Significance
CRISPR-Cas9 is a powerful site-specific genome-editing tool that has been used to genetically engineer many different systems. However, this tool has not yet been adopted for use in the baculovirus–insect cell system, which is an important recombinant protein production platform. Thus, we created new CRISPR-Cas9 vectors that can be used for genome editing in two relevant insect cell lines. This is significant because these tools will enable new efforts to enhance the capabilities and expand the utility of this important protein production platform.
Keywords: CRISPR-Cas9, baculovirus, insect cells, genome editing, glycoengineering
Abstract
The baculovirus–insect cell system (BICS) has been widely used to produce many different recombinant proteins for basic research and is being used to produce several biologics approved for use in human or veterinary medicine. Early BICS were technically complex and constrained by the relatively primordial nature of insect cell protein glycosylation pathways. Since then, recombination has been used to modify baculovirus vectors—which has simplified the system—and transform insect cells, which has enhanced its protein glycosylation capabilities. Now, CRISPR-Cas9 tools for site-specific genome editing are needed to facilitate further improvements in the BICS. Thus, in this study, we used various insect U6 promoters to construct CRISPR-Cas9 vectors and assessed their utility for site-specific genome editing in two insect cell lines commonly used as hosts in the BICS. We demonstrate the use of CRISPR-Cas9 to edit an endogenous insect cell gene and alter protein glycosylation in the BICS.
The baculovirus–insect cell system (BICS), first described in 1983 (1), has been used to produce thousands of different recombinant proteins for diverse areas of biomedical research. Since 2009, the BICS has also been used to produce several biologics approved for use in human or veterinary medicine (reviewed in ref. 2). Thus, the BICS is an important recombinant protein production platform that has had and will continue to have a large and broad impact on basic research, biotechnology, and medicine.
Two precedents suggest the BICS would have even more impact if it could be engineered to enhance its capabilities or extend its utility. In the 1980s, the isolation of baculovirus expression vectors was a highly inefficient, time-consuming, and frustrating process. However, by the early 1990s, efforts to engineer the baculoviral genome in various ways had greatly simplified this process (3–5). These refinements effectively converted a complex system created in highly specialized laboratories to a routine tool that could be easily used in many different laboratories. This was followed by efforts to enhance the BICS by engineering host protein N-glycosylation pathways (reviewed in ref. 6). However, host glycoengineering and other host improvement efforts have been limited to the use of nonhomologous recombination to knockin heterologous genes at random sites (reviewed in ref. 7). This is because there have been no tools for site-specific genome manipulation in the insect cell lines most commonly used as hosts in the BICS. These cell lines include Sf9 (8) and High Five (9), which are derived from the lepidopteran insects Spodoptera frugiperda (Sf) and Trichoplusia ni (Tn), respectively.
Sf9 and High Five cells clearly have the machinery required for protein N-glycosylation, but cannot synthesize the same end products as mammalian cells (reviewed in refs. 10–14). More specifically, both of these lepidopteran insect cell lines can transfer N-glycan precursors to nascent polypeptides and trim them to produce processing intermediates identical to those produced by mammalian cells. However, neither can process those intermediates through the additional steps needed to produce larger, mammalian-like structures with terminal sialic acids. Interestingly, we know insect cells encode the machinery needed to produce sialylated N-glycans and we also know some cells in whole insects naturally produce endogenously sialylated glycoproteins (15–17). Nevertheless, this genetic capacity is not expressed in Sf9 or High Five cells. Insect cells also have a trimming enzyme, absent in mammalian cells, which antagonizes N-glycan elongation (18–21). This enzyme, which is a specific, processing β-N-acetylglucosaminidase called fused lobes (FDL), removes a terminal N-acetylglucosamine residue from trimmed N-glycan–processing intermediates. This antagonizes elongation because it eliminates the N-glycan intermediates used as substrates for N-acetylglucosaminyltransferase II, which initiates the elongation process. The inability of the BICS to produce mammalian-type, elongated N-glycans is a major deficiency of this system because these structures are required for clinical efficacy in glycoprotein biologics (22). Because of its inability to synthesize these structures, it is widely believed that the BICS platform could never be used for glycoprotein biologics manufacturing.
As indicated above, this limitation has been addressed by using nonhomologous recombination to engineer insect cell N-glycosylation pathways for mammalian-type N-glycan biosynthesis (reviewed in refs. 12–14 and 23). These efforts have yielded new, transgenic insect cell lines that can be used to produce recombinant glycoproteins with fully elongated, mammalian-type N-glycans. However, further glycoengineering is needed to create host cell lines that can more efficiently process N-glycans in mammalian fashion and produce homogeneously glycosylated proteins. These more refined glycoengineering efforts will require tools for site-specific genome editing in the BICS, and fdl, which encodes an antagonistic function, will be a critically important target.
The CRISPR-Cas9 system is a relatively new and exceptionally powerful tool for site-specific genome editing (24–26). CRISPR-Cas9 vectors have been constructed for and used in many different biological systems, including insect cell systems. In fact, it has been shown that endogenous U6 promoters can be used to drive single-guide RNA (sgRNA) expression for CRISPR-Cas9 genome editing in S2R+, a cell line derived from the dipteran insect, Drosophila melanogaster (Dm) (27, 28), and BmN, a cell line derived from the lepidopteran insect, Bombyx mori (Bm) (29). These findings prompted us to attempt to adopt the CRISPR-Cas9 system for site-specific genome editing in the BICS. The broader purpose of this effort was to provide enabling technology for precise genetic modifications that will further enhance and expand the utility of this important recombinant protein production platform.
Results
Heterologous Insect U6 Promoters Fail to Support CRISPR-Cas9 Editing in Sf9 Cells.
When we undertook this effort, there were no known S. frugiperda or T. ni RNA polymerase III promoters. However, as noted above, there were DmU6 and BmU6 promoters with the known ability to drive sgRNA expression in D. melanogaster and B. mori cells, respectively (27–29). Thus, we chose to use the DmU6 and BmU6 promoters as potential surrogates for CRISPR-Cas9 genome editing in Sf9 and High Five cells, based on their ability to drive sgRNA expression in other insect cell systems. D. melanogaster is a dipteran and B. mori is a lepidopteran, so the former is relatively distantly and the latter more closely related to S. frugiperda and T. ni, from which Sf9 (8) and High Five (9) were derived.
