Significance
The steroid hormone 20-hydroxyecdysone (20E) promotes apoptosis during larval-to-adult metamorphosis in insects. However, the mechanisms governing this process are unclear. This work reveals that 20E up-regulates the expression of the protein kinase PKCδ isoform E. Overexpression of the catalytic domain of PKCδ is sufficient to increase caspase-3 activity and apoptosis. PKCδ directly phosphorylates a threonine residue at position 468 of the amino acid sequence of nuclear receptor EcRB1. The phosphorylation of EcRB1 is essential for apoptotic gene transcription. These results demonstrate the mechanism by which the steroid hormone 20E promotes PKCδ expression to regulate apoptosis.
Keywords: 20-hydroxyecdysone, protein kinase C delta, phosphorylation, ecdysone receptor B1, apoptosis
Abstract
The nuclear receptor EcRB1, which is activated by the insect steroid hormone 20-hydroxyecdysone (20E), is reportedly phosphorylated by a protein kinase after 20E induction. However, the protein kinase has not been identified, and the significance of EcRB1 phosphorylation is unclear. In this study, we identified a protein kinase C δ (PKCδ) isoform (the E isoform) that phosphorylates EcRB1 in the lepidopteran Helicoverpa armigera, a serious agricultural pest worldwide, to promote apoptotic gene expression and apoptosis during metamorphosis. Through activation of the EcRB1/USP1 transcription complex by 20E, PKCδ expression was up-regulated in several tissues during the metamorphic stage. Knockdown of PKCδ caused failure to transition from larvae to pupae, prevented tissues from undergoing programmed cell death (PCD), and down-regulated the expression of the transcription factor Brz-7 and the apoptosis executors caspase-3 and caspase-6. The threonine residue at position 1343 of PKCδ was phosphorylated and was critical for its proapoptotic function. Overexpression of the PKCδ catalytic domain was localized to the nuclei in HaEpi cells, which increased caspase-3 activity and apoptosis. PKCδ directly phosphorylated a threonine residue at position 468 in the amino acid sequence of EcRB1. The phosphorylation of EcRB1 was critical for its heterodimeric interaction with the USP1 protein and for binding to the ecdysone response element. The data suggested that 20E up-regulates PKCδ expression to regulate EcRB1 phosphorylation for EcRB1/USP1 transcription complex formation, apoptotic gene transcription, and apoptosis.
Protein kinas C δ (PKCδ) belongs to the serine/threonine protein kinase C family and has been shown to play an important role in apoptosis (1, 2). Human PKCδ contains two conserved regions (C1 and C2) and catalytic domains. In response to an apoptotic stimulus, PKCδ is cleaved by caspase-3 at the hinge region between C1 and the catalytic domain to generate a free and activated PKCδ catalytic fragment, which then translocates from the cytoplasm to the nucleus and induces apoptosis in humans (1). Phosphorylation at Ser359 increases PKCδ activity (3). Phosphorylation of Thr507 in the catalytic domain is required for in vivo activity, but not for in vitro activity in HEK293T cells. The unphosphorylated primary translational product is predicted to exhibit little or no activity in humans (4). The threonine residues in the catalytic domain of novel and atypical PKC (aPKC) isoforms are conserved, and mutations in these residues result in loss of catalytic competency (5). Overexpression of the PKCδ catalytic fragment leads to its localization in the nucleus and is sufficient to induce apoptosis (6). However, the factors acting upstream of PKCδ expression, especially concerning steroid hormones, are unclear.
The steroid hormone 20-hydroxyecdysone (20E) is produced in both insects (7) and plants (8). 20E promotes programmed cell death (PCD) during insect metamorphosis (9). In Drosophila melanogaster, 20E also stops stem cell proliferation (10). In Helicoverpa armigera, 20E causes a switch from autophagy to apoptosis (11), counteracts the action of insulin (12), promotes phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase (PTEN) expression (13), and promotes the Hippo pathway to induce apoptosis (14). Furthermore, 20E is used as a food additive to maintain the glucose–lipid balance in humans, without the side effects of mammalian hormones (15).
Through activation of the nuclear receptor complex (EcR/USP), 20E can initiate gene transcription. Protein phosphorylation is a key process in EcR/USP complex formation and binding of the complex to ecdysone response element (EcRE) for gene transcription. Some unknown PKCs are involved in the phosphorylation of the heterodimeric partner USP in 20E signaling, as demonstrated by inhibitor experiments in H. armigera (16) and in Drosophila (17). EcRB1 has also been shown to be phosphorylated in Bombyx mori (18) and H. armigera (19). However, it is unknown which isoform of PKC phosphorylates EcRB1.
To answer this question, we investigated the mechanism by which 20E regulates EcRB1 phosphorylation and the role played by PKCδ in this phosphorylation event. We demonstrated that 20E up-regulated PKCδ expression. PKCδ positively mediated larval tissue PCD and apoptosis-related gene expression. Phosphorylation of PKCδ at Thr1343 in the catalytic domain was necessary for its activation of caspase-3. In addition, PKCδ directly phosphorylated EcRB1 in the nucleus, which was critical for the formation of the EcRB1/USP1 transcriptional complex and its binding to the EcRE to induce gene transcription. These results suggest that 20E induces EcRB1 phosphorylation via up-regulation of PKCδ expression and subsequently mediates gene expression and apoptosis during insect metamorphosis.
Results
PKCδ Clone and Sequence Analysis.
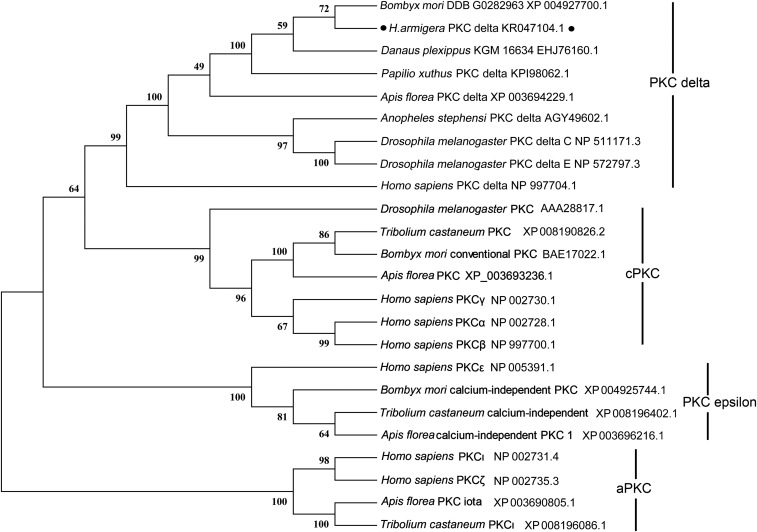
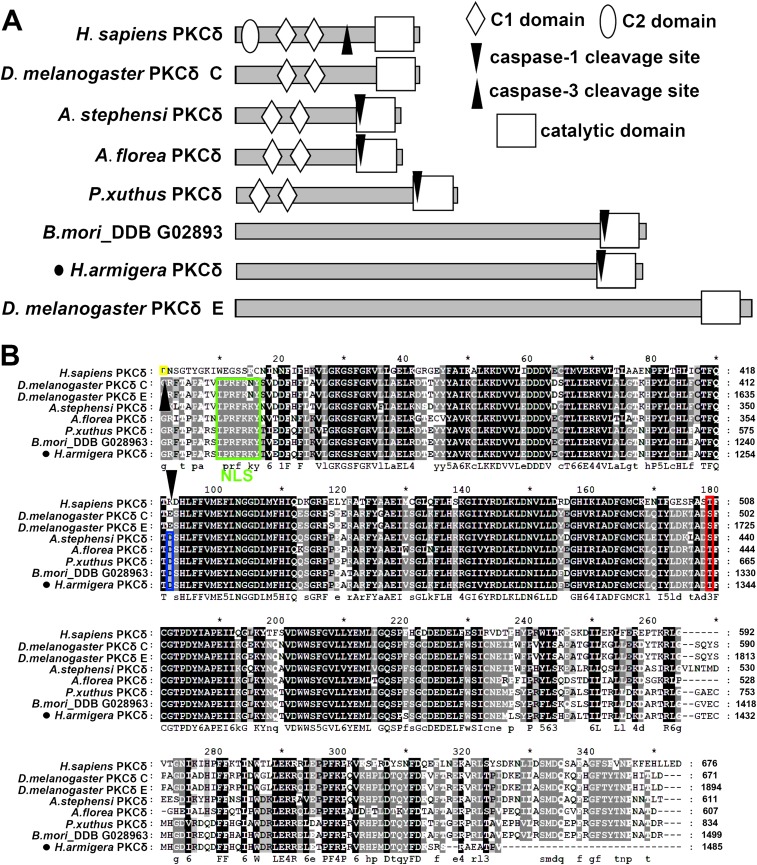
We cloned the full-length 5,145-bp cDNA of PKCδ from H. armigera, including a 55-bp 5′ untranslated region (UTR), a 4,458-bp ORF, and a 632-bp 3′ UTR. H. armigera PKCδ was identified as a PKCδ based on its short evolutionary distance from PKCδ of Homo sapiens and other insects, such as Papilio xuthus, Apis florea, Anopheles stephensi, and D. melanogaster (Fig. S1). Analysis using SMART software (smart.embl-heidelberg.de/) showed that the typical PKCδ of H. sapiens contains one C2 domain, two C1 domains, and the catalytic domain. The PKCδs of D. melanogaster (isoform C), A. florea, A. stephensi, and P. xuthus exhibit two C1 domains and the catalytic domain. The PKCδs with long sequences identified in D. melanogaster (isoform E), B. mori, and H. armigera have only a catalytic domain (Fig. S2A); thus, the H. armigera PKCδ was considered an E isoform. PeptideCutter (web.expasy.org/peptide_cutter/) analysis showed that the PKCδs of H. armigera and other insects, except D. melanogaster, exhibit a caspase-1 cleavage site, whereas H. sapiens PKCδ exhibits a caspase-3 cleavage site (Fig. S2A). The nuclear localization signal PRFRKYT was predicted at amino acids 1175–1181. Although PKCδs from different animals exhibit various structural characteristics, the catalytic domain is highly conserved from vertebrates to invertebrates according to alignment analysis (Fig. S2B).
