Significance
We have developed a bioinformatics tool that allows us to compare the sequences of all protein-coding genes of 36 sequenced mouse inbred strains with the reference mouse strain C57BL/6J. We also provide an estimate of the effect on protein function of each deviant protein sequence and have built a searchable database of all these sequences, giving researchers the opportunity to search for abnormal alleles of any protein coding gene across these strains. The database makes the enormous richness of variant alleles present in these 36 inbred strains visible, accessible, and useful to the whole mouse research community.
Keywords: mouse, genetics, sequence, polymorphisms, inbred strains
Abstract
Mouse inbred strains remain essential in science. We have analyzed the publicly available genome sequences of 36 popular inbred strains and provide lists for each strain of protein-coding genes that acquired sequence variations that cause premature STOP codons, loss of STOP codons and single nucleotide polymorphisms, and short in-frame insertions and deletions. Our data give an overview of predicted defective proteins, including predicted impact scores, of all these strains compared with the reference mouse genome of C57BL/6J. These data can also be retrieved via a searchable website (mousepost.be) and allow a global, better interpretation of genetic background effects and a source of naturally defective alleles in these 36 sequenced classical and high-priority mouse inbred strains.
The first inbred strains of mice were established more than a hundred years ago (1). Since then, mouse inbred lines have become essential in physiological, biomedical, and genetic research. Their importance and success reside in the stability of their homozygous genomes in both time and space. More than 500 inbred strains of mice are currently available, but the number of most frequently used strains does not exceed 40. For rather pragmatic reasons, the mouse strain C57BL/6J has become the standard mouse strain (2). These mice are well described, and their genome has been sequenced with the highest possible resolution (3). However, researchers have a number of good reasons to prefer to study a scientific question, or a mutant gene, in another inbred mouse strain background; for example, in the strain FVB/NJ or in BALB/cJ. Different mouse strains may have different degrees of susceptibility for a given pathology or challenge. For example, BALB/cJ mice easily develop plasmacytoma tumors (4), and DBA/1J mice are preferred for the induction of rheumatoid arthritis (5). Furthermore, there are many examples of the appearance of very different phenotypes, resulting from a targeted mutations (e.g., knockout allele), depending on the mouse inbred genetic background. One notorious example is the knockout mutation of the Apc gene, which is harmless in AKR/J mice but causes the appearance of thousands of colonic polyps in C57BL/6J mice (6). In such cases, the inbred genetic background determines the penetrance by which the mutant gene leads to a phenotype. Although these genetic background effects are fascinating, and may lead to the identification of important modulator genes, they may also be considered disturbing because the modulator genes may be very difficult to identify. Finally, certain inbred mouse strains have a very obvious phenotype, as a result of an inactivating sequence variation in a particular gene. C3H/HeJ mice, for example, are resistant to lethal shock induced by bacterial lipopolysaccharides, because they carry a missense mutation in the Tlr4 gene (7). Clearly, compared with the reference C57BL/6J genome, the different inbred strains contain a lot of interesting phenotypic characteristics. The Mouse Phenome Database provides a user-friendly overview of a multitude of those phenotypes (8), but an overview of the genetic variations in the genome of the most used, high-priority, inbred strains remain underexplored or poorly accessible to the broad community.
The mouse reference genome of C57BL/6J has been sequenced by the Genome Reference Consortium and is made available through several channels, including Ensembl (www.ensembl.org). The Wellcome Trust Sanger Institute has sequenced the genomes of 36 popular and/or important inbred strains (9). We recently reported the development of a bioinformatics tool that allows for the efficient and quick analysis of sequence variations of protein coding genes in the strains 129/SvImJ (10) and SPRET/Ei (11), starting from these genome sequences. On the basis of the obvious need for a user-friendly overview and searchable database, we decided to provide an overview of all protein-inactivating sequence variations of all these 36 strains compared with the reference sequence of C57BL/6J.
Results and Discussion
Comparative Genomics of the 36 Sequenced Inbred Strains and the C57BL/6J Reference Genome.
The Mouse Genomes Project (MGP), maintained by the Wellcome Trust Sanger Institute, is a collection of sequence data from 36 often-used laboratory mouse strains, not including the reference strain (C57BL/6J; www.sanger.ac.uk/science/data/mouse-genomes-project). The genomes of these mouse strains were analyzed by means of deep sequencing and genome assemblies for some strains, as well as single nucleotide polymorphisms (SNPs) and structural variation data for all 36 strains (9). For each of the 36 strains, 2 files are available on this MGP ftp server: one with small insertions and deletions (indels) and one with SNPs (Fig. 1). The files form the 2015 release, REL-1505, the most recent release available, which was downloaded from the ftp server (ftp://ftp-mouse.sanger.ac.uk/REL-1505-SNPs_Indels/strain_specific_vcfs/). These data were filtered so that only the variants with high confidence were retained. These processed data were stored in a sql database and indexed to allow fast searching. The Ensembl structural annotation of the C57BL/6J mm10 reference genome (the ensemble 86 release) was obtained from the ensemble website (www.ensembl.org) and used to extract the exon sequences of protein coding genes from the reference genome. To fully explore the consequences of all mutations present in a protein coding gene, an in-house script was used to process all genes and transcripts (Materials and Methods). This script constructed the coding sequence (CDS) of all transcripts based on the exon sequences and the information in the structural annotation gtf file. Using the coordinate information from the annotation file, the transcript CDS sequences were, for every strain, in silico mutated to the sequence that is present in that specific strain. The CDS of all references and all alternative transcripts were converted to the corresponding protein sequences, which were compared for classification.
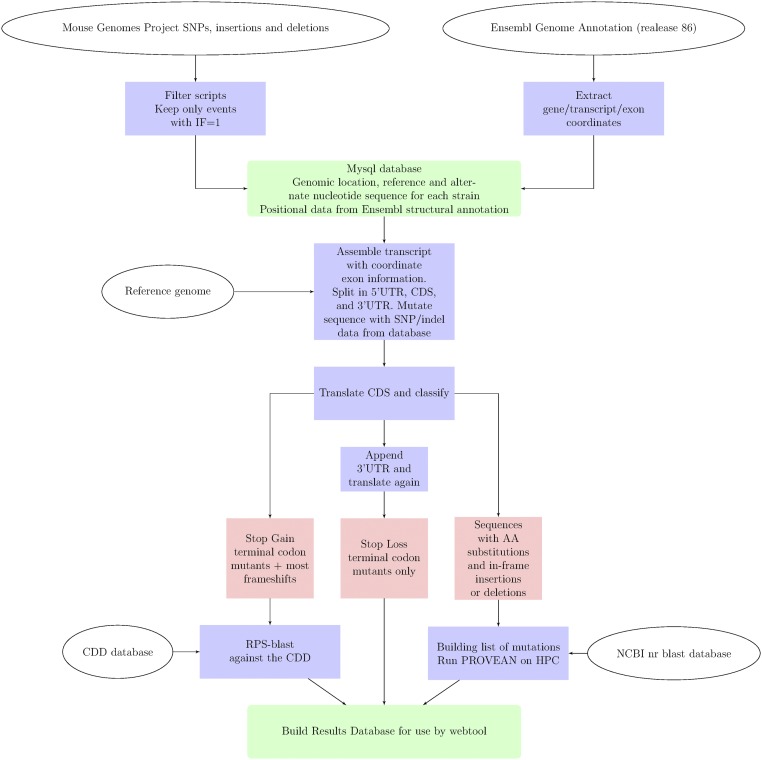
Fig. 1.
