Significance
Chemotherapy is a double-edged sword. It is anticancer because of its cytotoxicity. Paradoxically, by increasing chemoresistance and cancer metastasis, it is also pro-cancer. However, the mechanisms underlying chemotherapy-induced procancer activities are not well understood. Here we present data that provide mechanistic explanations for the ability of paclitaxel (PTX), a frontline chemotherapeutic agent, to exacerbate metastasis in mouse models of breast cancer. Importantly, Atf3, a stress-inducible gene, in the noncancer host cells is necessary for this PTX effect. Analyses of publicly available datasets suggest that our data from mouse models have relevance to human cancers. Thus, ATF3 links a chemotherapeutic agent—a stressor—to pro-metastatic changes in the host cells. Dampening the effect of ATF3 may improve the efficacy of chemotherapy.
Keywords: chemotherapy, metastasis, stress response, immune modulation, ATF3
Abstract
Chemotherapy is a double-edged sword. It is anticancer because of its cytotoxicity. Paradoxically, by increasing chemoresistance and cancer metastasis, it is also procancer. However, the underlying mechanisms for chemotherapy-induced procancer activities are not well understood. Here we describe the ability of paclitaxel (PTX), a frontline chemotherapeutic agent, to exacerbate metastasis in mouse models of breast cancer. We demonstrate that, despite the apparent benefit of reducing tumor size, PTX increased the circulating tumor cells in the blood and enhanced the metastatic burden at the lung. At the primary tumor, PTX increased the abundance of the tumor microenvironment of metastasis, a landmark microanatomical structure at the microvasculature where cancer cells enter the blood stream. At the metastatic lung, PTX improved the tissue microenvironment (the “soil”) for cancer cells (the “seeds”) to thrive; these changes include increased inflammatory monocytes and reduced cytotoxicity. Importantly, these changes in the primary tumor and the metastatic lung were all dependent on Atf3, a stress-inducible gene, in the noncancer host cells. Together, our data provide mechanistic insights into the procancer effect of chemotherapy, explaining its paradox in the context of the seed-and-soil theory. Analyses of public datasets suggest that our data may have relevance to human cancers. Thus, ATF3 in the host cells links a chemotherapeutic agent—a stressor—to immune modulation and cancer metastasis. Dampening the effect of ATF3 may improve the efficacy of chemotherapy.
Modern chemotherapy can reduce tumors to an undetectable level; however, in many cases the tumors relapse, with recurrence in the original, regional, or distant sites (1–3). The mechanisms for relapse are multifaceted and complex, including intrinsic changes in cancer cells and changes in the noncancer cells in the host—the organism carrying the cancer (4–9). Although the traditional concept is that chemotherapeutic drugs provide selection pressure for drug-resistant cancer cells to thrive, recent studies showed that chemotherapeutic drugs actually induce procancer changes (reviewed in refs. 4–9 and in the references cited below). Thus, chemotherapy is a double-edged sword: It is anticancer because of its cytotoxicity on cancer cells but also can be procancer by inducing changes in cancer and/or host cells. For cancer cell-intrinsic changes, chemotherapeutic drugs have been shown to induce the migration/invasion of cancer cells (10) and to up-regulate the expression of some antiapoptotic genes (11). For noncancer cells, chemotherapy theoretically can affect all host cells, because it is administered systemically. Advances in this nascent field have benefited greatly from the extensive literature on cancer–host interaction in the recent decades (5–7, 12). Although endothelial cells have been shown to play a role in mediating the procancer effect of chemotherapy (13–15), various reports also have identified myeloid-lineage cells (including macrophages and their precursor monocytes) as a key cell type with procancer effects that include protecting cancer cells from death, increasing cancer-initiating cells, and suppressing the anticancer immune response (16–23). Mechanistically, chemotherapeutic agents can increase the abundance of myeloid cells in the primary tumor (16, 17, 20, 21, 23) or alter their bioactivities (19). Functionally, inhibition of myeloid cells improved chemotherapy efficacy (16, 17, 19, 21, 24, 25).
The emerging picture is that chemotherapeutic drugs elicit procancer effects and thus counteract their own efficacy, contributing to chemoresistance and metastasis. However, many gaps in our understanding remain. Most papers focused on primary tumors; only a few demonstrated the effect of chemotherapy on the metastatic sties (15, 26, 27). Because successful metastasis requires not only the escape of cancer cells from the primary tumor but also their colonization at the distant site, to understand the impact of chemotherapy, it is essential to elucidate the mechanisms at both the primary tumor and the metastatic site. Thus, far, no reports have analyzed the mechanisms at both sites in the same mice. We addressed this gap by analyzing both the tumor and the metastatic site. In this study, we focus on the noncancer cells rather than on the intrinsic properties of cancer cells. Here we present evidence that the host cells play an important role in the exacerbation of breast cancer metastasis by paclitaxel (PTX), a frontline chemotherapeutic agent. Importantly, Atf3, a stress-inducible gene (28), is a necessary host factor for this PTX effect.
ATF3 is a member of the ATF/CREB family of transcription factors (29, 30). It is encoded by an immediate-early gene, whose expression is low in normal cells but is induced greatly and quickly (within 30 min to hours) by a wide spectrum of stress signals (28). As a transcription factor, ATF3 directly or indirectly alters the expression of a variety of genes, and one common characteristic for many of its target genes is their ability to regulate inflammation (31), a key modulator of cancer progression (32–35). Recently, we showed that Atf3 in the noncancer host cells (referred to hereafter as “host-Atf3”) is necessary for efficient metastasis, because cancer cells orthotopically injected into WT mice metastasized more efficiently than those injected into Atf3-knockout (Atf3-KO) mice (36). Because Atf3 is induced by various chemotherapeutic drugs, including PTX (37), cisplatin (38), and doxorubicin (39), we hypothesized that host-Atf3 plays an important role in the deleterious procancer effects of chemotherapy. Below, we present evidence supporting this hypothesis. Significantly, analyses of publicly available human datasets support the notion that our data from mouse models have potential relevance to human cancer.
Results
PTX Exacerbates Breast Cancer Metastasis in a Host-Atf3–Dependent Manner.
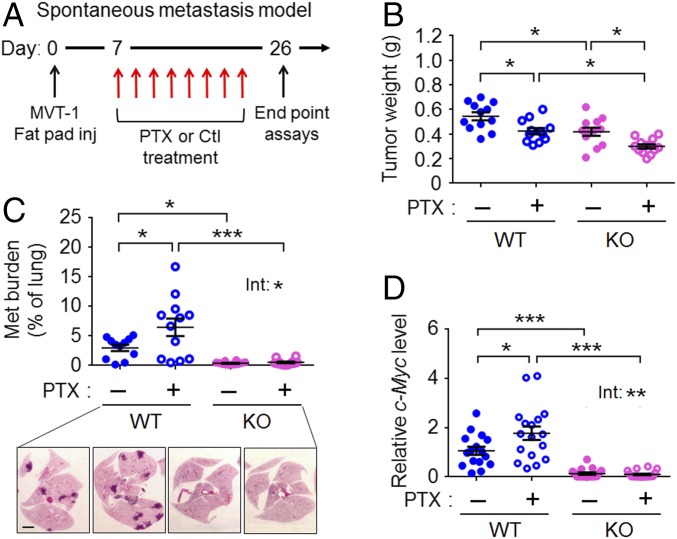
We used a spontaneous metastasis model to test the effect of PTX, a chemotherapeutic drug commonly used to treat breast cancer patients. As schematized in Fig. 1A, we injected MVT-1 breast cancer cells into immune-competent FVB/N mice at the orthotopic site (a fat pad), followed by control or PTX treatment (hereafter WT-Ctl and WT-PTX mice, respectively) and analyses of the primary tumor and lung (the metastatic site for MVT-1 cancer cells). By titration, we determined the highest PTX regimen without obvious toxicity to the mice (18 mg/kg, three times each week for a total of eight injections) (Methods). At this regimen, PTX reduced tumor size (Fig. 1B) but did not eradicate it. Area analysis of the lung showed that PTX exacerbated metastasis in the WT mice (Fig. 1C). Therefore, despite its benefit in reducing the size of primary tumor, PTX increased metastasis. This result is corroborated by the analysis of c-Myc, a transgene used as a readout of the cancer burden, in the MVT-1 cells (40): c-Myc was higher in WT-PTX than in WT-Ctl lungs (Fig. 1D). To test the role of host-Atf3, we compared WT and Atf3-KO mice. As shown in Fig. 1 C and D, the metastatic burden is much higher in the WT-Ctl mice than in their KO counterparts (KO-Ctl), indicating a prometastatic role for host-Atf3, as we reported before (36). Importantly, Atf3 deficiency in the host almost completely abolished the ability of PTX to exacerbate metastasis, indicating that this PTX effect is dependent on host-Atf3 (treatment–genotype interaction: P < 0.05, two-way ANOVA) (Fig. 1 C and D). We note that the same MVT-1 cancer cells were injected into the WT and Atf3-KO mice; only the host genotype was different. For convenience, we will refer to tumors from the WT mice as “WT tumors” and those from KO mice as “Atf3-KO tumors.”
Fig. 1.
