Significance
Living organisms perceive temperature using thermosensory molecules. In this study, we found that phototropin, a blue-light photoreceptor, perceives temperature via a mechanism based on the photoactivated chromophore’s lifetime to induce appropriate chloroplast positioning in plants. Our findings reveal that the chromophore of phototropin directs chloroplast positioning to optimize photosynthesis in plants by (i) sensing blue light and (ii) sensing temperature via a temperature-dependent lifetime mechanism. Because many photoreceptors in a range of organisms contain phtoactivatable chromophores with a thermodependent lifetime, the present study suggests that a common molecular principle underlies biological perception of temperature.
Keywords: chloroplast movement, dark reversion, photoreceptor, thermal reversion, thermosensor
Abstract
Living organisms detect changes in temperature using thermosensory molecules. However, these molecules and/or their mechanisms for sensing temperature differ among organisms. To identify thermosensory molecules in plants, we investigated chloroplast positioning in response to temperature changes and identified a blue-light photoreceptor, phototropin, that is an essential regulator of chloroplast positioning. Based on the biochemical properties of phototropin during the cellular response to light and temperature changes, we found that phototropin perceives temperature based on the temperature-dependent lifetime of the photoactivated chromophore. Our findings indicate that phototropin perceives both blue light and temperature and uses this information to arrange the chloroplasts for optimal photosynthesis. Because the photoactivated chromophore of many photoreceptors has a temperature-dependent lifetime, a similar temperature-sensing mechanism likely exists in other organisms. Thus, photoreceptors may have the potential to function as thermoreceptors.
Living organisms perceive temperature using thermosensory molecules. Various types of thermosensory molecules have been identified in microorganisms, animals, and plants, and these molecules perceive temperature via temperature-dependent intra- and intermolecular interactions (1–3). However, these thermosensory molecules and/or their mechanisms for temperature sensing differ among organisms (1).
As photosynthetic organelles, chloroplasts change their intracellular position in response to light and temperature (4–7). For example, under low light conditions at 22 °C, chloroplasts accumulate at the cell surface (along the periclinal cell wall) to maximize photosynthetic efficiency, in a phenomenon termed the accumulation response (4, 6). However, under the same light conditions, but at a temperature of 5 °C, the chloroplasts move to the cell periphery (along the anticlinal cell wall), possibly to avoid the light, in a phenomenon termed the cold-avoidance response or the cold-positioning response (5, 6). Because the cold-avoidance response is temperature dependent (5), the response is likely controlled by a thermosensory molecule. In a previous study, we found that the blue-light (BL) photoreceptor phototropin (phot) is responsible for the cold-avoidance response in the fern Adiantum capillus-veneris, which has three types of phot molecules (phot1, phot2, and neochrome1) (5, 8). In this fern, phot2 mediates the cold-avoidance response (5). Therefore, in the present study, we hypothesized that phot is involved in temperature perception and we further analyzed the cold-avoidance response in the liverwort Marchantia polymorpha, which has only a single copy of the PHOT gene (MpPHOT) (9).
Results and Discussion
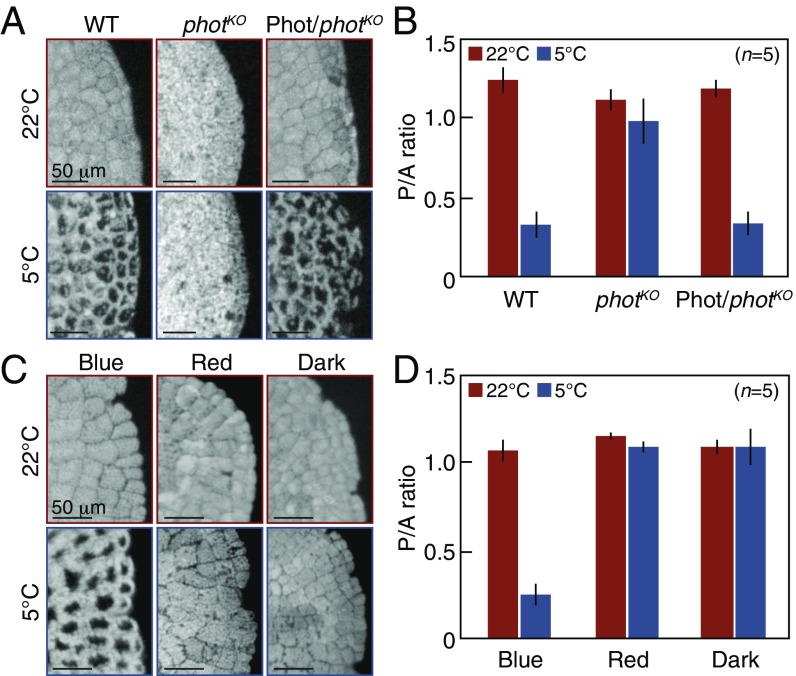
First, we examined whether MpPHOT is essential for the cold-avoidance response in M. polymorpha. When the wild type (WT), the knockout mutant (MpphotKO), and the complementation line (Mpphot/MpphotKO) (9) (Fig. S1) were treated with low white light at 5 °C for 24 h to induce the cold-avoidance response, all plants exhibited the response except for the MpphotKO mutant (Fig. 1A). To determine the position of the chloroplasts in the treated plants, we measured the intensities of chlorophyll autofluorescence along the periclinal (P) and anticlinal (A) cell walls, and the ratio of P/A autofluorescence was used as an arbitrary value to determine chloroplast position, according to previous reports (5, 6). The P/A ratio was lower at 5 °C than at 22 °C in the WT and the Mpphot/MpphotKO line, but the ratio was equivalent in the MpphotKO mutant at 5 °C and 22 °C (Fig. 1B). These results indicate that Mpphot is essential for the cold-avoidance response in M. polymorpha.
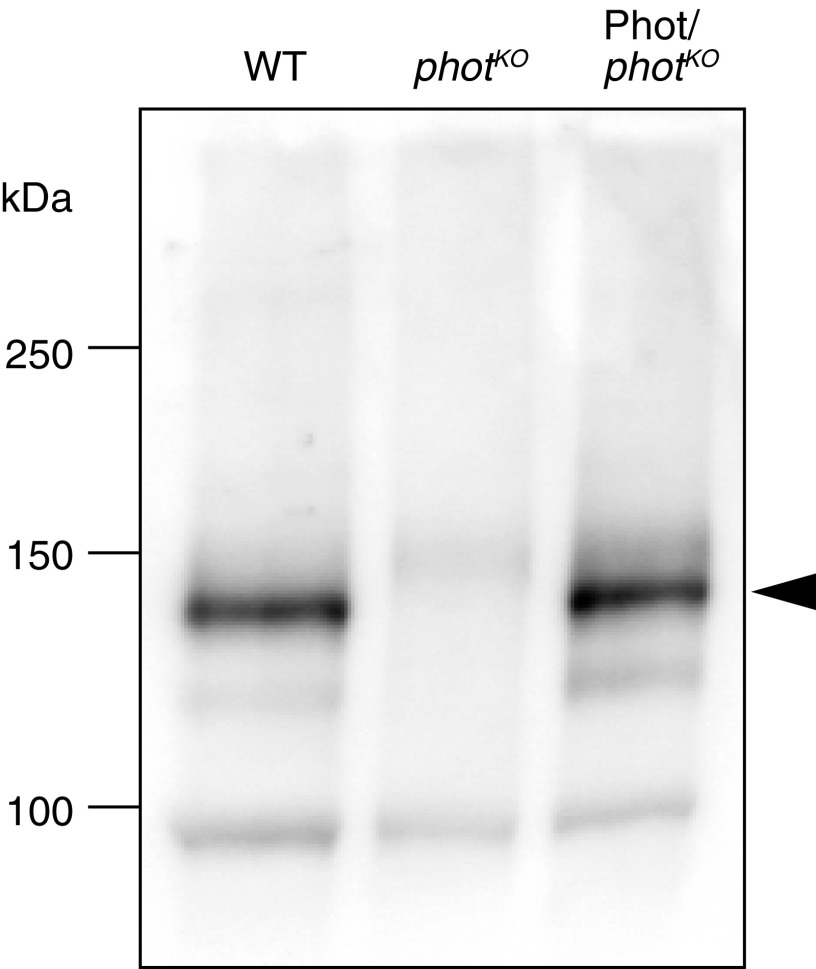
Fig. S1.