Schematic representation of the workflow used to process the sequences of the MGP of the Wellcome Trust Sanger Institute variation data to the final results, which can be queried by the web tool on mousepost.be. The MGP data were processed to only retain high-quality sequence hits, as described in Materials and Methods. Only the genomic location, reference sequence, and sequence in all other strains were kept. In parallel, the Ensembl Genome Annotation for mouse was processed into a table with the genomic coordinates of genes, transcripts and exons. Both the processed MGP and Ensembl tables were stored in a mysql database and indexed for fast access. The annotation tool uses this database in conjunction with the mm10 reference genome to construct the reference 5′ UTR, CDS, and 3′ UTR sequences and then change them to the actual sequences present in the strain of interest. CDS sequences are then translated to protein and compared with the reference sequence for classification. In the case of a SL, the 3′ UTR sequence is added to the canonical CDS to detect the new stop codon, if any. Stop gain genes are checked for lost protein domains by performing an RPS-blast against the conserved domain database from NCBI (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Mutated sequences have all changed positions scored by PROVEAN to predict the effect of the amino acid substitutions, insertions, or deletions. PROVEAN was run on a computer cluster using a local version of the NCBI database of nonredundant protein sequences. Finally, all results were placed in a mysql database (one table per class), which serves as the back-end for the web tool on mousepost.be.
We used three classes of protein sequence variations. The first is stop gain (SG), in which the protein is truncated compared with C57BL/6J. These variations only concern the occurrence of early stop codons, whereas length reductions resulting from in-frame deletions are not classified as SG variations. In the deleted parts of the transcripts, conserved domains were identified using the conserved domain database from the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). The reference protein sequences were located by searching this database with RPS-blast. These locations of missing conserved domains were thus defined. The second class was the stop loss (SL) variations, in which the normal stop codon has been lost so that the translation proceeds into the 3′ UTR. To determine the size of the extension, the 3′ UTR was added to the CDS and the in silico translation until a new stop codon was encountered, when it was performed again. Finally, the third class was mutated (MUT) variations. The transcripts placed in this group contain SNP mutations [leading to amino acid (aa) substitutions], in-frame insertions, and in-frame deletions.
In this MUT group of sequence variations, we attempted to predict the severity of the deviant sequences in terms of effect on the function of the protein. For this purpose, we used the Protein Variation Effect Analyzer (PROVEAN) software (12), which is a sequence homology-based prediction method. This software requires a sequence variation or mutation to be provided in the Human Genome Variation Society notation and can process multiple variations at once. For every transcript/protein, a file was constructed from the positional information obtained from the classification script and a global pairwise sequence alignment, containing all aa changes in the correct format for use with PROVEAN. The most recent version of the nonredundant protein blast database (the NCBI “nr” database, ftp://ftp.ncbi.nlm.nih.gov/blast/db/) was obtained and was used in combination with the reference sequences and the mutations file to predict the effect of the mutations on protein function. Prediction for all transcripts and strains was obtained by running this step on a computer cluster, and all results were stored in a mysql server that is able to be queried from our database website (mousepost.be). The standard cutoff for PROVEAN scores is set at −2.5. This corresponds to the maximal value suggested by the PROVEAN authors on the website to predict a mutation as deleterious. This cutoff is described as having the best balanced accuracy for the prediction, and this cutoff may be changed, with lower values being more specific and higher values being more sensitive. A score below the cutoff denotes a deleterious mutation, and the lower the score, the more severe the effect of the mutation.
A low PROVEAN score does not necessarily offer insight into the severity of the effect, because the number of supporting sequences used by PROVEAN to calculate the score also should be taken into account. The lower the number of supporting sequences, the less reliable the prediction; this becomes problematic when the number of supporting sequences drops below 50. This number of sequences is always reported in our database (mousepost.be), and sequences with fewer than 50 supporting sequences are not included.
Data Availability.
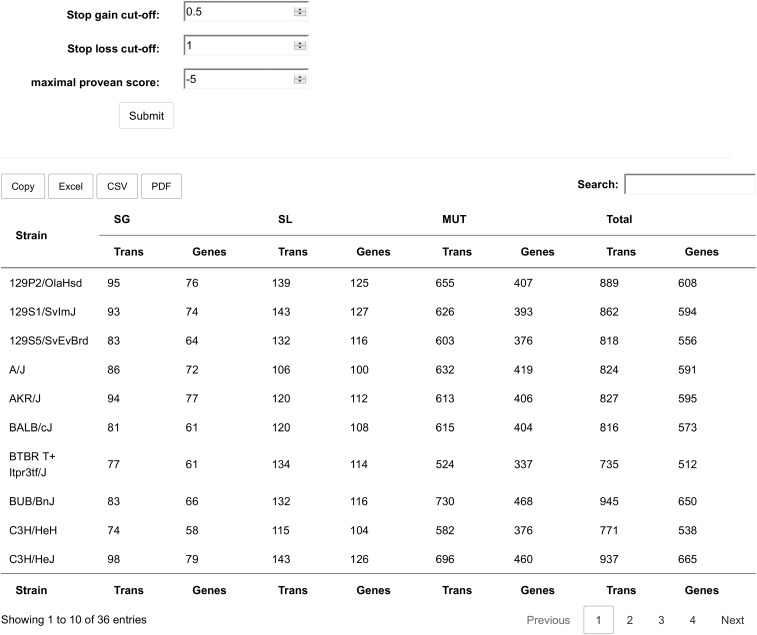
The numbers of protein coding transcripts that suffer from at least one aa truncation (SG) or extension (SL), as well as the number of transcripts leading to an aa change (MUT) with a PROVEAN impact score of −2.5 or less, are provided for each mouse strain in Table 1. This table essentially forms the database that is online, available, and searchable at mousepost.be. In addition to the tabular overview of all mouse strains, we also provide a detailed list of all affected transcripts (SG, SL, and MUT) for each individual strain. A search form allowing the user to search for a specific gene, or to search for a group of genes based on their GO terms, is also available on mousepost.be.
Table 1.
List of 36 mouse strains, sequenced by the Wellcome Trust Sanger Institute and addressed in this study
| Strain | SG | SL | MT | Total* | ||||
| Trans | Genes | Trans | Genes | Trans | Genes | Trans | Genes | |
| 129P2/OlaHsd | 227 | 179 | 139 | 125 | 2,155 | 1,298 | 2,521 | 1,602 |
| 129S1/SvImJ | 217 | 173 | 143 | 127 | 2,109 | 1,282 | 2,469 | 1,582 |
| 129S5/SvEvBrd | 202 | 159 | 132 | 116 | 2,068 | 1,246 | 2,402 | 1,521 |
| A/J | 229 | 185 | 106 | 100 | 2,036 | 1,263 | 2,371 | 1,548 |
| AKR/J | 224 | 181 | 120 | 112 | 2,052 | 1,248 | 2,396 | 1,541 |
| BALB/cJ | 205 | 158 | 120 | 108 | 1,925 | 1,199 | 2,250 | 1,465 |
| BTBR T+ Itpr3tf/J | 200 | 147 | 134 | 114 | 1,744 | 1,073 | 2,078 | 1,334 |
| BUB/BnJ | 213 | 163 | 132 | 116 | 2,233 | 1,352 | 2,578 | 1,631 |
| C3H/HeH | 185 | 144 | 115 | 104 | 1,773 | 1,082 | 2,073 | 1,330 |
| C3H/HeJ | 234 | 188 | 143 | 126 | 2,170 | 1,328 | 2,547 | 1,642 |
| C57BL/10J | 29 | 21 | 59 | 54 | 190 | 114 | 278 | 189 |
| C57BL/6NJ | 17 | 12 | 52 | 47 | 23 | 17 | 92 | 76 |
| C57BR/cdJ | 132 | 96 | 88 | 76 | 970 | 615 | 1,190 | 787 |
| C57L/J | 111 | 85 | 84 | 71 | 902 | 566 | 1,097 | 722 |
| C58/J | 121 | 99 | 100 | 90 | 1,273 | 764 | 1,494 | 953 |
| CAST/EiJ | 634 | 515 | 317 | 258 | 6,001 | 3,626 | 6,952 | 4,399 |
| CBA/J | 181 | 149 | 121 | 103 | 1,667 | 1,050 | 1,969 | 1,302 |
| DBA/1J | 230 | 180 | 133 | 119 | 2,189 | 1,335 | 2,552 | 1,634 |
| DBA/2J | 240 | 194 | 136 | 119 | 2,279 | 1,408 | 2,655 | 1,721 |
| FVB/NJ | 242 | 183 | 129 | 114 | 2,100 | 1,261 | 2,471 | 1,558 |
| I/LnJ | 267 | 206 | 135 | 122 | 2,220 | 1,384 | 2,622 | 1,712 |
| KK/HiJ | 234 | 192 | 147 | 125 | 2,128 | 1,365 | 2,509 | 1,682 |
| LEWES/EiJ | 299 | 230 | 161 | 139 | 3,112 | 1,881 | 3,572 | 2,250 |
| LP/J | 260 | 204 | 152 | 131 | 2,202 | 1,377 | 2,614 | 1,712 |
| MOLF/EiJ | 653 | 528 | 328 | 266 | 5,962 | 3,661 | 6,943 | 4,455 |
| NOD/ShiLtJ | 241 | 184 | 129 | 115 | 2,175 | 1,346 | 2,545 | 1,645 |
| NZB/BlNJ | 199 | 162 | 127 | 112 | 2,087 | 1,278 | 2,413 | 1,552 |
| NZO/HlLtJ | 217 | 176 | 129 | 112 | 2,106 | 1,316 | 2,452 | 1,604 |
| NZW/LacJ | 229 | 193 | 140 | 121 | 2,309 | 1,419 | 2,678 | 1,733 |
| PWK/PhJ | 694 | 583 | 350 | 278 | 6,273 | 3,824 | 7,317 | 4,685 |
| RF/J | 224 | 174 | 121 | 110 | 2,210 | 1,358 | 2,555 | 1,642 |
| SEA/GnJ | 224 | 177 | 126 | 115 | 2,067 | 1,303 | 2,417 | 1,595 |
| SPRET/EiJ | 1,342 | 1,055 | 556 | 459 | 10,235 | 6,107 | 12,133 | 7,621 |
| ST/bJ | 246 | 178 | 128 | 115 | 2,011 | 1,233 | 2,385 | 1,526 |
| WSB/EiJ | 326 | 257 | 170 | 144 | 3,186 | 1,951 | 3,682 | 2,352 |
| ZALENDE/EiJ | 359 | 291 | 177 | 156 | 3,457 | 2,193 | 3,993 | 2,640 |
For each strain, the number of protein-coding transcripts and genes with a SG, SL, or short indel or single amino acid sequence variation (MUT) compared with C57BL/6J, is given. Only deviant sequences with a PROVEAN score of −2.5 or less are given.