PTX exacerbates breast cancer metastasis in a host-Atf3–dependent manner. (A) A schematic of the spontaneous metastasis model with PTX or control (Ctl) treatment starting on day 7 after cancer cell injection (inj). (B) Primary tumor weight on day 26 (n = 12 from two independent experiments). (C, Upper) Lung metastasis (met) burden on day 26 as percent of total lung area. Shown are representative data of more than 12 experiments (n = 12 from two independent experiments). (Lower) Representative images H&E staining. (Scale bar, 2 mm.) (D) Lung cancer burden on day 26 as assayed by the mRNA level of c-Myc, a transgene, in MVT-1 cancer cells. The RT-qPCR signals were standardized against that of actin, and the average c-Myc level in the WT-Ctl group was arbitrarily defined as 1 (n = 16–18 from three independent experiments). Bars indicate mean ± SEM; two-way ANOVA with post hoc Bonferroni test; *P < 0.05; ***P < 0.001. Int, treatment–genotype interaction.
PTX Affects the Vasculature Properties and Increases Cancer Cell Escape from the Primary Tumor in a Host-Atf3–Dependent Manner.
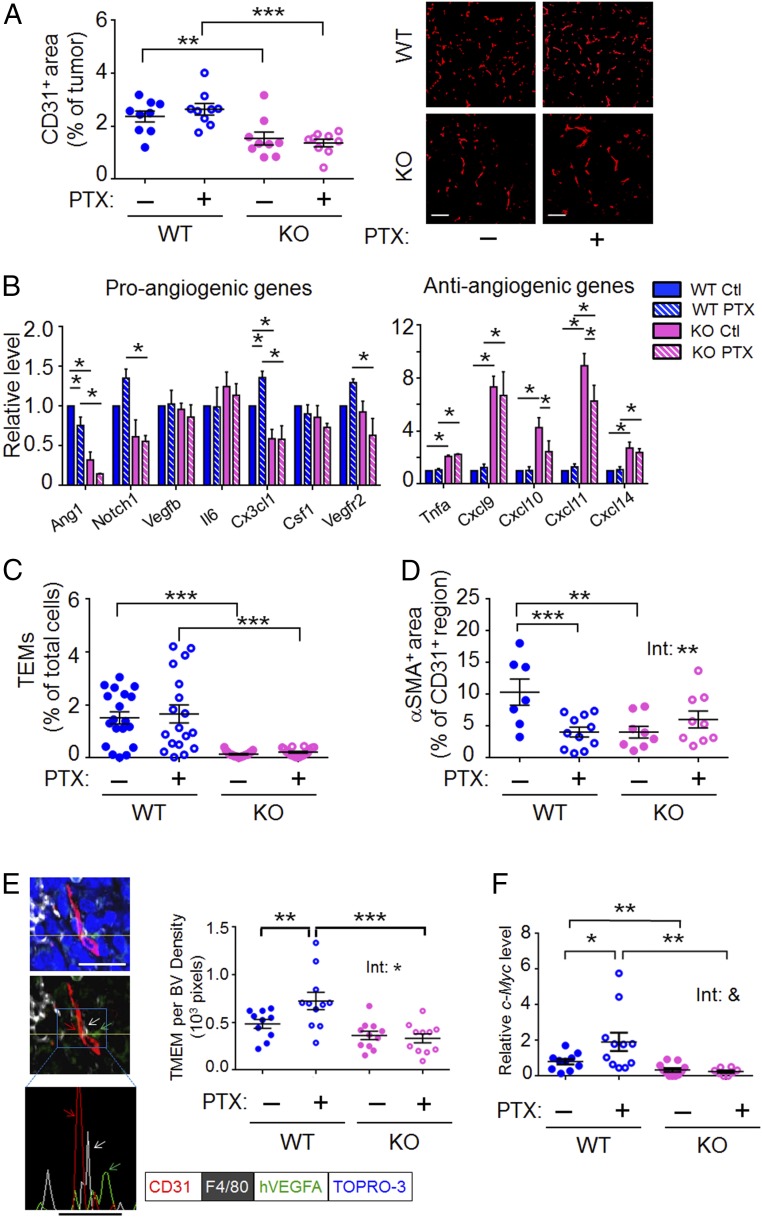
Blood vessels are a key route for cancer cells to escape primary tumors. We examined vessel density by CD31 staining and found that WT groups (both WT-Ctl and WT-PTX) had much higher microvascular density than their KO counterparts on day 15 and day 26 after cancer cell injection (Fig. 2A and SI Appendix, Fig. S1A). Analyses of gene expression by RT-qPCR showed that WT tumors had higher expression of proangiogenic genes (Ang1, Notch1, and Cx3cl1) but lower expression of the antiangiogenic genes (Cxcl9, 10, 11, and 14) than their KO counterparts (Fig. 2B). Furthermore, the WT tumors had higher abundance of Tie2-expressing monocytes/macrophages (TEMs; markers CD11+, F4/80+, TIE2+), a subset of macrophages that is proangiogenic (41), than did their Atf3-KO counterparts (Fig. 2C). SI Appendix, Fig. S1B shows that the higher abundance of TEMs in WT than in KO tumors was not caused by a higher macrophage abundance in general, because the numbers of CD11+, F4/80+ cells were similar in all four groups. Taken together, the evidence shows that WT tumors had a more proangiogenic tumor microenvironment than KO tumors, as assayed by vessel density, gene expression, and TEM abundance. Interestingly, PTX had no effect on vessel density or TEM abundance (Fig. 2 A and C). Its effect on gene expression is not statistically significant, with some exceptions (within WT for Ang1 and Cx3cl1, and within KO for Cxcl10 and Cxcl11) (Fig. 2B). We next considered the possibility that PTX may affect the properties of blood vessel and/or TEMs. We found that PTX reduced pericyte coverage on the vessel in WT but not in Atf3-KO tumors (treatment–genotype interaction; P < 0.01, two-way ANOVA) (Fig. 2D and SI Appendix, Fig. S1C). We tested the effects of PTX on TEM properties by a coculture assay using a Matrigel-coated Boyden chamber. As shown in SI Appendix, Fig. S1D, TEMs isolated from the WT-PTX tumors stimulated cancer cell invasion more efficiently than did TEMs isolated from the WT-Ctl tumors, indicating that PTX affects the activity of TEMs. We did not examine the TEMs from the Atf3-KO tumors because of their low abundance.
Fig. 2.
Host-Atf3 promotes a proangiogenic microenvironment, and PTX increases the abundance of TMEM and CTCs in a host-Atf3–dependent manner. (A) The microvascular density as assayed by immunofluorescence for CD31+ cells. (Left) The percent of CD31+ tumor area in a FOV from day-26 tumors was averaged from at least five FOVs; each dot represents one tumor (n = 9 from two independent experiments). See Methods for the details of image analysis. (Right) Representative images. (Scale bars, 100 μm.) (B) The relative mRNA level for the indicated genes from day-26 tumors. The average level for each gene in the WT-Ctl group was arbitrarily defined as 1 (n = 12 from four independent experiments). (C) TEMs (CD11b+ F4/80+ TIE2+) in day-26 tumors as the percent of total cells in the tumors (n = 18 from six independent experiments). (D) Pericyte coverage of day-22 tumors. The percent of CD31+ area also positive for αSMA in each FOV, averaged from at least five FOVs per tumor; each dot represents one tumor (n = 7–11 from three independent experiments). See Methods for detailed imaging analysis. (E, Left) A representative image of day-26 WT tumors analyzed by coimmunofluorescence for the three cell types in a TMEM: macrophage (white, F4/80), endothelium (red, CD31), and cancer cell (green, hVEGFA, a transgene in MVT-1 cells), with nuclei stained by TOPRO-3 (blue). See Methods for details of TMEM identification. The yellow line denotes the plane for the histogram in the Bottom panel. Nuclear signal (blue) was removed from the Middle and Bottom panels for clarity, and the arrows indicate the three cell types in TMEM. (Scale bars, 20 μm.) (Right) The number of TMEMs per blood vessel (BV) density (1,000 pixels) averaged from 10 FOVs for each tumor; each dot represents one tumor (n = 10–12 from three independent experiments). More than four hundred images were scrambled from all four groups of mice and analyzed in a blind fashion (see Methods for details). (F) CTCs as indicated by the relative mRNA levels of c-Myc (a transgene in the MVT-1 cancer cells) in the blood cells on day 26 after cancer cell injection. The RT-qPCR signals were standardized against that of actin, and the average level in the WT-Ctl group was arbitrarily defined as 1 (n = 8–11 mice from four independent experiments). Bars indicate mean ± SEM; two-way ANOVA with post hoc Bonferroni test; *P < 0.05; **P < 0.01; ***P < 0.001; &, P = 0.056. Int, treatment–genotype interaction.
Recently, intravital imaging of mouse breast tumors has revealed an intriguing phenomenon: Cancer cells enter the blood vessels (intravasate) at sites with a microanatomical landmark called “tumor microenvironment of metastasis” (TMEM), a structure composed of a perivascular macrophage and a cancer cell in close proximity (42–44). Because PTX increased metastasis in the WT mice (see above and Fig. 1 C and D), we tested whether PTX increased the abundance of TMEMs, sites for intravasation. We carried out coimmunofluorescence assays to detect TMEMs using selective markers: (i) CD31 for endothelium, (ii) F4/80 for macrophage, and (iii) human VEGFA (hVEGFA), encoded by a transgene in MVT-1, for cancer cells. We identified TMEM as detailed in Methods; Fig. 2E, Left shows a representative image of a TMEM. To avoid bias, we randomized more than 400 images from four groups of mice (n = 9–12 mice per group, 10 images per tumor), and analyzed them in a blind fashion. As shown in Fig. 2E, Right, PTX increased TMEM (per blood vessel density) in the WT tumors. Importantly, this PTX effect was not observed in the Atf3-KO tumors, indicating its dependence on host-Atf3 (treatment–genotype interaction, P < 0.05). We also carried out another coimmunofluorescence assay by identifying cancer cells using antibody against MENA rather than hVEGFA. MENA is a protein in the Invasive signature (45–47) and was previously used as a marker to identify cancer cells in TMEMs (44). SI Appendix, Fig. S1E shows a similar trend, corroborating the result shown in Fig. 2E that PTX increases TMEMs in a host-Atf3–dependent manner.