Mpphot amounts in WT, MpphotKO, and Mpphot/MpphotKO plants. Immunoblot analysis using anti-Mpphot antibodies. Mpphot was not detected in MpphotKO, but was detected in Mpphot/MpphotKO at the same level as in the WT. Arrowhead indicates Mpphot.
Fig. 1.
The cold-avoidance response is dependent on Mpphot and BL. (A) Cold-avoidance response in the wild type (shown as WT), the MpphotKO mutant (shown as photKO), and the Mpphot/MpphotKO complementation line (shown as Phot/photKO) (Fig. S1) under white-light conditions. Chlorophyll fluorescence images at 22 °C and 5 °C under weak white light (140 µmol photons m−2⋅s−1) are shown. To induce the cold-avoidance response, the samples were incubated at 5 °C for 24 h. (B) Quantitative analysis of the samples shown in A using the P/A ratio. (C) Cold-avoidance response of WT cells in different light conditions, including blue (25 µmol photons m−2 ⋅s−1) and red (35 µmol photons m−2⋅s−1) light, and in darkness. The samples were incubated at 5 °C for 24 h. (D) Quantitative analysis of the samples in C using the P/A ratio. (B and D) Error bars represent SDs.
Next, we examined whether BL is required by Mpphot for the cold-avoidance response. Following the induction of the accumulation response at 22 °C with low light, the WT cells were treated with weak BL and two control conditions (red light and darkness) at 5 °C. The cold-avoidance response was observed in the BL-treated plants, but not in those subjected to control conditions (Fig. 1 C and D). To confirm the BL-dependent response, we constructed a temperature-controlled microscope (10), equipped with a BL microbeam irradiation system, and performed single-cell analysis. When the WT cells were irradiated with weak BL at 22 °C, the chloroplasts accumulated in the irradiated area (Movie S1). However, when the cells were irradiated at 5 °C, only the chloroplasts inside the irradiated area exhibited the cold-avoidance response and moved to the outside of the cell to avoid the light (Movie S2). These results indicate that the cold-avoidance response requires BL, suggesting that BL perception by Mpphot is essential.
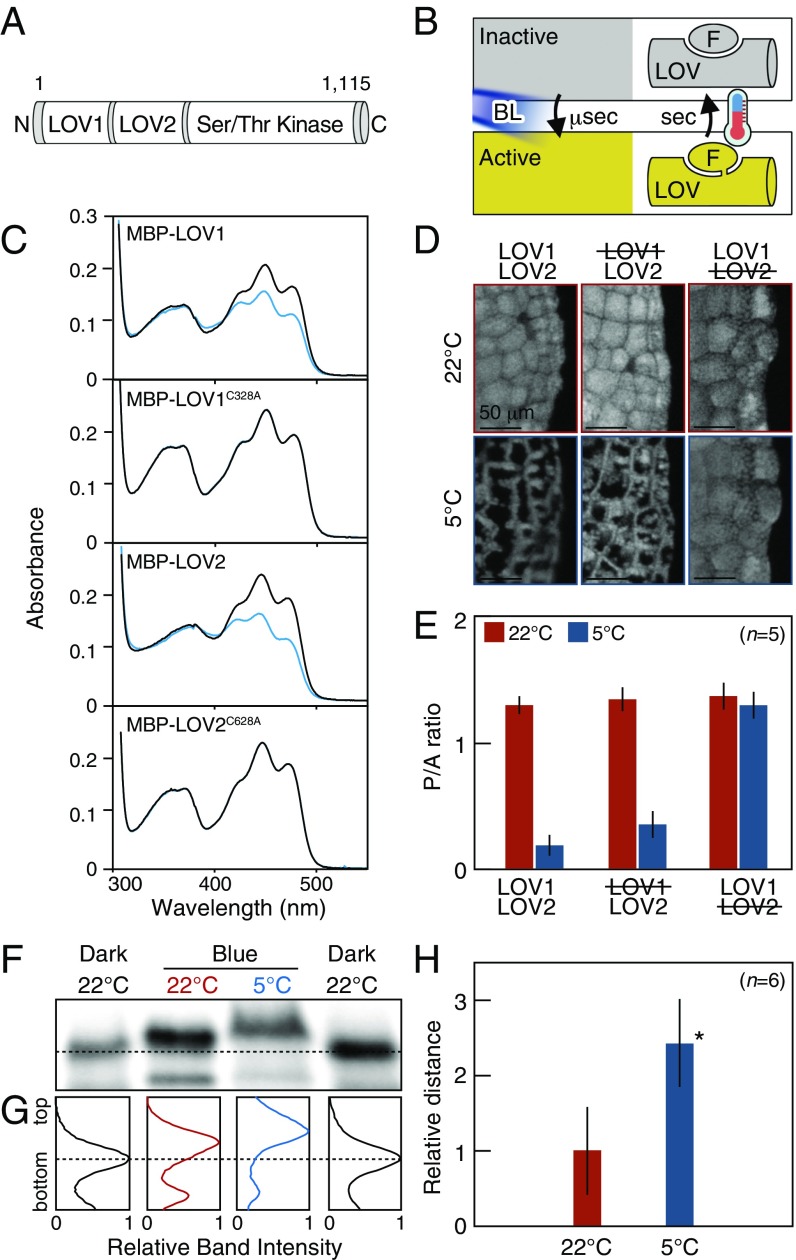
BL perception by phot is carried out by two photosensory light/oxygen/voltage (LOV1 and LOV2) domains at the N-terminal region (Fig. 2A), which have active and inactive states determined by a bond with a flavin mononucleotide (FMN) (11). In darkness, the LOV domain noncovalently binds to the FMN conferring the inactive state (Fig. 2B). Upon BL excitation, on a microsecond scale, the LOV domain covalently binds to the FMN at a conserved cysteine residue conferring the active state (Fig. 2B). The state change is thermoreversible; a temperature increase causes the active LOV domain to disconnect from the FMN and to return to the inactive state on a second scale (Fig. 2B). Because the thermodependent reversion can be experimentally observed under dark conditions, this phenomenon is termed dark reversion or thermal reversion. This state change regulates the activity of a serine/threonine kinase at the C-terminal region of phot (Fig. 2A). The kinase-mediated signaling of phot induces the BL-dependent movement of the chloroplasts (11).
Fig. 2.