Fig. S1.
Identification of H. armigera PKCδ via phylogenetic tree analysis. MEGA 5.0 was used to conduct phylogenetic tree analysis with the corresponding amino acid sequences of PKC isoforms obtained from NCBI.
Fig. S2.
Structural characteristics and alignment of PKCδ catalytic domain. (A) The SMART tool was used to analyze the PKCδ domains of humans and various insects included in the phylogenetic tree shown in Fig. S1. (B) Alignment of the catalytic domains was performed with MEGA 5.0 and GENEDOC based on the structural analysis described in A. The yellow mark indicates the caspase-3 cleavage site (after the amino acid) in H. sapiens. The sequence highlighted in the green box is the predicted nuclear localization signal. The blue box denotes the predicted caspase-1 cleavage sites (after the amino acid). The red box indicates the phosphorylation site in H. armigera PKCδ at Thr1343 that was identified in this work.
PKCδ Is Highly Expressed During Metamorphosis via 20E Regulation in Parts of Tissues.
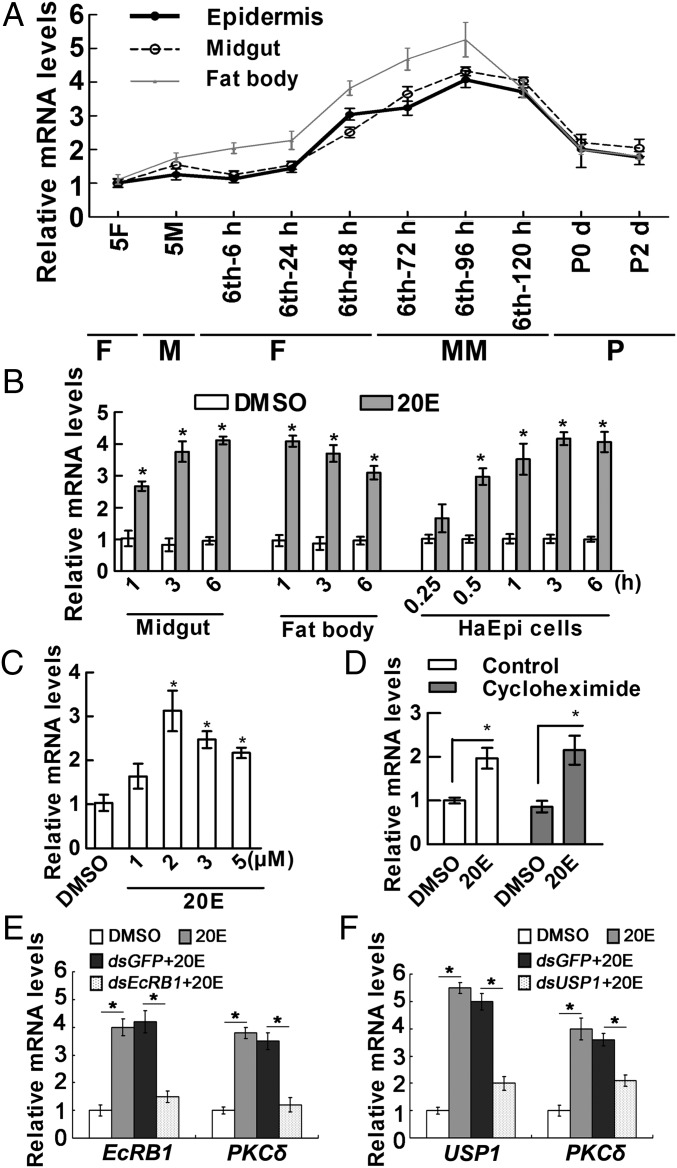
The transcript levels of PKCδ in different tissues at various developmental stages were detected using qRT-PCR to investigate the function of PKCδ in insect development. PKCδ displayed increased expression in the epidermis, midguts, and fat bodies in the whole wandering stage from sixth-72 h larval to sixth-120 h larval stages during metamorphosis (Fig. 1A). These findings suggested a potential role of PKCδ during insect metamorphosis.
Fig. 1.
qRT-PCR showing the 20E-regulated expression of PKCδ. (A) Expression profiles of PKCδ in tissues during larval development. Sixth-6, sixth-24, sixth-48, sixth-72, sixth-96, and sixth-120 represent sixth instar larvae at the corresponding hours. P0 d and P2 d indicate 0- and 2-d-old pupae. 5F, fifth instar feeding larvae; 5M, fifth instar molting larvae; F, feeding; M, molting; MM, metamorphic molting; P, pupae. (B) The transcript levels of PKCδ in the midgut, fat body (injection of 500 ng of 20E/larva), and HaEpi cells (2 μM 20E). Equal diluted DMSO was used as a solvent control. (C) Expression of PKCδ in HaEpi cells after being treated with different concentrations of 20E for 3 h. Equal diluted DMSO was used as a control. (D) Expression of PKCδ when protein synthesis was inhibited by cycloheximide. The control experimental cells were treated with DMSO or 2 μM 20E for 3 h. Other cells were incubated with 50 μM cycloheximide for 30 min and then treated with 2 μM 20E for 3 h. (E and F) Knockdown of EcRB1 or USP1 using dsEcRB1 or dsUSP1 (2 μg/mL for 48 h) in HaEpi cells, followed by treatment with 20E (2 μM) for 6 h. dsGFP and DMSO were used as negative controls. All of the relative mRNA levels were calculated by 2–∆∆CT, and the bars indicate the mean ± SD. Significant differences were calculated using Student’s t test (*P < 0.05) according to three biological replicates and three technical replicates in all qRT-PCRs.
Given the up-regulated expression of PKCδ during metamorphosis, we detected the effect of 20E on PKCδ expression by injecting 20E into sixth instar larvae at 6 h or by incubating the H. armigera epidermal cell line (HaEpi) with 20E for different times to examine whether 20E induces PKCδ expression. The mRNA levels of PKCδ increased quickly from 0.25 to 6 h in the midgut, fat body, and HaEpi cells after 2 μM 20E treatment compared with that in the dimethyl sulfoxide (DMSO) solvent control (Fig. 1B). The expression of PKCδ was significantly up-regulated by 2 μM to 5 μM of 20E treatment in HaEpi cells (Fig. 1C). To detect whether the expression of PKCδ was directly induced by 20E, we inhibited the protein synthesis using the protein synthesis inhibitor cycloheximide. 20E up-regulated expression of PKCδ when cycloheximide was added (Fig. 1D). These results suggested that 20E directly up-regulates the expression of PKCδ in these tissues and HaEpi cells.
The 20E nuclear receptor EcRB1 and its heterodimeric partner USP1 were depleted in HaEpi cells via RNA interference (RNAi) to investigate the pathway by which 20E regulates PKCδ expression. 20E could not up-regulate PKCδ mRNA after the depletion of EcRB1 or USP1 compared with the dsGFP control (Fig. 1 E and F), indicating that 20E mediates PKCδ expression through EcRB1 and USP1.
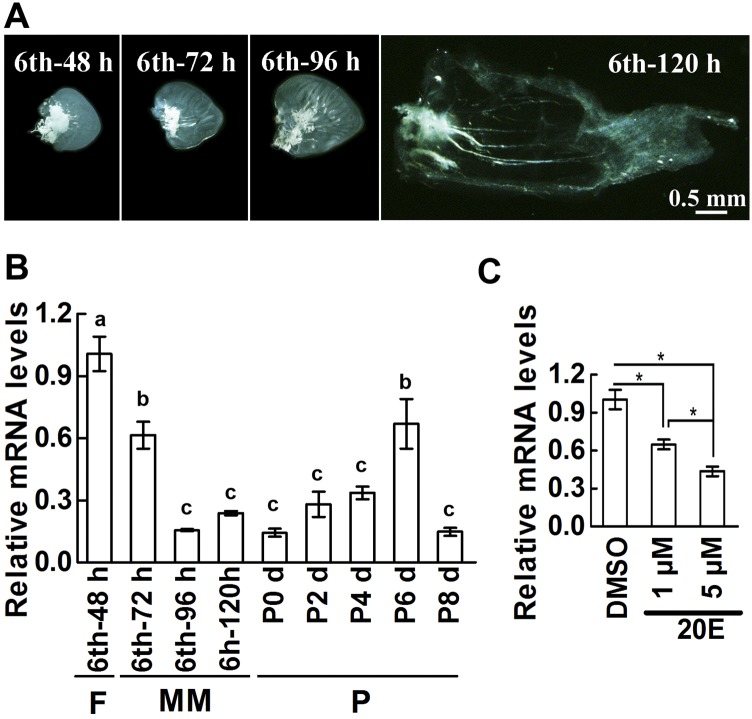
To examine the expression of PKCδ in nonapoptotic tissues, the fore wing imaginal disks were dissected out from the sixth instar 48 h (sixth-48 h) feeding larvae to sixth instar 120 h (sixth-120 h) metamorphic larvae. The disks were growing rapidly after larvae entered the wandering stage from sixth-72 h to sixth-120 h (Fig. S3A). The expression of PKCδ was high at the feeding stage at sixth-48 h; however, it was gradually decreased at the wandering stage from sixth-72 h to sixth-120 h and at the pupal stage from P0 to P8 d (Fig. S3B). To examine the repression of 20E on PKCδ expression, the fore wing imaginal disks from sixth-48 h larvae were dissected out and cultured in vitro. The results showed that the expression of PKCδ was repressed by 1 μM and 5 μM 20E treatment (Fig. S3C). The data suggested that the expression of PKCδ has a negative correlation with the concentration of 20E and wing disk growth.
Fig. S3.