The features described above—vessel density, vessel property, and TMEM abundance (per blood vessel density)—can all affect the likelihood of cancer cells’ escape. We examined the circulating tumor cells (CTCs) by analyzing the blood cells for c-Myc, a transgene, in MVT-1 cancer cells. As shown in Fig. 2F, CTCs were much higher in WT than in Atf3-KO mice, indicating that host-Atf3 facilitates cancer cell escape, a genotype effect we reported previously (36). PTX further increased CTCs in WT but not in Atf3-KO mice (Fig. 2F).
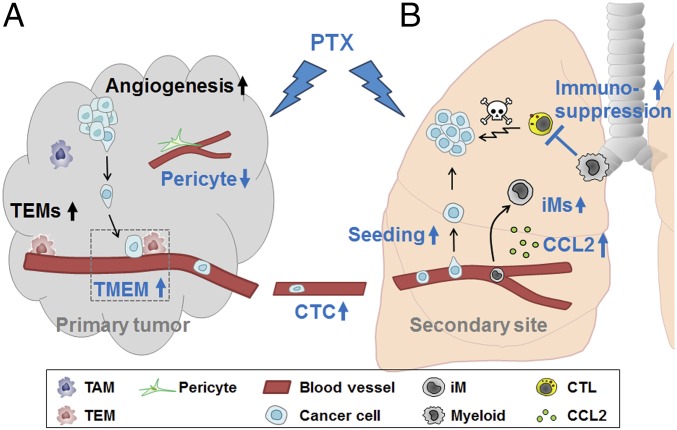
We summarize the data presented above as follows (Fig. 3A). On the one hand, the genotype (host-Atf3) facilitated a proangiogenic microenvironment (as shown by gene expression, TEM abundance, and vessel density). On the other hand, the treatment (PTX) affected the property of blood vessels (pericyte coverage), the property of TEMs (shown in the in vitro assay), and the abundance of TMEM (per vessel density). The PTX effects in vivo (pericyte coverage and TMEM) were dependent on host-Atf3 status, with a statistically significant treatment–genotype interaction. The overall consequence was higher CTC numbers in WT than in Atf3-KO mice and the exacerbation by PTX in WT but not in KO mice. Because the same cancer cell (MVT-1) was used, and only the host-Atf3 status was different, PTX exerted its effect on cancer cells in our models indirectly through the host, at least in part via ATF3-regulated events.
Fig. 3.
A model showing how host-Atf3 and PTX affect multiple steps in the metastatic cascade at both the primary tumor site (A) and the metastatic site (B). Black text: genotype effect only with no PTX effect; blue text: PTX effect, all dependent on host-Atf3 (statistically significant treatment–genotype interaction). CTC, circulating tumor cell; CTL, cytotoxic T lymphocyte; down-arrow, decrease; iM, inflammatory monocyte; TAM, tumor-associated macrophage; TEM, Tie2-expressing monocyte/macrophage; TMEM, tumor microenvironment metastasis; up-arrow, increase. See text for details.
PTX Exacerbates Lung Colonization by Cancer Cells in a Host-Atf3–Dependent Manner.
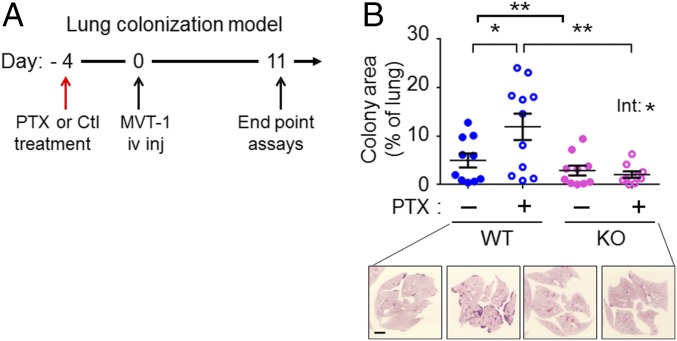
To establish metastatic nodules, the escaped cancer cells must colonize the distant site (reviewed in ref. 48). Experimentally, colonization of the lung (the metastatic site in our model) can be tested by the lung colonization (or experimental metastasis) model, which injects cancer cells i.v. into the circulation. To test whether PTX affects lung colonization, we used a pretreatment design (Fig. 4A) in which the mice were treated with PTX on day 4 before cancer cell injection. Because PTX is cleared from mice within 24–48 h of injection (49), this schedule minimizes the direct killing of cancer cells by PTX. Any effect of PTX on lung colonization would be the result of its effect on the host cells, which in turn affect cancer cells. As shown in Fig. 4B, PTX exacerbated lung colonization in the WT host (assayed at 11 d after injection), indicating that PTX changes the lung environment in a way that is favorable for cancer cells. Importantly, this effect is dependent on host-Atf3 status, because PTX showed no exacerbation in the Atf3-KO mice (treatment–genotype interaction P < 0.05) (Fig. 4B). Because this model does not require a fat pad for injection, we also examined the effect of PTX on lung colonization in male mice. It is worth testing the male mice, because men also develop breast cancer, albeit at a much lower frequency (50). SI Appendix, Fig. S2A shows similar results in male mice. We note that the overall cancer burden was lower in male lungs than in female lungs (compare the y axes of Fig. 4B and SI Appendix, Fig. S2A), in congruence with the lower propensity of men to develop breast cancer. Together, data from the spontaneous metastasis and lung colonization models indicate that PTX can enhance the ability of cancer cells to escape from tumors and to colonize the second site.
Fig. 4.
PTX exacerbates lung colonization in a host-Atf3–dependent manner. (A) A schematic of the lung colonization model with PTX or control (Ctl) treatment before i.v. injection (iv inj) of cancer cells. (B, Upper) Cancer burden in lungs from female mice on day 11 as the percent of total lung area (n = 9–11 from three independent experiments). Bars indicate mean ± SEM; two-way ANOVA with post hoc Bonferroni test; *P < 0.05; **P < 0.01. Int, treatment–genotype interaction. (Lower) Representative images of H&E staining. (Scale bar, 2 mm.)
PTX Increases Cancer Cell Seeding but Decreases the Cytotoxic Program in the Lung in a Host-Atf3–Dependent Manner.
To colonize the lung, the CTCs need to extravasate, survive, and proliferate in the lung. Using the pretreatment design described above, we studied seeding by analyzing the lung shortly (3 d) after i.v. injection of cancer cells. We used turbo GFP (tGFP)-labeled MVT-1 cells to facilitate the counting of single cells or microcolonies. As shown in Fig. 5A and SI Appendix, Fig. S2B, the host-Atf3 genotype facilitated seeding, and PTX increased seeding in WT but not in Atf3-KO mice. This result was observed in both female and male mice, with males showing a more robust PTX effect than females (compare the fold difference and P values in Fig. 5A and SI Appendix, Fig. S2B).
Fig. 5.
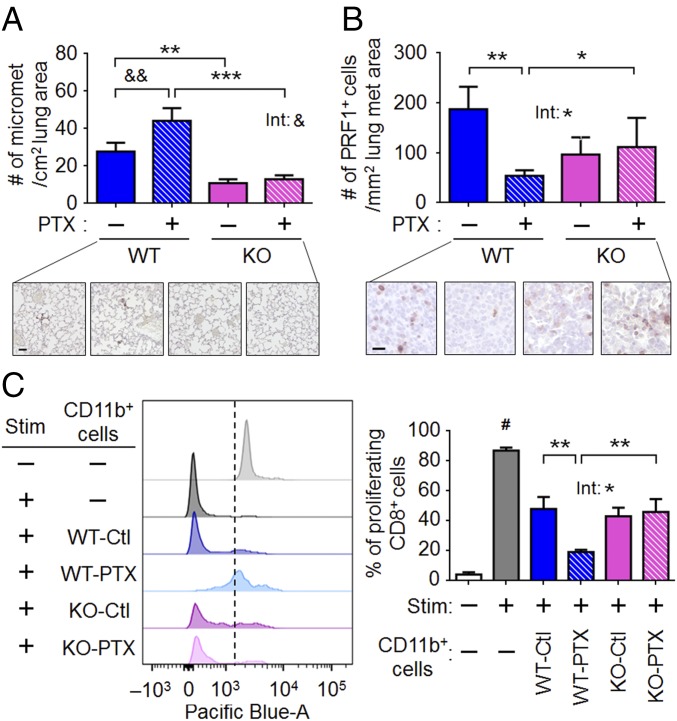
PTX increases cancer cell seeding but decreases the cytotoxic program in the lung in a host-Atf3–dependent manner. (A) Cancer cell seeding on day 3 after injection in the lung colonization model. (Upper) Average numbers of micrometastases (micromets) per square centimeter of lung area. A single cell or cluster of cells (fewer than five cells) was counted as one micromet (n = 12–14 from three independent experiments). (Lower) Representative images of the tGFP-labeled cells (by immunohistochemistry). (Scale bar, 40 μm.) (B) Analysis of PRF1+ cells in day-26 lungs from the spontaneous metastasis model. (Upper) Average numbers of PRF1+ cells per square millimeter of lung metastasis (met) area (n = 7–12 from three independent experiments). (Scale bar, 20 μm.) (Lower) Representative images. Brown: PRF1 signal. (C) T-cell–suppression assay using CD11b+ cells isolated from day-26 lungs from the spontaneous metastasis model. (Left) Representative histograms of the proliferation dye (Pacific Blue-A) for CD8+ T cells, without (−) or with (+) stimulation (Stim) by anti-CD3ε and anti-CD28 antibodies. Nonproliferating cells are on the right of the dotted line; proliferating cells are on the left of the dotted line. Signal from the proliferating cells was divided by the total signal to obtain the percent of proliferating CD8+ T cells. (Right) Percent of proliferating T cells from unstimulated (Stim: −) or stimulated (Stim: +) splenocytes incubated without (−) or with the indicated CD11b+ cells (n = 11 or 12 mice from three independent experiments). Bars indicate mean ± SEM; two-way ANOVA with post hoc Bonferroni test (except for those indicated by #); *P < 0.05; **P < 0.01; ***P < 0.001; &, P = 0.128; &&, P = 0.064; #P < 0.01 between the indicated bar and the other bars in C analyzed by t test (two-sided). Int, treatment-genotype interaction.
