below threshold. Using this method, we could visually map true binding events. It should be noted that this method was solely used for visualization purposes and was not used to produce the calibration curve.
LOD Calculation.
We computed the LOD for the assay as follows:
where LOB (limit of blank) is obtained by
in which the average signal of the blank (the sample with 0 nM thrombin) is added to the 5% false-negative rate (1.645*SD).
Based on our calibration curve, LOB = 0.059 nM and LOD = 0.0858 nM.
Conclusions
We describe a sensitive, specific method for quantitative target detection that combines the ELISA dual affinity reagent sandwich architecture with SERS. The SERS-based technique uses a pair of affinity reagents, each labeled with a Raman reporter; one is conjugated to AuNPs, whereas the other is bound to a gold surface. The simultaneous binding of the protein to the two affinity reagents produces a SERS hot spot for each binding event. The data are analyzed by using CLS analysis to quantify the results and eliminate false positives. In an initial demonstration with thrombin as a target, we achieved a LOD of 86 pM, and we further show that our assay can be applied to other targets, such as TNF-α. CLS analysis of these spots made it possible to distinguish true-positive SERS spectra produced by both reporters from false positives produced by only a single reporter. In this initial iteration our thrombin assay delivered a LOD that was higher than that of ELISA (4 pM), which we attribute primarily to the fact that ELISA has an amplification step (through HRP) whereas our method is a direct readout of a single-particle binding event. However, our method is capable of detecting physiologically relevant target concentrations in complex mixtures and has a powerful capability to distinguish between true and false binding. Furthermore, our assay is considerably faster than ELISA, and we have demonstrated a fourfold shorter assay time in this work. This is because it requires fewer washing steps and, as noted above, is less prone to nonspecific binding. Finally, we have demonstrated that higher sensitivity can be obtained by either increasing incubation time to induce more AuNP–target complexes to bind, and thereby shifting the monotonic region toward the lower concentration, or increasing the area interrogated to reduce the deviation. This produces a lower LOD while still resulting in a faster assay than ELISA with the capacity to discriminate between true and false binding. In this way, we anticipate that future iterations of our assay will deliver both greater speed and higher sensitivity.
Materials and Methods
Materials.
AuNP assembly.
All reagents were purchased from Sigma Aldrich and used as received except where specified. For nanoparticle assembly, we used bis(p-sulfonatophenyl)phenylphosphine dihydratedipotassium salt (phosphine ligand, 99%), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (500 mM), NBT (80%), 6-mercapto-1-hexanol (MCH) (99%), 3,7-diamino-5-phenothiazinium acetate (thionine) (dye content >85%), 60 nm citrate-capped AuNPs (2.6 × 1010 particles/mL; Ted Pella), methanol, thrombin-binding aptamer (TBA-29; Integrated DNA Technologies), and TNF-α binding aptamer (Integrated DNA Technologies).
Film assembly.
We used thrombin-binding aptamer (MBA-TBA-15; Biosearch Technologies), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (99%), N-hydroxysuccinimide (NHS) (97%), DTNB (98%; Molecular Probes), BSA, TNF-α antibody (MAb11; eBioscience), acetone, isopropanol, silicon wafer (University Wafer), and a 96-well plate (Corning).
Buffers were diluted with ultrapure water at 18.2 MΩ·cm (EMD Millipore). The buffers used include 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (Hepes) (1 M), Tris-EDTA (TE) (pH 8.0; Invitrogen), 20× saline sodium citrate buffer (SSC) (3 M NaCl, 300 mM sodium citrate, pH 7.0; Invitrogen), 7.5 M ammonium acetate solution, and 1× phosphate-buffered saline (PBS) (pH 7.0; Invitrogen).
Prepared assay buffer (50 mM Trizma, 140 mM sodium chloride, 100 mM potassium chloride, and 1 mM magnesium chloride, pH 7.2), 50 mM potassium phosphate buffer (KPB), human serum (male, AB type), 2-(N-morpholino)ethanesulfonic acid hydrate (Mes) (pH 5), human α-thrombin (Haematologic Technologies), and TNF-α (R&D Systems) were diluted in assay buffer.
Oligonucleotide Deprotection.