The expression of PKCδ was repressed by 20E in wing imaginal disk. (A) Development of fore wing imaginal disk of H. armigera from sixth-48 h to sixth-120 h larvae. All the disks were the same magnification. (B) Relative mRNA levels of PKCδ in fore wing imaginal disk during larval development. Sixth-48 h, sixth-72 h, sixth-96 h, and sixth-120 h represent sixth instar larvae at the corresponding hours. P0 to P8 indicate 0- to 8-d-old pupae. F, feeding; M, molting; MM, metamorphic molting; P, pupa. The values are the mean ± SD based on 10 disks. Significant differences were calculated using ANOVA, and different letters (a, b, c) represent significant differences. (C) 20E suppresses PKCδ expression in fore wing disk. The mRNA was extracted after wing disks cultured with 20E (1 μM or 5 μM) for 48 h. DMSO served as a negative control. The relative mRNA of PKCδ was analyzed by qRT-PCR. Bars represent mean ± SD based on 12 disks. Significant differences were calculated using Student’s t test (*P < 0.05). Three biological replicates and three technical replicates were performed for the experiments.
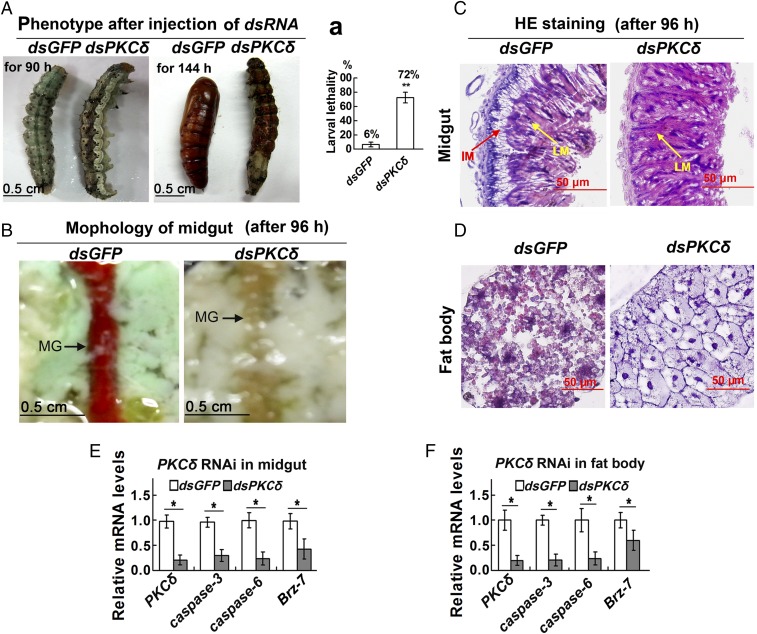
PKCδ Depletion in Larvae via RNAi Suppresses Metamorphosis, Tissue Degradation, and Apoptotic Gene Expression.
Given the high expression of PKCδ during metamorphosis, we investigated its function in the larval–pupal transition by injecting PKCδ dsRNA into larval hemocoel. Phenotype analysis showed that the injection of dsPKCδ resulted in slower larval growth, failure to pupate, and larval death compared with larvae injected with dsGFP (Fig. 2A). The lethality of dsPKCδ-injected larvae was 12 times that of dsGFP-injected larvae (Fig. 2 A, a). The midguts of larvae in the dsGFP control group exhibited characteristics of degradation, showing red color of the midgut, at 96 h post-second dsRNA injection, whereas dsPKCδ-treated larval midguts did not appear red (Fig. 2B). Histochemical analysis further revealed that the larval midgut separated from the imaginal midgut in the dsGFP control, whereas no clear separation was observed in the dsPKCδ-treated larval midgut (Fig. 2C). Similarly, the fat body of the dsGFP control larvae showed degradation, while the fat body cells of the PKCδ knockdown larvae were still closely arranged and did not exhibit degradation (Fig. 2D). qRT-PCR analysis demonstrated that the apoptosis executors caspase-3 and caspase-6 as well as the metamorphosis-regulating transcription factor Brz-7 were decreased in the midgut and fat body after PKCδ knockdown in larvae (Fig. 2 E and F). These data suggested that PKCδ participates in the degradation of the midgut and the fat body during metamorphosis via regulation of apoptosis-related gene expression.
Fig. 2.
PKCδ knockdown via RNAi in larvae restrains larval pupation and tissue PCD and suppresses apoptotic gene expression. (A) Insect phenotypes after PKCδ knockdown via two injections of dsPKCδ or dsGFP (1–2 μg) into the larval hemocoel (the first in fifth instar-12 h larvae and the second in sixth instar-6 h larvae). Phenotypes were observed at 90 h and 144 h after the second injection. (a) Statistics of larval lethality after injection of dsRNA by Student’s t test from three biological repeats. Every 30 larvae were injected with dsRNA in each experiment, and the dead larvae were counted when all of the larvae had become pupae in the dsGFP control. The values are the mean ± SD. (B) Midgut phenotypes observed at 96 h after the second dsPKCδ or dsGFP injection. MG, midgut. (C and D) Hematoxylin and eosin (HE) staining showing the tissue histology of the midgut at 96 h after the second dsPKCδ or dsGFP injection. Tissues were collected during the observations indicated in B. IM, imaginal midgut; LM, larval midgut. (E and F) Detection of apoptosis-related genes using qRT-PCR in the midgut and fat body tissue after the second dsPKCδ or dsGFP injection for 96 h. The values are the mean ± SD. Significant differences were calculated using Student’s t test from three biological repeats (*P < 0.05, **P < 0.01).
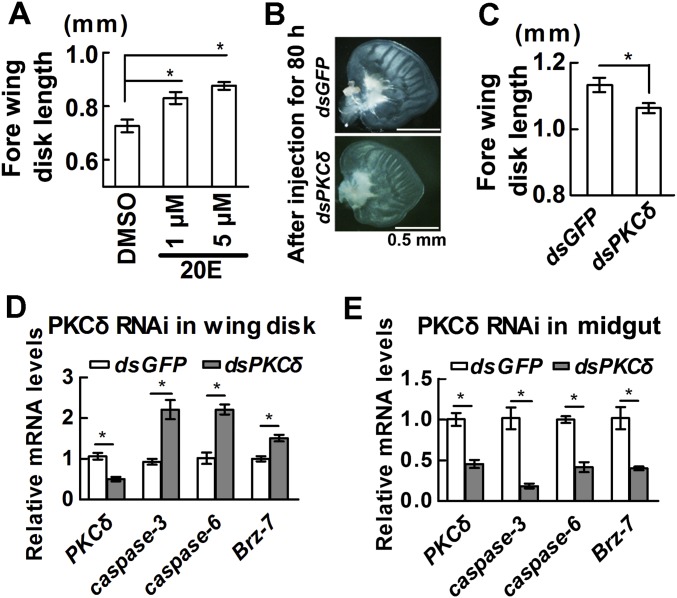
To confirm 20E promoting wing disk growth during metamorphosis, the fore wing imaginal disks from sixth-48 h larvae were dissected out and cultured in vitro with 20E treatment. 20E indeed promotes wing disk growth (Fig. S4A), which was consistent with the results obtained in Manduca sexta (20). In the dsPKCδ-injected larvae, the wing disk growth was suppressed compared with dsGFP-injected larvae (Fig. S4 B and C). However, the expression of caspases and Brz-7 was increased in the wing disk after PKCδ was knocked down (Fig. S4D), which was opposite to the gene expression in midgut by dsPKCδ injection (Fig. S4E). The involved mechanism needs further study.
Fig. S4.
PKCδ knockdown via RNAi in larvae restrains wing imaginal disk growth and promotes apoptotic gene expression in wing disk. (A) 20E promotes fore wing disk growth. Fore wing imaginal disks were separated and cultured in Grace’s medium with 20E for 48 h. The wing disk length was measured using ocular micrometer. 20E used a lower concentration (1 μM) and a higher concentration (5 μM). DMSO was used as a solvent control. Each bar represents the mean of 10 disks. Bars are mean ± SD. (B) Morphology of fore wing imaginal disk after injection of dsRNA. The fresh fore wing disks were separated under microscope for observation after two injections of dsRNA for 80 h. dsGFP served as a control experiment. (C) The length of wing imaginal disk after PKCδ knockdown. The length of fore wing disks was measured using ocular micrometer after the disks were separated. Each bar represents the mean of six disks. Significant differences were calculated using Student’s t test (*P < 0.05). (D and E) Detection of apoptosis-related genes using qRT-PCR in the wing disk and midgut after the second dsPKCδ or dsGFP injection for 80 h. The wing disk and midgut were separated from the same individual. The values are the mean ± SD. Significant differences were calculated using Student’s t test (*P < 0.05). Three biological replicates and three technical replicates were performed for the qRT-PCR.
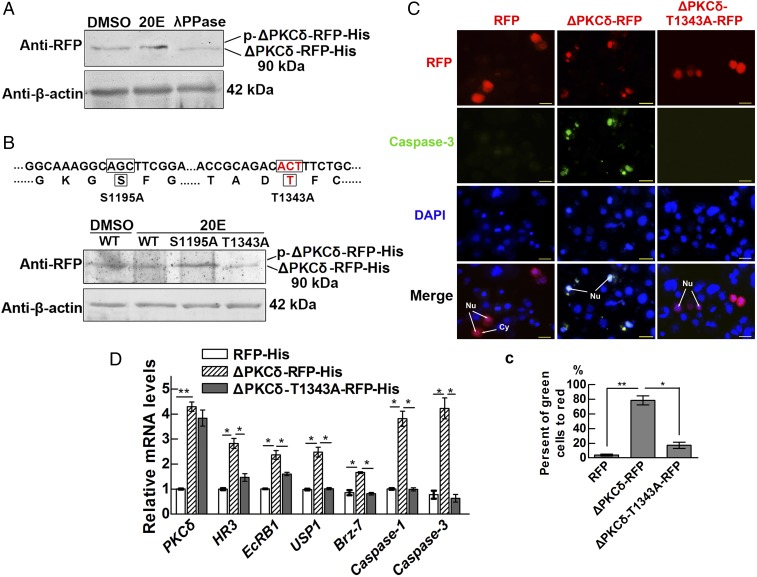
Overexpression of the Catalytic Domain of PKCδ (ΔPKCδ) Is Sufficient to Induce Apoptosis Without 20E Induction.