After seeding, cancer cells need to proliferate and evade death to become detectable nodules. We examined these processes by immunohistochemical analyses of specific markers: phospho-histone H3 for mitosis and activated caspase 3 for apoptosis. We used the lung sections from the spontaneous metastasis model and found no statistically significant difference between any groups in either assay (SI Appendix, Fig. S2 C and D). Because activated caspase 3 reflects only a subset of cell death, we examined perforin 1 (PRF1), a marker for the activated cytotoxic lymphocytes (CD8+ T and NK cells) that induce cancer cells to die via caspase 3-dependent and -independent pathways (51). Fig. 5B shows that PTX reduced the number of Prf1-expressing cells in the WT lung nodule but had no effect on the Atf3-KO counterparts, indicating that PTX reduces the cytotoxic microenvironment in a host-Atf3–dependent manner. We note that WT-Ctl and KO-Ctl mice had no statistically significant difference in their PRF1+ cells. This result may appear surprising, because the WT-Ctl lung had a higher cancer burden than the KO-Ctl lung. One explanation is that the higher seeding in the WT-Ctl than in KO-Ctl lung would contribute to the higher cancer burden in the WT-Ctl lung.
To investigate further the ability of PTX to reduce cytotoxicity, we carried out a T-cell–suppression assay. A hallmark of cancer progression is the ability of cancer cells to reprogram myeloid cells (52), which in turn suppress the PRF1-secreting cytotoxic immune cells (CD8+ T and NK cells) (53). We isolated myeloid cells (CD11b+) from the lungs derived from the spontaneous metastasis model and tested their ability to suppress T cells in vitro. As shown by the fluorescent dye dilution assay, WT myeloid cells suppressed T cells (Fig. 5C); PTX further enhanced this immunosuppression. Analysis of the median fluorescent intensity (MFI) showed similar results (SI Appendix, Fig. S2E). Importantly, this PTX effect was not observed in the Atf3-KO myeloid cells (Fig. 5C and SI Appendix, Fig. S2E), again indicating the dependence of PTX on host-Atf3. Consistent with the perforin assay described above, myeloid cells from WT-Ctl and KO-Ctl lungs showed a similar ability to suppress T cells; that is, the Atf3 genotype did not affect T-cell suppression. Taken together these results show that the host-Atf3 genotype facilitated seeding but did not affect the cytotoxic microenvironment. PTX treatment, however, enhanced both seeding and immunosuppression in a host-Atf3–dependent manner. Together, these data provide a mechanistic explanation for the higher cancer burden in WT than in KO hosts and the further exacerbation by PTX in WT but not in KO hosts.
PTX Increases the Abundance of Inflammatory Monocytes in the Lung in a Host-Atf3–Dependent Manner.
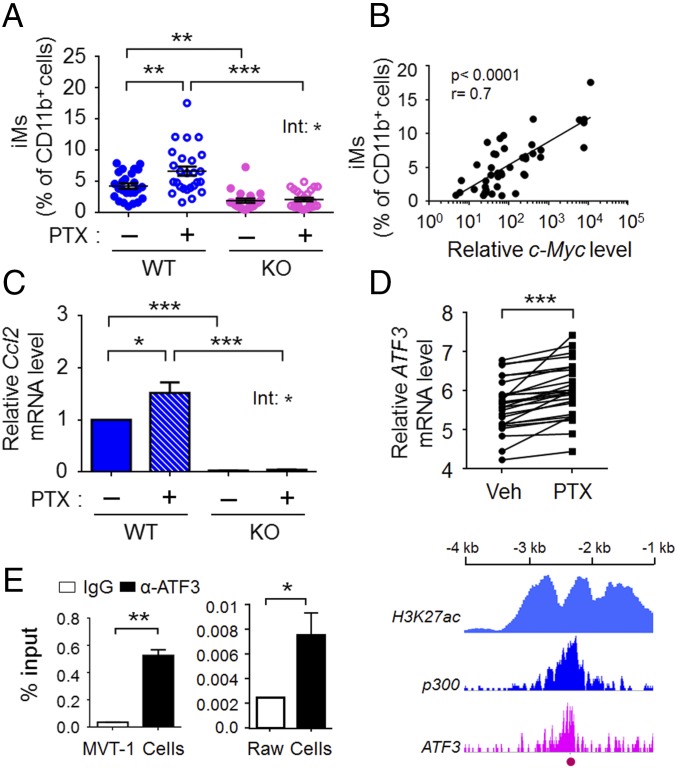
Inflammatory monocytes (iMs), a subset of myeloid-lineage of cells (54), were recently shown to be recruited to the lung and to differentiate into macrophages that facilitate cancer cell metastasis and lung colonization (55). In light of this finding, we tested whether their abundance is affected by PTX and/or host-Atf3 status. Flow cytometry analyses of lungs from the spontaneous metastasis model showed that iM (CD11b+ F4/80+ CCR2+ LY6C+) numbers were higher in WT than in Atf3-KO mice (Fig. 6A), indicating that host-Atf3 promotes a lung microenvironment more conducive to iMs. Importantly, PTX increased the abundance of iMs in the WT lungs but not in the Atf3-KO lungs (treatment–genotype interaction, P < 0.05) (Fig. 6A). We note that the abundance of CD11b+ cells in the lung did not vary among the groups (SI Appendix, Fig. S3A). Therefore, the difference in iM abundance is not caused by a difference in the abundance of myeloid cells in general. Analyses of the lungs from the lung colonization model (day 11 after cancer cell injection) showed similar results: higher iM abundance in WT than in Atf3-KO lungs, with PTX further increasing iM numbers in WT but not in KO lungs (SI Appendix, Fig. S3B). We note that the level of iMs correlated with cancer burden in the lung in both the spontaneous metastasis model (see Fig. 1C for cancer burden and Fig. 6A for iMs) and the lung colonization model (see Fig. 4B for cancer burden and SI Appendix, Fig. S3B for iMs). In those experiments, separate cohorts of mice were used to assay iM (by flow analysis of single-cell suspensions) versus cancer burden (by area analysis of lung sections). We repeated the experiments using a cohort of mice and analyzed single-cell suspensions of their lungs side-by-side for the percent of iMs and for the levels of c-Myc, a transgene, in the cancer cells. Fig. 6B shows a linear correlation between iM percentages and c-Myc levels (Pearson correlation, P < 0.0001, r = 0.7).
Fig. 6.
PTX increases iMs in the lung in a host-Atf3–dependent manner, and CCL2, a monocyte recruitment factor, is a potential target gene of ATF3. (A) iMs (CD11b+ F4/80+ Ly6C+ CCR2+) expressed as percent of CD11b+ cells using day-26 lungs from the spontaneous metastasis model (n = 21–26 from nine independent experiments). (B) Correlation between the abundance of iMs (as percent of CD11b+ cells) and c-Myc mRNA level (an indicator for the metastatic burden) in day-26 lungs from the spontaneous metastasis model (n = 8–10 from four independent experiments). The RT-qPCR signals of c-Myc were standardized against that of actin, and the standardized c-Myc in the normal healthy lungs was arbitrarily defined as 1. The x axis shows log base 10 values of relative c-Myc levels. (C) Ccl2 gene-expression data in the day-26 lungs from the spontaneous metastasis model. The RT-qPCR signals were standardized against that of actin, and the average number in the WT-Ctl group was arbitrarily defined as 1 (n = 11 or 12 from four independent experiments). (D) The relative ATF3 mRNA levels in 27 human breast cancer cell lines treated with vehicle (Veh) or PTX (at IC50 for each cell line, based on antiproliferative activity) for 24 h (data were extracted from GEO accession no. GSE50811). The y axis shows log base 2 values of spot intensity on the microarray chip after subtracting background signals and normalization. (E, Left) ChIP signals, as percent of input in MVT-1 cells and Raw264.7 macrophages using IgG or ATF3 antibody to examine the occupancy of ATF3 on the Ccl2 promoter at the site −2.3-kb from the transcriptional start site. Shown is a representative of three independent experiments. (Right) Data extracted from a genome-wide ChIP-seq dataset from The ENCODE Project Consortium (61). The ATF3, p300, and H3K27ac ChIP peaks on the CCL2 promoter are shown. Numbers indicate the distance from the transcriptional start site. The red dot corresponds to the −2.3-kb site. Bars in C and E indicate mean ± SEM; two-ANOVA with post hoc Bonferroni test; Int, treatment–genotype interaction; (B) Pearson correlation for P and r values, linear regression for the line of best fit. (E) Student t test (two-sided); *P < 0.05; **P < 0.01; ***P < 0.001.