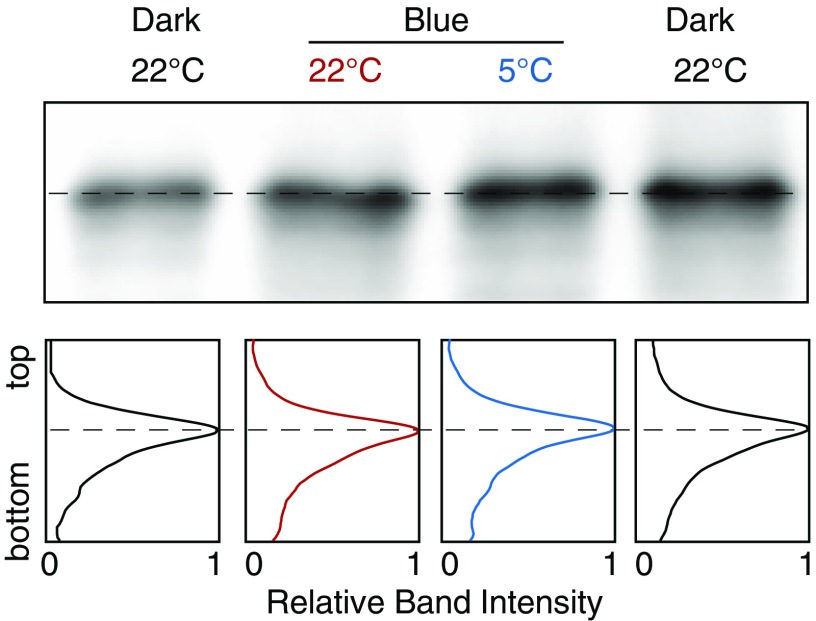
Active LOV2 mediates the cold-avoidance response. (A) Schematic illustration of Mpphot. The number of amino acids is shown above the image. N and C indicate the N and C termini, respectively. (B) Schematic illustration of the state change of the LOV domain. F indicates FMN. (C) Elimination of the active LOV1 and LOV2 domains of Mpphot. Bacterially expressed MBP–LOV1, MBP–LOV1C328A, MBP–LOV2, and MBP–LOV2C628A were purified and then analyzed using a double-beam spectrophotometer equipped with a temperature-controlled cuvette cell. (D) Disruption of the active LOV domain by the cold-avoidance response at 5 °C for 24 h. Crossed-out text shows disruption of the indicated LOV domain. Mpphot/MpphotKO, MpphotC328A/MpphotKO (line 1, Fig. S2), and MpphotC628A/MpphotKO (line 2, Fig. S2) plants are shown as LOV1LOV2, LOV1LOV2, and LOV1LOV2, respectively. (E) Quantitative analysis of the samples in D. (F) Detection of autophosphorylated Mpphot by the electrophoretic mobility shift assay. Dark-adapted plants were incubated under weak BL (25 µmol photons m−2⋅s−1) at 22 °C or 5 °C for 24 h. The dashed line indicates the basal band position of the dark sample. (G) Band intensity histograms of the samples in F. The dashed line indicates the basal band position of the dark sample. (H) Quantitative analysis of the mobility shift of the samples shown in F. Relative distance from the band position of the dark sample was measured. Student’s t test was performed for statistical evaluation (*P < 0.001). (E and H) Error bars represent SDs.
To determine whether the state change of the LOV domain is involved in the cold-avoidance response, the conserved cysteine residue (9) was substituted with alanine (C328A and C628A for LOV1 and LOV2, respectively) to block the covalent binding with FMN and eliminate the active state of the LOV domains (Fig. 2C). The absorbance spectra of recombinant LOV1 and LOV2 proteins fused with maltose-binding protein (MBP–LOV1 and MBP–LOV2, respectively) were dramatically changed by pulse BL irradiation (200 µmol photons m−2⋅s−1) for 30 s (blue line) and were restored after dark treatment (5 min) (black line). By contrast, BL irradiation did not affect the absorbance spectra in MBP–LOV1C328A and MBP–LOV2C628A, indicating complete elimination of the active state (Fig. 2C). When transgenic plants lacking active LOV1 (MpphotC328A/MpphotKO) or LOV2 (MpphotC628A/MpphotKO) were produced (Fig. S2), MpphotC328A could induce the cold-avoidance response, whereas MpphotC628A could not (Fig. 2 D and E). Thus, active LOV2 mediates the cold-avoidance response.
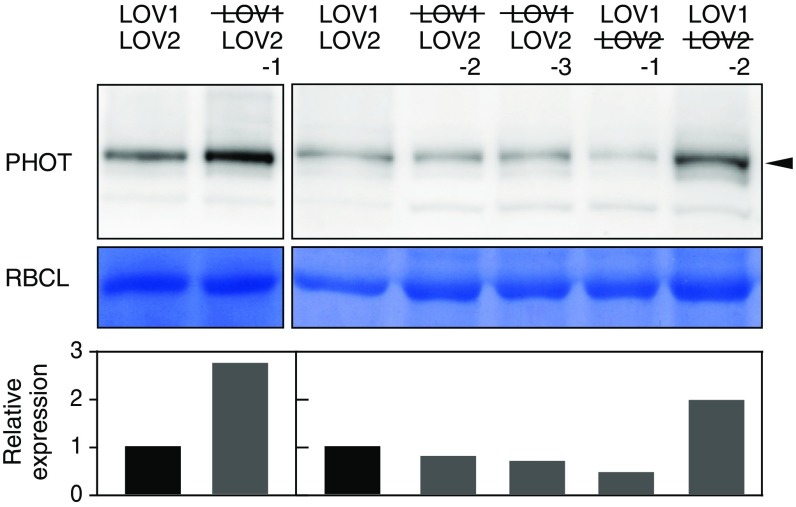
Fig. S2.
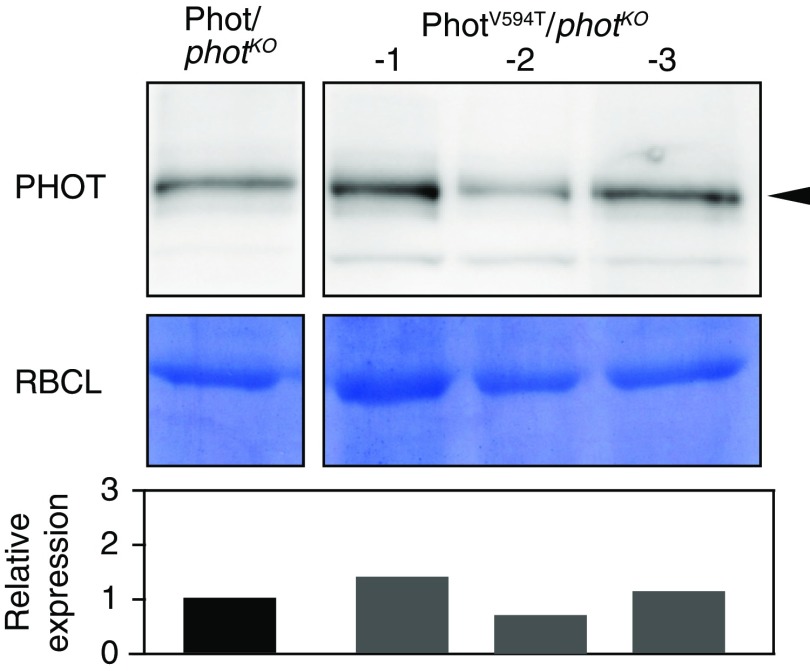
The amount of Mpphot in the MpphotC328A/MpphotKO and MpphotC628A/MpphotKO plants. The amount of Mpphot in Mppot/MpphotKO (shown as LOV1LOV2), MpphotC328A/MpphotKO (shown as LOV1LOV2), and MpphotC628A/MpphotKO (shown as LOV1LOV2) plants was detected by immunoblot analysis using anti-Mpphot antibodies. Arrowhead indicates Mpphot. To calculate the relative expression, the band intensity of Mpphot was divided by that of the large subunit of ribulose-1,5-bisphosphate carboxylase (RBCL). The band intensities were measured using ImageJ software.
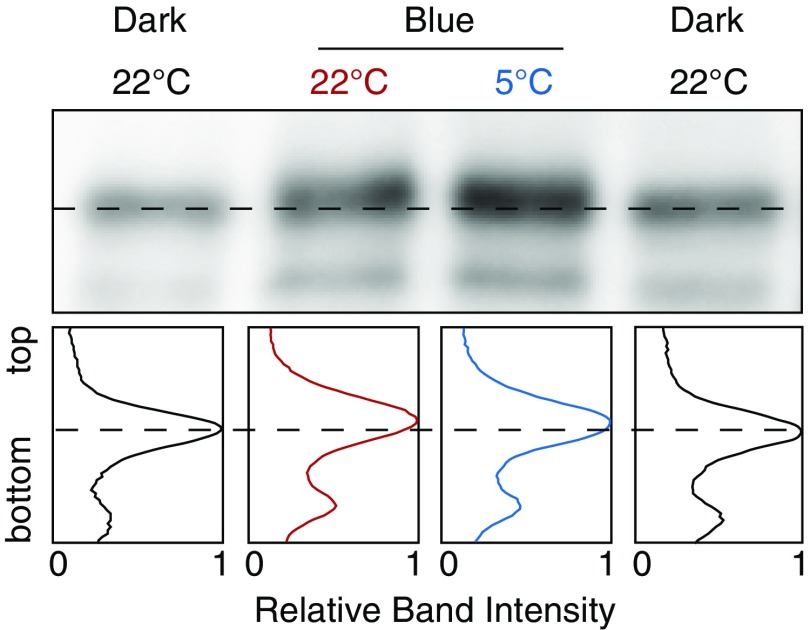
Because active LOV2 induces autophosphorylation of phot via the C-terminal kinase (12) (Fig. 2A), the level of autophosphorylation reflects the amount of active LOV2 (12). To determine the amount of active LOV2 in WT cells during the cold-avoidance response, we analyzed autophosphorylation of Mpphot by the electrophoretic mobility shift assay. The autophosphorylation level under weak BL was significantly higher at 5 °C than at 22 °C (Fig. 2 F–H and Fig. S3), suggesting that levels of active LOV2 increase during the cold-avoidance response.