Transcripts and genes are given only once; that is, a transcript with a SG or a SL and a MUT will appear in the SG or SL list respectively.
All the known sequence variations and mutations in mouse inbred strains are confirmed in our database. Two well-known examples are the Lpsd (Tlr4P712H) mutation in the LPS-resistant mouse strain C3H/HeJ (7), which receives a PROVEAN score of −7.833, and the albino (TyrC103S) mutation (PROVEAN score −9.738), leading to the albino phenotype of BALB/c mice (13) (Table 2). By searching the database, it is found that exactly the same mutation in the Tyr gene is found in 10 mouse strains closely related to BALB/c, all of which are albino (e.g., A/J and AKR/J, as well as FVB/NJ).
Table 2.
Examples of known mutations, validated by mousepost.be, and new interstrain gene variations, as described in this report
| Known mutation/gene | Variation found by mousepost.be | PROVEAN score | Mouse strain | (Expected) phenotype and reference |
| Lpsd/Tlr4 | Tlr4P712H | −7.833 | C3H/HeJ | Resistance to LPS (7) |
| Albino/Tyr | TyrC103S | −9.738 | BALB/cJ | Albinism (13) |
| CyfipM1N/Cyfip2 | Cyfip2S968F | −5.251 | C57BL/6NJ | Retinal degeneration (17) |
| Interstrain gene variants/gene | ||||
| Rd8/Crb1 | Crb1R1161G | NA: SG: 14% shorter protein | C57BL/6NJ | Response to cocaine and methamphetamine (18) |
| Adamts12 | Adamts12C1518F | −7.131 | C57BL/6NJ | Cancer phenotype (19) |
| Ugt genes | R > S | −4.545 to −4.791 | C57BL/6NJ | Poor detoxification |
| Adamts4 | Adamts4L17F | NA: SG: 96% shorter protein | FVB/NJ | Resistance to atherosclerosis (22) |
| Ccr5 | Ccr5P185L | −9.103 | FVB/NJ | Resistance to acetaminophen (23–25) |
| Brca1 | Brca1N623S | −3.369 | CAST/EiJ | Breast cancer susceptibility |
| Brca2 | Brca2L1495del | −12.166 | CAST/EiJ | Breast cancer susceptibility |
| Nlrp3 | Nlrp3P214A | −7.090 | CAST/EiJ | Deficient NLRP3 inflammasome function |
| Tnfrsf1b | Tnfrsf1bP431L | −7.325 | 129 strains | Resistance to TNF-mediated inflammation |
| BTBR T+ Itpr3tf/J | ||||
| LP/J | ||||
| Ripk3 | Ripk3T166K | −5.114 | BTBR T+ Itpr3tf/J | Resistance to necroptosis |
| DBA/2J | ||||
| IL1a | IL1aY118_T119del | −11.218 | C3H/HeN | Resistance to IL1α-mediated inflammation |
| C3H/HeJ | ||||
| Il1r1 | Il1r1E500G | −6.401 | PWK/PhJ | Resistance to IL1-mediated inflammation |
The genetic characterization of C57BL/6NJ is of critical importance, as the International Knockout Mouse Consortium has decided to use embryonic stem cells derived from this strain (14, 15). The C57BL/6NJ strain has been established, starting from C57BL/6J mice (derived from the Jackson Laboratories in 1951) at NIH. Now, 66 y later, compared with the reference C57BL/6J, the strain C57BL/6NJ is still closely related, but no longer identical. A comparison between C57BL/6J and C57BL/6NJ was performed in the past (16). Using our tool, only 17 transcripts were shown to contain an SG variation, some of which might, however, be important (Table 2); for example, the gene Crb1, which appears to have a 14% shorter protein and is cause for retinal degeneration in this strain (17). Also in these mice, only a few MUT changes have been described; for example, the Cyfip2S968F mutation, which we find in our database with a PROVEAN score −5.251, and which leads to an unstable protein, the CyfipM1N allele (18), which was linked to a reduced acute and sensitized response to cocaine and methamphetamine (18). The point mutation in the Adamts12 gene (Adamts12C1518F with PROVEAN score of −7.131) might lead to a specific cancer phenotype in these mice, as the knockout allele of this gene leads to increased tumor angiogenesis and invasion (19). An increased susceptibility for development of colon cancer in these C57BL/6NJ mice compared with C57BL/6J has been described (20). Finally, several genes of the UDP glucuronosyltransferase 1 family, responsible for the glucuronidation of hydrophobic substrates, are mutated in this strain: Ugt1a1, Ugt1a6a, Ugt1a7c, Ugt1a5, Ugt1a9, Ugt1a2, and Ugt1a10. All these mutations are exactly the same missense mutation (R > S; PROVEAN score, −4.545 to −4.791), as these different genes all share the affected exon (Fig. 2).
Fig. 2.
Overview of the Ugt1a genomic locus on mouse chromosome 1, and the sequence variation in the exon shared by multiple Ugt1a genes. The genes that were found mutated in the C57BL/6NJ strain, Ugt1a1, Ugt1a6a, Ugt1a7c, Ugt1a5, Ugt1a9, Ugt1a2, and Ugt1a10, all make use of a common exon. In this exon, the affected codon leading to the mutation (R > S) is found, and is enlarged in the figure and labeled in red. For each gene, this relevant sequence change leads to a specific mutation; for example, for Ugt1a10 and Ugt1a10R439S.