We further characterized these iMs by LY6G, a granulocyte marker, and found that the majority (>99%) of them are negative (SI Appendix, Fig. S4A), indicating that the iMs presented here have a monocytic rather than a granulocytic nature. Previously, a VEGFR1+ subset of bone marrow-derived cells was shown to initiate the premetastatic niche (56). In addition, a distinct VEGFR1+ population of macrophages was shown to mediate metastatic growth in the lung (57). Interestingly, 64–83% of the iMs we studied here were VEGFR1+ (SI Appendix, Fig. S4B). Although more analyses are required to clarify how the iMs described here relate to the VEGFR1+ cells in previous studies, our data support the notion that VEGFR1 is likely to be important in a subset of myeloid cells. Because iMs also can be recruited to the primary tumor (55) (albeit with lower efficiency than to the lung), we analyzed the primary tumor for the abundance of iMs. SI Appendix, Fig. S5A shows that they did not vary among the four groups of mice. Thus, in contrast to the situation in the lung, iMs are not modulated by host-Atf3 or PTX in the primary tumor. In a reciprocal experiment, we examined whether TEMs, which are modulated by host-Atf3 in the primary tumor (as described above), are modulated by host-Atf3 or PTX in the lung. SI Appendix, Fig. S5B shows that TEMs in the lung did not vary among the four groups of mice. Together, our data implicate distinct subsets of myeloid cells at two sites: TEMs modulated by host-Atf3 in the primary tumor and iMs modulated by PTX and host-Atf3 in the metastatic lung.
Host-Atf3 Plays a Necessary Role for PTX to Up-Regulate the Expression of Chemokine (C-C Motif) Ligand 2.
Chemokine (C-C motif) ligand 2 (CCL2) is a key recruitment factor for monocytes, and the CCL2–CCR2 axis plays an important role in cancer development (58). Recently, blockade of iMs by CCL2 antibody or CCR2 inhibitor was shown to abrogate their prometastatic effects (55, 59). We tested whether Ccl2 gene expression is modulated by host-Atf3 and/or PTX. As shown in Fig. 6C, Ccl2 mRNA level was much higher in WT than in Atf3-KO lungs (from the spontaneous metastasis model), indicating that its expression is up-regulated directly or indirectly by ATF3. PTX further increased the Ccl2 mRNA level in a host-Atf3–dependent manner. The pattern of Ccl2 expression parallels that of iM abundance and is consistent with the role of CCL2 as a recruitment factor for monocytes. Analysis of a publicly available microarray dataset derived from 27 breast cancer cell lines showed the induction of Atf3 by PTX (Fig. 6D) and a positive correlation between the mRNA levels of Atf3 and Ccl2 (SI Appendix, Fig. S3C).
The Ccl2 promoter contains potential ATF3-binding sites (SI Appendix, Fig. S3D), i.e., ATF/CRE, AP-1, and closely related sites (29, 60). We tested whether ATF3 binds to the Ccl2 promoter by ChIP assay and found consistent binding at the −2.3-kb site (Fig. 6E, Left). This result is corroborated by a publicly available ChIP-sequencing (ChIP-seq) dataset, the ENCODE Project Consortium (61), which shows an ATF3-binding peak at the −2.3-kb region on the Ccl2 promoter. Intriguingly, this peak overlaps with the p300-binding peak and is in a region with high levels of histone H3K27ac (Fig. 6E, Right) (61). Because p300 binding and H3K27ac are associated with transcriptionally active areas (62, 63), these ChIP data, in combination with the higher Ccl2 mRNA level in WT than in Atf3-KO lungs, strongly suggest that ATF3 (a transcription factor) up-regulates the Ccl2 promoter.
Taking our findings together, we summarize the data as follows (Fig. 3B): PTX, as a stress signal, increases lung iM abundance, cancer cell seeding, and lung colonization and reduces the anticancer immune microenvironment, all in a manner dependent on host-Atf3. Importantly, ATF3 up-regulates the expression of Ccl2, which encodes a recruitment factor for iMs. These findings, together with reports in the literature on the roles of CCL2 and iM in cancer progression, support the notion that ATF3-mediated up-regulation of Ccl2 expression and iM abundance in the lung contributes to the ability of PTX to exacerbate cancer burden.
The Potential Relevance of Data from Mouse Models for Human Cancers.
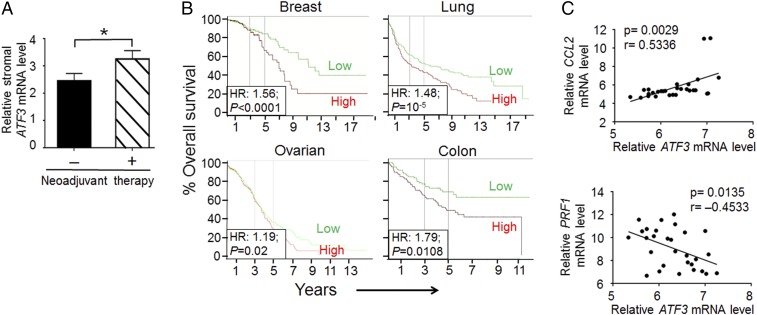
To test whether the data from mouse models reported above have any relevance to human cancers, we analyzed publicly available microarray datasets (64). As shown in Fig. 7A, ATF3 expression was higher in the breast tumor stroma from patients who had undergone chemotherapy than in those who had not. Interestingly, high coexpression of ATF3 and TEK, the human ortholog of Tie2, correlated with reduced overall survival in breast, lung, ovarian, and colon cancers (Fig. 7B). This coexpression is a more robust predictor than the expression of either gene alone (SI Appendix, Fig. S6A). We note that microarray datasets do not distinguish the cell source for differential gene expression. However, the worse outcome data are consistent with our mouse data that WT tumors had higher Tie2 gene expression in their macrophages than their Atf3-KO counterparts (SI Appendix, Fig. S6B) and had higher cancer escape and metastasis (as described above).
Fig. 7.
Analyses of human cancer specimens. (A) Induction of ATF3 by chemotherapy in the human breast tumor stroma. ATF3 gene expression in the tumor stroma from breast cancer patients with (n = 20) or without (n = 27) chemotherapy was extracted from Boersma et al. (64) and analyzed on the Oncomine website. Bars indicate mean ± SEM; Student t test (one-sided); *P < 0.05. (B) Coexpression of high ATF3 and high TEK (the human ortholog of mouse Tie2) correlates with worse outcome in human cancers. Patients with the indicted cancer types from publicly available microarray datasets were arbitrarily classified into a ATF3-high and TEK-high (both above median, red trace) group versus the ATF3-low and TEK-low (both below median, green trace) group. Breast cancer and ovarian cancer data are from the TCGA database; lung cancer and colon cancer data are from the GEO database (accession nos. GSE30219 and GSE17536, respectively). The online tool PROGgene V2 was used, and the Kaplan–Meier curves of survival are shown. Log-rank test; P values and hazard ratios (HR) are indicated. (C) The correlation between the mRNA levels of ATF3 and the indicated genes. Data were extracted from Foukakis et al. (76) (GEO accession GSE54323), which were derived from breast cancer metastatic sites (including lymph node, bone, and brain, 15 patients, 29 samples). The x and y axes are log base 2 values of spot intensity on the microarray chip after subtracting background signals and normalization. Pearson correlation; P and r values are indicated; linear regression for the line of best fit.
We also analyzed microarray datasets derived from metastatic sites from human breast cancer patients. The ATF3 mRNA level correlated positively with the CCL2 but negatively with the PRF1 mRNA level (Fig. 7C). Furthermore, analysis of other markers indicated that ATF3 expression correlated with higher CCL7 and CCL8 levels (encoding two other monocyte recruitment factors) but with lower cytotoxic immune cell markers, such as CD8, various granzymes, and natural killer (NK) cell markers (SI Appendix, Fig. S7). Thus, data from patient samples are consistent with the notion that ATF3 promotes a microenvironment at the metastatic sites that favors monocyte recruitment and immunosuppression. The data from patient samples not only support our data but also lend credence to our mouse models.
Discussion
Here we describe the effect of chemotherapy on both the primary tumor and metastatic sites using samples from the same mice. The conceptual advance is twofold. (i) It demonstrates that, despite the apparent therapeutic benefit of reducing tumor size, PTX enhances the dissemination of cancer cells (the seeds) from primary tumors and facilitates the preparation of the lung microenvironment (the soil) to be more hospitable to cancer cells, thereby explaining the paradoxical procancer effect of chemotherapy in the context of the seed-and-soil theory (65). (ii) The report delineates a pathway from chemotherapy to stress response, to immune modulation and metastasis, with Atf3, a stress gene, as an essential link in this pathway. Significantly, a recent study showed that residual cancer burden after neoadjuvant chemotherapy is associated with worse clinical outcome (66). Thus, elucidating the mechanisms by which chemotherapy exacerbates cancer progression has clinical relevance.