We overexpressed the PKCδ catalytic domain (ΔPKCδ) in HaEpi cells by fusing it with red fluorescence protein (RFP) and His tag (ΔPKCδ-RFP-His) to determine the role of this domain in apoptosis. The results showed that the ΔPKCδ-RFP-His displayed the same molecular mass under DMSO or 20E induction. Lambda protein phosphatase (λPPase) treatment decreased the molecular mass of ΔPKCδ-RFP-His, indicating that ΔPKCδ-RFP-His retained its phosphorylation (Fig. 3A). According to the reported phosphorylation sites in the PKCδ catalytic domain of humans (3, 4), we identified two conserved Ser/Thr residues (S1195 and T1343, conserved with human S369 and T507, respectively) in ΔPKCδ. Overexpression of the wild-type (WT) or single alanine substitution mutants (S1195A or T1343A) of ΔPKCδ showed that T1343A displayed a lower molecular mass, indicating that ΔPKCδ was phosphorylated at Thr1343 (Fig. 3B). Both WT ΔPKCδ-RFP and the ΔPKCδ-T1343A-RFP mutant were localized to the nucleus; however, only ΔPKCδ-RFP significantly increased the activity of caspase-3 (Fig. 3 C, c). The transcript levels of genes responding to 20E (HR3, EcRB1, USP1, Brz-7) and apoptosis (caspase-1, caspase-3) were enhanced when ΔPKCδ-RFP-His was overexpressed compared with overexpression of either RFP-His or mutant T1343A, according to qRT-PCR analysis (Fig. 3D). These results indicated that the ability of PKCδ to promote apoptosis depends on the overexpression and phosphorylation of its catalytic domain.
Fig. 3.
Overexpression of the PKCδ catalytic domain in HaEpi cells induces apoptosis and 20E-responsive gene expression. (A) Western blot analysis detecting the phosphorylation of ΔPKCδ-RFP-His using λPPase degradation. ΔPKCδ-RFP-His was overexpressed in HaEpi cells for 48 h, and the cells were then treated with 20E (2 μM) for 30 min. DMSO was used as the solvent control, and a 7.5% gel concentration was used for SDS/PAGE. A mouse monoclonal antibody against RFP was used for Western blot analysis. β-Actin was used as the protein quantity control. (B) Identification of the phosphorylation site in ΔPKCδ through overexpression of ΔPKCδ-RFP-His, ΔPKCδS1195A-RFP-His, or ΔPKCδT1343A-RFP-His in HaEpi cells, which were then treated with 20E (2 μM) for 30 min. DMSO was used as the solvent control. Western blot analysis was performed as described in A. (C) Detection of the activity of caspase-3 in RFP-, ΔPKCδ-RFP–, or ΔPKCδT1343A-RFP–overexpressing cells for 48 h after transfection. Red fluorescence indicates RFP, ΔPKCδ-RFP, or ΔPKCδT1343A-RFP. Green fluorescence indicates activity of caspase-3 using NucView 488 caspsae-3 assay kit. The blue signals indicate the cell nuclei stained by DAPI. (Scale bar, 20 μm at 40× magnification.) (c) Statistical analysis of the data in C by Student’s t test from three biological repeats (*P < 0.05; **P < 0.01). The values are the mean ± SD. (D) Relative mRNA levels of 20E- and apoptosis-related genes determined via qRT-PCR. Analysis of significance differences was performed with Student’s t test from three biological repeats and three technical replicates (*P < 0.05; **P < 0.01). The values are the mean ± SD.
PKCδ Phosphorylates EcRB1.
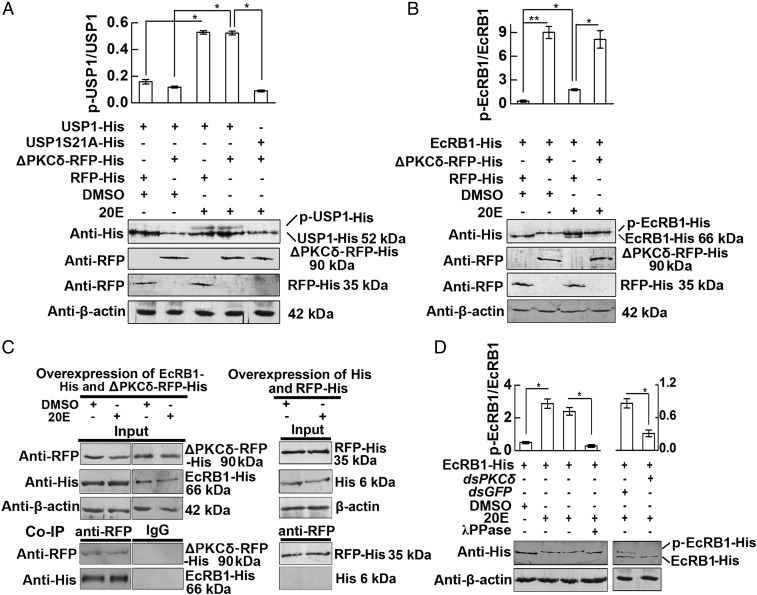
We investigated the effects of PKCδ on the 20E nuclear receptor EcRB1 and its partner protein USP1 to address the mechanism by which PKCδ promotes apoptosis. Overexpression of ΔPKCδ-RFP-His did not induce the phosphorylation of overexpressed USP1-His unless with treatment of 20E, compared with the control DMSO treatment. The phosphorylation site mutant USP1S21A-His was used as a nonphosphorylated USP1 control (Fig. 4A). This meant that the 20E-induced phosphorylation of USP1 is independent from the expression of PKCδ. However, overexpression of ΔPKCδ-RFP-His not only induced the phosphorylation of overexpressed EcRB1 under DMSO treatment but also enhanced the partial EcRB1 phosphorylation induced by 20E (Fig. 4B). The coimmunoprecipitation (co-IP) experiment showed that the anti-RFP antibody precipitated overexpressed ΔPKCδ-RFP-His, together with overexpressed EcRB1-His, with or without 20E induction, which was controlled with the lgG and overexpressed His and RFP-His tags (Fig. 4C). These data suggested that PKCδ directly interacts with EcRB1 to induce EcRB1 phosphorylation.
Fig. 4.
ΔPKCδ regulates the phosphorylation of EcRB1. (A) Overexpression of ΔPKCδ-RFP-His has no effect on the phosphorylation of USP1 by Western blot analysis. After cotransfection with USP1-His, USP1-S21A-His (phosphorylation site mutation), and ΔPKCδ-RFP-His or RFP-His for 48 h, the cells were treated with 20E (2 μM) for 30 min. DMSO was used as a negative control. Anti-His and anti-RFP mAbs were used for detection of the corresponding proteins following 7.5% SDS/PAGE. β-Actin was used as a loading control. Statistical analyses of the density Western blotting bands were performed with Quantity One software, based on three independent biological experiments. The values are the mean ± SD from three independent experiments (*P < 0.05). (B) Overexpression of ΔPKCδ-RFP induced EcRB1 phosphorylation. After cotransfection with EcRB1-His and ΔPKCδ-RFP or RFP for 48 h, the cells were treated with 20E (2 μM) for 30 min. DMSO was used as a negative control. Western blot analysis was performed as described in A. (C) ΔPKCδ-RFP-His combined with EcRB1-His with or without 20E induction. HaEpi cells were treated with 2 μM 20E or an equivalent amount of DMSO for 30 min after cotransfection with ΔPKCδ-RFP-His and EcRB1-His for 48 h. The protein expression levels of EcRB1-His, ΔPKCδ-RFP-His, RFP-His, and β-actin in HaEpi cells were detected via Western blot following 7.5% SDS/PAGE. For co-IP analysis, ΔPKCδ-RFP-His was immunoprecipitated with a mAb against RFP, and the coprecipitated EcRB1-His was detected via Western blot analysis using an anti-His mAb. Mouse lgG was used as a negative control of the antibody. His tag and RFP-His were overexpressed in HaEpi cells using the pIEx-4-His and pIEx-4-RFP-His plasmids as tag controls. The cells were treated and proteins were detected using the same methods as described in B. The pictures represent three repeats. (D) PKCδ knockdown via RNAi repressed EcRB1 phosphorylation. HaEpi cells were transfected with pIEx-4-EcRB1-His for 48 h, then incubated with dsPKδ or dsGFP (2 µg/mL) for 24 h, and induced with 2 µM 20E or an equivalent amount of DMSO for 0.5 h. λPPase treatment of the proteins was performed for 0.5 h, and EcRB1 and β-actin were detected via Western blot analysis following 7.5% SDS/PAGE. Statistical analyses of the density of Western blot bands were performed by Quantity One software based on three independent biological experiments. The values are the mean ± SD from three independent experiments, and statistical analyses were performed using Student’s t test (*P < 0.05).
To confirm that PKCδ induces EcRB1 phosphorylation, PKCδ was knocked down via RNAi in HaEpi cells. Knockdown of PKCδ significantly suppressed 20E-induced EcRB1 phosphorylation. The molecular mass of the upper band was degraded to that of the lower band by λPPase (Fig. 4D). These results confirmed that PKCδ is involved in 20E-induced EcRB1 phosphorylation.
EcRB1 Phosphorylation Determines the Formation of the EcRB1/USP1 Complex and Its Binding to EcRE to Regulate Gene Transcription.
The phosphorylation site of EcRB1 (GenBank accession no. EU526831.1) was determined to confirm the 20E-induced EcRB1 phosphorylation. Thr176, Thr468, and Ser510 of EcRB1 were predicted as possible PKC phosphorylation sites using tools provided in KinasePhos (kinasephos.mbc.nctu.edu.tw/) and NetPhos (www.cbs.dtu.dk/services/NetPhos/). These residues of EcRB1 were mutated to alanines (T176A, T468A, and S510A), followed by overexpression in HaEpi cells. The phosphorylation levels of WT and mutant EcRB1 were detected using a phosphoprotein phosphate estimation assay kit. The number of moles of phosphorus per mole of EcRB1 was increased under 20E induction for the T176A and S510A mutants compared with that seen under DMSO treatment, while the level in the T468 mutant was not increased (Fig. 5A). This result indicated that T468 is the phosphorylation site of EcRB1 by 20E induction. To further verify the phosphorylation site of EcRB1, EcRB1-His and EcRB1-T468A-His were analyzed by Western blotting. EcRB1-His showed a higher molecular mass band after 20E treatment than after DMSO treatment, but T468A-His did not (Fig. 5B). Therefore, EcRB1 is phosphorylated at residue Thr468 under 20E induction.