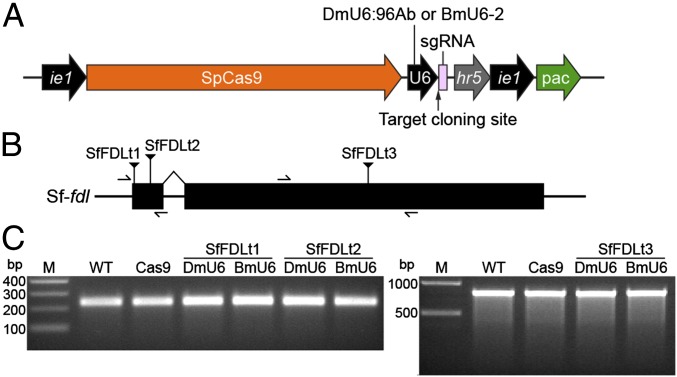
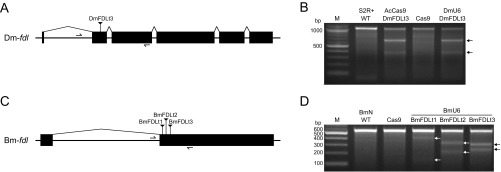
We initially designed generic CRISPR-Cas9 vectors (Fig. 1A) that included an S. frugiperda codon-optimized Streptococcus pyogenes (Sp) Cas9 coding sequence under the control of a baculovirus ie1 promoter, which provides constitutive transcription in a wide variety of organisms (30), followed by either the DmU6:96Ab or BmU6-2 promoter for sgRNA expression and a targeting sequence cloning site. These vectors also included a puromycin-resistance marker (puromycin acetyl transferase, pac) under the control of baculovirus hr5 enhancer and ie1 promoter elements (Fig. 1A). After constructing, mapping, and sequencing the generic DmU6:96Ab and BmU6-2 CRISPR-Cas9 vectors, we designed, synthesized, and inserted targeting sequences (Table S1) for the D. melanogaster (Fig. S1A) or B. mori (Fig. S1C) fdl genes. We then examined the editing capacities of the products by transfecting D. melanogaster (S2R+) or B. mori (BmN) cell lines, respectively, and performing CEL-I nuclease assays on puromycin-resistant derivatives. The results of this control experiment showed the Dm-fdl gene was efficiently edited in S2R+ cells transfected with the DmU6 vector encoding the Dm-fdl–specific sgRNA and in S2R+ cells transfected with AcCas9DmFDLt3, a previously described CRISPR-Cas9 vector encoding a Dm-fdl–specific sgRNA (28), but not in S2R+ cells transfected with a vector encoding Cas9 alone (Fig. S1B). Similarly, the Bm-fdl gene was efficiently edited in BmN cells transfected with each of three BmU6-2 vectors encoding different Bm-fdl–specific sgRNAs, but not in BmN cells transfected with a vector encoding Cas9 alone (Fig. S1D). These results indicated our CRISPR-Cas9 vectors produced functional Cas9 under ie1 promoter control, functional sgRNAs under DmU6:96Ab and BmU6-2 promoter control, and also showed they could be used for efficient CRISPR-Cas9 editing of endogenous gene targets in cells from the homologous species.
Fig. 1.
D. melanogaster and B. mori U6 promoters do not support CRISPR-Cas9 editing in Sf9 cells. (A) Diagram showing generic CRISPR-Cas9 vectors encoding, (Left to Right) SpCas9 under the control of a baculovirus ie1 promoter, an sgRNA expression cassette that includes an insect species-specific U6 promoter and a targeting sequence cloning site consisting of two SapI recognition sites, and a puromycin-resistance marker under the control of baculovirus hr5 enhancer and ie1 promoter elements. (B) Diagram showing Sf-fdl gene structure and highlighting specific Cas9 targeting sequences (Table S1) and PCR primer sites. (C) CEL-I nuclease assay results obtained using genomic DNA from Sf9 cells edited with CRISPR-Cas9 vectors encoding various Sf-fdl targeting sequences (SfFDLt1, SfFDLt2, and SfFDLt3) (Table S1) under the control of either the DmU6:96Ab or the BmU6-2 promoter.
Table S1.
sgRNA targeting sequences used in this study
| Name of target site | Target gene | Sequence (5′ to 3′) |
| DmFDLt3 | Dm-fdl | gcgccatattcatcctga |
| SfFDLt1 | Sf-fdl | ggcagtgcgatgaagtgg |
| SfFDLt2 | Sf-fdl | gccgcggcgctgctgtac |
| SfFDLt3 | Sf-fdl | gaagtgtcggaacgttgc |
| TnFDLt | Tn-fdl | gaagtgtccgagcgctgc |
| BmFDLt1 | Bm-fdl | gcgagaggtatcaagcat |
| BmFDLt2 | Bm-fdl | gctctggccacagccgac |
| BmFDLt3 | Bm-fdl | ggcctgtcagcctcgcat |
| EGFPt | EGFP | gggcgaggagctgttcac |
Fig. S1.
CRISPR-Cas9 editing of fdl in S2R+ and BmN cells. The figure shows diagrams of the (A) Drosophila melanogaster and (C) Bombyx mori fdl genes and CEL-I nuclease assay results demonstrating CRISPR-Cas9 editing of the (B) D. melanogaster and (D) B. mori fdl genes.
Therefore, we constructed DmU6:96Ab and BmU6-2 CRISPR-Cas9 vectors encoding sgRNAs with three different Sf-fdl targeting sequences (Fig. 1B and Table S1) and used them to transfect Sf9 cells in an effort to edit the Sf-fdl gene. However, CEL-I nuclease assays revealed no evidence of Sf-fdl indels in the resulting puromycin-resistant Sf9 derivatives (Fig. 1C). Because the results obtained with D. melanogaster and B. mori cells indicated these vectors induced adequate Cas9 and pac expression, this result suggested the DmU6 and BmU6 promoters were unable to support adequate sgRNA expression in Sf9 cells, which are derived from a heterologous insect species. Therefore, we concluded we needed to identify an endogenous SfU6 promoter to induce sgRNA expression in Sf9 cells.
An Identified SfU6 Promoter Supports CRISPR-Cas9 Editing in Sf9 Cells.