Fig. S3.
Phosphorylation-dependent mobility shift of Mpphot. After desalting of the lysates of blue light (25 µmol photons m−2⋅s−1)-irradiated Mpphot/MpphotKO using an Amicon Ultra 30K device (EMD Millipore), 15 units of FastAP thermosensitive alkaline phosphatase (Thermo Fisher Scientific) was treated for 1 h at 37 °C. The band intensity histograms are shown. The light- and cold-dependent phosphorylations of Mpphot (Fig. 2 F and G) completely disappeared by the phosphatase treatment.
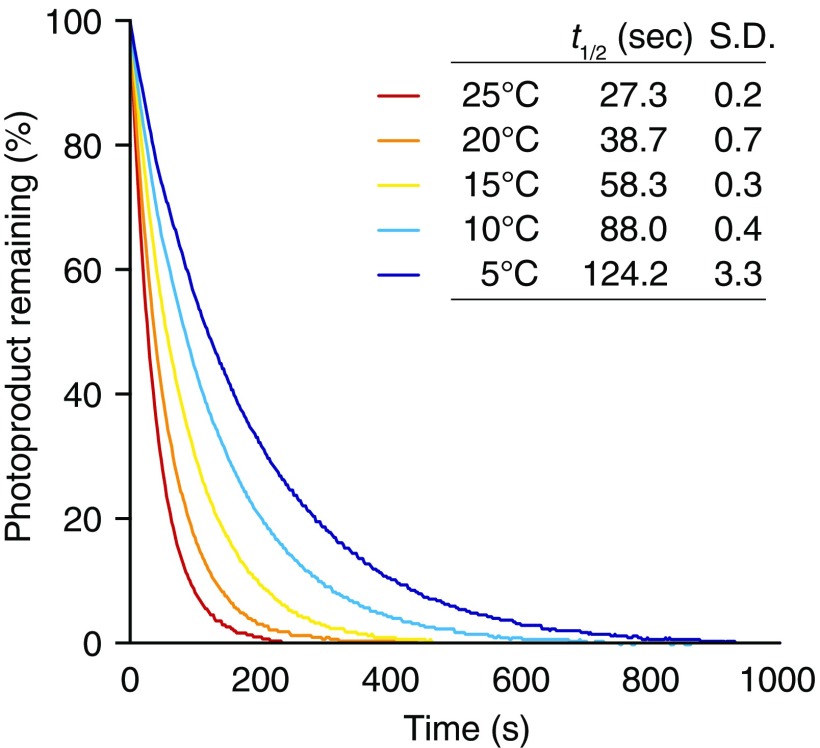
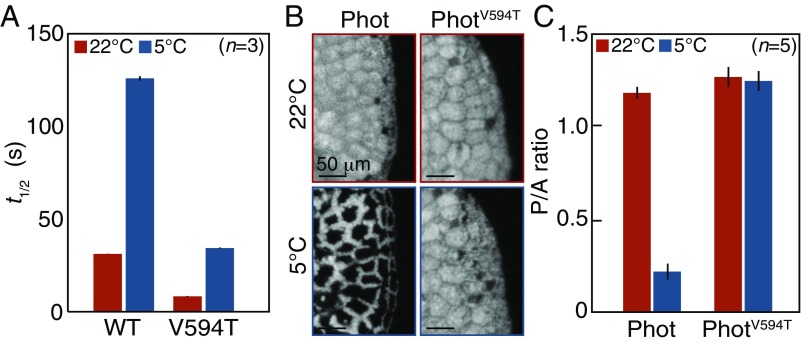
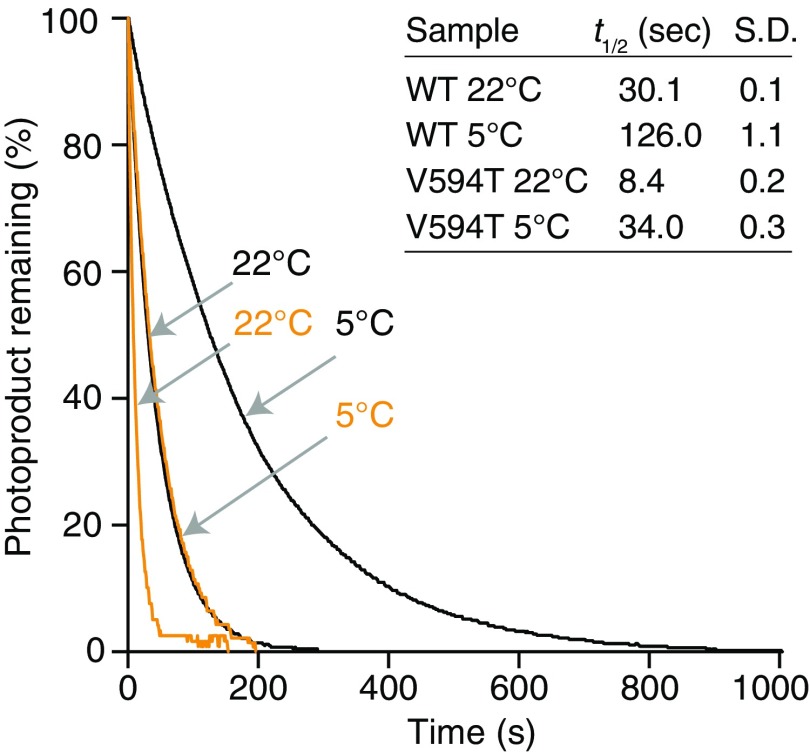
To increase the amount of active LOV2 during the cold-avoidance response, the lifetime of active LOV2 should be prolonged by slow thermal reversion under low temperatures (Fig. 2B). Using LOV2 from Mpphot, we confirmed that this molecule undergoes thermodependent reversion from the active to inactive state (Fig. S4) and found that the lifetime of active LOV2 at 5 °C [half-life (t1/2) ∼ 120 s] was fourfold longer than that at 22 °C (t1/2 ∼ 30 s) (Fig. 3A and Fig. S5). Given the temperature-dependent change of the lifetime, we predicted that Mpphot perceives temperature using the lifetime of active LOV2 to selectively induce the accumulation response or the cold-avoidance response. To test this possibility, we used an amino acid substitution (V594T) (13) (Fig. S6) that accelerates the speed of thermal reversion fourfold and confirmed that the lifetime of active LOV2V594T at 5 °C (t1/2 ∼ 30 s) is comparable to that of active LOV2 at 22 °C (t1/2 ∼ 30 s) (Fig. 3A and Fig. S5). We hypothesized that LOV2V594T would misperceive the 5 °C temperature as 22 °C. When the transgenic plants (MpphotV594T/MpphotKO) containing LOV2V594T (Fig. S7) were treated with weak BL at 5 °C to induce the cold-avoidance response, MpphotV594T misperceived the 5 °C temperature as 22 °C and thereby induced the accumulation response, but not the cold-avoidance response (Fig. 3 B and C). In the transgenic plants (MpphotV594T/MpphotKO), the autophosphorylation level of MpphotV594T (i.e., the amount of active LOV2V594T) under weak BL was confirmed to be comparable at 22 °C and 5 °C (Fig. S8). These results indicate that Mpphot perceives temperature using the lifetime of active LOV2, which then determines the accumulation response or the cold-avoidance response.