Fig. 5.
Phosphorylation of EcRB1 promotes an interaction between EcRB1 and USP1 as well as gene expression in HaEpi cells. (A) Identification of the phosphorylation site in EcRB1 by determining the number of moles of phosphorus per mole of EcRB1-His or its mutants (T176A, T468A, and S510A) overexpressed in cells. EcRB1-His and site mutants of the protein were overexpressed in cells for 48 h, followed by treatment with 20E or an equivalent amount of DMSO. (B) Western blot analysis confirmed that the site of phosphorylation in EcRB1 was Thr468, under the conditions described in A. β-Actin was used as a loading control. Statistical analyses of the density of Western blotting bands were performed with Quantity One software. (C) ChIP analyses of the EcRE-binding abilities of EcRB1-RFP-His and EcRB1-T468A-RFP-His in the HR3 promoter region via anti-RFP precipitation and qRT-PCR analysis. 20E (2 µM) or DMSO treated the transfected cells for 3 h. Results are presented as percentage of enrichment relative to input. Input, sample before immunoprecipitation. Enrichment relative to input = (sample precipitated by antibody – sample without antibody)/input × %. lgG, nonspecific mouse lgG. Primer EcRE, primers targeted to EcRE. Primer HR3, primers targeted to HR3 ORF. (D) Phosphorylation of EcRB1 T468 promoted the interaction of EcRB1 with USP1 under 20E induction. The proteins were extracted from transfected cells after treatment with 2 µM 20E or DMSO for 3 h. The input showed the levels of the USP1-GFP-His, EcRB1-RFP-His, and EcRB1-T468A-RFP-His proteins expressed in the cells. β-Actin was used as a loading control, and the proteins were detected via Western blot analysis following SDS/PAGE using gels with a concentration of 7.5%. The phosphorylation of USP1 and EcRB1 was not detected due to the GFP and RFP tags increasing the molecular mass. Nonspecific mouse lgG as a negative control. (E) Comparison of HR3 expression levels in RFP-His–, EcRB1-RFP-His–, and EcRB1-T468A-RFP-His–overexpressing cells under 20E induction. pIEx-4-RFP-His, pIEx-4-EcRB1-RFP-His, and pIEx-4-EcRB1-T468A-RFP-His plasmid-transfected cells were treated with 20E (2 µM) for 6 h. An equivalent amount of diluted DMSO was added as a control. Statistical analyses were performed using Student’s t test (*P < 0.05, ***P < 0.001), and the values are the mean ± SD. Three biological replicates and three technical replicates were performed for A–E.
To address the significance of EcRB1 phosphorylation in the 20E pathway, we examined the binding of EcRB1-RFP-His and EcRB1-T468A-RFP-His to EcRE via a chromatin immunoprecipitation (ChIP) assay. EcRB1-RFP-His bound more EcRE DNA fragments than EcRB1-T468A-RFP-His under 20E treatment (Fig. 5C), suggesting that EcRB1 phosphorylation is necessary for its binding to the EcRE. In addition, a co-IP assay using anti-GFP detected an interaction between EcRB1-RFP-His and USP1-GFP-His but not between EcRB1-T468A-RFP-His and USP1-GFP-His under 20E induction when USP1-GFP-His and EcRB1-RFP-His were equally expressed (Fig. 5D). This suggested that EcRB1 phosphorylation is necessary for the interaction between EcRB1-RFP-His and USP1-GFP-His. The expression level of HR3 was significantly increased in EcRB1-RFP-His–overexpressing cells under 20E induction compared with that in EcRB1-T468A-RFP-His–overexpressing cells (Fig. 5E), suggesting that EcRB1 phosphorylation is necessary for 20E-induced gene expression. Therefore, in the 20E pathway, EcRB1-Thr468 phosphorylation is required for the formation of the EcRB1-USP1 transcriptional complex and the binding of this complex to EcRE to promote 20E-induced gene expression.
Discussion
PKCδ has been shown to play an important role in apoptosis in mammals (1, 2), and the steroid hormone 20E promotes apoptosis in insects (16, 21). However, the role of PKCδ in 20E-induced apoptosis is unclear. Here, we reveal that 20E up-regulates the expression of a novel PKCδ isoform. This PKCδ isoform is phosphorylated upon its expression and subsequently functions in the regulation of apoptosis. PKCδ phosphorylates EcRB1 at Thr468 to form the EcRB1/USP1 transcriptional complex to promote apoptotic gene expression and apoptosis.
Characteristics of the Novel PKCδ of H. armigera.
Based on the structural characteristics and modes of activation of PKCs, they are subclassified into three groups: conventional PKCs (cPKCs, α, βI, βII, and γ), novel PKCs (nPKCs, ε, θ, δ, and η), and atypical PKCs (aPKCs, ι, ζ, and λ) (22). All PKC isoforms contain an N-terminal regulatory domain for maintaining an inactive conformation and a C-terminal catalytic domain (kinase domain) to phosphorylate substrates. All isoforms of PKC also contain a C1 domain in the regulatory domain for DAG or phorbol ester binding, with the exception of aPKCs, which cannot bind DAG or phorbol esters. Conventional and novel isoforms of PKC contain a C2 domain as well, which acts as a Ca2+ sensor but functions only in the conventional isoform (23).
This work describes the function of PKCδ isoform E. Because of the absence of the C1 and C2 domains in PKCδ isoform E, the activity of PKCδ-E does not theoretically rely on calcium or DAG. When the catalytic domain is overexpressed, it localizes to the nucleus, where it shows phosphorylation and proapoptotic activity in 20E-free conditions. Therefore, up-regulation of the expression levels of PKCδ isoform E is a key step to control its function in apoptosis. The cleavage of PKCδ by caspase-1 was not investigated due to the lack of a specific antibody and the difficulty of overexpressing the large PKCδ protein. Nevertheless, overexpression of the PKCδ catalytic domain promotes caspase-1 expression in the HaEpi cells (Fig. 3D), and 20E promotes caspase-1 expression in hemocytes in H. armigera (24), which might cleave PKCδ in vivo.
20E Up-Regulates PKCδ Expression to Promote Apoptosis During Metamorphosis.
The high expression levels of PKCδ in H. armigera during metamorphosis were correlated with PCD in the tissue and were in accord with the higher 20E titer observed in lepidopteran insects (9). Via EcRB1/USP1, 20E up-regulates PKCδ expression in larval tissues and HaEpi cells, confirming that 20E is one of the upstream positive regulators of PKCδ. This finding is consistent with the major function of 20E at a high concentration, inducing apoptosis (25).
We previously revealed that a serine/threonine protein kinase with 56% identity to testis-specific serine/threonine protein kinase 3-like of D. melanogaster promotes midgut and fat body apoptosis in H. armigera (26). Now, we report that PKCδ is involved in 20E-induced apoptosis in HaEpi cells and H. armigera larvae, which is consistent with the function of PKCδ in mammalian apoptosis (6, 27, 28). In humans, PKCδ acts as a tumor suppressor (29). Although many studies have shown that PKCδ mediates cell apoptosis, this study shows that PKCδ induces apoptosis in insects.
We demonstrated that phosphorylation at Thr1343 is required for the proapoptotic activity of PKCδ in H. armigera. This residue is highly conserved from vertebrates to invertebrates, as demonstrated by alignment analysis, and can also be phosphorylated in humans (30, 31). In our study, phosphorylation was found to occur once the catalytic domain of PKCδ was overexpressed, and we therefore propose that PKCδ is autophosphorylated; however, this hypothesis requires further study in future work.
Studies have shown that the growth and differentiation of the wing imaginal disk in M. sexta has a 20E concentration dependence (20). 20E also induced cell proliferation in the Bombyx wing disk (32). However, our study showed that the expression of PKCδ was negatively correlated with the growth of wing disks during metamorphosis in H. armigera. In addition, 20E repressed PKCδ expression in the wing disk. Although knockdown of PKCδ repressed 20E-promoted wing disk growth, the gene expression profiles quite differently in wing disk and in midgut after knockdown of PKCδ. A possible mechanism is the differentially expressed genes in the apoptotic tissues and the growing tissues. 20E might use different pathways to induce midgut apoptosis and wing disk growth, which needs further study in the future work.
PKCδ Phosphorylates EcRB1 to Regulate Gene Transcription.
Both EcRB1 and USP1 are members of the nuclear receptor superfamily and are ligand-activated transcription factors. The activities of nuclear receptors are regulated not only by transcriptional/translational mechanisms but also by posttranslational mechanisms, such as phosphorylation. The regulation of USP phosphorylation by 20E through PKC has been demonstrated in D. melanogaster (17), M. sexta (33), Tenebrio molitor (34), and H. armigera (16), although the PKC was not identified yet until now. Furthermore, EcR has been demonstrated to be a phosphoprotein in adult B. mori (18), Drosophila larvae (35), and H. armigera (19). However, these studies only demonstrated the phosphorylation of EcR and did not identify its phosphorylation sites or the specific PKC involved. To our surprise, we found that when overexpressed, the PKCδ catalytic domain can directly phosphorylate EcRB1 at Thr468 without 20E induction, although it has no significant effect on USP1 phosphorylation. The phosphorylation of EcRB1 is required for the formation of the EcRB1/USP1 complex and for binding of the complex to the EcRE for subsequent gene transcription. This finding reveals one mechanism of 20E-regulated gene expression.
The overexpressed catalytic domain of PKCδ localizes to the nucleus. Full-length PKCδ was also predicted to contain nuclear localization signals (at amino acids 716–723 and 1175–1181) and to localize to the nucleus, based on analysis using the PSORT website (www.genscript.com/psort.html). Thus, we infer that the phosphorylation of EcRB1 occurs in the nucleus.