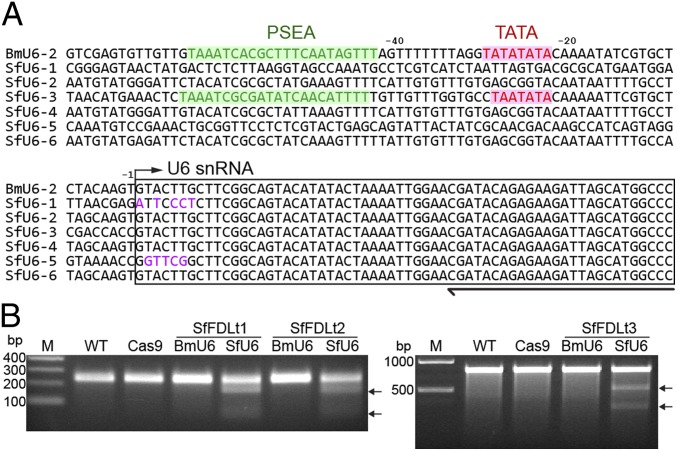
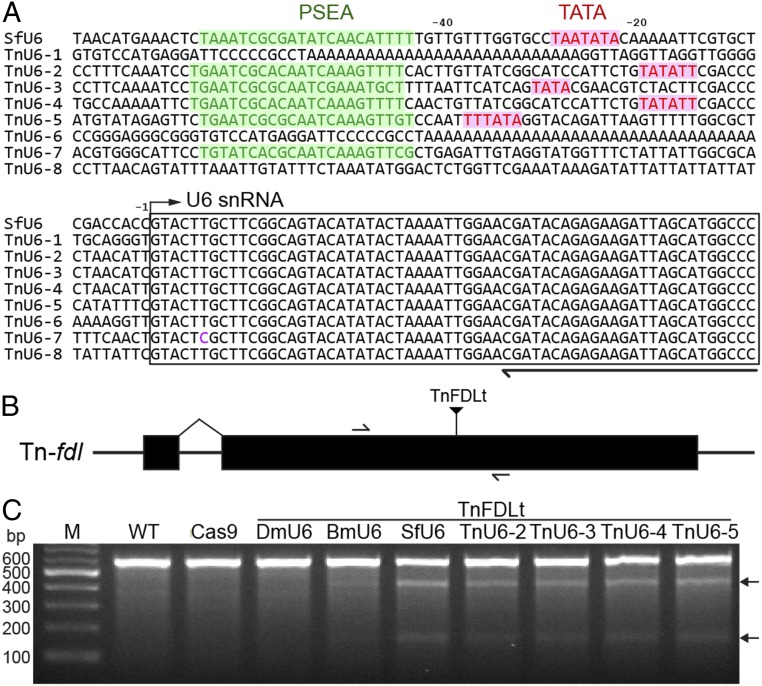
Using the BmU6-2 snRNA sequence (31) as a query to search the S. frugiperda draft genome sequence (32), we found only one putative SfU6 snRNA coding sequence. We had no confidence in this hit because insect snRNA sequences are often derived from pseudogenes (31). Thus, we used splinkerette PCR (33) in an attempt to experimentally isolate SfU6 promoter candidates from Sf9 genomic DNA. This approach yielded six unique U6 snRNA upstream sequences (Fig. 2A), including the one (SfU6-3) identified using bioinformatics. Additional bioinformatics showed only SfU6-3 included the proximal sequence element A (PSEA) and TATA box required for insect U6 promoter function (Fig. 2A).
Fig. 2.
Identification of putative SfU6 promoters and successful CRISPR-Cas9 editing of Sf-fdl. (A) Multiple sequence alignment of BmU6-2 promoter and SfU6 promoter candidates. (B) CEL-I nuclease assay results obtained using genomic DNA from Sf9 cells edited with CRISPR-Cas9 vectors encoding Sf-fdl targeting sequences (Table S1) under the control of the BmU6-2 or SfU6-3 promoters.
Based on these results, we used SfU6-3 to construct a generic CRISPR-Cas9 vector (Fig. 1A) and then constructed three derivatives using the Sf-fdl targeting sequences previously inserted into the DmU6 and BmU6 CRISPR-Cas9 vectors (Fig. 1B and Table S1). We used each construct to transfect Sf9 cells, selected puromycin-resistant derivatives, and then performed CEL-I nuclease assays with genomic DNAs from those cells. The results showed all three SfU6-3–based CRISPR-Cas9 vectors produced Sf-fdl indels (Fig. 2B) and this was confirmed by PCR and sequencing (Fig. S2). These results demonstrated the SfU6-3 promoter, but not the DmU6:96Ab or BmU6-2 promoters, can be used for CRISPR-Cas9 editing in Sf9 cells.
Fig. S2.
Sequences of Sf-fdl amplification products from Sf9 cells or Sf9 cells transfected with SfU6 CRISPR-Cas vectors encoding (A) SfFDLt1, (B) SfFDLt2, or (C) SfFDLt3 and selected for puromycin resistance.
Identified TnU6 Promoters Support CRISPR-Cas9 Editing in T. ni Cells.
We extended these results by using splinkerette PCR to identify eight putative TnU6 promoters as potential tools for CRISPR-Cas9 editing of High Five cells (Fig. 3A). We then used TnU6-2, -3, -4, and -5, all of which had PSEA and TATA elements, to construct generic CRISPR-Cas9 vectors. Finally, we inserted a Tn-fdl–specific targeting sequence (Fig. 3B and Table S1), transfected High Five cells with the resulting constructs, selected for puromycin resistance, and examined the cellular Tn-fdl genes using CEL-I nuclease assays. The results indicated the Tn-fdl gene was edited in each case, demonstrating that TnU6-2, -3, -4, and -5 are all effective as promoters for CRISPR-Cas9 editing in High Five cells (Fig. 3C). Interestingly, the CEL-I nuclease assays also indicated the SfU6-3, BmU6-2, and DmU6:96Ab CRISPR-Cas9 vectors encoding the Tn-fdl–specific sgRNA produced efficient, inefficient, and no detectable Tn-fdl gene editing in High Five cells, respectively (Fig. 3C). These results showed that TnU6-2, -3, -4, -5, and SfU6-3 promoters can all be used for CRISPR-Cas9 editing in High Five cells.
Fig. 3.
Identification of putative TnU6 promoters and successful CRISPR-Cas9 editing of Tn-fdl. (A) Multiple sequence alignment of SfU6 promoter and TnU6 promoter candidates. (B) Diagram showing Tn-fdl gene structure and highlighting specific Cas9 targeting sequences and PCR primer sites. (C) CEL-I nuclease assay results obtained using genomic DNA from High Five cells edited with CRISPR-Cas9 vectors encoding a Tn-fdl targeting sequence (Table S1) under the control of the DmU6:96Ab, BmU6-2, SfU6, or TnU6 promoters.