Fig. S4.
Thermoreversibility of photoactivated LOV2 from Mpphot. Reversion from the active to inactive states was analyzed using bacterially expressed MBP–LOV2 under different temperature conditions (5, 10, 15, 20, and 25 °C) and a double-beam spectrophotometer equipped with a temperature-controlled cuvette cell.
Fig. 3.
Mpphot perceives temperature using the lifetime of active LOV2. (A) Lifetime of active LOV2 and LOV2V594T at 22 °C and 5 °C. The graph shows the half-life (t1/2) in seconds. (B) Cold-avoidance response in Mpphot/MpphotKO (shown as Phot) (Fig. S1) and the accumulation response in MpphotV594T/MpphotKO (shown as PhotV594T) (line 3, Fig. S7) at 5 °C for 24 h. (C) Quantitative analysis of the samples shown in B. (A and C) Error bars represent SDs.
Fig. S5.
Measurement of the lifetime of active LOV2 at 22 °C and 5 °C. The half-life (t1/2) indicated in Fig. 3A was obtained from these measurements.
Fig. S6.
Alignment of the amino acid sequences of Asphot1 and Mpphot. The valine residue positioned at 416 in Avena sativa phot1 (13) is conserved at position 594 in Mpphot. Red arrowhead indicates the conserved valine residue. The V416T mutation of Asphot1 accelerates thermal reversion (13).
Fig. S7.
The amount of Mpphot in MpphotV594T/MpphotKO plants. The amount of Mpphot in Mppot/MpphotKO and MpphotV594T/MpphotKO plants was detected by immunoblot analysis using anti-Mpphot antibodies. The images were captured from a single membrane, and this figure was prepared without any change in brightness or contrast. Arrowhead indicates Mpphot. To calculate the relative expression, the band intensity of Mpphot was divided by that of the large subunit of ribulose-1,5-bisphosphate carboxylase (RBCL). The band intensities were measured using ImageJ software.
Fig. S8.
Detection of autophosphorylated MpphotV594T by an electrophoretic mobility shift assay. Dark-adapted transgenic plants (MpphotV594T/MpphotKO) were incubated under weak BL (25 µmol photons m−2⋅s−1) at 22 °C or 5 °C for 24 h. The dashed line indicates the basal band position of the dark sample. The band intensity histograms are shown. MpphotV594T was autophosphorylated by the BL irradiation, and the autophosphorylation level was comparable at 22 °C and 5 °C.
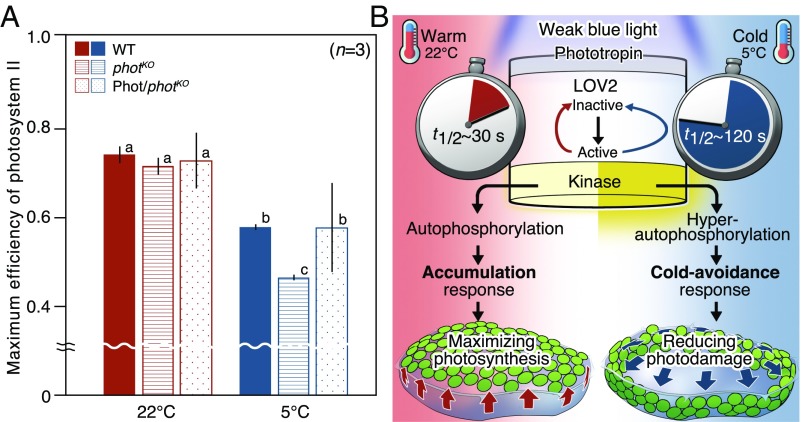
The accumulation response induced by Mpphot-mediated perception of normal temperatures (e.g., 22 °C) for plant growth and development contributes to the maximum photosynthetic efficiency (4), whereas the physiological significance of the cold-avoidance response by the perception of low temperature (e.g., 5 °C) remains to be determined. In a previous study, we observed the cold-avoidance response in evergreen ferns, but not in summer-green ferns, suggesting that the cold-avoidance response may be involved in maintaining photosynthesis at low temperature (5). To examine this possibility, we determined the impact of the cold-avoidance response on photosynthesis. When the maximum efficiency of photosystem II was measured after induction of the cold-avoidance response in the WT, the MpphotKO mutant, and the Mpphot/MpphotKO complementation line (Fig. S1), the efficiency was notably reduced in the MpphotKO mutant (Fig. 4A). This result suggests that the Mpphot-mediated cold-avoidance response may serve to reduce photodamage of the chloroplasts to maintain photosynthesis at low temperatures.
Fig. 4.
Physiological significance of the cold-avoidance response and a suggested model. (A) Comparison of the maximum efficiency of photosystem II between the wild type (shown as WT), MpphotKO (shown as photKO), and Mpphot/MpphotKO (shown as Phot/photKO) (Fig. S1). The plants were incubated under weak white light (70 µmol photons m−2⋅s−1) at 22 °C or 5 °C for 24 h. Error bars represent SDs. Means with different letters show significant differences (Tukey’s HSD, P < 0.001). (B) Schematic illustration of a suggested model of phot induction and its functions.
Based on these findings, we concluded that Mpphot perceives temperature using the lifetime of active LOV2 and thereby optimizes photosynthesis under different temperatures (Fig. 4B). The same cold-avoidance response mediated by phot has also been found in different clades of plants, including the fern Adiantum (5) and the angiosperm Arabidopsis (14) (Movies S3–S5), and the response may be conserved in many plant species. Based on our biochemical and physiological experiments, LOV2 plays a central role to perceive temperature, and the lifetime of active LOV2 determines appropriate chloroplast positioning at different temperatures. However, temperature perception by phot may be more complicated in vivo, because the lifetime (or the kinetics of dark reversion) of active LOV2 is altered in LOV1–LOV2 fusion and full-length phot proteins of Arabidopsis (15). In this context, the lifetime of active LOV2 seems to be affected by intramolecular interactions with other domains such as LOV1 and/or kinase. Further experiments such as in vivo measurement of the lifetime will be necessary to elucidate the phot-mediated perception of temperature in plants. As observed for chloroplasts, relocations of the nucleus and peroxisome under cold conditions (6) were impaired in the MpphotKO mutant (Fig. S9). Thus, multiple organelle relocations under cold conditions are mediated by phot. Recently, the red/far-red light photoreceptor, phytochrome (phy), in Arabidopsis was reported to perceive temperature (2, 3). Because the bioactive form of phy, which has a red-light–activated chromophore, thermally reverts to the inactive form, the molecular mechanism underlying temperature perception was inferred to be the temperature-dependent lifetime of the bioactive form (2, 3). Given that the lifetime of phy is relatively long (t1/2 = over 30 min) (16), phot, which has a short lifetime (t1/2 = ≈30 s), may be better able to cope with sudden changes of temperature in plants. Although both phot and phy require light to perceive temperature, their light-absorbing spectra (blue and red/far-red, respectively) predominate in sunlight at different times of day (morning and evening, respectively). Taken together, these findings suggest that plants use various thermosensory photoreceptors, such as phot and phy, to perceive ambient conditions. Furthermore, like phot and phy in plants, many photoreceptors (e.g., cryptochrome and rhodopsin) in other organisms have a photoactivatable chromophore that exhibits a temperature-dependent lifetime (i.e., the lifetime of the photoactivated form of the chromophore is determined by a thermal reversion mechanism) (17). Thus, photoreceptors may have the potential to function as thermoreceptors, and the lifetime-mediated mechanism for perceiving temperature may be a common molecular principle in living organisms.
Fig. S9.