Because of the big size of their oocytes and zygotes, FVB/NJ mice have been used as the preferential strain for transgenic overexpression by injection of DNA in zygote pronuclei. Therefore, many biological systems have been studied in these mice. We found that 242 transcripts of these mice have a SG, that is, a nonsense mutation, compared with the reference genome, comprising a long list of very important genes, such as Adamts4. This gene encodes a protein of 648 aa, but in FVB/NJ, only 27 aa. Adamts4 knockout mice are resistant to high-fat-diet–induced atherosclerosis (21), a trait also described in FVB/NJ mice (22). Among the 2,100 MUT variations, many interesting sequence variations with impressively low PROVEAN score are found; for example, in the gene coding for the important chemokine receptor CCR5, Ccr5, a P185L variation, is found, leading to a PROVEAN score of −9.103. As CCR5 knockout mice were found to be resistant to acetaminophen (23, 24), this mutant version found in FVB/NJ might explain their resistance to this inducer of hepatitis (25).
Mouse strains that are, from an evolutionary point of view, very distant from C57BL/6J, for example, SPRET/EiJ and CAST/EiJ, display many thousands of potentially important sequence variations. CAST/EiJ, a strain generated from the Mus musculus castaneus subspecies, shows 634 SG, 317 SL, and 6,001 MUT transcripts. These mice, for example, carry an exceptional SL mutation in the Ahr gene, leading to a 43-aa-longer protein, but also they have a MUT in Brca1 (Brca1N623S) with PROVEAN score of −3.369 and 10 sequence variations (with PROVEAN scores of −2.5 or less) in the Brca2 gene, the most severe one (PROVEAN score −12.166) being a single-aa-deletion Brca2L1495del. Their Nlrp3 gene (coding for a major inflammasome protein) has a single MUT leading to Nlrp3P214A (PROVEAN score of −7.09). In fact, these mice have severe MUT versions in most of their Nlrp genes. By studying the variant alleles of these mice, their value as a reservoir of interesting alleles becomes apparent.
Exploring and Exploiting the Full Richness of the Mouse Sequence Variations.
To explore the full richness of the gene variations in mice for functional research, deviant versions of proteins can be searched across all 36 mouse strains, using the search functions of the online tool (mousepost.be; Figs. S1–S9 for a user manual). The search function of this web tool allows a gene-by-gene investigation of polymorphisms across the 36 mouse strains, and the reports include links to the University of California, Santa Cruz; Ensembl; and PubMed websites. This way, a variant form of the TNF receptor 2 (encoded by the Tnfrsf1b gene) is found in five mouse strains, namely in all three 129 strains, the BTBR strain and the LP/J strain (all with a PROVIAN score of −7.325). Similarly, BTBR and DBA/2J mice appear to express a variant form of the essential protein for necroptosis RIPK3 (26) (encoded by Ripk3 gene, PROVIAN score −5.114), and both C3H strains express a severely attenuated form of the important cytokine interleukin 1 alpha (IL1α, encoded by Il1a gene, PROVIAN score −11.218) and the PWK/PhJ strain a mutant form of the IL1R1 protein (PROVIAN score −6.401). Finally, the new database provides a fast view on candidate polymorphic protein-coding sequences within a critical chromosomal region, which was defined by a linkage analysis. For example, the TNF resistance locus found on distal chromosome 12 (104.3 Mb) in DBA/2J mice (27) can now be studied in the context of the variations in sequences in the Serpin genes, found on this locus.
Fig. S1.
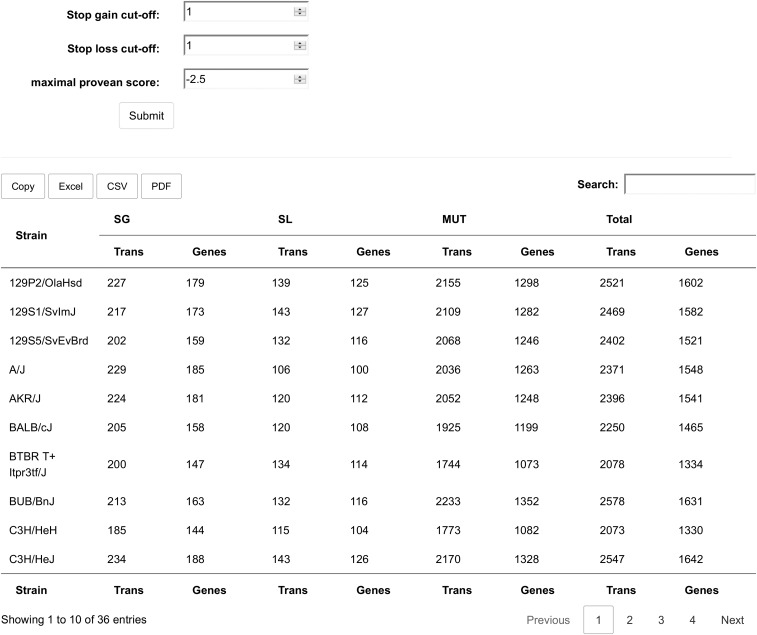
The first page a user sees in the web tool homepage. This page provides an overview of the number of affected genes per strain in all 3 classes using the cutoffs set at the top of the page. The 3 cutoff fields can be set by the user, and on submitting the changes, the table will be updated with the new cutoffs. The default cutoffs for SG and SL are 1, which means not to use any minimal deletion or extension size. The PROVEAN cutoff is set at −2.5, as suggested by the authors of the PROVEAN software as a good tradeoff between specificity and sensitivity. Lower values are more specific, but at the cost of finding less true positives; higher scores are more sensitive, but with the disadvantage of picking up more false-positives.
Fig. S9.
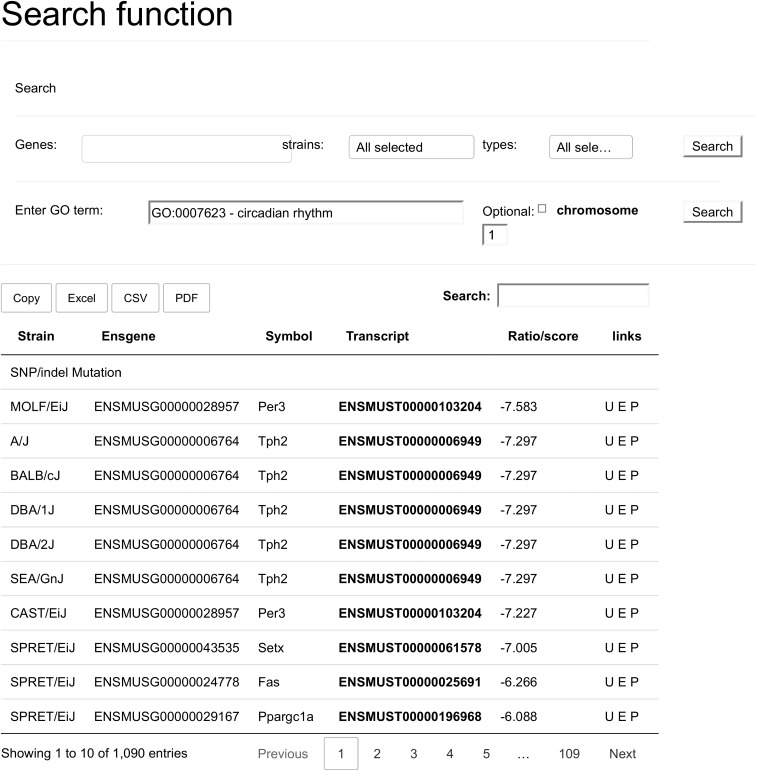
A search on the GO term circadian rhythm returns all genes that belong to this terms and that are affected by one of the events. These results are grouped by type and sorted by strain and gene fields.
Fig. S2.
After changing the cutoffs, the table is updated to show the number of genes that pass the new filters. The table can be exported to the clipboard, an excel file, a csv file of a pdf file using the buttons above the table. The search field may be used to quickly select a strain of interest. Each number in the table is a link to a different part of the website in which a list of the actual genes affected for each strain an class may be obtained. The filter settings entered here will be taken into account when generating those lists.
Fig. S3.