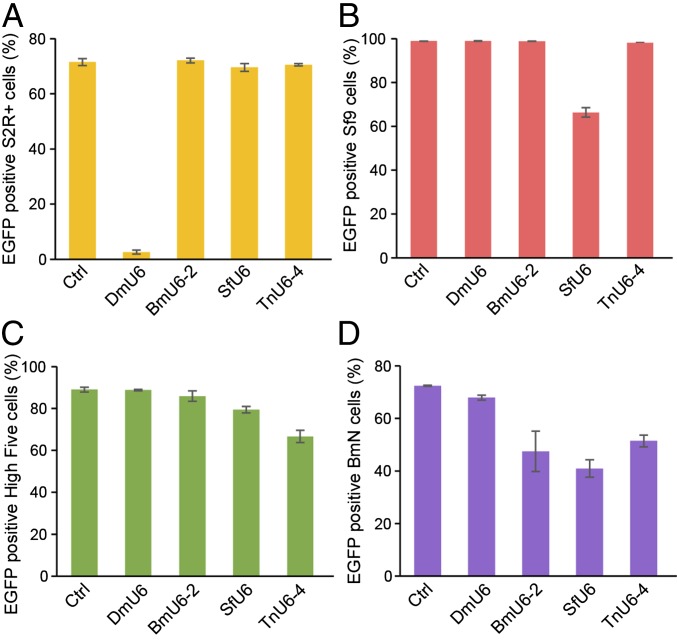
CRISPR-Cas9 Editing Efficiencies Mediated by Various Insect U6 Promoters in Various Insect Cell Lines.
Considering the U6 promoters derived from T. ni and S. frugiperda both mediated Tn-fdl gene editing in High Five cells, we chose to more quantitatively document the efficiencies of CRISPR-Cas9 editing provided by various insect U6 promoters in the various insect cell lines used in this study. First, we transformed S2R+, Sf9, High Five, and BmN cells with an EGFP expression plasmid. Then, we transfected each transformed derivative with CRISPR-Cas9 vectors encoding an EGFP-specific sgRNA under the control of D. melanogaster, B. mori, S. frugiperda, or T. ni U6 promoters and measured cellular fluorescence (Fig. 4). The results showed only the homologous U6 CRISPR-Cas9 vectors significantly reduced fluorescence in S2R+ and Sf9 cells (Fig. 4 A and B), whereas the U6 promoters from several species reduced fluorescence in T. ni and B. mori cells (Fig. 4 C and D). Overall, among those tested, the DmU6:96Ab, SfU6-3, and TnU6-4 promoters would be the best choices for CRISPR-Cas9 editing in S2R+, Sf9, and High Five cells, respectively. In contrast, the BmU6-2, SfU6-3, and TnU6-4 promoters all provided about the same efficiencies and the heterologous SfU6-3 promoter would likely be the best choice for CRISPR-Cas9 editing in BmN cells.
Fig. 4.
CRISPR-Cas9 editing efficiencies by various insect U6 promoters in various insect cell lines. (A) S2R-EGFP, (B) Sf9-EGFP, (C) High Five-EGFP, and (D) BmN-EGFP cells were transfected with DmU6:96Ab, SfU6, TnU6-4, and BmU6-2 CRISPR-Cas9 vectors encoding an EGFP-specific sgRNA, selected for puromycin resistance, and EGFP was measured by flow cytometry (the bars show mean fluorescence ± SD, n = 3 per group).
Phenotypic Impact of Gene Editing with SfU6-3-SfFDLt1 CRISPR-Cas9 Vector in Sf9 Cells.
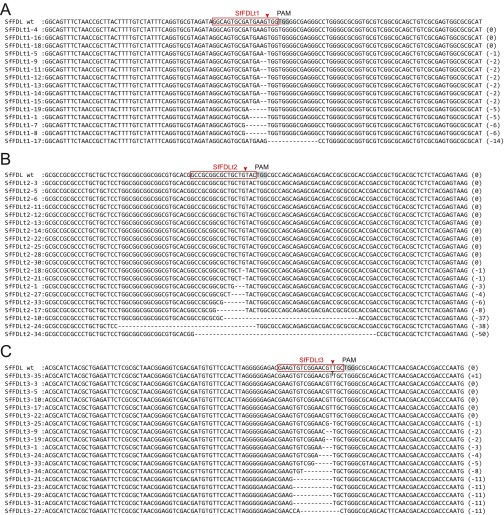
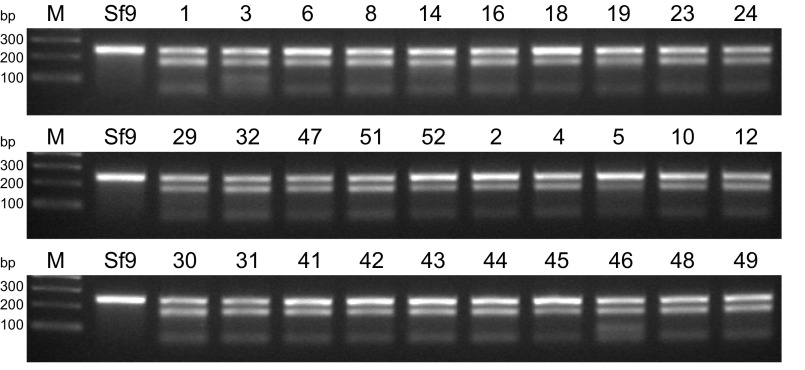
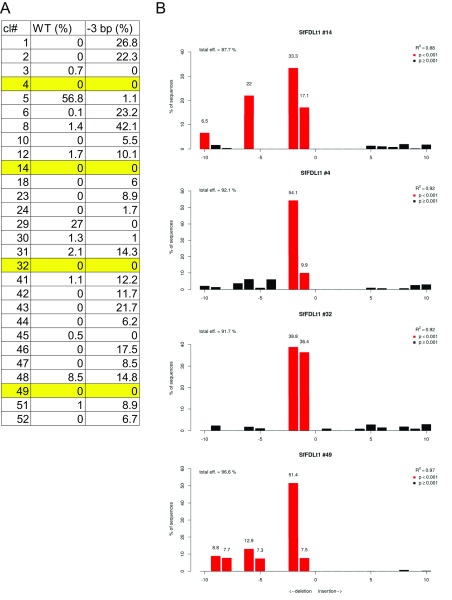
Finally, we assessed the phenotypic impact of gene editing using one of the CRISPR-Cas9 tools created in this study. Sf9 cells were transfected with the CRISPR-Cas9 vector encoding the Sf-FDLt1 sgRNA under SfU6-3 promoter control, puromycin-selected, and the resulting polyclonal cell population (SfFDLt1) was used to isolate 30 single-cell clones. The Sf-fdl sequences in the parental Sf9, polyclonal SfFDLt1, and SfFDLt1 clones were then examined by CEL-I nuclease assays and TIDE analysis, as described in Materials and Methods. The CEL-I nuclease assay results indicated all 30 clones had Sf-fdl indels (Fig. S3) and TIDE analysis revealed four clones had no wild-type Sf-fdl sequences or potentially functional in-frame deletions (Fig. S4 and Table S2).
Fig. S3.
CEL-I nuclease assays demonstrating Sf-fdl indels in SfFDLt1 clones.
Fig. S4.