To test whether the exacerbation of metastasis by PTX is limited to MVT-1 cells (which have c-Myc and VEGF as the oncogenic drivers), we used Met-1 breast cancer cells (which have the Polyoma virus-middle T antigen as the oncogenic driver) (67). As shown in SI Appendix, Fig. S8A, PTX also exacerbated Met-1 lung colonization in a host-Atf3–dependent manner. In addition, PTX showed a similar trend in a different cancer (SI Appendix, Fig. S8B), the lung cancer model, in which Lewis lung carcinoma (LLC) cells were injected into C57BL/6 mice, which have a different genetic background from the FVB/N mice used in all the experiments described above. Although our study focused on PTX, we found that cyclophosphamide (CTX), another frontline chemotherapeutic drug, also exacerbated lung colonization in both the MVT-1 (SI Appendix, Fig. S8C) and Met-1 (SI Appendix, Fig. S8D) breast cancer models in a host-Atf3–dependent manner. Thus, the phenotype—the exacerbation of lung colonization by chemotherapy in a host-Atf3–dependent manner—is not limited to PTX, nor is it limited to one cancer cell line or to one mouse genetic background. However, it is not clear whether the underlying mechanisms for the phenotype are the same in these models. Further investigation is required to address this issue.
Several features of ATF3 provide important perspectives for our findings. (i) The Atf3 gene is expressed at a low basal level, but its expression is greatly increased by a wide spectrum of stress signals (28). Intriguingly, by global analysis of ATF3-binding sites using ChIP-seq, Yan and colleagues (60) showed that ATF3 binds to a surprisingly large number of sites on the genome; furthermore, their data supported the idea that ATF3 “bookmarks” sites on the genome to allow rapid transcriptional response upon stress stimulation. (ii) Atf3 is an immediate-early gene (31), which has a hallmark of encoding transcription factors that regulate the expression of other transcription factor genes, resulting in a cascade of changes in transcriptional profile. (iii) Many ATF3 target genes encode cytokines and chemokines irrespective of the signals that induce it, prompting the idea that one unifying function of ATF3 is to modulate the immune response (31). Combining these features of ATF3 with our data, which are derived from mice with host-Atf3 status, we propose that the induction of Atf3 in the noncancer host cells represents an early cellular stress response and links PTX (and likely other chemotherapeutic drugs) to an altered transcriptome, resulting in a modified immune response and enhanced cancer metastasis. The stress-inducible nature of Atf3 has particular relevance to the following idea regarding chemoresistance: that the cellular stress response has evolved to promote tissue repair but has been co-opted to help cancer cell survival and progression (8, 12). That is, chemotherapy, as a stressor, can counteract its own efficacy by inducing a stress response. Our data suggest that Atf3 in the noncancer host cells is a key gene mediating this process. Interestingly, peptidyl-prolyl cis-trans isomerase A (PPIA), a stress-response chaperone in cancer cells, was shown to play an important role in the ability of cancer cells to resist PTX (68). Thus, a stress response in both noncancer cells (our study) and cancer cells (the PPIA study) can counteract chemotherapy.
Our report provides several important findings. First, we demonstrated that PTX decreases pericyte coverage but increases TMEM, CTCs, seeding, iMs, and immune-suppression, all in a host-Atf3–dependent manner. None of these effects has been reported before. Although cisplatin, a different chemotherapeutic drug, has been shown to increase seeding (15), it is not clear whether host-Atf3 plays a role. Second, the increase of TMEM by PTX is of particular interest. TMEM is a microanatomical landmark for intravasation (42, 43, 69). One implication of this finding is that cancer cells escape the primary tumor via an “active” process through a specific structure. Importantly, high TMEM density in human breast cancer was shown to be associated with higher metastasis, independent of tumor grade or lymph node status (44, 70). Thus, our finding that PTX increases TMEM is a timely finding that provides a mechanistic explanation for the emerging procancer effect of chemotherapy. It would be interesting to test whether chemotherapy increases TMEM abundance in human tumors (as we found in the mouse models). Third, our data show a parallel pattern among Ccl2 expression, iM abundance, and the metastatic burden in the lung: all were higher in WT than in Atf3-KO mice and were further exacerbated by PTX. These parallels, combined with the reports that CCL2 recruits iMs and that iMs promote metastasis (21, 55, 59), suggest that iMs may be a contributing factor for the phenotypes we observed. Although iMs have been shown to enhance metastasis (55), they have not been implicated in the context of chemotherapy. Fourth, the iM abundance in the primary tumor did not vary among the four groups of mice (SI Appendix, Fig. S5A). Thus, iM levels in the primary tumor are not a relevant factor for the phenotypes we observed. Conversely, TEMs, the cell type whose abundance in the primary tumor was affected by host-Atf3 status (being higher in WT than in KO mice), did not show this genotype difference in the lung (SI Appendix, Fig. S5B). Taken together our findings identified two subsets of myeloid cells: (i) TEMs modulated by the host-Atf3 genotype in the primary tumor but not in the metastatic lung; and (ii) iMs modulated by PTX and the host-Atf3 genotype in the metastatic lung but not in the primary tumor. Although it has been postulated that different subsets of myeloid cells are functionally important at primary tumor versus metastatic sites, their nature is not well understood (6, 7). Our data provide an insight into this knowledge gap. Fifth, although host-Atf3 was previously shown to be prometastatic (36), its role in the effect of chemotherapy on cancer progression has never been reported. Furthermore, the ability of host-Atf3 to increase vessel density, TEM abundance, and iMs has not been demonstrated. Thus, our study provides insights into the mechanisms by which host-Atf3 may promote metastasis.
Our study has limitations and caveats. Here we discuss a few of them. (i) In our models, the cancer cells are the same (with endogenous Atf3 gene), but the host genotype is different. However, the differences we observed between WT and Atf3-KO mice reflect the combined effect of cancer–host interactions, not only the host cells per se. (ii) Because we used traditional KO mice, our data do not delineate the roles of specific host cells. Further studies are required to distinguish the roles of Atf3 in different cell types, such as myeloid versus endothelial cells. (iii) The lymphatic vessel is another route by which cancer cells can escape primary tumors (71). It would be interesting to test whether Atf3 or PTX affects its density or properties. (iv) Our data indicate that PTX does not affect the abundance of tumor-associated macrophages (TAMs) (SI Appendix, Fig. S1B). However, others have shown an increase in TAMs induced by chemotherapeutic agents such as PTX and cyclophosphamide (16, 17, 23). This apparent discrepancy may reflect differences in mouse strain, cancer cells, the chemotherapy regimen, and the timing of sample collection relative to the last treatment. (v) Previously, PTX was shown to increase blood vessel density (27) in primary tumors. However, we did not observe this effect. One possibility is that the MVT-1 cancer cells express human VEGFA as a transgene (40). Because VEGFA is highly proangiogenic (72), the tumor microenvironment may already be saturated with angiogenic signals, making it difficult to detect the effect of PTX in increasing vessel density. (vi) To address the impact of chemotherapy and the stromal expression of ATF3, further experiments are required, including immunohistochemical or immunofluorescent analyses of samples from patients with or without chemotherapy.
In conclusion, PTX enhances multiple steps in the metastatic cascade, and host-Atf3 is necessary for these deleterious effects of PTX. Significantly, bioinformatics analyses suggest that our data from the mouse models are relevant to human cancer. Because chemotherapy is an important treatment for cancer patients, dampening the effect of ATF3 may help improve the efficacy of chemotherapy.
Materials and Methods
Animal Studies.
Unless otherwise indicated, age-matched FVB/N WT (ATF3+/+) and KO (ATF3−/−) female mice (6–8 wk of age) were used. For the spontaneous metastasis model, MVT-1 cancer cells (2 × 105 cells, in DMEM, 10% FBS:Matrigel, 1:1, in 30 μL) were injected into the fourth mammary fat pad, followed by PTX (described below) or control saline treatment and final analyses (Fig. 1A). Analyses of primary tumors and lung nodules were carried out as previously reported (36). PTX (Sigma) in a 1:1:2 ratio of cremophore EL:ethanol:PBS (18 mg/kg body weight in 100–150 μL) was i.p. injected on day 7 after cancer cell injection and then was injected three times each week for a total of eight injections. We note that this dosage is slightly below the maximum tolerable dose. When we increased the dose by ∼10% (20 mg/kg three times each week for a total of eight injections), about 30% of the mice displayed symptoms that met the early removal criteria, including weight loss (reaching 15%), lethargy, and hunched back. In postmortem analysis, these mice showed extremely distended intestine and shrunken spleen. Data from pilot experiments using different doses and frequencies of PTX injection are shown in SI Appendix, Fig. S9. For the lung colonization model, PTX (18 mg/kg body weight, unless otherwise indicated) or saline was injected i.p. on day 4 before MVT-1 cancer cell injection via the tail vein (2 × 106 cells in 200 μL of PBS), and lung colonies were analyzed on day 11 after cancer injection (Fig. 4A), except for the seeding experiment, in which lung colonies were analyzed on day 3.
General Cell Culture Work and Isolation of Cells from the Blood, Primary Tumors, and Lungs for Assays.
MVT-1 cells were cultured and blood cells were isolated as previously described (36). Single-cell suspensions from primary tumors and the lungs were prepared using the gentleMACS Dissociator (Miltenyi Biotec) according to the manufacturer’s instructions. CD11b+ cells from the lungs for the T-cell–suppression assay and gene expression analysis were isolated by microbeads (Miltenyi Biotec) bound with the CD11b antibodies as detailed before (36). Macrophages (F4/80+) from primary tumors were enriched by MACS beads as described previously (36). Antibodies are listed in SI Appendix, Table S1. The purity of the isolated cells (usually ≥80–90%) was estimated by flow analysis on a BD LSR II flow cytometer (BD Bioscience). For activity assays, the cells were used within 4–6 h after isolation; for RNA analyses, cells were put into TRIzol immediately.