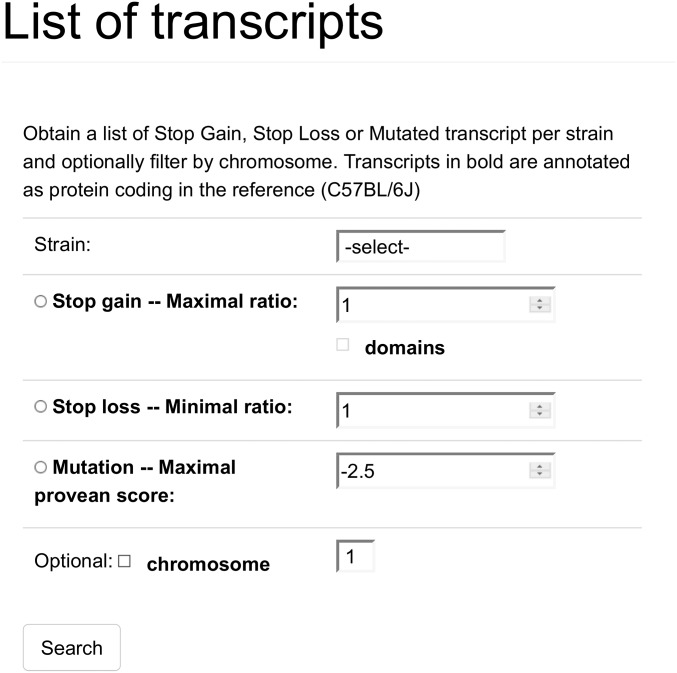
When selecting “Lists” from the menu, this is the page that is displayed. Here a user must select a strain of interest and one event type. In addition, a cutoff value must also be set, and optionally, the user may restrict the search to only one chromosome. The domains selector for SG events is also optional and, if selected, will include information about any lost conserved protein domains in the output, which is not by default included. In the case that the user clicked one of the numbers in the table of the homepage, he will also be redirected to this page, but the strain, event type, and cutoff will be already selected, and results will be immediately displayed.
Fig. S4.
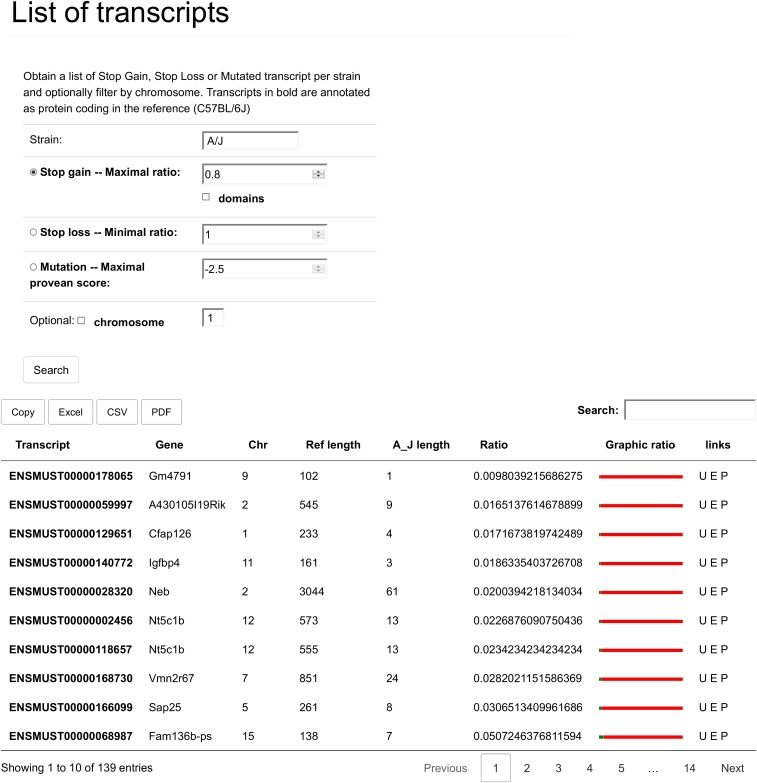
This is the output that is generated for SG in the strain A/J, with a minimal loss of 20% of the reference sequence. There are eight columns. The first two specify the affected transcript and gene, respectively, and link to a details page, where all the information about this gene may be found. Then there is the chromosome where the gene is located, and the following four columns have to do with the length and length difference between the reference C57BL/6J strain and the strain of interest (here A/J). The “Ref length” shows how long the protein encoded by this transcript is in the reference strain. The “A/J length” column shows how long the protein is in the strain of interest, taking the early stop codon into account. The “ratio” column shows the ratio between “A/J length” and “Ref length.” By default, the table is also ordered on this column from largest difference to smallest. The “Graphic ration” shows an image-based representation of the length difference, where the red color stands for sequence lost. The final column is a set of links to Ensembl (E); University of California, Santa Cruz, genome browser (U); and PubMed (P). These link to ensemble transcript information, a view of the transcript on the genome browser, and a PubMed search of the gene, respectively. The data can also be exported, but because of limitations of the export library, the “Graphic ratio” and “Links” are not included when doing so. In the case that SL is selected, the table has the same fields as the one show here. The difference is only in the sorting (from largest relative extension to smallest) and in the interpretation of the “Graphic ratio,” where the red then becomes the amount of sequence added to the normal protein length.
Fig. S5.
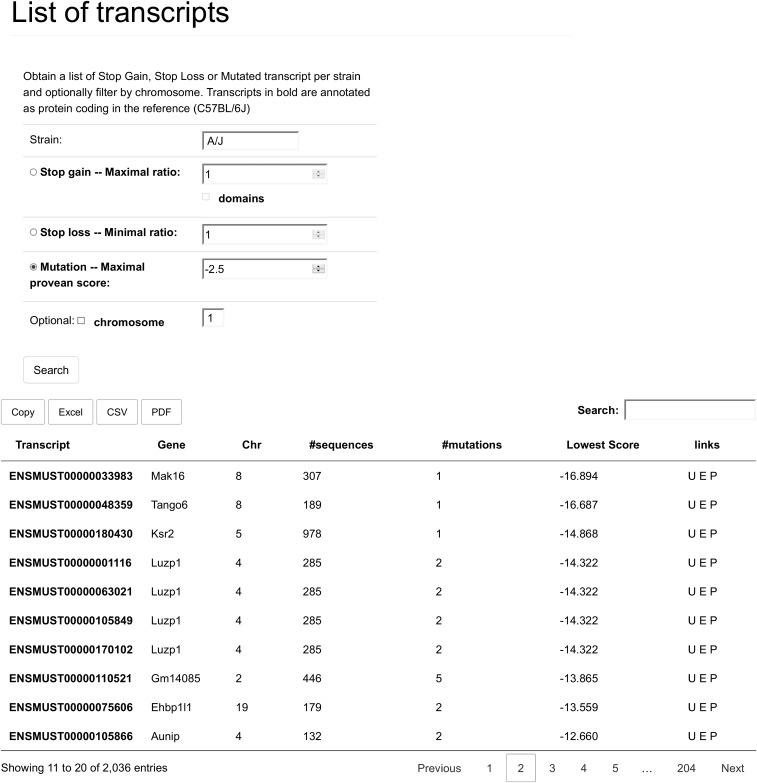
The table obtained when listing the MUT events differs lightly from the SL and SG tables. The “Transcrip,” “Gene,” “Chr,” and “links” columns are the same; see Fig. S4 for the explanation. The fourth column here is the “#sequences” column. This shows the number of supporting sequences PROVEAN used to calculate it’s score. This is a quality metric for the prediction, and values lower than 50 in this field indicate that the PROVEAN score may not be reliable. “#mutations” indicates how many mutations are present in the transcript. “Lowes Scores” shows the score of the mutation with the lowest PROVEAN score in the protein encoded by this transcript. The table is also sorted on this field.
Fig. S6.