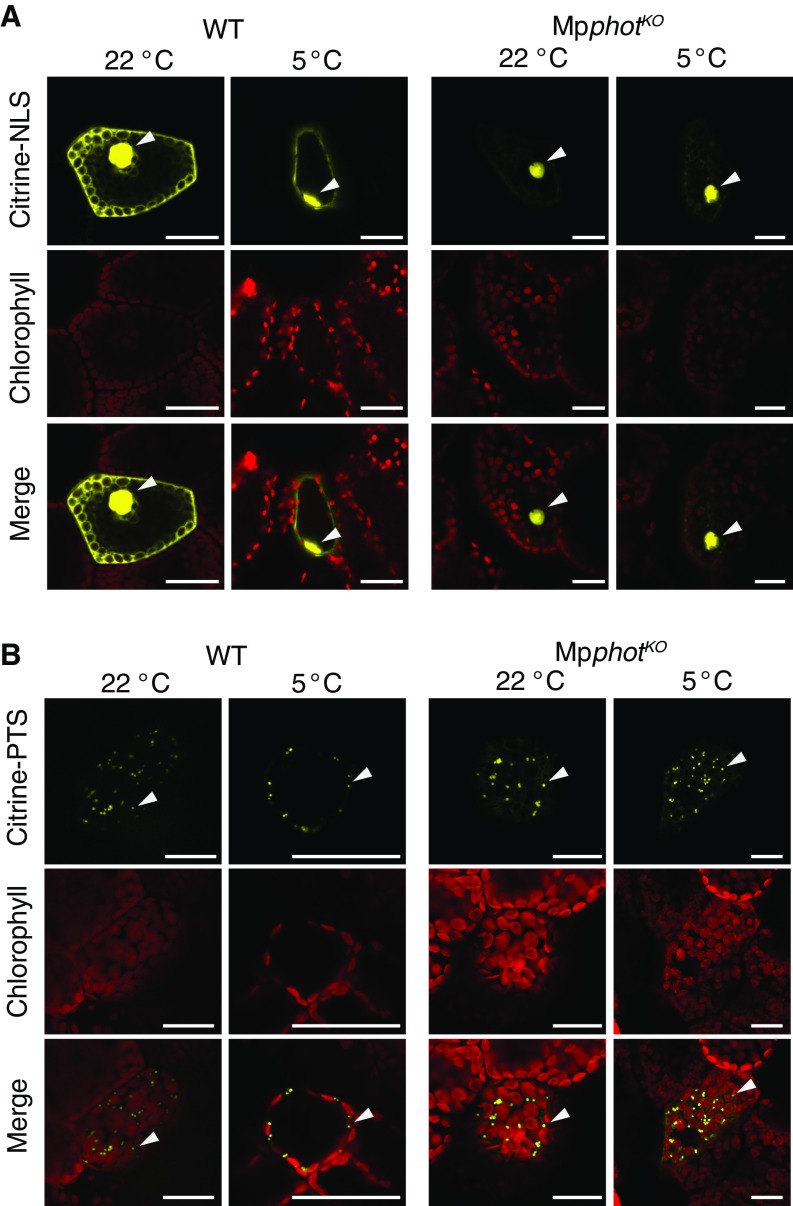
Cold-induced relocation of the nucleus and peroxisome in the MpphotKO mutant. The nucleus (A) and peroxisome (B) were visualized using yellow fluorescent proteins (Citrine–NLS and Citrine–PTS, respectively). The bombarded cells (Citrine–NLS/WT, Citrine–NLS/MpphotKO, Citrine–PTS/WT, and Citrine–PTS/MpphotKO) were incubated under weak BL (25 µmol photons m−2⋅s−1) at 22 °C or 5 °C for 24 h and observed by confocal laser scanning microscopy (SP8X, Leica Microsystems) with the time-gating method (26). For detection of Citrine fluorescence, a 513-nm laser line and a range of 520–560 nm were used for excitation and emission, respectively. (Scale bar, 25 µm.) Arrowheads indicate fluorescent nuclei (A) and peroxisomes (B).
Materials and Methods
Plant Materials and Culture Conditions.
The liverwort M. polymorpha was grown in a culture room (temperature: 22 °C, humidity: ≈40%) and asexually maintained on M51C or one-half-strength B5 medium with 1% agar under 75 µmol photons m−2⋅s−1 continuous white light (FL40SW, NEC Corporation) as described previously (18–20). The male accession Takaragaike-1 (Tak1), which was used as the WT, and MpphotKO and Mpphot/MpphotKO plants were kindly provided by Takayuki Kohchi, Kyoto University, Kyoto; the MpphotKO and the Mpphot/MpphotKO plants were previously produced by Aino Komatsu, Kimitsune Ishizaki, Noriyuki Suetsugu, Masamitsu Wada, and Takayuki Kohchi (9). Arabidopsis thaliana (gl line as the WT, phot1-5, and phot2-1) were cultured on one-half-strength MS medium in the same culture room. A. thaliana seeds were kindly provided by Masamitsu Wada, Tokyo Metropolitan University, Tokyo. The other culture conditions for A. thaliana were previously reported (21, 22).
Light and Temperature Treatments.
Light and temperature treatments for M. polymorpha were previously described (6). Ten-day-old sporelings (thalli grown from spores) or 1-d-old gemmalings (thalli grown from gemmae) in Petri dishes were incubated at 22 °C or 5 °C in temperature-controlled incubators (IJ100 and IJ101, Yamato Scientific). To induce the cold-avoidance response, an illuminator with white- or blue-colored light-emitting diodes (LEDs) (OptoSupply) was set to 140 µmol photons m−2⋅s−1 or 25 µmol photons m−2⋅s−1 in the incubator. The LEDs were controlled by a direct-current stabilized power supply (AD-8723D, A&D Company). In some experiments (Fig. 1 C and D), blue (25 µmol photons m−2⋅s−1) and red (35 µmol photons m−2⋅s−1) lights were obtained from white-colored LEDs through plastic films (catalog no. 72 for blue and no. 21 for red, Tokyo Butai Showmei Holdings) (5). The light intensity was measured with a light meter LI-250A (LI-COR Bioscience).
Analysis of Chloroplast Position with Chlorophyll Fluorescence.
Chloroplast positions were observed by monitoring chlorophyll fluorescence using a Leica MZ16F stereo fluorescence microscope (Leica Microsystems) equipped with an excitation filter at 480/40 nm and a long-pass barrier filter at 510 nm. Fluorescence images were acquired as TIFF (tagged image file format) files of red/green/blue (RGB) digital images using a DP73 digital camera (Olympus). Image processing was carried out using ImageJ software (https://imagej.nih.gov/ij/). The chloroplast position in thalli cells of M. polymorpha was evaluated using the P/A ratio method, as previously reported (6). The P/A ratio (i.e., the brightness ratio of chlorophyll fluorescence emitted from chloroplasts along the periclinal and anticlinal cell walls) is a quantification method used to determine chloroplast position. The P/A ratios with a SD were obtained as an average of experiments repeated five times.
Time-Lapse Observations.
In a previous study, we constructed a temperature-regulated microscope system for observing chloroplasts under cold conditions (10). In this system, an inverted light microscope DM IL LED (Leica Microsystems) was used. In the present study, to equip the microscope with a microbeam irradiation system, a light-guiding optics module was used instead of a conventional fluorescence module. A blue-colored LED fiber at 450.8 nm (FOLS-01, Pi Photonics, Inc.) was used as the light source, and the beam diameter was determined with the diaphragm of the optical system. The intensity of the microbeam was measured using a power meter 1918-R (Newport Corporation) with a silicon detector 918D-SL-OD1. Chloroplast movement was tracked using a time-lapse video-recording system with a complementary metal oxide semiconductor (CMOS) camera, Moticam2000 (Shimadzu Rika Corporation), and analyzed with ImageJ software.
Plasmid Construction.