All oligonucleotides (Integrated DNA Technologies and Biosearch Technologies) were purchased at the 1-μM scale in lyophilized form. The 5′ protection groups were removed by reacting with 1 mL 100 mM TCEP in pH 8.0 TE buffer overnight at room temperature (RT), followed by purification through NAP-10 columns (GE Healthcare). The purified oligo solution was concentrated to 250 μM with Amicon Ultra-4 centrifugal filters (EMD Millipore) in TE buffer for long-term storage at −20 °C. Before AuNP or gold film conjugation, the oligo aliquots were annealed in 10 mM TCEP at 95 °C for 5 min to reduce potential disulfide bonds and to break up secondary, hydrogen-bonded structures that the aptamers may have adopted during storage.
Oligonucleotide sequences are shown in Table S1. MB-TBA-15 and TBA-29 were used in the assays for the detection of thrombin. MB-TBA-15 aptamers were immobilized onto gold film, and TBA-29 aptamers were immobilized on AuNP. TNF-α aptamers were immobilized onto AuNP for the TNF-α assays.
Table S1.
Aptamer sequences used in the assays
| Aptamer | Conjugation substrate | Sequence |
| MB-TBA-15 | Gold film | 5′ S-(CH2)6-/MB/iSp18/TTTTTG GTT GGT GTG GTT GG -3′ |
| iSp18 is a hexa-ethyleneglycol spacer for increasing flexibility and density of aptamer on the gold film (24) | ||
| TBA-29 | 60 nm AuNP | 5′ S-(CH2)6-AGT CCG TGG TAG GGC AGG TTG GGG TGA CT-3′ |
| TNF-α | 60 nm AuNP | 5′ S-(CH2)6-ATC CAG AGT GAC GCA GCATGC TTA AGG GGG GGG CGG GTT AAG GGA GTG GGG AGG GAG CTG GTG TGG ACA CGGTGG CTT AGT -3′ |
Oligonucleotide sequences are shown 5′–3′. MB-TBA-15 and TBA-29 were used in the assays for the detection of thrombin. MB-TBA-15 aptamers were immobilized onto gold film, and TBA-29 aptamers were immobilized on AuNP. TNF-α aptamers were immobilized onto AuNP for the TNF-α assays.
Preparation of Aptamer–Nanoparticle Conjugates.
All incubations under rotation were performed at RT on an Eppendorf rotisserie rotator (6 rpm). All centrifugation (Eppendorf minispin) steps were performed at 4,300 relative centrifugal force (rcf) for 15 min. AuNP concentrations were verified using UV-vis spectroscopy, calculated with absorbance at 540 nm and an extinction coefficient of 3.5 × 1010 L⋅mol−1⋅cm−1 (20). The aptamer–AuNP conjugation was conducted as follows.
We performed ligand exchange on purchased citrate-capped AuNPs to stabilize the particles during aptamer immobilization. A total of 1 mL 60 nm AuNPs from Ted Pella stock was centrifuged to remove storage buffer, resuspended in 1 mL aqueous solution of 0.9 mM phosphine ligand, and rotated overnight. We linked the thiol-conjugated aptamer to AuNPs following a modified protocol based on the procedure described by Zhang et al. (21). Briefly, the resulting phosphine-capped AuNPs were concentrated to 2 × 1011 particles/mL (100 µL) by centrifugation, to which we added 3.2 µL (1.2 µL TBA-29 or TNF-α taken from 250 µM stock without dilution and 2 µL of 500 mM TCEP) of the annealed solution to achieve a final aptamer concentration of 3 μM. To encourage DNA binding, the solution pH was adjusted to 3.0, using sodium citrate (final 10 mM NaCit). After 20 min rotation, the pH was neutralized to pH 7.0 with 1 M Hepes buffer to a final concentration of 130 mM Hepes. To maximize the quantity of aptamer bound to the surface, the particles were salt aged by adding 20× SSC in 300-mM increments every 20 min to a final NaCl concentration of ∼1 M. Along with the first 300-mM SSC addition, we added 50 mM KPB to help the aptamers form G-quadruplex structures and minimize overpacking (15, 22). The AuNPs were sonicated in a bath (Branson Ultrasonic Bath) as needed to prevent aggregation. Finally, the AuNPs were put on the rotator to age overnight in the salt solution. We used Raman label (NBT for thrombin experiments and thionine for TNF-α experiments) and MCH to label and passivate, respectively, the remaining, uncoated Au surface. This was done by adding NBT in methanol (1 mM stock) and MCH (1 mM stock) in ethanol simultaneously to the Au colloidal solution to a final concentration of 100 µM and 5 μM, respectively, after which we rotate the solution for 2 h at RT. For TNF-α experiments, we used an identical labeling procedure but using thionine dissolved in water (1 mM stock) rather than NBT in methanol. After 2 h, the AuNP conjugates were isolated by centrifugation and washed in KPB three times to ensure complete removal of unbound reagents. Finally, the concentration of particles was adjusted to 350 pM in assay buffer, and this mixture was then used immediately. We noted that aptamers were displaced from the particles over the course of storage (Fig. S5), and particles were therefore prepared fresh for each experiment.