A previous study showed that USP phosphorylation is necessary for it interacting with EcRB1 in H. armigera (16). In addition, acetylation of USP1 is also necessary for it interacting with EcRB1 (36). 20E induces phosphorylation of cyclin-dependent kinase 10 to promote EcRB1/USP1 complex formation (21). This work further found that 20E induces EcRB1 phosphorylation and it was important for the EcRB1/USP1 complex formation. All of the data suggested that the posttranslational modification is an important mechanism for the EcRB1/USP1 transcription complex formation.
In Conclusion
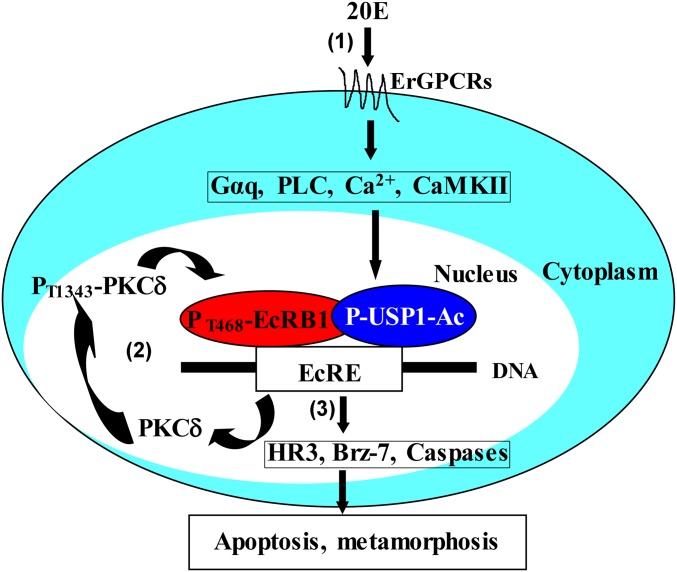
Previous work has shown that 20E via ErGPCR1 and ErGPCR2, Gαq, PLC, calcium ion, and CaMKII signaling triggers USP1 phosphorylation and acetylation (P-USP1-Ac) to modulate USP1 binding to EcRE for subsequent gene transcription (16, 37). In this work, we further revealed that 20E rapidly up-regulates PKCδ expression via EcRB1 and USP1. The overexpressed PKCδ catalytic domain was autophosphorylated and localized in the nucleus to phosphorylate EcRB1 at T468 directly. The phosphorylated EcRB1 interacted with P-USP1-Ac to form the EcRB1/USP1 transcriptional complex to promote apoptotic gene expression and apoptosis. Through a positive feedback mechanism, 20E via PKCδ promotes midgut apoptosis during insect metamorphosis (Fig. 6).
Fig. 6.
A schematic representation of the mechanism by which 20E regulates PKCδ expression and EcRB1 phosphorylation. (1) 20E via ErGPCR1 and ErGPCR2, Gαq, PLC, calcium ion, and CaMKII signaling triggers USP1 phosphorylation and acetylation to modulate formation of EcRB1/USP1 transcription complex to bind on EcRE for 20E pathway gene transcription based on our previous studies (16, 36). (2) 20E rapidly up-regulates PKCδ expression via EcRB1 and USP1. The overexpressed PKCδ catalytic domain was autophosphorylated and localized to the nucleus to phosphorylate EcRB1 at T468. (3) The phosphorylated EcRB1 interacts with P-USP1-Ac to form the EcRB1/USP1 transcriptional complex to promote apoptotic gene expression and apoptosis. Through this positive feedback mechanism, 20E promotes apoptosis during insect metamorphosis.
Materials and Methods
Insects.
Cotton boll worms were cultured in our laboratory at 27 ± 1 °C under a photoperiod of 14 h light/10 h dark. The larvae were reared on a previously described artificial diet (38).
Cell Culture.
The Helicoverpa epidermal cell line (HaEpi) was established from the H. armigera integument and has previously been well characterized (39). HaEpi cells were developed as a loosely attached monolayer and were maintained at 27 °C with Grace’s insect cell culture medium containing 10% FBS (Gibco).
20E Induction in Larvae and in HaEpi Cells.
20E was first dissolved at a concentration of 10 mg/mL (20 mM) in DMSO and then diluted 1:100 in sterile PBS (140 mM NaCl, 10 mM sodium phosphate, pH 7.4). Next, 500 ng/5 μL of 20E was injected into sixth instar-6 h larva, and an equal diluted DMSO was injected as a control. Total RNA was isolated from the fat body and midgut of the injected larvae (3–5 larvae in each panel) after 3, 6, or 12 h of induction by 20E and DMSO. The relative expression of PKCδ mRNA was then analyzed via qRT-PCR. HaEpi cells were treated with 2 μM 20E for 0.25, 0.5, 1, 3, and 6 h to confirm the response of PKCδ to 20E induction, and equal diluted DMSO was used as a solvent control. HaEpi cells were treated with various doses of 20E for 3 h.
DsRNA Synthesis.
Some laboratories had difficulty with knocking down genes in lepidopteran larvae in the previous year (40). However, 100 peer-reviewed papers have described successful RNAi experiments in 10 families and 25 species of moth in recent years (41). We describe the detailed method here as a reference for people doing RNAi experiments in insects. About 500 bp cDNA of target gene was amplified by a single PCR with the T7 promoter appended to both PCR primers (Table S1) as the template to synthesize dsRNA. The long dsRNAs specifically suppress expression of a target gene in worms (42) after being broken down into smaller fragments in vivo (43). Both strands of the dsRNA were synthesized in one reaction according to the method described in MEGAscript RNAi kit (Ambion). The purity of dsRNA is critical for the efficacy of RNAi in larvae. We used the phenol-chloroform method to purify the dsRNA. After DNaseI digestion to remove the DNA template, the dsRNA was filled with nuclease-free water to 200 μL, and an equal volume water saturated with phenol-chloroform was added. After mixing gently and centrifugation (9,000 × g) at 4 °C, the upper aqueous phase was mixed with chloroform and centrifuged again. The upper aqueous was added with 1/10th volume of 3M sodium acetate (pH 4.0) and 2.5 volume of 100% ethanol, mixed, and kept at –20 °C overnight. The dsRNA was collected by centrifugation. The dsRNA pellet was washed with 75% ethanol, air-dried, and suspended in 20 μL nuclease-free water. The quality of dsRNA was examined by 1% agrose gel electrophoresis and quantified by microvolume spectrophotometers.
Table S1.
Oligonucleotide sequences of PCR primers
| Primer name | Sequence, 5′–3′ |
| qRT-PCR | |
| PKCδ-RTF | acgccgtgaagtgtttga |
| PKCδ-RTR | cagaggtagggatggttagtg |
| EcRB1-RTF | aattgcccgtcagtacga |
| EcRB1-RTR | tgagcttctcattgagga |
| USP1-RTF | ggtcctgacagcaatgtt |
| USP1-RTR | ttccagctccagctgactgaag |
| HR3-RTF | tcaagcacctcaacagcagcccta |
| HR3-RTR | gactttgctgatgtcaccctccgc |
| Brz-7-RTF | ggtgactgtccttactgcggcat |
| Brz-7-RTR | ttaattcctttgaccatgact |
| caspase-1 RTF | gagttgggaatgttgtatgct |
| caspase-1 RTR | gatgcagacgatgatccatca |
| caspase-3 RTF | ggagacaagggtggagaa |
| caspase-3 RTR | ggaaggcgtgtatgtggt |
| caspase-6 RTF | gctgtgatcagtgctacggat |
| caspase-6 RTR | ccgaatcagctgcatacactt |
| β-actin RTF | cctggtattgctgaccgtatgc |
| β-actin RTR | ctgttggaaggtggagagggaa |
| CHIP F | atattcgaatcgttggcg |
| CHIP R | cagtgtaattaagagaca |
| RNAi | |
| PKCδRNAiF | gcgtaatacgactcactatagggatcgatggacttgtcacggta |
| PKCδRNAiR | gcgtaatacgactcactatagggaatttgccggtttagatgcac |
| EcRB1RNAiF | gcgtaatacgactcactatagggacgctggtataacaacggagga |
| EcRB1RNAiR | gcgtaatacgactcactatagggaagctggagacaactcctcacg |
| USP1RNAiF | gcgtaatacgactcactatagggacgaaccatcccctaagtggttc |
| USP1RNAiR | gcgtaatacgactcactatagggaccttgatgagcaggatctggtc |
| GFPRNAiF | gcgtaatacgactcactatagggatggtcccaattctcgtggaac |
| GFPRNAiR | gcgtaatacgactcactatagggacttgaagttgaccttgatgcc |
| Overexpression: | |
| ΔPKCδoexF | tcgttaacacgtcaagagaaggccgtgagacctt |
| ΔPKCδoexR | tgggatccgcgagctcgacgggggtcgcctccgccc |
| EcRB1oexF | taacacgtcaagagctcatgagacgccgctggtataac |
| EcRB1oexR | gcaggcgcgccgagatctggagcgccggcgagtccgcca |
| Clone the full-length | |
| PKCδ F | acgcgctccgatgagcgac |
| PKCδ R | ctgcgcctgctaaatgtaac |
RNAi in HaEpi Cells.
For RNAi in HaEpi cells, 2 μg of dsRNA was transfected into the cells using the QuickShuttle-enhanced transfection reagent (Biogragon-Immunotech) in 1 mL of Grace’s medium, according to the manufacturer’s instructions and our previous work (11), while the control group received an equivalent amount of dsGFP. After 48 h of dsRNA treatment, 2 μM 20E or an equivalent amount of DMSO was added to the cell culture media, followed by an additional 6 h of incubation. Total RNA was then extracted from the cells for qRT-PCR analysis.
RNAi in Larvae.