Immunophenotyping.
Single-cell suspensions from primary tumors and lungs were prepared as described above. Cells were stained as previously described (36) using the indicated antibodies (SI Appendix, Table S1) and were analyzed on a BD LSR II flow cytometer (BD Bioscience). Forward scatter (FSC) and side scatter (SSC) were used to exclude small cell debris or large aggregates; unstained and single stained cells were used for gating controls. Examples of gating (for TEMs and iMs) are shown in SI Appendix, Fig. S10 A and B.
Immunohistochemistry, Immunofluorescent Staining, and Imaging Analyses.
Staining was performed on formalin-fixed paraffin-embedded tissue sections as described previously (36, 73) using the antibodies listed in SI Appendix, Table S1. After immunohistochemistry, the slides were counterstained with hematoxylin and captured on a Nikon Diaphot 300 microscope (Nikon). After immunofluorescence, the slides were counterstained with Topro-3 (Molecular Probes) and captured on an Olympus FV 1,000 Filter confocal microscope (Olympus) or a Leica TCS SL confocal microscope (Leica Microsystems). For seeding, PRF1, phospho-histone H3, and activated caspase 3 experiments, cells with positive signals from the entire section or within the lung cancer nodules (as indicated) were counted and standardized against either the lung area or lung metastasis area as specified in the figure legends. For vessel density, at least five fields were imaged for each tumor; the percent of CD31+ areas in each field of view (FOV) was determined by ImageJ, and the average number for each tumor was calculated. SI Appendix, Fig. S10C shows an example of CD31 analysis. For pericyte coverage, CD31 was used as the endothelial marker, and αSMA was used as the pericyte marker. At least five FOVs were imaged for each section and analyzed by ImageJ to obtain the percent of CD31+ areas that were αSMA+. SI Appendix, Fig. S10D shows an example. For TMEM analyses, two sets of markers were used to stain the cells: (i) CD31 for endothelial cells, F4/80 for macrophages, and hVEGFA for cancer cells; (ii) CD31 for endothelial cells and F4/80 for macrophages, but with MENA replacing hVEGFA for cancer cells. The following criteria were used to identify TMEM: (a) The distance between any two cells within this tripartite structure (macrophage, endothelium, cancer cell) had to be ≤20 μm. (b) If any two signals completely overlapped with each other (based on the histogram), they were considered signals from a single cell stained by more than one antibody, because of the coexpression of markers or potential cross-reaction. As such, any hVEGFA+ or MENA+ cells that were also stained by the F4/80 or CD31 antibody were not considered cancer cells. We note that, according to the manufacturer (Santa Cruz), the hVEGFA antibody is selective against hVEGF with low cross-activity to the mouse VEGF. As a control, we tested whether this antibody can detect pericyte, which may express mouse VEGF. Because of its perivascular location, the hVEGFA+ pericyte will compromise the assay by appearing as the cancer cell in the tripartite TMEM structure. Analysis of >150 pericytes in the WT tumors indicated that less than 1.5% of the pericytes were detected by this antibody. This low background, combined with criteria a and b, helps make this assay a robust gauge to estimate TMEM abundance. Five to ten FOVs were imaged from each section. To avoid bias, the image files from all four groups (WT-Ctl, WT-PTX, KO-Ctl, and KO-PTX) were combined, coded, and reshuffled (manually or using the Random Names script). Thus the scorer did not have cues about the potential identity of any image resulting from the cluster it fell in. The images were scored by one blinded individual (J.D.M.) to maintain internal consistency, even though the numbers from two investigators (J.D.M. and S.P.J.) were close in test trials. The files were decoded, and the average number of TMEMs per vessel density (1,000 pixels) was calculated for each tumor. The number of mice and total images analyzed are indicated in the figure legends.
RNA Extraction, RT-qPCR, and ChIP Analysis.
Total RNA was prepared using TRIzol (Invitrogen) according to the manufacturer’s instructions, and RT-qPCR was performed as previously described (74) using actin as an internal control. ChIP analysis was described previously (75). All primers are listed in SI Appendix, Table S2.
T-Cell–Suppression Assay.
Spleens from WT healthy mice were squeezed between two sterile glass slides and filtered through a 100-μm strainer. Red blood cells were lysed in BD Pharm Lyse Buffer (BD Biosciences), and the remaining cells were gently washed twice in ice-cold PBS, filtered through a 40-μm strainer, kept on ice, and used within 4–6 h. To reduce biological variation, splenocytes from three mice were pooled. The cells (at 2 × 107/mL, counted by hemocytometer) then were mixed with an equal volume of eFluor 450 (Pacific Blue-A) cell proliferation dye (eBioscience) at 200 μM in PBS and were incubated at room temperature in the dark for 20 min before the addition of ice-cold growth medium (DMEM, 10% FBS, 5× volume) and incubation on ice for 5 min to stop the reaction. Splenocytes then were washed three times in ice-cold PBS and were resuspended in growth medium (at 2 × 105 cells/100 μL) before being added to a round-bottomed 96-well plate precoated with anti-CD3ε antibody (100 μg/mL in PBS, 50 μL per well), incubated at 37 °C for 2–3 h, and washed twice in PBS. To each well of splenocytes (100 μL), anti-mouse CD28 antibody (1 μg in 1 μL) was added to stimulate T cells, with the immediate addition of CD11b+ cells (2 × 105/100 μL of medium) isolated from the lung by the Miltenyi microbeads (as described above) or growth medium without CD11b+ cells as a control. The plates were incubated at 37 °C in the CO2 incubator for 5 d before staining by CD8-PE antibody to detect CD8+ T cells and flow cytometry analysis.
Analyses of Publicly Available Datasets Derived from Human Samples.
For primary tumors, the online tool PROGgeneV2 (watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene) was used to analyze the correlation between patient outcome and gene expression of ATF3 and/or TEK (the human ortholog of mouse Tie2). Human microarray datasets from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) or the National Center for Biotechnology Information Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) database are specified in the figure legends. For the metastatic sites, data were extracted from Foukakis et al. (76) (GEO accession no. GSE54323). The correlation between the mRNA levels of Atf3 and those of the indicated genes was analyzed using the Pearson correlation, and the line of best fit was generated by linear regression.
Statistics.
Unless otherwise indicated, statistic methods were two-way ANOVA followed by the Bonferroni post hoc analysis using the Sigma Plot software. The Pearson correlation analysis, linear regression, log-rank test, and Student t test (two-sided, unless otherwise indicated) were also used as indicated in the figure legends. Data represent the mean ± SEM; P < 0.05 was considered statistically significant.
Study Approval.
All mouse studies were approval by the Ohio State University Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank the imaging facility at Rightmire Hall supported by NIH Grant P30-NS045758 and the Ohio State University Comprehensive Cancer Center for core facilities (the Campus Microscopy and Imaging Facility, the Comparative Pathology and Mouse Phenotyping Shared Resource, and the Analytical Cytometry Shared Resource); the ENCODE Project Consortium and the ENCODE production laboratories (Dr. R. M. Myers at the HudsonAlpha Institute for Biotechnology and Dr. B. E. Bernstein at the Broad Institute) for the ChIP-seq data (https://genome.ucsc.edu/); G. Gadre for the script to automate the analyses of the CD31 area and pericyte coverage; Dr. J. Jontes for helpful comments; and Dr. C. C. Wolford and J. L. Dominick for initial analyses. This work was supported in part by Department of Defense Grant W81XWH-14-1-0179 (to T.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700455114/-/DCSupplemental.