Fig. S5.
Percentage of coverage of TBA-29 aptamer on 60 nm AuNPs as a function of time. When particles are freshly functionalized with TBA-29 and NBT, there is an average of 1,830 ± 335 TBA-29 strands per particle. The theoretical maximum coverage is ≈1,880 strands of TBA-29 per 60-nm particle, assuming the footprint of the TBA-29 G-quadruplex to be 3 × 2 nm2. As particles are stored, the coverage decreases from full coverage to around 50% by week 4. For consistency, particles are always made fresh and used immediately for the assay.
Preparation of MB-TBA-15 Modified Monolayer Au Film.
All incubations under rotation were performed at RT on an orbital rotator in a 96-well plate. As with AuNPs, to prevent oligo displacement over storage, aptamer immobilization was therefore prepared fresh for each experiment.
The MB-TBA-15 aptamers were immobilized onto the film through thiol-gold conjugation. Gold films were prepared by electron-beam deposition (Temescal) on a 4-inch silicon wafer. The wafer was cleaned (bath sonication in acetone, isopropanol, and water for 3 min each) and ozone/plasma scrubbed (Novascan) for 3 min. A 30-nm titanium film was deposited as an adhesion layer at the following rates: 0.05 nm/s to 3 nm and 0.1 nm/s to 30 nm. A total of 300 nm of Au were layered on top of the titanium layer at the following rates: 0.1 nm/s to 10 nm, 0.2 nm/s to 30 nm, and 0.4 nm/s to 300 nm. The wafer was diced using a dicing saw (Advanced Dicing Technologies) into 4 × 4-mm squares and stored under nitrogen. Immediately before use, the Au films were cleaned by sonication in isopropanol for 3 min and UV-ozone treated for 20 min. For every substrate prepared, 50 µL 2 μM MB-TBA-15 was annealed in 10 mM TCEP at 95 °C for 10 min. The annealed oligos were added to the cleaned gold substrates in individual 96-well plates. After 10 min of incubation, 1 µL 500 mM sodium citrate, pH 3.0, was added to the well to lower the pH for 20 min, followed by neutralization with 6.6 µL of 1 M Hepes to reach a final concentration of 130 mM. Finally, 57.6 µL 20× SSC was added to the same well to encourage maximum packing of MB-TBA-15 on the gold films, resulting in a final solution concentration of 1 μM MB-TBA-15 and 1.5 M NaCl. The films were salt aged under these conditions on a covered plate at RT overnight on an orbital rotator. The next morning, the films were washed three times with 300 µL of 50 mM KPB, delivered by pipette, followed by 300 µL of 30 mM NaOH for 3 min to remove nonspecifically adhered aptamers. For each wash, substrates were moved to a new well. Finally, the films were washed with water and submerged in assay buffer for 10 min to prime the films for use in the assay.
Preparation of Mab11 Modified Au Film.
For TNF-α detection, the Mab11 films are prepared using the same 4-mm × 4-mm diced Au chips described above. After cleaning the Au films by sonication in isopropanol and UV-ozone treatment, the films were immediately submerged in 200 µL of 5 mM DTNB in absolute ethanol in a well on a 96-well plate and reacted overnight. The next day, the films were washed three times with ethanol, followed by two washes in 0.1 M Mes buffer, pH 5. A total of 200 µL of freshly prepared EDC and NHS (400 µM and 100 µM, respectively, in 0.1 M Mes buffer) was added to the films to activate the carboxylic acid functional group for 1 h. The films were washed once with 0.1 M Mes buffer, after which 100 µL of 5 µg/mL MAb11 in PBS was added to each film and incubated for 2 h. The substrates were washed with PBS three times. Finally, to block uncoated regions of the gold film, 200 µL of 1% BSA in PBS was added and stored for at least 12 h at 4 °C for use the following day.