The newly synthesized dsRNA was diluted to the appropriate concentration with RNase-free PBS. Larvae were placed on ice for 15 min until they did not move. The sterile dsRNA was injected into the larval hemocoel from the side of the front abdomen, with the needle direction backward. Injection was careful to not puncture the midgut. The larvae need to be injected twice with 5 μL dsRNA. The first time, 1 μg of dsRNA was injected into the fifth instar-12 h larvae, and the second time, 2 μg of dsRNA was injected into the sixth instar-6 h feeding larvae. Each experiment group contained 30 larvae, and the experiments were repeated three times. Injection of dsGFP was used as control.
Overexpression of Protein in HaEpi Cells.
The cDNA fragment encoding the catalytic domain (ΔPKCδ, 1064–1485 aa) of PKCδ was amplified using the corresponding primers (Table S1) and then inserted into an insect cell-specific overexpression vector (pIEx-4-RFP-His) fused with RFP (RFP-His) to construct the pIEx-4-ΔPKCδ-RFP-His plasmid, as described in our previous work (44). The recombinant vector was verified via direct DNA sequencing. Also, other recombinant vectors had been prepared, such as pIEx-4-EcRB1-His, pIEx-4-EcRB1-RFP-His, pIEx-4-USP1-His, pIEx-4-USP1-GFP-His, and other mutants. HaEpi cells were maintained at 80% confluence under normal growth conditions, as previously described by Shao et al. (39). Five micrograms of plasmid was transfected into the HaEpi cells using the QuickShuttle-enhanced transfection reagent (Biogragon-Immunotech). After 48 h of transfection, total protein was extracted for Western blot analysis or other experiments. Total RNA was isolated for detection of the transcripts of genes via qRT-PCR.
λPPase Treatment.
The pIEx-4-ΔPKCδ-RFP-His plasmid or pIEx-4-EcRB1-His plasmid (5 μg) was transfected into HaEpi cells for 48 h. The cells were then treated with 2 μM 20E for 30 min, and protein was extracted from the cells using radio immunoprecipitation assay buffer (RIPA, 50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.1% SDS, and 1 mM PMSF). A 40-µL sample of the protein (2 mg/mL) was subsequently incubated with 0.5 μL of λPP, 5 μL of buffer, and 5 μL of MnCl2 at 30 °C for 30 min, according to the manufacturer’s specifications (New England Biolabs). Next, the proteins were subjected to SDS polyacrylamide gel electrophoresis (SDS/PAGE) in 7.5% low-concentration gels, followed by Western blot analysis. Protein phosphorylation was examined based on variations in molecular mass. β-Actin was used as the protein control using antiserum against β-actin in H. armigera.
Western Blot.
The total protein was extracted from cells using 40 mM Tris·HCl, pH 7.5, with 1 mM phenylmethanesulfonyl fluoride (PMSF), followed by centrifugation at 10,000 × g at 4 °C for 10 min. The supernatant was then collected, and Bradford’s method (45) was used to determine the protein concentration. The details are in SI Materials and Methods.
Caspase-3 Activity Assay.
The activity of caspase-3 was detected in pIEx-4-ΔPKCδ-RFP–overexpressing HaEpi cells with the NucView 488 caspase-3 assay kit (NO. 30029 Biotium) via immunocytochemistry, according to the manufacturer’s instructions. After 48 h of overexpression of ΔPKCδ-RFP-His, the NucView 488 Caspase-3 Substrate (2 μM) was added to the cell culture media, followed by 0.5 h of incubation at room temperature. Finally, the nuclei were stained with 1 μg/mL 4', 6-diamidino-2-phenylindole (DAPI) for 10 min, and the resulting signals were observed using a Zeiss LSM 700 laser confocal microscope (Zeiss) or Olympus BX51 fluorescence microscope (Shinjuku-ku, Tokyo).
Co-IP.
HaEpi cells were seeded into a six-well plate. When the cell density was ∼80% in each well, the reconstructed plasmids were cotransfected into the cells. After 48 h, the cells were treated with 2 μM 20E for 1 h, and DMSO was used as the control. Protein was subsequently extracted according to the RIPA buffer assay, and the supernatant was harvested via centrifugation at 12,000 × g for 10 min (4 °C). The supernatant was added to protein A resin to eliminate nonspecific binding and harvested through centrifugation (1,000 × g for 2 min). The supernatant was then incubated with the protein A resin–monoclonal antibody complex for 2–4 h with gentle shaking at 4 °C. The resin was subsequently harvested via centrifugation and washed with RIPA buffer three times. Finally, the resin was treated with SDS/PAGE loading buffer and boiled for 10 min, and the samples were subjected to SDS/PAGE, followed by Western blot analysis with mouse monoclonal antibodies against RFP, GFP, and His (Sungenebiotech) to detect the target proteins.
Detection of Phosphorylation Levels.
EcRB1-His and the mutant plasmids (EcRB1-T176A-His, EcRB1-T468A-His, or EcRB1-S510A-His) were transfected into HaEpi cells for 72 h. Protein was then purified using a Ni2+-NTA affinity column to detect phosphorylation levels after different treatments. The number of moles of phosphate per mole of purified protein was determined using the Phosphoprotein Phosphate Estimation Assay Kit (Sangon Biotech), based on the alkaline hydrolysis of phosphate from Ser and Thr residues in phosphoproteins. The released phosphates were then quantified using malachite green and ammonium molybdate, in accordance with the manufacturer’s instructions.
ChIP Assay.
The EcRB1-RFP and EcRB1-T468A-RFP plasmids were transfected into cells with a 70% density in six-well plates for 72 h. Subsequently, the cells were treated with 20E for 3 h. DMSO treatment was used as the control. A monoclonal antibody against RFP was used to link EcRB1-RFP with the EcRE complex. The EcRE fragments in the H. armigera HR3 promoter were purified using a ChIP Assay Kit (Beyotime) according to the manufacturer’s instructions. Then, the DNA was analyzed via qRT-PCR using the ChIP F/ChIP R primers (target to EcRE) shown in the Table S1.
SI Materials and Methods
Cloning of the Full-Length cDNA of PKCδ and Analysis of Its Sequence Information.
The 3′ terminal cDNA sequence was obtained from H. armigera transcriptomic data, and the 5′ sequence was provided by Zhen Zou, Institute of Zoology, Chinese Academy of Sciences, Beijing. Using BLASTX (https://www.ncbi.nlm.nih.gov/), these sequences were suggested to be the 5′ termini and 3′ termini of the PKCδ gene. The full-length cDNA was cloned via PCR using the terminal primers PKCδ-F and PKCδ-R (Table S1) and verified through direct sequencing. The ORF was determined using the ExPASy tool (web.expasy.org/translate/). Protein domain predictions were performed with SMART software (smart.embl-heidelberg.de/). Sequence alignments and phylogenetic trees were obtained using MEGA 5.0 and GENEDOC.
Analysis of mRNA Levels via qRT-PCR.
Total RNA was extracted using the TranZol reagent (TransGen), and the first strand of cDNA was synthesized using a first-strand cDNA synthesis kit (TianGen) according to the manufacturer’s instructions. Relative transcript levels were measured by using the 2× SYBR RT-PCR premixture (BioTeke Corp.) and the CFX96 real-time system (Bio-Rad) with the corresponding primers (Table S1) in a final volume of 10 μL. The following steps were included in the amplification program: 95 °C initial denaturation for 15 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s, with plate reading at 78 °C for 2 s and melting curve analysis from 65 °C to 95 °C. 2–∆∆CT was used to calculate the relative mRNA levels (39). Data were analyzed using the formula: R = 2−[ΔCt sample − ΔCt control], where R is the relative expression level, ΔCt sample is the difference between the Ct of the gene and the average β-actin in the experimental sample, and ΔCt control is the difference between the Ct of the gene and the average of β-actin in the control sample. Three biological replicates and three technical replicates were performed for all qRT-PCRs in the manuscript.
HE Staining of Tissues.
For HE staining, the larval midgut and fat body were isolated, fixed with 4% paraformaldehyde at 4 °C overnight, and then gradient dehydrated. Next, the tissues were embedded in paraffin, sliced into 7-μm sections with a paraffin slicing machine, and adhered to gelatin-coated glass slides. The slides were dried at 42 °C overnight and subsequently dewaxed, gradient rehydrated, and digested with 20 mM proteinase K at 37 °C for 10 min. The assay was finally conducted following the method described in our previous work (42). Positive signals were observed using an Olympus BX51 fluorescence microscope (Shinjuku-ku).
Western Blot.
The total protein was extracted from cells using 40 mM Tris·HCl, pH 7.5, with 1 mM PMSF, followed by centrifugation at 10,000 × g at 4 °C for 10 min. The supernatant was then collected, and Bradford’s method (44) was used to determine the protein concentration. Next, 50 μg of protein from each sample was subjected to 7.5–12.5% SDS/PAGE and then electrophoretically transferred onto nitrocellulose membranes. The membranes were incubated at room temperature in blocking buffer (5% fat-free powdered milk in Tris-buffered saline—10 mM Tris·HCl, 150 mM NaCl, pH 7.5) for 1 h. The primary antibodies used for these assays were diluted with blocking buffer, followed by overnight incubation at 4 °C. The membrane was subsequently washed three times with TBST (0.02% Tween in TBS) for 10 min each time, followed by the addition of the alkaline phosphatase-conjugated AffiniPure horse anti-rabbit/anti-mouse IgG secondary antibody, diluted 1:5,000 with the same blocking buffer. The protein signal was observed in 10 mL of a Tribuffered saline solution combined with 5% p-nitro-blue tetrazolium chloride (NBT) and 5% 5-bromo-4-chloro-3-indolyl phosphate (BCIP) in the dark for 20 min.
Wing Disk Culture and Measure.
The sixth instar-48 h larvae were placed on ice for 15 min until they did not move. Then larvae were surface-sterilized by immersion in 75% ethanol for 2–3 min and dissected in sterile solution of PBS in culture dish. Wing disks were cultured in 24-well plates in 300 μL Grace’s medium (Gibco, Invitrogen) and supplemented with 10% FBS (Scitecher) and two antibiotics, ampicillin (125 U/mL) and streptomycin sulfate (100 μg/mL) (Sangon Biotech) under 27 °C for 48 h. Pairs of wing disks from the same individuals were cultured, using one wing as a control and the other wing as the experimental. The length of fore wing disks was measured using ocular micrometer under microscope after the disks were separated.