Gateway Cloning Technology (Invitrogen) was used to construct plasmids according to the manufacturer’s instructions. To introduce the mutations (C328A, V594T, and C628A) into the genomic fragment of MpPHOT (gMpPHOT), pENTR–gMpPHOT (9) (a donor vector containing gMpPHOT) was used as a template for the following PCR experiments. pENTR–gMpPHOT was a gift from Takayuki Kohchi, Kyoto University, Kyoto. For the C328A mutation, DpnI-mediated site-directed mutagenesis (23) was performed using the primers 5′-GAGGTCATCGGAAGAAACGCGTACGCATTC-3′ and 5′-GAATGCGTACGCGTTTCTTCCGATGACCTC-3′. The V594T and C628A mutations were introduced by overlap extension PCR (24) with complementary primers (for V594T: 5′-CGAATTGAGAAGAATTTCACGATTACAGATCCTCG-3′ and 5′-CGAGGATCTGTAATCGTGAAATTCTTCTCAATTCG-3′, and for C628A: 5′-ATTATCGGAAGAAATGCTCGGTAAGTGGCG-3′ and 5′-CGCCACTTACCGAGCATTTCTTCCGATAAT-3′), 5′-end primer (5′-AGTACCGGTTGATGGGGGATCAAGAATGA-3′), and 3′-end primer (5′-GAGGCCTAGGAAAGACAACACGAGAGTGATTCGCC-3′). The mutated DNA fragments were replaced with the corresponding sequences in pENTR–gMpPHOT using the unique restriction enzyme sites, AgeI/AvrII. The resulting plasmids were mixed with a destination vector, pMpGWB301 (25), and the LR reaction was performed to produce pMpGWB301–MpPHOTC328A, pMpGWB301–MpPHOTV594T, and pMpGWB301–MpPHOTC628A.
DNA fragments for the LOV1 and LOV2 domains of Mpphot were amplified with MpPHOT cDNA as template (26) by PCR with primers (for LOV1: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCAGCAGACCTTTGTGGTC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCATGTGTATTTGCTGACC-3′, and for LOV2: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAGAGTATTGATGACAGT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAGTGCTCACTTCCATCG-3′) and cloned into the pDONR207 vector by the BP reaction. The C328A mutation for LOV1 and the C628A or V594T mutation for LOV2 were introduced by DpnI-mediated site-directed mutagenesis with the following primers (for C328A: 5′-ATCGGAAGAAACGCTCGATTTCTCCAGGGA-3′ and 5′-TCCCTGGAGAAATCGAGCGTTTCTTCCGAT-3′, for C628A: 5′-ATCGGAAGAAATGCTCGGTTTCTGCAAGGG-3′ and 5′-CCCTTGCAGAAACCGAGCATTTCTTCCGAT-3′, and for V594T: 5′-CGAATTGAGAAGAATTTCACGATTACAGATCCTCG-3′ and 5′-CGAGGATCTGTAATCGTGAAATTCTTCTCAATTCG-3′). The resulting plasmid was mixed with pKM839 (27), a destination vector for the N-terminal maltose-binding protein (MBP)-tag fusion and bacterial expression, and the LR reaction was performed to produce pKM839–LOV1, pKM839–LOV1C328A, pKM839–LOV2, pKM839–LOV2C628A, and pKM839–LOV2V594T. pKM839 (Addgene plasmid 8838) was a gift from David Waugh, National Cancer Institute, NIH.
Transformation of M. polymorpha.
For transformation of M. polymorpha, Agrobacterium-mediated transformation methods (AgarTrap methods) were used (18–20). The resulting M. polymorpha (T1) transformants were cultivated for about 1 mo and then G1 gemmae were obtained. Subsequently, the G1 gemmae were cultivated for 1 mo and then G2 gemmae were obtained. Transgenic G2 gemmae were used for all experiments.
Immunoblot Analysis.
Four-day-old gemmalings incubated on one-half-strength B5 medium with 1% agar were used for immunoblot analysis of Mpphot. The gemmalings were frozen in liquid nitrogen and kept at −80 °C until use. The frozen samples were homogenized in a mortar and mixed in lysis buffer [150 mM NaCl, 1% Triton X-100, and 50 mM Tris⋅HCl (pH 8.0)] containing protease inhibitor mixture cOmplete Mini (Roche). The homogenates were mixed with 2× sample buffer [4% (wt/vol) SDS, 20% (vol/vol) glycerol, 0.01% (wt/vol) bromophenol blue, 10% (vol/vol) 2-mercaptoethanol, and 0.5 M Tris⋅HCl (pH 6.8)] and then incubated at 95 °C for 5 min. After centrifugation at 14,000 × g for 10 min at 4 °C, the total protein amount was quantified using an XL-Bradford Kit (APRO Science). Then, 15 µg of protein was subjected to SDS-polyacrylamide gel electrophoresis (SDS/PAGE) using an 8% (wt/vol) polyacrylamide gel. After electrophoresis, the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane. Mpphot protein was detected with rabbit antibodies (anti-MpPHOT) as the primary antibody raised against a synthetic peptide (C+VDERAPPSKGSAKE) (custom-made antibodies, Eurofins Genomics). Horseradish peroxidase-conjugated anti-rabbit IgG (catalog no. 32460, Thermo Scientific) was used as the secondary antibody. The primary and the secondary antibodies were used at dilutions of 1:3,000 and 1:2,000, respectively. Subsequently, chemiluminescent signal was detected using ECL-Select (GE Healthcare) and Light Capture (ATTO).
Recombinant Protein.
The appropriate plasmid (pKM839–LOV1, pKM839–LOV1C328A, pKM839–LOV2, pKM839–LOV2C628A, or pKM839–LOV2V594T) was transformed into Escherichia coli strain Rosetta2 (Novagen) and recombinant protein was bacterially expressed under white light (200 µmol photons m−2⋅s−1) at 20 °C for LOV1, LOV1C328A, LOV2, and LOV2C628A, or at 14 °C for LOV2V594T. After centrifugation at 18,000 × g for 20 min at 4 °C, the bacterial cells were frozen at −20 °C, thawed on ice, and resuspended in lysis buffer [50 mM Tris HCl (pH 8.0), 150 mM NaCl, and 2 mM CaCl2] with 200 µg of ml−1 lysozyme and 0.2% Triton X-100. The cells were lysed by sonication on ice and the MBP-tagged recombinant proteins were purified using Amylose Resin High Flow (New England Biolabs) and Poly-Prep Chromatography Columns (Bio-Rad) according to the manufacturers’ protocols. The MBP–LOV proteins were concentrated with Amicon Ultra Centrifucal Filters (Millipore) and diluted in 20 mM Tris⋅HCl (pH 7.8) buffer containing 200 mM NaCl and 10% glycerol.
Analysis of Phosphorylated Mpphot.
Four-day-old gemmalings were incubated in darkness at 22 °C for 3 d. The dark-adapted gemmalings were then incubated for 24 h at 22 °C or 5 °C under weak BL (25 µmol photons m−2⋅s−1) or in darkness. Each sample was collected, frozen in liquid nitrogen, and stored at −80 °C until use. The dark-treated gemmalings were collected under red-light conditions. The frozen samples were homogenized in 1× sample buffer [2% SDS, 10% glycerol, 5% 2-mercaptoethanol, and 0.25 M Tris⋅HCl (pH 6.8)] using a mortar and then incubated at 95 °C for 5 min under red light. The phosphorylated Mpphot was separated by SDS/PAGE using an 8% gel (acrylamide: N,N′-methylenebisacrylamide = 29.9:0.1), and detected by immunoblot analysis as described above. The migration distance was determined using ImageJ software. Pairwise comparison of the means (n = 6) was performed using a Student’s t test in R statistical software.
Photochemical Analysis.
The procedure for photochemical analysis was previously reported (12). The photochemical reaction of the LOV domains was recorded using a U-2810 spectrophotometer (Hitachi Hitec). The temperature of the cuvette was controlled by a water circulated cell holder (catalog no. 210–2111, Hitachi Hitec) with a NCB-1200 chiller unit (EYELA). The samples were excited in the cuvette under an illuminator with blue-colored LEDs (OptoSupply).
Photosynthetic Analysis.