TIDE analysis of Sf-fdl indels in SfFDLt1 clones. (A) Proportions of wild-type and in-frame (−3 bp) deletions in monoclonal SfFDLt1 cell lines as determined using the TIDE program. Four clones with no wild-type sequences or potentially functional in-frame deletions are highlighted in yellow. (B) Raw TIDE data for the four clones highlighted in A.
Table S2.
Indels found in SfFDLt1 monoclonal cell lines
| Indels, bp | SfFDLt1#4 | SfFDLt1 #14 | SfFDLt1 #32 | SfFDLt1 #49 |
| −1 | 2 | 1 | 5 | 1 |
| −2 | 8 | 3 | 2 | 8 |
| − | 1 | |||
| −95 | 3 | |||
| +76 | 3 |
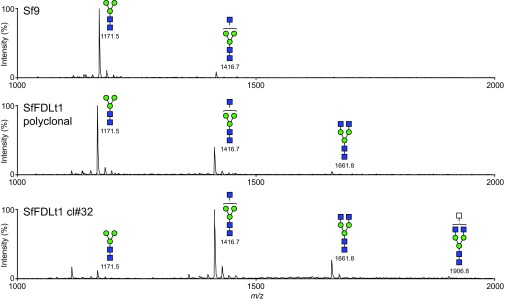
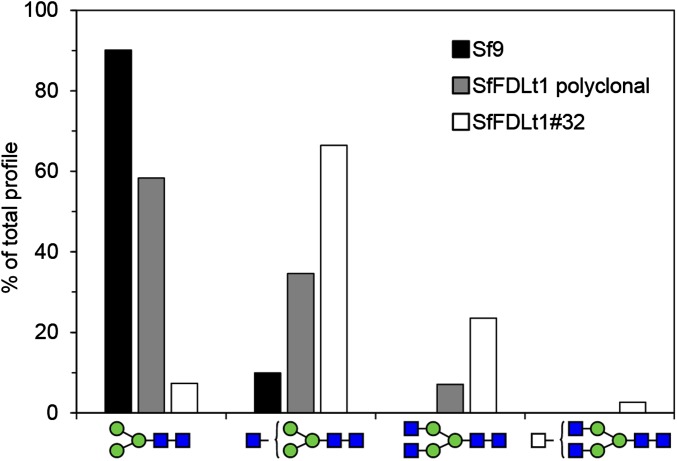
We subsequently infected one of those clones (#32), as well as Sf9 cells and the polyclonal SfFDLt1 cell population, with a recombinant baculovirus encoding an affinity-tagged version of human erythropoietin (hEPO) and purified the secreted product from each culture, as described in Materials and Methods. We then enzymatically released the N-glycans from each purified protein preparation and analyzed the permethylated glycan structures by MALDI-TOF-MS, as described in Materials and Methods. The spectra showed the major N-glycan on hEPO from Sf9 and SfFDLt1 (polyclonal) cells was Man3GlcNAc2, whereas the major N-glycan on hEPO from SfFDLt1 #32 was GlcNAcMan3GlcNAc2 (Fig. S5). A quantitative analysis showed Man3GlcNAc2 represented about 90%, 60%, and 8% of the total N-glycans on hEPO from Sf9, SfFDLt1 (polyclonal), and SfFDLt1 clone #32, respectively (Fig. 5). Reciprocally, GlcNAcMan3GlcNAc2 represented about 10%, 30%, and 65% of total N-glycans on hEPO from Sf9, SfFDLt1 (polyclonal), and SfFDLt1 clone #32, respectively (Fig. 5). Finally, GlcNAc2Man3GlcNAc2 was only detected on hEPO from SfFDLt1 (polyclonal), and SfFDLt1 #32 (Fig. 5).
Fig. S5.
Impact of Sf-fdl editing on N-glycan processing. N-glycans were isolated from hEPO produced by Sf9, SfFDLt1 polyclonal, or SfFDLt1 monoclonal cl#32 cells, derivatized, and profiled by MALDI-TOF-MS, as described in Materials and Methods. All molecular ions were detected as [M + Na]+, assigned and annotated using the standard cartoon symbolic representations.
Fig. 5.
CRISPR-Cas9–mediated Sf-fdl gene editing for host engineering in the BICS. The bar graph shows the relative proportions of different N-glycan structures released from hEPO produced by Sf9, SfFDLt1 polyclonal population, and SfFDLt1 clone #32. These data are derived from the MALDI-TOF-MS profiles shown in Fig. S5 and represent the relative percentages of each N-glycan shown along the bottom of the figure as a percentage of total.
These results clearly demonstrate the phenotypic impact of genome editing with the SfU6-3-SfFDLt1 CRISPR-Cas9 vector in Sf9 cells. Specifically, the structures of the N-glycans observed in the Sf9 cells treated with this vector reveal a partial (polyclonal) and nearly complete (clone #32) loss of FDL function resulting from fdl editing with this vector. We conclude that the CRISPR-Cas tools created in this study can be used to engineer host pathways in efforts to enhance and expand the capabilities of the BICS.
Discussion
The major outcome of this study was successful creation of CRISPR-Cas9 tools that can be used for site-specific genome editing in the BICS. These tools will enable far more sophisticated host-cell engineering efforts, which to date have been limited to using nonhomologous recombination to knockin genes at random sites in the insect cell genome. Thus, these tools will enable new efforts to enhance and expand the utility of the BICS as a recombinant protein production platform.
In our efforts to achieve this goal, we initially tested D. melanogaster and B. mori U6 promoters that were previously shown to direct effective sgRNA expression and CRISPR-Cas9–mediated genome editing in dipteran and lepidopteran insect cells (27–29). We assumed these promoters might drive these same functions in S. frugiperda and T. ni cells, which would have allowed us to quickly produce CRISPR-Cas9 vectors for the BICS.
In fact, CRISPR-Cas9 vectors encoding Dm- or Bm-fdl–specific targeting sequences under D. melanogaster or B. mori U6 promoter control produced indels in cell lines from homologous species (Fig. S2). However, CRISPR-Cas9 vectors with these same D. melanogaster or B. mori U6 promoters encoding sgRNAs with Sf- or Tn-fdl–specific targeting sequences failed to produce any detectable indels in S. frugiperda (Fig. 1) or T. ni cells (Fig. 3), respectively. This forced us to identify putative S. frugiperda and T. ni U6 promoters, which we then used to produce CRISPR-Cas9 vectors encoding sgRNAs with the same Sf- or Tn-fdl–specific targeting sequences. We found CRISPR-Cas9 vectors with the homologous U6 promoters efficiently produced indels in S. frugiperda (Fig. 2) and T. ni (Fig. 3) cells, respectively.