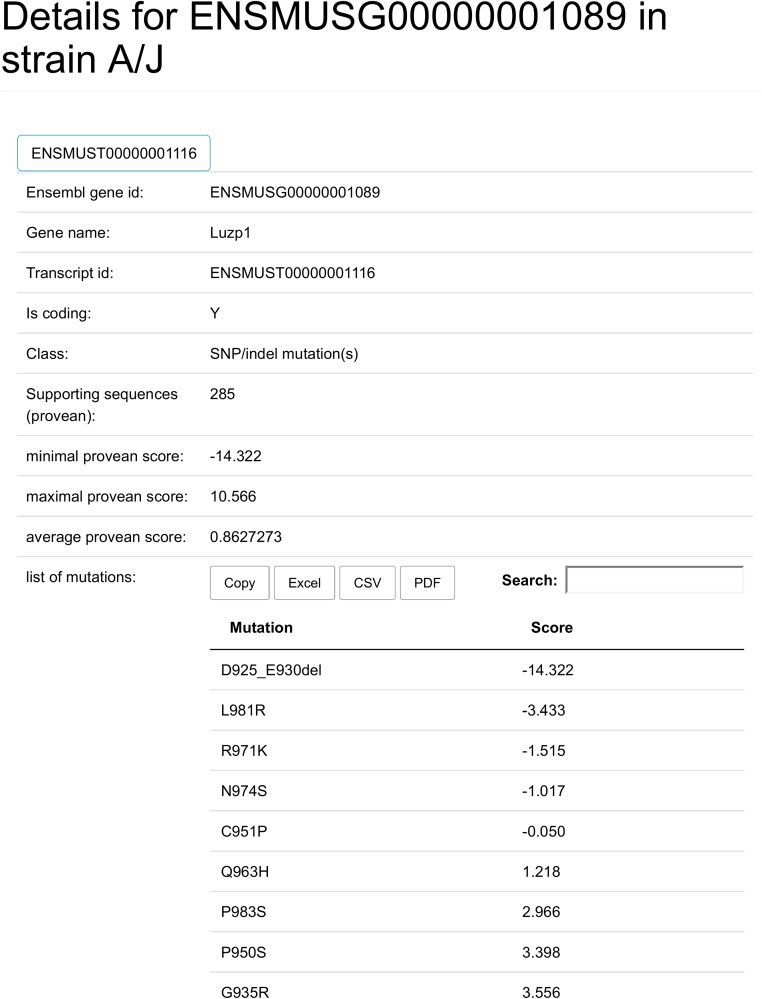
A details page, this is where the links on the Gene and Transcript table fields point to. In this case, the details page for Luzp1 in the A/J strain is shown (in part). This contains all the information from the tables previously described. In addition, it also contains a list of all PROVEAN scored mutations, including those that do not meet the cutoff. The page also includes the GO annotation and the protein sequence in the reference train and strain of interest (SG and SL).
Fig. S7.
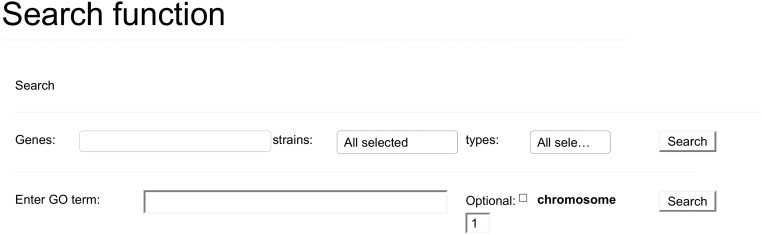
The tool also contains a search function. There are 2 types of search that may be performed. The first, and most simple, search is searching a gene. After starting to type a gene name, autocomplete options will be offered, and the search may be restricted to a certain strain or event type. This restriction is useful for answering questions such as, “is gene X affected by a mutation in strain Y?” (strain restriction), or “is there a strain where gene X is truncated due to a SG mutation?” (event-type restriction). The second type of search is a search on function. One may enter a GO term (term identifier or term description) and the search function will select for a group of genes all having this GO term. The results returned may optionally be restricted to a selected chromosome. This field also offers autocomplete suggestions, and because this search is rather demanding, it can only be run once a valid GO term has been entered into the search field.
Fig. S8.
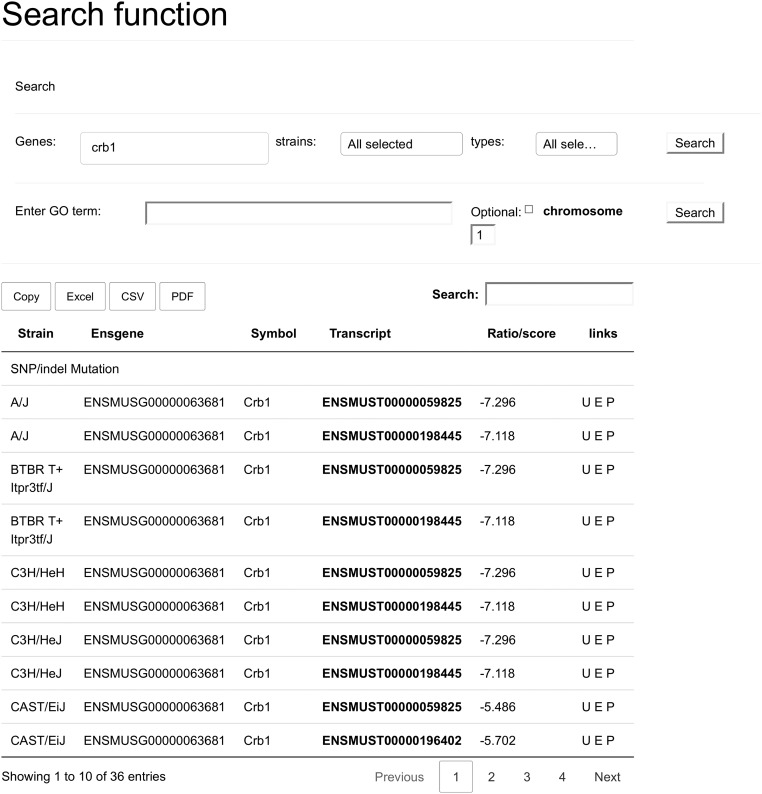
A search for the Crb1 gene without restrictions. This returns a table with all hits for the gene in all event types, the table is sorted by strain and grouped by event type. The search field of the table can be used to narrow down the returned set further on transcript or strain.
In conclusion, an easily accessible and searchable online repository (which will be updated twice per year) of variant alleles of protein coding genes is now available and will lead to the full exploration and exploitation of the naturally occurring mutant variants fixed in the 36 sequenced mouse strains. Obviously, our analysis and database concerns sequence variations in protein-coding genes only. An extension of this study toward noncoding RNA sequences, as well as a link toward mRNA expression levels, might be considered in the future. In these days of fast and efficient mutagenesis using CRISPR/Cas, the availability of naturally occurring sequence variations in these 36 mouse strains is a really good start to identify potentially function-compromising mutations.
Materials and Methods
Sequence Variation Data.
We obtained the sequence variation data (SNPs, insertions, and deletions) of 36 often-used laboratory strains of mice from the ftp site of the mouse genomes project (9). We made use of the strain-specific files, one with SNPs and one with indels per strain, from the REL-1505-SNPs-Indels version of the data (ftp://ftp-mouse.sanger.ac.uk/REL-1505-SNPs_Indels/strain_specific_vcfs). The variants in each downloaded file were filtered on the FI tag, so that only high-quality events (FI = 1) were retained. All files were processed into a single mysql table, which contains the DNA sequence for every position in which a variant was called in every strain, including the reference (C57BL/6J).
Reference Annotation.
We used the GRCm38.p4 version of the mouse genome reference strain (C57BL/6J), which we obtained from the ensemble ftp site (ftp://ftp.ensembl.org/pub/release-86). The structural annotation from this version, in gtf format, was also obtained from the same source. This file was processed to allow searches on features important for the analysis (locations, transcripts, and exons).
Transcript Classification.
We developed a perl script to assess the combined effect of all mutations on each transcript in each strain. The script iterated over all 36 strains and all transcripts. Only transcripts with at least one sequence variation were subject to further processing: the cDNA sequences was constructed using positional information and the reference sequences and split up in the 5′ UTR, CDS, and 3′ UTR. The sequence variations were applied to the CDS, followed by translating it to the aa sequence, using the standard vertebrate coding table. Classification was performed by comparing the reference and alternate aa sequences into three classes. Although the database is focused on protein coding genes, the rare occurrence of pseudogene sequences is not excluded.
Basic Local Alignment Search Databases.