References
- 1.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Holen KD, Saltz LB. New therapies, new directions: Advances in the systemic treatment of metastatic colorectal cancer. Lancet Oncol. 2001;2:290–297. doi: 10.1016/S1470-2045(00)00324-7. [DOI] [PubMed] [Google Scholar]
- 3.Corradini P, Ladetto M, Pileri A, Tarella C. Clinical relevance of minimal residual disease monitoring in non-Hodgkin’s lymphomas: A critical reappraisal of molecular strategies. Leukemia. 1999;13:1691–1695. doi: 10.1038/sj.leu.2401559. [DOI] [PubMed] [Google Scholar]
- 4.Ran S. The role of TLR4 in chemotherapy-driven metastasis. Cancer Res. 2015;75:2405–2410. doi: 10.1158/0008-5472.CAN-14-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiao SL, Ganesan AP, Rugo HS, Coussens LM. Immune microenvironments in solid tumors: New targets for therapy. Genes Dev. 2011;25:2559–2572. doi: 10.1101/gad.169029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Palma M, Lewis CE. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013;23:277–286. doi: 10.1016/j.ccr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Emmenegger U, Kerbel RS. Cancer: Chemotherapy counteracted. Nature. 2010;468:637–638. doi: 10.1038/468637a. [DOI] [PubMed] [Google Scholar]
- 9.Shaked Y. Balancing efficacy of and host immune responses to cancer therapy: The yin and yang effects. Nat Rev Clin Oncol. 2016;13:611–626. doi: 10.1038/nrclinonc.2016.57. [DOI] [PubMed] [Google Scholar]
- 10.Quintavalle M, Elia L, Price JH, Heynen-Genel S, Courtneidge SA. A cell-based high-content screening assay reveals activators and inhibitors of cancer cell invasion. Sci Signal. 2011;4:ra49. doi: 10.1126/scisignal.2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang AC, Su QB, Wu FX, Zhang XL, Liu PS. Role of TLR4 for paclitaxel chemotherapy in human epithelial ovarian cancer cells. Eur J Clin Invest. 2009;39:157–164. doi: 10.1111/j.1365-2362.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert LA, Hemann MT. Chemotherapeutic resistance: Surviving stressful situations. Cancer Res. 2011;71:5062–5066. doi: 10.1158/0008-5472.CAN-11-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaked Y, et al. Rapid chemotherapy-induced acute endothelial progenitor cell mobilization: Implications for antiangiogenic drugs as chemosensitizing agents. Cancer Cell. 2008;14:263–273. doi: 10.1016/j.ccr.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–366. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daenen LG, et al. Chemotherapy enhances metastasis formation via VEGFR-1-expressing endothelial cells. Cancer Res. 2011;71:6976–6985. doi: 10.1158/0008-5472.CAN-11-0627. [DOI] [PubMed] [Google Scholar]
- 16.Shree T, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25:2465–2479. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeNardo DG, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinushi M, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc Natl Acad Sci USA. 2011;108:12425–12430. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruchard M, et al. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat Med. 2013;19:57–64. doi: 10.1038/nm.2999. [DOI] [PubMed] [Google Scholar]
- 20.Nakasone ES, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21:488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchem JB, et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013;73:1128–1141. doi: 10.1158/0008-5472.CAN-12-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding ZC, et al. Immunosuppressive myeloid cells induced by chemotherapy attenuate antitumor CD4+ T-cell responses through the PD-1-PD-L1 axis. Cancer Res. 2014;74:3441–3453. doi: 10.1158/0008-5472.CAN-13-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes R, et al. Perivascular M2 macrophages stimulate tumor relapse after chemotherapy. Cancer Res. 2015;75:3479–3491. doi: 10.1158/0008-5472.CAN-14-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulus P, Stanley ER, Schäfer R, Abraham D, Aharinejad S. Colony-stimulating factor-1 antibody reverses chemoresistance in human MCF-7 breast cancer xenografts. Cancer Res. 2006;66:4349–4356. doi: 10.1158/0008-5472.CAN-05-3523. [DOI] [PubMed] [Google Scholar]
- 25.Ruffell B, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingis-Velitski S, et al. Host response to short-term, single-agent chemotherapy induces matrix metalloproteinase-9 expression and accelerates metastasis in mice. Cancer Res. 2011;71:6986–6996. doi: 10.1158/0008-5472.CAN-11-0629. [DOI] [PubMed] [Google Scholar]
- 27.Volk-Draper L, et al. Paclitaxel therapy promotes breast cancer metastasis in a TLR4-dependent manner. Cancer Res. 2014;74:5421–5434. doi: 10.1158/0008-5472.CAN-14-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 29.Hai T, Hartman MG. The molecular biology and nomenclature of the ATF/CREB family of transcription factors: ATF proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 30.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 31.Hai T, Wolford CC, Chang YS. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: Is modulation of inflammation a unifying component? Gene Expr. 2010;15:1–11. doi: 10.3727/105221610x12819686555015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balkwill F, Mantovani A. Inflammation and cancer: Back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 33.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 35.Tan TT, Coussens LM. Humoral immunity, inflammation and cancer. Curr Opin Immunol. 2007;19:209–216. doi: 10.1016/j.coi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Wolford CC, et al. Transcription factor ATF3 links host adaptive response to breast cancer metastasis. J Clin Invest. 2013;123:2893–2906. doi: 10.1172/JCI64410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh YK, et al. Role of activating transcription factor 3 on TAp73 stability and apoptosis in paclitaxel-treated cervical cancer cells. Mol Cancer Res. 2008;6:1232–1249. doi: 10.1158/1541-7786.MCR-07-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St Germain C, et al. Cisplatin induces cytotoxicity through the mitogen-activated protein kinase pathways and activating transcription factor 3. Neoplasia. 2010;12:527–538. doi: 10.1593/neo.92048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park EJ, Kwon HK, Choi YM, Shin HJ, Choi S. Doxorubicin induces cytotoxicity through upregulation of pERK-dependent ATF3. PLoS One. 2012;7:e44990. doi: 10.1371/journal.pone.0044990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei XF, et al. Explant-cell culture of primary mammary tumors from MMTV-c-Myc transgenic mice. In Vitro Cell Dev Biol Anim. 2004;40:14–21. doi: 10.1290/1543-706X(2004)40<14:ECOPMT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 41.De Palma M, et al. Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell. 2005;8:211–226. doi: 10.1016/j.ccr.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Wyckoff JB, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 43.Harney AS, et al. Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA. Cancer Discov. 2015;5:932–943. doi: 10.1158/2159-8290.CD-15-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson BD, et al. Tumor microenvironment of metastasis in human breast carcinoma: A potential prognostic marker linked to hematogenous dissemination. Clin Cancer Res. 2009;15:2433–2441. doi: 10.1158/1078-0432.CCR-08-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 46.Wang W, et al. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67:3505–3511. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- 47.Di Modugno F, et al. The cytoskeleton regulatory protein hMena (ENAH) is overexpressed in human benign breast lesions with high risk of transformation and human epidermal growth factor receptor-2-positive/hormonal receptor-negative tumors. Clin Cancer Res. 2006;12:1470–1478. doi: 10.1158/1078-0432.CCR-05-2027. [DOI] [PubMed] [Google Scholar]
- 48.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sparreboom A, van Tellingen O, Nooijen WJ, Beijnen JH. Preclinical pharmacokinetics of paclitaxel and docetaxel. Anticancer Drugs. 1998;9:1–17. doi: 10.1097/00001813-199801000-00001. [DOI] [PubMed] [Google Scholar]
- 50.Anderson WF, Jatoi I, Tse J, Rosenberg PS. Male breast cancer: A population-based comparison with female breast cancer. J Clin Oncol. 2010;28:232–239. doi: 10.1200/JCO.2009.23.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barry M, Bleackley RC. Cytotoxic T lymphocytes: All roads lead to death. Nat Rev Immunol. 2002;2:401–409. doi: 10.1038/nri819. [DOI] [PubMed] [Google Scholar]
- 52.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Noy R, Pollard JW. Tumor-associated macrophages: From mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 55.Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaplan RN, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian B, et al. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One. 2009;4:e6562. doi: 10.1371/journal.pone.0006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panni RZ, Linehan DC, DeNardo DG. Targeting tumor-infiltrating macrophages to combat cancer. Immunotherapy. 2013;5:1075–1087. doi: 10.2217/imt.13.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanford DE, et al. Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: A role for targeting the CCL2/CCR2 axis. Clin Cancer Res. 2013;19:3404–3415. doi: 10.1158/1078-0432.CCR-13-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao J, Li X, Guo M, Yu J, Yan C. The common stress responsive transcription factor ATF3 binds genomic sites enriched with p300 and H3K27ac for transcriptional regulation. BMC Genomics. 2016;17:335. doi: 10.1186/s12864-016-2664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boersma BJ, et al. A stromal gene signature associated with inflammatory breast cancer. Int J Cancer. 2008;122:1324–1332. doi: 10.1002/ijc.23237. [DOI] [PubMed] [Google Scholar]
- 65.Fokas E, Engenhart-Cabillic R, Daniilidis K, Rose F, An HX. Metastasis: The seed and soil theory gains identity. Cancer Metastasis Rev. 2007;26:705–715. doi: 10.1007/s10555-007-9088-5. [DOI] [PubMed] [Google Scholar]
- 66.Symmans WF, et al. Long-term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J Clin Oncol. 2017;35:1049–1060. doi: 10.1200/JCO.2015.63.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borowsky AD, et al. Syngeneic mouse mammary carcinoma cell lines: Two closely related cell lines with divergent metastatic behavior. Clin Exp Metastasis. 2005;22:47–59. doi: 10.1007/s10585-005-2908-5. [DOI] [PubMed] [Google Scholar]
- 68.Fujioka H, et al. Comparative proteomic analysis of paclitaxel resistance-related proteins in human breast cancer cell lines. Oncol Lett. 2017;13:289–295. doi: 10.3892/ol.2016.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roh-Johnson M, et al. Macrophage contact induces RhoA GTPase signaling to trigger tumor cell intravasation. Oncogene. 2014;33:4203–4212. doi: 10.1038/onc.2013.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rohan TE, et al. Tumor microenvironment of metastasis and risk of distant metastasis of breast cancer. J Natl Cancer Inst. 2014;106:dju136. doi: 10.1093/jnci/dju136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stacker SA, et al. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat Rev Cancer. 2014;14:159–172. doi: 10.1038/nrc3677. [DOI] [PubMed] [Google Scholar]
- 72.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 73.Hai T, Jalgaonkar S, Wolford CC, Yin X. 2011. Immunohistochemical detection of activating transcription factor 3, a hub of the cellular adaptive-response network. Methods in Enzymology, Unfolded Protein Response and Cellular Stress, Part B, ed Conn MP (Elsevier Inc. Academic, Burlington, MA), Vol 490, pp 175–194. [DOI] [PMC free article] [PubMed]
- 74.Yin X, Dewille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene. 2008;27:2118–2127. doi: 10.1038/sj.onc.1210861. [DOI] [PubMed] [Google Scholar]
- 75.Lu D, Wolfgang CD, Hai T. Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J Biol Chem. 2006;281:10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 76.Foukakis T, et al. Gene expression profiling of sequential metastatic biopsies for biomarker discovery in breast cancer. Mol Oncol. 2015;9:1384–1391. doi: 10.1016/j.molonc.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.