To determine the maximal quantum efficiency of photosystem II, chlorophyll fluorescence was measured with a Junior-PAM pulse amplitude-modulated fluorometer according to the manufacturer’s instructions (Walz). Gemmalings were cultured on a sterilized black cloth rested on one-half-strength B5 medium to reduce scattered light. After the incubation under weak white light (70 µmol photons m−2⋅s−1) at 22 °C or 5 °C for 24 h, the minimal chlorophyll fluorescence at the open photosystem II center (Fo) was determined by measuring light at 460 nm. A saturating pulse at 460 nm was applied to determine the maximal chlorophyll fluorescence at the closed photosystem II center (Fm). The maximal quantum efficiency of photosystem II (termed as Fv/Fm) was calculated using the following equation: (Fm − Fo)/Fm (28). Tukey’s honest significant difference (HSD) test was performed using R statistical software.
Observation of the Nucleus and Peroxisome.
To visualize the nucleus and peroxisomes in MpphotKO cells, pMpGWB102–Citrine–NLS (nuclear localization signal) and pMpGWB102–Citrine–PTS (peroxisome targeting signal) plasmids (6) were introduced into the cells by particle bombardment, according to a previously described method (29). The bombarded cells were observed by confocal laser scanning microscopy (SP8X, Leica Microsystems) with the time-gating method (26) for blocking chlorophyll autofluorescence.
Supplementary Material
Acknowledgments
We thank D. Inomata (Suana Science) for the illustrations; T. Matsushita (Kyushu University), Y. Tada (Nagoya University), and M. Matsuda (Utsunomiya University) for critiquing the manuscript; T. Kohchi (Kyoto University) for the M. polymorpha plants and the pENTR–gMpPHOT vector; K. Ishizaki (Kobe University) for various information on handling M. polymorpha; M. Wada (Tokyo Metropolitan University) for A. thaliana plants; D. Waugh (National Cancer Institute) for the pKM839 vector; W. Yamori (University of Tokyo) for valuable comments; K. Numata (RIKEN) for valuable information; N. Habu (Utsunomiya University) for the use of the spectrophotometer; and members of the Y.K. laboratory for various support. This work was supported by the Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (KAKENHI 26840088 to Y.K.), the Exploratory Research for Advanced Technology (ERATO) from the Japan Science and Technology Agency (JST) (Numata Organelle Reaction Cluster, JPMJER 1602 to Y.K.), the Plant Transgenic Design Initiative (PTraD) of University of Tsukuba (Y.K.), and the Center of Excellence (UU-COE) (N.K., S.H., Y.H., and Y.K.) and the Creative Department of Innovation (CDI) of Utsunomiya University (Y.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704462114/-/DCSupplemental.
References
- 1.Sengupta P, Garrity P. Sensing temperature. Curr Biol. 2013;23:R304–R307. doi: 10.1016/j.cub.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung JH, et al. Phytochromes function as thermosensors in Arabidopsis. Science. 2016;354:886–889. doi: 10.1126/science.aaf6005. [DOI] [PubMed] [Google Scholar]
- 3.Legris M, et al. Phytochrome B integrates light and temperature signals in Arabidopsis. Science. 2016;354:897–900. doi: 10.1126/science.aaf5656. [DOI] [PubMed] [Google Scholar]
- 4.Wada M. Chloroplast movement. Plant Sci. 2013;210:177–182. doi: 10.1016/j.plantsci.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Kodama Y, Tsuboi H, Kagawa T, Wada M. Low temperature-induced chloroplast relocation mediated by a blue light receptor, phototropin 2, in fern gametophytes. J Plant Res. 2008;121:441–448. doi: 10.1007/s10265-008-0165-9. [DOI] [PubMed] [Google Scholar]
- 6.Ogasawara Y, Ishizaki K, Kohchi T, Kodama Y. Cold-induced organelle relocation in the liverwort Marchantia polymorpha L. Plant Cell Environ. 2013;36:1520–1528. doi: 10.1111/pce.12085. [DOI] [PubMed] [Google Scholar]
- 7.Kimura S, Kodama Y. Actin-dependence of the chloroplast cold positioning response in the liverwort Marchantia polymorpha L. PeerJ. 2016;4:e2513. doi: 10.7717/peerj.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wada M. The fern as a model system to study photomorphogenesis. J Plant Res. 2007;120:3–16. doi: 10.1007/s10265-006-0064-x. [DOI] [PubMed] [Google Scholar]
- 9.Komatsu A, et al. Phototropin encoded by a single-copy gene mediates chloroplast photorelocation movements in the liverwort Marchantia polymorpha. Plant Physiol. 2014;166:411–427. doi: 10.1104/pp.114.245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka H, et al. Chloroplast aggregation during the cold positioning response in the liverwort Marchantia polymorpha L. J Plant Res. 2017 doi: 10.1007/s10265-017-0958-9. [DOI] [PubMed] [Google Scholar]
- 11.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 12.Okajima K, Kashojiya S, Tokutomi S. Photosensitivity of kinase activation by blue light involves the lifetime of a cysteinyl-flavin adduct intermediate, S390, in the photoreaction cycle of the LOV2 domain in phototropin, a plant blue light receptor. J Biol Chem. 2012;287:40972–40981. doi: 10.1074/jbc.M112.406512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawano F, Aono Y, Suzuki H, Sato M. Fluorescence imaging-based high-throughput screening of fast- and slow-cycling LOV proteins. PLoS One. 2013;8:e82693. doi: 10.1371/journal.pone.0082693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Łabuz J, Hermanowicz P, Gabryś H. The impact of temperature on blue light induced chloroplast movements in Arabidopsis thaliana. Plant Sci. 2015;239:238–249. doi: 10.1016/j.plantsci.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 15.Kasahara M, et al. Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 2002;129:762–773. doi: 10.1104/pp.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eichenberg K, Hennig L, Martin A, Schäfer E. Variation in dynamics of phytochrome A in Arabidopsis ecotypes and mutants. Plant Cell Environ. 2000;23:311–319. [Google Scholar]
- 17.Briggs RW, Spudich LJ. Handbook of Photosensory Receptors. Wiley VCH; Weinheim, Germany: 2005. [Google Scholar]
- 18.Tsuboyama S, Kodama Y. AgarTrap: A simplified Agrobacterium-mediated transformation method for sporelings of the liverwort Marchantia polymorpha L. Plant Cell Physiol. 2014;55:229–236. doi: 10.1093/pcp/pct168. [DOI] [PubMed] [Google Scholar]
- 19.Tsuboyama-Tanaka S, Kodama Y. AgarTrap-mediated genetic transformation using intact gemmae/gemmalings of the liverwort Marchantia polymorpha L. J Plant Res. 2015;128:337–344. doi: 10.1007/s10265-014-0695-2. [DOI] [PubMed] [Google Scholar]
- 20.Tsuboyama-Tanaka S, Nonaka S, Kodama Y. A highly efficient agarTrap method for genetic transformation of mature thalli of the liverwort Marchantia polymorpha L. Plant Biotechnol. 2015;32:333–336. [Google Scholar]
- 21.Kodama Y, Suetsugu N, Kong SG, Wada M. Two interacting coiled-coil proteins, WEB1 and PMI2, maintain the chloroplast photorelocation movement velocity in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:19591–19596. doi: 10.1073/pnas.1007836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii Y, Kodama Y. In planta comparative analysis of improved green fluorescent proteins with reference to fluorescence intensity and bimolecular fluorescence complementation ability. Plant Biotechnol. 2015;32:81–87. [Google Scholar]
- 23.Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–571, 574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi R, Krummel B, Saiki RKA. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizaki K, et al. Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One. 2015;10:e0138876. doi: 10.1371/journal.pone.0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kodama Y. Time gating of chloroplast autofluorescence allows clearer fluorescence imaging in planta. PLoS One. 2016;11:e0152484. doi: 10.1371/journal.pone.0152484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox JD, Routzahn KM, Bucher MH, Waugh DS. Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett. 2003;537:53–57. doi: 10.1016/s0014-5793(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell K, Johnson GN. Chlorophyll fluorescence: A practical guide. J Exp Bot. 2000;51:659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- 29.Kodama Y. A bright green-colored bimolecular fluorescence complementation assay in living plant cells. Plant Biotechnol. 2011;28:95–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.