Databases for the blast tools were obtained from the NCBI ftp site. The database with nonredundant protein sequences for use with PROVEAN was downloaded November 16, 2016 (ftp://ftp.ncbi.nlm.nih.gov/blast/db/). The second database that we used was the conserved domains database (CDD) (release from June 28, 2016; ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/).
Searching for Lost Domains.
Using RPS-blast, the references protein sequences were used to query the conserved domain database (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Hits were filtered on the E-value field, and only those with E-values < 0.01 were retained. In a second step of filtering, we removed the hits that did not overlap the truncated part of the sequence.
PROVEAN.
Several programs have been developed to estimate the effect of a given sequence variation on the function of the protein. Because PROVEAN is able to interpret small insertions and deletions (12), this tool was selected. First, a file for each transcript in each strain was constructed with the mutated positions. For this, a global pairwise sequence alignment was constructed between the reference and alternative transcript with needle (EMBOSS tools) (28). This alignment file, along with the positional data from the classification step, was processed with a perl script that was specifically created to build these files. To minimize running time, the PROVEAN tool was run on a high-performance computing (HPC) cluster for each strain in sequence; in this way, we could save and reuse the supported sequence sets. The score provided by PROVEAN depends in part on the number of available sequences for a given transcript. We have followed the suggestions of the authors of the PROVEAN tool (12) and have applied a cutoff of 50 sequences as the minimum reliable amount of sequences. In their paper, the authors of PROVEAN show an overview in which the balanced accuracy is determined in the function of the number of supporting sequences. For 51+ sequences, this balanced accuracy remains higher than 73%. However, it is shown that the balanced accuracy decreases when the number of supporting sequences drops to 50 or lower. For this reason, we used 50 supporting sequences as a cutoff, as from this point the accuracy of the prediction drops. We did not exclude cases with fewer than 50 supporting sequences from the database, but they are not reported in the web tool mousepost.be because this does not mean that the prediction are not correct, but they are more likely to be wrong. As suggested by Choi et al. (12), a PROVEAN cutoff score of −2.5 is applied as the default cutoff in the mousepost.be web tool. This cutoff has been motivated by these authors. However, the user of the mousepost.be has the option to set the cutoff at another level, according to desire.
Gene Ontology.
The gene ontology (GO) annotation for was downloaded the gene ontology consortium website (www.geneontology.org/). We processed this file to obtain the GO terms for all genes in our dataset to allow the GO search functionality in the web tool.
Figs. S1–S9 provide a user manual for the web tool mousepost.be, which allows us to search for sequence variations of protein coding genes across the 36 sequenced mouse strains compared with the reference genome of C57BL/6J.
Acknowledgments
This work was supported by the Belgian Science Policy (BELSPO) Interuniversity Attraction Poles program (IAP-VI-18), University Ghent (BOF13/GOA/005 project), Flanders Institute for Biotechnology (basic funding), and the Research Foundation-Flanders (G.0.005.10.N.10 project).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706168114/-/DCSupplemental.
References
- 1.Silver LM. Mouse genetics: Concepts and applications. Oxford University Press; New York: 1995. p. xiii. [Google Scholar]
- 2.Battey J, Jordan E, Cox D, Dove W. An action plan for mouse genomics. Nat Genet. 1999;21:73–75. doi: 10.1038/5012. [DOI] [PubMed] [Google Scholar]
- 3.Church DM, et al. Mouse Genome Sequencing Consortium Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva S, et al. Spontaneous development of plasmacytomas in a selected subline of BALB/cJ mice. Eur J Cancer. 1997;33:479–485. doi: 10.1016/s0959-8049(97)89025-9. [DOI] [PubMed] [Google Scholar]
- 5.Myers LK, Rosloniec EF, Cremer MA, Kang AH. Collagen-induced arthritis, an animal model of autoimmunity. Life Sci. 1997;61:1861–1878. doi: 10.1016/s0024-3205(97)00480-3. [DOI] [PubMed] [Google Scholar]
- 6.Moser AR, Dove WF, Roth KA, Gordon JI. The Min (multiple intestinal neoplasia) mutation: Its effect on gut epithelial cell differentiation and interaction with a modifier system. J Cell Biol. 1992;116:1517–1526. doi: 10.1083/jcb.116.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 8.Grubb SC, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2014;42:D825–D834. doi: 10.1093/nar/gkt1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanden Berghe T, et al. Passenger mutations confound interpretation of all genetically modified congenic mice. Immunity. 2015;43:200–209. doi: 10.1016/j.immuni.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steeland S, et al. Efficient analysis of mouse genome sequences reveal many nonsense variants. Proc Natl Acad Sci USA. 2016;113:5670–5675. doi: 10.1073/pnas.1605076113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibahara S, et al. A point mutation in the tyrosinase gene of BALB/c albino mouse causing the cysteine––serine substitution at position 85. Eur J Biochem. 1990;189:455–461. doi: 10.1111/j.1432-1033.1990.tb15510.x. [DOI] [PubMed] [Google Scholar]
- 14.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettitt SJ, et al. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon MM, et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013;14:R82. doi: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mekada K, et al. Genetic differences among C57BL/6 substrains. Exp Anim. 2009;58:141–149. doi: 10.1538/expanim.58.141. [DOI] [PubMed] [Google Scholar]
- 18.Kumar V, et al. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science. 2013;342:1508–1512. doi: 10.1126/science.1245503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Hour M, et al. Higher sensitivity of Adamts12-deficient mice to tumor growth and angiogenesis. Oncogene. 2010;29:3025–3032. doi: 10.1038/onc.2010.49. [DOI] [PubMed] [Google Scholar]
- 20.Diwan BA, Blackman KE. Differential susceptibility of 3 sublines of C57BL/6 mice to the induction of colorectal tumors by 1,2-dimethylhydrazine. Cancer Lett. 1980;9:111–115. doi: 10.1016/0304-3835(80)90114-7. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, et al. Loss of ADAMTS4 reduces high fat diet-induced atherosclerosis and enhances plaque stability in ApoE(-/-) mice. Sci Rep. 2016;6:31130. doi: 10.1038/srep31130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sontag TJ, et al. Apolipoprotein A-I protection against atherosclerosis is dependent on genetic background. Arterioscler Thromb Vasc Biol. 2014;34:262–269. doi: 10.1161/ATVBAHA.113.302831. [DOI] [PubMed] [Google Scholar]
- 23.Choi DY, Ban JO, Kim SC, Hong JT. CCR5 knockout mice with C57BL6 background are resistant to acetaminophen-mediated hepatotoxicity due to decreased macrophages migration into the liver. Arch Toxicol. 2015;89:211–220. doi: 10.1007/s00204-014-1253-3. [DOI] [PubMed] [Google Scholar]
- 24.Jaeschke H. Commentary to Choi et al. (2015): CCR5 knockout mice with C57BL6 background are resistant to acetaminophen-mediated hepatotoxicity due to decreased macrophages migration into the liver. Arch Toxicol. 2015;89:807–808. doi: 10.1007/s00204-015-1499-4. [DOI] [PubMed] [Google Scholar]
- 25.Weerasinghe SVW, Park MJ, Portney DA, Omary MB. Mouse genetic background contributes to hepatocyte susceptibility to Fas-mediated apoptosis. Mol Biol Cell. 2016;27:3005–3012. doi: 10.1091/mbc.E15-06-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newton K, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 27.Libert C, et al. Identification of a locus on distal mouse chromosome 12 that controls resistance to tumor necrosis factor-induced lethal shock. Genomics. 1999;55:284–289. doi: 10.1006/geno.1998.5677. [DOI] [PubMed] [Google Scholar]
- 28.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The numbers of protein coding transcripts that suffer from at least one aa truncation (SG) or extension (SL), as well as the number of transcripts leading to an aa change (MUT) with a PROVEAN impact score of −2.5 or less, are provided for each mouse strain in Table 1. This table essentially forms the database that is online, available, and searchable at mousepost.be. In addition to the tabular overview of all mouse strains, we also provide a detailed list of all affected transcripts (SG, SL, and MUT) for each individual strain. A search form allowing the user to search for a specific gene, or to search for a group of genes based on their GO terms, is also available on mousepost.be.