Acknowledgments
We thank Dr. Zhen Zou from the Institute of Zoology, Chinese Academy of Sciences for providing 5′ terminal sequence information of H. armigera PKCδ. This work was supported by National Natural Science Foundation of China Grants 31230067 and 31572328.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KR047104.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704999114/-/DCSupplemental.
References
- 1.Reyland ME. Protein kinase Cdelta and apoptosis. Biochem Soc Trans. 2007;35:1001–1004. doi: 10.1042/BST0351001. [DOI] [PubMed] [Google Scholar]
- 2.Zhao M, Xia L, Chen GQ. Protein kinase cδ in apoptosis: A brief overview. Arch Immunol Ther Exp (Warsz) 2012;60:361–372. doi: 10.1007/s00005-012-0188-8. [DOI] [PubMed] [Google Scholar]
- 3.Gong J, et al. The C2 domain and altered ATP-binding loop phosphorylation at Ser(3)(5)(9) mediate the redox-dependent increase in protein kinase C-delta activity. Mol Cell Biol. 2015;35:1727–1740. doi: 10.1128/MCB.01436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Belkina NV, Graham C, Shaw S. Independence of protein kinase C-delta activity from activation loop phosphorylation: Structural basis and altered functions in cells. J Biol Chem. 2006;281:12102–12111. doi: 10.1074/jbc.M600508200. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Paramio P, Cabrerizo Y, Bornancin F, Parker PJ. The broad specificity of dominant inhibitory protein kinase C mutants infers a common step in phosphorylation. Biochem J. 1998;333:631–636. doi: 10.1042/bj3330631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeVries TA, Neville MC, Reyland ME. Nuclear import of PKCdelta is required for apoptosis: Identification of a novel nuclear import sequence. EMBO J. 2002;21:6050–6060. doi: 10.1093/emboj/cdf606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert LI. Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol Cell Endocrinol. 2004;215:1–10. doi: 10.1016/j.mce.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Adler JH, Grebenok RJ. Biosynthesis and distribution of insect-molting hormones in plants––A review. Lipids. 1995;30:257–262. doi: 10.1007/BF02537830. [DOI] [PubMed] [Google Scholar]
- 9.Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem Mol Biol. 2003;33:1327–1338. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Homem CC, et al. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014;158:874–888. doi: 10.1016/j.cell.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 11.Li YB, Li XR, Yang T, Wang JX, Zhao XF. The steroid hormone 20-hydroxyecdysone promotes switching from autophagy to apoptosis by increasing intracellular calcium levels. Insect Biochem Mol Biol. 2016;79:73–86. doi: 10.1016/j.ibmb.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Liu CY, Zhao WL, Wang JX, Zhao XF. Cyclin-dependent kinase regulatory subunit 1 promotes cell proliferation by insulin regulation. Cell Cycle. 2015;14:3045–3057. doi: 10.1080/15384101.2015.1053664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai MJ, et al. 20-Hydroxyecdysone activates Forkhead box O to promote proteolysis during Helicoverpa armigera molting. Development. 2016;143:1005–1015. doi: 10.1242/dev.128694. [DOI] [PubMed] [Google Scholar]
- 14.Wang D, et al. The steroid hormone 20-hydroxyecdysone promotes the cytoplasmic localization of yorkie to suppress cell proliferation and induce apoptosis. J Biol Chem. 2016;291:21761–21770. doi: 10.1074/jbc.M116.719856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunyadi A, et al. Ecdysteroid-containing food supplements from Cyanotis arachnoidea on the European market: Evidence for spinach product counterfeiting. Sci Rep. 2016;6:37322. doi: 10.1038/srep37322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Cai MJ, Zheng CC, Wang JX, Zhao XF. Phospholipase Cγ1 connects the cell membrane pathway to the nuclear receptor pathway in insect steroid hormone signaling. J Biol Chem. 2014;289:13026–13041. doi: 10.1074/jbc.M113.547018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Q, Sun X, Jin XY. 20E-regulated USP expression and phosphorylation in Drosophila melanogaster. Insect Biochem Mol Biol. 2003;33:1211–1218. doi: 10.1016/j.ibmb.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Sridhara S. Ecdysone receptor and ultraspiracle proteins are tyrosine phosphorylated during adult development of silkmoths. Insect Biochem Mol Biol. 2012;42:91–101. doi: 10.1016/j.ibmb.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Ren J, et al. G-protein αq participates in the steroid hormone 20-hydroxyecdysone nongenomic signal transduction. J Steroid Biochem Mol Biol. 2014;144:313–323. doi: 10.1016/j.jsbmb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Nijhout HF, et al. The control of growth and differentiation of the wing imaginal disks of Manduca sexta. Dev Biol. 2007;302:569–576. doi: 10.1016/j.ydbio.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Cai MJ, Wang JX, Zhao XF. In a nongenomic action, steroid hormone 20-hydroxyecdysone induces phosphorylation of cyclin-dependent kinase 10 to promote gene transcription. Endocrinology. 2014;155:1738–1750. doi: 10.1210/en.2013-2020. [DOI] [PubMed] [Google Scholar]
- 22.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webb BL, Hirst SJ, Giembycz MA. Protein kinase C isoenzymes: A review of their structure, regulation and role in regulating airways smooth muscle tone and mitogenesis. Br J Pharmacol. 2000;130:1433–1452. doi: 10.1038/sj.bjp.0703452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D, Chai L, Wang J, Zhao X. Molecular cloning and characterization of Hearm caspase-1 from Helicoverpa armigera. Mol Biol Rep. 2008;35:405–412. doi: 10.1007/s11033-007-9100-8. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Pei XY, Zhao WL, Zhao XF. Steroid hormone 20-hydroxyecdysone promotes higher calcium mobilization to induce apoptosis. Cell Calcium. 2016;60:1–12. doi: 10.1016/j.ceca.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Liu CY, Liu W, Zhao WL, Wang JX, Zhao XF. Upregulation of the expression of prodeath serine/threonine protein kinase for programmed cell death by steroid hormone 20-hydroxyecdysone. Apoptosis. 2013;18:171–187. doi: 10.1007/s10495-012-0784-4. [DOI] [PubMed] [Google Scholar]
- 27.Leverrier S, Vallentin A, Joubert D. Positive feedback of protein kinase C proteolytic activation during apoptosis. Biochem J. 2002;368:905–913. doi: 10.1042/BJ20021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao M, et al. Protein kinase Cdelta stimulates proteasome-dependent degradation of C/EBPalpha during apoptosis induction of leukemic cells. PLoS One. 2009;4:e6552. doi: 10.1371/journal.pone.0006552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyland ME. Protein kinase C isoforms: Multi-functional regulators of cell life and death. Front Biosci (Landmark Ed) 2009;14:2386–2399. doi: 10.2741/3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parekh DB, et al. Beta1-integrin and PTEN control the phosphorylation of protein kinase C. Biochem J. 2000;352:425–433. [PMC free article] [PubMed] [Google Scholar]
- 31.Konishi H, et al. Phosphorylation sites of protein kinase C delta in H2O2-treated cells and its activation by tyrosine kinase in vitro. Proc Natl Acad Sci USA. 2001;98:6587–6592. doi: 10.1073/pnas.111158798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriyama M, et al. Ecdysteroid promotes cell cycle progression in the Bombyx wing disc through activation of c-Myc. Insect Biochem Mol Biol. 2016;70:1–9. doi: 10.1016/j.ibmb.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Song Q, Gilbert LI. Alterations in ultraspiracle (USP) content and phosphorylation state accompany feedback regulation of ecdysone synthesis in the insect prothoracic gland. Insect Biochem Mol Biol. 1998;28:849–860. doi: 10.1016/s0965-1748(98)00075-7. [DOI] [PubMed] [Google Scholar]
- 34.Nicolaï M, Bouhin H, Quennedey B, Delachambre J. Molecular cloning and expression of Tenebrio molitor ultraspiracle during metamorphosis and in vivo induction of its phosphorylation by 20-hydroxyecdysone. Insect Mol Biol. 2000;9:241–249. doi: 10.1046/j.1365-2583.2000.00181.x. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y, An S, Henrich VC, Sun X, Song Q. Proteomic identification of PKC-mediated expression of 20E-induced protein in Drosophila melanogaster. J Proteome Res. 2007;6:4478–4488. doi: 10.1021/pr0705183. [DOI] [PubMed] [Google Scholar]
- 36.Jing YP, et al. The steroid hormone 20-Hydroxyecdysone enhances gene transcription through the cAMP response element-binding protein (CREB) signaling pathway. J Biol Chem. 2016;291:12771–12785. doi: 10.1074/jbc.M115.706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jing YP, Liu W, Wang JX, Zhao XF. The steroid hormone 20-hydroxyecdysone via nongenomic pathway activates Ca2+/calmodulin-dependent protein kinase II to regulate gene expression. J Biol Chem. 2015;290:8469–8481. doi: 10.1074/jbc.M114.622696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao XF, Wang JX, Wang YC. Purification and characterization of a cysteine proteinase from eggs of the cotton boll worm, Helicoverpa armigera. (Translated from English) Insect Biochem Molec. 1998;28:259–264. [Google Scholar]
- 39.Shao HL, et al. Establishment of a new cell line from lepidopteran epidermis and hormonal regulation on the genes. PLoS One. 2008;3:e3127. doi: 10.1371/journal.pone.0003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terenius O, et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Xu J, et al. RNA interference in moths: Mechanisms, applications, and progress. Genes (Basel) 2016;7:E88. doi: 10.3390/genes7100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 43.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 44.Cai MJ, et al. Heat shock protein 90 maintains the stability and function of transcription factor Broad Z7 by interacting with its Broad-Complex-Tramtrack-Bric-a-brac domain. Insect Mol Biol. 2014;23:720–732. doi: 10.1111/imb.12118. [DOI] [PubMed] [Google Scholar]
- 45.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]