Protein Sandwich Assay.
The modified AuNPs were added to solutions containing 0 nM, 0.1 nM, 0.2 nM, 0.5 nM, 1 nM, 5 nM, and 10 nM thrombin (or 0 nM, 0.07 nM, 0.14 nM, 0.29 nM, 0.57 nM, and 1.14 nM TNF-α) as well as either 5 µM albumin in assay buffer or 1% human serum to a final colloidal concentration of 50 pM and a final volume of 100 µL. The solution was mixed for 30 min at RT on an orbital rotator to allow the AuNPs to capture the target in solution. The 100-µL modified AuNP and protein mixtures were added to the gold films in fresh wells and incubated on an orbital shaker at RT for 30 min. After three washes with assay buffer, followed by two washes with KPB to remove unbound molecules, the films were quickly submerged in 300 mM ammonium acetate to remove excess salts that may hinder microscopic imaging. The film was dried under a nitrogen stream. Raman data were collected as described below, followed by SEM imaging.
Obtaining Reference Spectra.
Reference spectra shown in Fig. 3A were obtained by adding 1 µM Raman tag to 100 µL of 100 pM AuNPs after the phosphine ligand exchange step. The particles were incubated for 1 h, after which the tagged particles were dropcasted onto a clean gold film and allowed to dry. Reference SERS spectra were obtained by measuring the AuNP aggregates at the “coffee-ring” edges.
SERS Measurements.
SERS measurements were performed on a LabRam Aramis system (Horiba Jobin-Yvon) equipped with a CCD detector thermoelectrically cooled to −70 °C. The sample was excited with the 633-nm HeNe laser (Melles Griot) through a 50× objective lens with a 600-μm hole, a 400-μm slit width, and a 600-lines/mm grating. The laser beam spot size was roughly 1 μm2, with 0.87 mW laser irradiation power. The exposure was set to 1 s per spot.
Scanning Electron Microscopy.
Electron micrographs were obtained from representative Au film post assay and SERS measurement on an FEI XL40 Sirion FEG digital scanning microscope. Images were taken using 10.0 kV voltage at 10,000× magnification, using ultrahigh resolution.
Thrombin ELISA.
The thrombin ELISA (Abnova; KA0511) was performed per manufacturer instructions on 1% human serum to determine the concentration of endogenous thrombin in the sample. Briefly, a 50-µL thrombin standard was prepared at 20 ng/mL. A twofold serial dilution was performed using the provided EIA dilution buffer to obtain a total of seven standard points (0.313–20 ng/mL) and a blank containing only the EIA dilution buffer. A total of 50 µL of the standards, seven individually prepared 1% human serum samples, and 5 µM albumin (negative control) were added to the provided 96-well plate and allowed to incubate for 2 h at RT. After washing the wells five times with a wash buffer, 50 µL of biotinylated thrombin antibody was added per well and incubated for 1 h. The wells were washed five times before 50 µL of an SP conjugate was added per well and incubated for 30 min. The wells were again washed five times before adding 50 µL of chromogen substrate per well and incubated for 10 min. Finally, the chromogenic reaction was stopped using a stop solution. The wells were read at 450 nm immediately afterward, using the TECAN Infinite M1000. The data were fitted with a four-parameter logistic curve fit per manufacturer’s suggestion.
Statistical/Data Analysis.
We used classical least-squares analysis from the PLS_toolbox software (Eigenvector Research Inc.), accessed within the MATLAB computational environment, to analyze the spectral data. NBT and MB reference spectra were used to construct the model. Reference spectra were baselined and normalized by area within the software, resulting in a model that was used to assess spectra for relative contributions of the two reporters.
Acknowledgments
We thank Dr. Binghui Wu for his insightful critique of the manuscript and Brian Evanko for advice on presenting heat map results. Funding was provided by the Institute for Collaborative Biotechnologies through Contract W911NF-09-D-0001 from the US Army Research Office, by Grant N66001-14-2-4055 from the Defense Advanced Research Projects Agency, and by Contract W911NF-10-2-0114 from the US Army Medical Research and Material Command, Defense Medical Research and Army Research Office. Additional funding from Omnis Global Technologies is acknowledged and greatly appreciated. H.T.S. is a Chan Zuckerberg Biohub Investigator. The content of the information does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700317114/-/DCSupplemental.