We subsequently established an EGFP reduction assay, which could be used to more quantitatively measure the relative efficiencies of editing by CRISPR-Cas9 vectors encoding a GFP-specific sgRNA under the control of various insect U6 promoters in different insect cell species, as described in Materials and Methods. The results (Fig. 4) indicated only the CRISPR-Cas9 vectors with homologous U6 promoters significantly reduced GFP expression in D. melanogaster and S. frugiperda cells. In contrast, while the homologous U6 promoter provided the highest CRISPR-Cas9 editing efficiency in High Five cells, SfU6-3 also provided a reasonable efficiency and the B. mori, S. frugiperda, and T. ni promoters all provided about the same efficiencies of CRISPR-Cas9 editing in BmN cells. These results indicate SfU6-3 has the broadest—while DmU6:96Ab has the narrowest—host range among the insect U6 promoters tested in the insect cell lines used in this study. Additional studies will be required to extend these findings to include a broader range of insect U6 promoters and cell lines and, ultimately, determine their underlying host-range determinants at the molecular level.
In a previous study, a recombinant baculovirus designed to express Cas9 and sgRNAs under the control of mammalian promoters was used as a transducing vector to induce CRISPR-Cas9 genome editing in mammalian cells (34). In contrast, the present study yielded new CRISPR-Cas9 tools designed to express Cas9 and sgRNAs under the control of baculovirus and insect cell promoters and examined their relative utility for genome editing in the BICS. Finally, our study provided proof-of-concept that these tools can be used for host cell engineering in the BICS. In our example, we targeted fdl, which encodes a key enzyme that distinguishes insect and mammalian cell protein N-glycosylation pathways by antagonizing N-glycan elongation. As such, fdl has been a high-priority target for knockout, as this would facilitate efforts to glycoengineer the BICS and other insect-based recombinant protein production platforms for high-efficiency mammalian-type protein N-glycosylation. Several previous publications demonstrated various RNAi approaches can reduce FDL activity, but with little or no phenotypic impact on N-glycan processing (20, 35–37). We previously used existing CRISPR-Cas9 tools to knockout Dm-fdl in S2R+ cells and demonstrate this had the expected impact on N-glycan processing (28). However, we were unable to knockout Sf-fdl or Tn-fdl until we created the tools needed for site-specific gene editing in the BICS. We then used a CRISPR-Cas9 vector encoding an Sf-fdl–specific sgRNA under the control of the SfU6-3 promoter to produce polyclonal and monoclonal Sf9 cell derivatives. CEL-I nuclease assays and TIDE analysis indicated this CRISPR-Cas9 vector directed efficient editing of the Sf-fdl gene (Fig. S4 and Table S2). Finally, we documented the phenotypic impact of these genotypic changes by analyzing the N-glycans isolated from recombinant hEPO produced by polyclonal and monoclonal Sf-fdl knockout cells described in this study. As expected, we observed reduced proportions of paucimannose (Man3GlcNAc2) and increased proportions of terminally GlcNAcylated (GlcNAc1–2Man3GlcNAc2) structures on hEPO produced by SfFDLt1 cells, compared with Sf9 cells (Fig. 5 and Fig. S5). Thus, in summary, this study presents CRISPR-Cas9 vectors for site-specific genome editing and clearly demonstrates they can be used successfully for host cell engineering in the BICS.
Materials and Methods
Cells.
S2R+ cells (38) were maintained at 28 °C as adherent cultures in Schneider’s Drosophila medium (Life Technologies) containing 10% (vol/vol) FBS (Atlanta Biologics). Sf9, High Five, and BmN cells were maintained at 28 °C as adherent cultures in TNM-FH medium containing 10% (vol/vol) FBS. Sf9 cells were transfected using a modified calcium phosphate method (8) and S2R+, High Five, and BmN cells were transfected with polyethyleneimine, as described previously (28). S2R-EGFP, Sf9-EGFP, Tn-EGFP, and BmN-EGFP cells are transgenic derivatives of S2R+, Sf9, High Five, and BmN cells, respectively, produced by transfecting each parental cell line with pIE1-EGFP-Bla and selecting for blasticidin resistance. Blasticidin-resistant cells expressing EGFP in the top quartile were isolated using a MoFlo Legacy Cell Sorter (Beckman Coulter) and enriched cell subpopulations were maintained under the same growth conditions as the parental cell lines.
Plasmid Constructions.
All CRISPR-Cas9 constructs were generically designed to include three distinct cassettes for expression of Cas9, an sgRNA, and a puromycin resistance marker. The Cas9 expression cassette consists of a S. pyogenes Cas9 sequence codon optimized for S. frugiperda and assembled with the AcMNPV ie1 promoter and p10 polyadenylation signal using the Golden Gate method. The sgRNA expression cassettes consist of DmU6:96Ab, BmU6-2, SfU6-3, TnU6-2, TnU6-3, TnU6-4, or TnU6-5 promoters assembled with various downstream sgRNA sequences. The targeting sequences incorporated into various sgRNAs are given in Table S1. A targeting sequence cloning site consisting of two SapI recognition sites was inserted between the U6 promoter and sgRNA in each CRISPR-Cas9 plasmid. Finally, the puromycin-resistance marker was codon optimized for S. frugiperda and assembled with the AcMNPV ie1 promoter and p10 polyadenylation signal. The generic CRISPR-Cas9 plasmid and specific target-sgRNA sequences are given in SI Appendix.
Splinkerette PCR.
Splinkerette PCR was performed as described previously (39). Briefly, Sf9 or High Five genomic DNA was digested with BamHI, BglII, BstYI, HindIII, SalI, SpeI, or XbaI, and ligated with splinkerette adaptors complementary to the resulting overhangs. Primary and secondary PCRs were performed with Splink1 and SfU6-Rv1 and Splink2 and SfU6-Rv2 as the primer pairs, respectively (primer sequences are given in Table S3). The resulting amplimers were cloned into pGEM-T (Promega) and three independent clones were sequenced to determine the consensus.
Table S3.
Primers used for splinkerette PCR
| Primer name | Sequence (5′ to 3′) |
| Splink-GATC-TOP | gatcccactagtgtcgacaccagtctctaattttttttttcaaaaaaa |
| Splink-CTAG-TOP | ctagccactagtgtcgacaccagtctctaattttttttttcaaaaaaa |
| Splink-TCGA-TOP | tcgaccactagtgtcgacaccagtctctaattttttttttcaaaaaaa |
| Splink-AGCT-TOP | agctccactagtgtcgacaccagtctctaattttttttttcaaaaaaa |
| Splink-bottom | cgaagagtaaccgttgctaggagagaccgtggctgaatgagactggtgtcgacactagtgg |
| Splink1 | cgaagagtaaccgttgctaggagagacc |
| Splink2 | gtggctgaatgagactggtgtcgac |
| SfU6-Rv1 | gcttcacgattttgcgtgtcatccttg |
| SfU6-Rv2 | gggccatgctaatcttctctgtatcg |
Genomic DNA Isolation and CEL-I Nuclease Assays.
Genomic DNA was extracted from Sf9, High Five, BmN, and S2R+ cells using the Wizard genomic DNA extraction kit (Promega) according to the manufacturer’s instructions. CEL-I nuclease assays were performed as described previously (28). The sequences of the primers used to amplify various target loci are given in Table S4.
Table S4.
Primers used to amplify sequences surrounding target sites
| Primer name | Target site | Sequence (5′ to 3′) |
| DmFDLsurv-Fw | DmFDLt3 | acaggcctggtggtggtgtc |
| DmFDLsurv-Rv | DmFDLt3 | aaagttaagatccccggatttgagcac |
| SfFDLt12-Fw | SfFDLt1, SfFDLt2 | ggcagtttctaaccgcttacttttg |
| SfFDLt12-Rv | SfFDLt1, SfFDLt2 | cttactcgtagagagcgtgcagc |
| SfFDLt3-Fw | SfFDLt3 | cgcggacttctccttgacacag |
| SfFDLt3-Rv | SfFDLt3 | cgaacccgcagtccaggtac |
| TnFDLsurv-Fw | TnFDLt | atgaagtggtggggcga |
| TnFDLsurv-Rv | TnFDLt | gccacagctgtgtcgagtc |
| BmFDLsurv-Fw | BmFDLt1, BmFDLt2, BmFDLt3 | cttttatttatcgattcgggc |
| BmFDLsurv-Rv | BmFDLt1, BmFDLt2, BmFDLt3 | gaatgcgctgtgatgtctac |
TIDE Analysis.
We performed TIDE analysis as described previously (40). Briefly, we directly sequenced the PCR products amplified from Sf9 and SfFDLt1 monoclonals’ genomic DNA as described above and used the sequencing results as queries for a TIDE web program (https://tide-calculator.nki.nl/). All analyses were performed with a default setting.
EGFP Reduction Assay.
S2R-EGFP, Sf9-EGFP, Tn-EGFP, and BmN-EGFP cells were transfected with various CRISPR-Cas9 vectors targeting EGFP or a control vector encoding no sgRNA and selected for puromycin resistance. Puromycin-resistant survivors were analyzed using a Guava easyCyte HT flow cytometer (Millipore) and EGFP+ cell populations were quantified using FlowJo software.
Expression and Purification of hEPO.
We isolated AcRMD2-p6.9-hEPO, a recombinant baculovirus encoding an N-terminally affinity-tagged version of hEPO, in two steps. First, we recombined a gene encoding the Pseudomonas aeruginosa GDP-4-dehydro-6-deoxy-d-mannose reductase (rmd) cds under the control of the AcMNPV ie1 promoter into the chi-cth locus of a baculovirus vector called BacPAK6-p6.9-GUS (41) to produce AcRMD2. Second, we recombined a honey bee melittin signal peptide, 8XHIS-tag, Strep II-tag, tobacco etch virus recognition site, and mature hEPO cds under the control of the AcMNPV p6.9 promoter into the polh locus of AcRMD2. hEPO was expressed and purified as described previously (42).
Isolation and Characterization of Monoclonal SfFDLKO Cell Lines.
Single cell clones were isolated from the polyclonal SfFDLt1 cell population, as described previously (43). Indels were analyzed by CEL-I nuclease assays and the Sf-fdl gene sequences in clones 4, 14, 32, and 49 were amplified, sequenced, and the sequences were analyzed by TIDE, as described previously (28, 40).
Mass Spectrometry.
N-glycans were enzymatically released from purified hEPO and derivatized, as described previously (42), then analyzed by MALDI-TOF-MS using an Applied Biosystems SCIEX TOF/TOF 5800 (SCIEX), with 400 shots accumulated in reflectron positive-ion mode. Structures were manually assigned to peaks based on knowledge of the insect cell N-glycan processing pathway. Quantification involved dividing the peak intensities of permethylated N-glycan structures by the total intensity of all annotated N-glycan peaks having >1% of total intensities.
Supplementary Material
Acknowledgments
We thank Dr. Kostas Iatrou of the Institute of Biosciences & Applications, National Center for Scientific Research “Demokritos,” Greece, for providing BmN cells; and Leslie Armstrong-Lea, of the Research Cores Facilities Program, Colorado State University, for performing Moflo Flow Cytometry-based cell sorting services. This work was supported by Award R43 GM102982 from the National Institute of General Medical Sciences, NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the NIH.
Footnotes
Conflict of interest statement: D.L.J. is the President of and H.M.-A. was a consultant for GlycoBac, LLC. D.L.J. and H.M.-A. are coinventors on a US provisional patent application based on the work described herein.
This article is a PNAS Direct Submission. V.M.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705836114/-/DCSupplemental.
References
- 1.Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1983;3:2156–2165. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felberbaum RS. The baculovirus expression vector system: A commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnol J. 2015;10:702–714. doi: 10.1002/biot.201400438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitts PA, Ayres MD, Possee RD. Linearization of baculovirus DNA enhances the recovery of recombinant virus expression vectors. Nucleic Acids Res. 1990;18:5667–5672. doi: 10.1093/nar/18.19.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitts PA, Possee RD. A method for producing recombinant baculovirus expression vectors at high frequency. Biotechniques. 1993;14:810–817. [PubMed] [Google Scholar]
- 5.Luckow VA, Lee SC, Barry GF, Olins PO. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler C, Mabashi-Asazuma H, Jarvis DL. An overview and history of glyco-engineering in insect expression systems. Methods Mol Biol. 2015;1321:131–152. doi: 10.1007/978-1-4939-2760-9_10. [DOI] [PubMed] [Google Scholar]
- 7.Harrison RL, Jarvis DL. Transforming lepidopteran insect cells for improved protein processing and expression. Methods Mol Biol. 2016;1350:359–379. doi: 10.1007/978-1-4939-3043-2_18. [DOI] [PubMed] [Google Scholar]
- 8.Summers MD, Smith GE. 1987. A Manual of Methods for Baculovirus Vectors and Insect Cell Culture Procedures Tex Agric Exp Stn Bull 1555:1–57.