References
- 1.Gervais L, de Rooij N, Delamarche E. Microfluidic chips for point-of-care immunodiagnostics. Adv Mater. 2011;23:H151–H176. doi: 10.1002/adma.201100464. [DOI] [PubMed] [Google Scholar]
- 2.Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 3.de la Rica R, Stevens MM. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat Nanotechnol. 2012;7:821–824. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- 4.Tsai CT, Robinson PV, Spencer CA, Bertozzi CR. Ultrasensitive antibody detection by agglutination-PCR (ADAP) ACS Cent Sci. 2016;2:139–147. doi: 10.1021/acscentsci.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon J, et al. Highly sensitive detection of thrombin using SERS-based magnetic aptasensors. Biosens Bioelectron. 2013;47:62–67. doi: 10.1016/j.bios.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Femtomolar detection of prostate-specific antigen: An immunoassay based on surface-enhanced Raman scattering and immunogold labels. Anal Chem. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 7.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogt RF, Jr, Phillips DL, Henderson LO, Whitfield W, Spierto FW. Quantitative differences among various proteins as blocking agents for ELISA microtiter plates. J Immunol Methods. 1987;101:43–50. doi: 10.1016/0022-1759(87)90214-6. [DOI] [PubMed] [Google Scholar]
- 9.Song Y, et al. Point-of-care technologies for molecular diagnostics using a drop of blood. Trends Biotechnol. 2014;32:132–139. doi: 10.1016/j.tibtech.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang K, Hu Y, Dong N. A novel biosensor based on competitive SERS immunoassay and magnetic separation for accurate and sensitive detection of chloramphenicol. Biosens Bioelectron. 2016;80:373–377. doi: 10.1016/j.bios.2016.01.064. [DOI] [PubMed] [Google Scholar]
- 11.Baniukevic J, et al. Magnetic gold nanoparticles in SERS-based sandwich immunoassay for antigen detection by well oriented antibodies. Biosens Bioelectron. 2013;43:281–288. doi: 10.1016/j.bios.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Deng B, et al. Aptamer binding assays for proteins: The thrombin example–a review. Anal Chim Acta. 2014;837:1–15. doi: 10.1016/j.aca.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 13.Jayasena SD. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 14.Chiu TC, Huang CC. Aptamer-functionalized nano-biosensors. Sensors. 2009;9:10356–10388. doi: 10.3390/s91210356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 16.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 17.Olmsted IR, et al. Measurement of aptamer-protein interactions with back-scattering interferometry. Anal Chem. 2011;83:8867–8870. doi: 10.1021/ac202823m. [DOI] [PubMed] [Google Scholar]
- 18.Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29:S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford AC, Skuratovsky A, Porter MD. Sampling error: Impact on the quantitative analysis of nanoparticle-based surface-enhanced Raman scattering immunoassays. Anal Chem. 2016;88:6515–6522. doi: 10.1021/acs.analchem.6b01263. [DOI] [PubMed] [Google Scholar]
- 20.Mulvaney P. Surface plasmon spectroscopy of nanosized metal particles. Langmuir. 1996;12:788–800. [Google Scholar]
- 21.Zhang X, Servos MR, Liu J. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J Am Chem Soc. 2012;134:7266–7269. doi: 10.1021/ja3014055. [DOI] [PubMed] [Google Scholar]
- 22.Nagatoishi S, Tanaka Y, Tsumoto K. Circular dichroism spectra demonstrate formation of the thrombin-binding DNA aptamer G-quadruplex under stabilizing-cation-deficient conditions. Biochem Biophys Res Commun. 2007;352:812–817. doi: 10.1016/j.bbrc.2006.11.088. [DOI] [PubMed] [Google Scholar]
- 23.Singhal GS, Rabinowitch E. Changes in the absorption spectrum of methylene blue with pH. J Phys Chem. 1967;71:3347–3349. [Google Scholar]
- 24.Balamurugan S, Obubuafo A, Soper SA, McCarley RL, Spivak DA. Designing highly specific biosensing surfaces using aptamer monolayers on gold. Langmuir. 2006;22:6446–6453. doi: 10.1021/la060222w. [DOI] [PubMed] [Google Scholar]