- 9.Wickham TJ, Davis T, Granados RR, Shuler ML, Wood HA. Screening of insect cell lines for the production of recombinant proteins and infectious virus in the baculovirus expression system. Biotechnol Prog. 1992;8:391–396. doi: 10.1021/bp00017a003. [DOI] [PubMed] [Google Scholar]
- 10.Marz L, Altmann F, Staudacher E, Kubelka V. Protein glycosylation in insects. In: Montreuil J, Vliegenthart JFG, Schachter H, editors. Glycoproteins. Vol 29a. Elsevier; Amsterdam: 1995. pp. 543–563. [Google Scholar]
- 11.Marchal I, Jarvis DL, Cacan R, Verbert A. Glycoproteins from insect cells: Sialylated or not? Biol Chem. 2001;382:151–159. doi: 10.1515/BC.2001.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison RL, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell expression system and engineering of insect cells to produce “mammalianized” recombinant glycoproteins. Adv Virus Res. 2006;68:159–191. doi: 10.1016/S0065-3527(06)68005-6. [DOI] [PubMed] [Google Scholar]
- 13.Geisler C, Jarvis DL. Insect cell glycosylation patterns in the context of biopharmaceuticals. In: Walsh B, editor. Post-Translational Modification of Protein Biopharmaceuticals. Wiley-Blackwell; Weinheim, Germany: 2009. pp. 165–191. [Google Scholar]
- 14.Shi X, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell system. Curr Drug Targets. 2007;8:1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koles K, Irvine KD, Panin VM. Functional characterization of Drosophila sialyltransferase. J Biol Chem. 2004;279:4346–4357. doi: 10.1074/jbc.M309912200. [DOI] [PubMed] [Google Scholar]
- 16.Koles K, et al. Identification of N-glycosylated proteins from the central nervous system of Drosophila melanogaster. Glycobiology. 2007;17:1388–1403. doi: 10.1093/glycob/cwm097. [DOI] [PubMed] [Google Scholar]
- 17.Aoki K, et al. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 18.Altmann F, Schwihla H, Staudacher E, Glössl J, März L. Insect cells contain an unusual, membrane-bound β-N-acetylglucosaminidase probably involved in the processing of protein N-glycans. J Biol Chem. 1995;270:17344–17349. doi: 10.1074/jbc.270.29.17344. [DOI] [PubMed] [Google Scholar]
- 19.Léonard R, et al. The Drosophila fused lobes gene encodes an N-acetylglucosaminidase involved in N-glycan processing. J Biol Chem. 2006;281:4867–4875. doi: 10.1074/jbc.M511023200. [DOI] [PubMed] [Google Scholar]
- 20.Geisler C, Aumiller JJ, Jarvis DL. A fused lobes gene encodes the processing beta-N-acetylglucosaminidase in Sf9 cells. J Biol Chem. 2008;283:11330–11339. doi: 10.1074/jbc.M710279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler C, Jarvis DL. Identification of genes encoding N-glycan processing beta-N-acetylglucosaminidases in Trichoplusia ni and Bombyx mori: Implications for glycoengineering of baculovirus expression systems. Biotechnol Prog. 2010;26:34–44. doi: 10.1002/btpr.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solá RJ, Griebenow K. Glycosylation of therapeutic proteins: An effective strategy to optimize efficacy. BioDrugs. 2010;24:9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvis DL. Baculovirus-insect cell expression systems. Methods Enzymol. 2009;463:191–222. doi: 10.1016/S0076-6879(09)63014-7. [DOI] [PubMed] [Google Scholar]
- 24.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 27.Bassett AR, Tibbit C, Ponting CP, Liu JL. Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol Open. 2014;3:42–49. doi: 10.1242/bio.20137120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mabashi-Asazuma H, Kuo CW, Khoo KH, Jarvis DL. Modifying an insect cell N-glycan processing pathway using CRISPR-Cas technology. ACS Chem Biol. 2015;10:2199–2208. doi: 10.1021/acschembio.5b00340. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Highly efficient multiplex targeted mutagenesis and genomic structure variation in Bombyx mori cells using CRISPR/Cas9. Insect Biochem Mol Biol. 2014;49:35–42. doi: 10.1016/j.ibmb.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Fujita R, Ono C, Ono I, Asano S, Bando H. Analysis of the Bombyx mori nucleopolyhedrovirus ie-1 promoter in insect, mammalian, plant, and bacterial cells. Biochem Biophys Res Commun. 2015;464:1297–1301. doi: 10.1016/j.bbrc.2015.07.126. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez G, Jr, Valafar F, Stumph WE. Insect small nuclear RNA gene promoters evolve rapidly yet retain conserved features involved in determining promoter activity and RNA polymerase specificity. Nucleic Acids Res. 2007;35:21–34. doi: 10.1093/nar/gkl982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kakumani PK, Malhotra P, Mukherjee SK, Bhatnagar RK. A draft genome assembly of the army worm, Spodoptera frugiperda. Genomics. 2014;104:134–143. doi: 10.1016/j.ygeno.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Devon RS, Porteous DJ, Brookes AJ. Splinkerettes—Improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 1995;23:1644–1645. doi: 10.1093/nar/23.9.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansouri M, et al. Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat Commun. 2016;7:11529. doi: 10.1038/ncomms11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nomura T, et al. Improvement of glycosylation structure by suppression of β-N-acetylglucosaminidases in silkworm. J Biosci Bioeng. 2015;119:131–136. doi: 10.1016/j.jbiosc.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 36.Kim NY, et al. Short-hairpin RNA-mediated gene expression interference in Trichoplusia ni cells. J Microbiol Biotechnol. 2012;22:190–198. doi: 10.4014/jmb.1108.08045. [DOI] [PubMed] [Google Scholar]
- 37.Kim YK, et al. Suppression of beta-N-acetylglucosaminidase in the N-glycosylation pathway for complex glycoprotein formation in Drosophila S2 cells. Glycobiology. 2009;19:301–308. doi: 10.1093/glycob/cwn138. [DOI] [PubMed] [Google Scholar]
- 38.Yanagawa S, Lee JS, Ishimoto A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem. 1998;273:32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- 39.Mabashi-Asazuma H, et al. Targeted glycoengineering extends the protein N-glycosylation pathway in the silkworm silk gland. Insect Biochem Mol Biol. 2015;65:20–27. doi: 10.1016/j.ibmb.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maghodia AB, Geisler C, Jarvis DL. Characterization of an Sf-rhabdovirus-negative Spodoptera frugiperda cell line as an alternative host for recombinant protein production in the baculovirus-insect cell system. Protein Expr Purif. 2016;122:45–55. doi: 10.1016/j.pep.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geisler C, Mabashi-Asazuma H, Kuo CW, Khoo KH, Jarvis DL. Engineering β1,4-galactosyltransferase I to reduce secretion and enhance N-glycan elongation in insect cells. J Biotechnol. 2015;193:52–65. doi: 10.1016/j.jbiotec.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrison RL, Jarvis DL. Transforming lepidopteran insect cells for continuous recombinant protein expression. Methods Mol Biol. 2016;1350:329–348. doi: 10.1007/978-1-4939-3043-2_16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.