Table 1.
List of 36 mouse strains, sequenced by the Wellcome Trust Sanger Institute and addressed in this study
| Strain | SG | SL | MT | Total* | ||||
| Trans | Genes | Trans | Genes | Trans | Genes | Trans | Genes | |
| 129P2/OlaHsd | 227 | 179 | 139 | 125 | 2,155 | 1,298 | 2,521 | 1,602 |
| 129S1/SvImJ | 217 | 173 | 143 | 127 | 2,109 | 1,282 | 2,469 | 1,582 |
| 129S5/SvEvBrd | 202 | 159 | 132 | 116 | 2,068 | 1,246 | 2,402 | 1,521 |
| A/J | 229 | 185 | 106 | 100 | 2,036 | 1,263 | 2,371 | 1,548 |
| AKR/J | 224 | 181 | 120 | 112 | 2,052 | 1,248 | 2,396 | 1,541 |
| BALB/cJ | 205 | 158 | 120 | 108 | 1,925 | 1,199 | 2,250 | 1,465 |
| BTBR T+ Itpr3tf/J | 200 | 147 | 134 | 114 | 1,744 | 1,073 | 2,078 | 1,334 |
| BUB/BnJ | 213 | 163 | 132 | 116 | 2,233 | 1,352 | 2,578 | 1,631 |
| C3H/HeH | 185 | 144 | 115 | 104 | 1,773 | 1,082 | 2,073 | 1,330 |
| C3H/HeJ | 234 | 188 | 143 | 126 | 2,170 | 1,328 | 2,547 | 1,642 |
| C57BL/10J | 29 | 21 | 59 | 54 | 190 | 114 | 278 | 189 |
| C57BL/6NJ | 17 | 12 | 52 | 47 | 23 | 17 | 92 | 76 |
| C57BR/cdJ | 132 | 96 | 88 | 76 | 970 | 615 | 1,190 | 787 |
| C57L/J | 111 | 85 | 84 | 71 | 902 | 566 | 1,097 | 722 |
| C58/J | 121 | 99 | 100 | 90 | 1,273 | 764 | 1,494 | 953 |
| CAST/EiJ | 634 | 515 | 317 | 258 | 6,001 | 3,626 | 6,952 | 4,399 |
| CBA/J | 181 | 149 | 121 | 103 | 1,667 | 1,050 | 1,969 | 1,302 |
| DBA/1J | 230 | 180 | 133 | 119 | 2,189 | 1,335 | 2,552 | 1,634 |
| DBA/2J | 240 | 194 | 136 | 119 | 2,279 | 1,408 | 2,655 | 1,721 |
| FVB/NJ | 242 | 183 | 129 | 114 | 2,100 | 1,261 | 2,471 | 1,558 |
| I/LnJ | 267 | 206 | 135 | 122 | 2,220 | 1,384 | 2,622 | 1,712 |
| KK/HiJ | 234 | 192 | 147 | 125 | 2,128 | 1,365 | 2,509 | 1,682 |
| LEWES/EiJ | 299 | 230 | 161 | 139 | 3,112 | 1,881 | 3,572 | 2,250 |
| LP/J | 260 | 204 | 152 | 131 | 2,202 | 1,377 | 2,614 | 1,712 |
| MOLF/EiJ | 653 | 528 | 328 | 266 | 5,962 | 3,661 | 6,943 | 4,455 |
| NOD/ShiLtJ | 241 | 184 | 129 | 115 | 2,175 | 1,346 | 2,545 | 1,645 |
| NZB/BlNJ | 199 | 162 | 127 | 112 | 2,087 | 1,278 | 2,413 | 1,552 |
| NZO/HlLtJ | 217 | 176 | 129 | 112 | 2,106 | 1,316 | 2,452 | 1,604 |
| NZW/LacJ | 229 | 193 | 140 | 121 | 2,309 | 1,419 | 2,678 | 1,733 |
| PWK/PhJ | 694 | 583 | 350 | 278 | 6,273 | 3,824 | 7,317 | 4,685 |
| RF/J | 224 | 174 | 121 | 110 | 2,210 | 1,358 | 2,555 | 1,642 |
| SEA/GnJ | 224 | 177 | 126 | 115 | 2,067 | 1,303 | 2,417 | 1,595 |
| SPRET/EiJ | 1,342 | 1,055 | 556 | 459 | 10,235 | 6,107 | 12,133 | 7,621 |
| ST/bJ | 246 | 178 | 128 | 115 | 2,011 | 1,233 | 2,385 | 1,526 |
| WSB/EiJ | 326 | 257 | 170 | 144 | 3,186 | 1,951 | 3,682 | 2,352 |
| ZALENDE/EiJ | 359 | 291 | 177 | 156 | 3,457 | 2,193 | 3,993 | 2,640 |
For each strain, the number of protein-coding transcripts and genes with a SG, SL, or short indel or single amino acid sequence variation (MUT) compared with C57BL/6J, is given. Only deviant sequences with a PROVEAN score of −2.5 or less are given.
Transcripts and genes are given only once; that is, a transcript with a SG or a SL and a MUT will appear in the SG or SL list respectively.
All the known sequence variations and mutations in mouse inbred strains are confirmed in our database. Two well-known examples are the Lpsd (Tlr4P712H) mutation in the LPS-resistant mouse strain C3H/HeJ (7), which receives a PROVEAN score of −7.833, and the albino (TyrC103S) mutation (PROVEAN score −9.738), leading to the albino phenotype of BALB/c mice (13) (Table 2). By searching the database, it is found that exactly the same mutation in the Tyr gene is found in 10 mouse strains closely related to BALB/c, all of which are albino (e.g., A/J and AKR/J, as well as FVB/NJ).
Table 2.
Examples of known mutations, validated by mousepost.be, and new interstrain gene variations, as described in this report
| Known mutation/gene | Variation found by mousepost.be | PROVEAN score | Mouse strain | (Expected) phenotype and reference |
| Lpsd/Tlr4 | Tlr4P712H | −7.833 | C3H/HeJ | Resistance to LPS (7) |
| Albino/Tyr | TyrC103S | −9.738 | BALB/cJ | Albinism (13) |
| CyfipM1N/Cyfip2 | Cyfip2S968F | −5.251 | C57BL/6NJ | Retinal degeneration (17) |
| Interstrain gene variants/gene | ||||
| Rd8/Crb1 | Crb1R1161G | NA: SG: 14% shorter protein | C57BL/6NJ | Response to cocaine and methamphetamine (18) |
| Adamts12 | Adamts12C1518F | −7.131 | C57BL/6NJ | Cancer phenotype (19) |
| Ugt genes | R > S | −4.545 to −4.791 | C57BL/6NJ | Poor detoxification |
| Adamts4 | Adamts4L17F | NA: SG: 96% shorter protein | FVB/NJ | Resistance to atherosclerosis (22) |
| Ccr5 | Ccr5P185L | −9.103 | FVB/NJ | Resistance to acetaminophen (23–25) |
| Brca1 | Brca1N623S | −3.369 | CAST/EiJ | Breast cancer susceptibility |
| Brca2 | Brca2L1495del | −12.166 | CAST/EiJ | Breast cancer susceptibility |
| Nlrp3 | Nlrp3P214A | −7.090 | CAST/EiJ | Deficient NLRP3 inflammasome function |
| Tnfrsf1b | Tnfrsf1bP431L | −7.325 | 129 strains | Resistance to TNF-mediated inflammation |
| BTBR T+ Itpr3tf/J | ||||
| LP/J | ||||
| Ripk3 | Ripk3T166K | −5.114 | BTBR T+ Itpr3tf/J | Resistance to necroptosis |
| DBA/2J | ||||
| IL1a | IL1aY118_T119del | −11.218 | C3H/HeN | Resistance to IL1α-mediated inflammation |
| C3H/HeJ | ||||
| Il1r1 | Il1r1E500G | −6.401 | PWK/PhJ | Resistance to IL1-mediated inflammation |
The genetic characterization of C57BL/6NJ is of critical importance, as the International Knockout Mouse Consortium has decided to use embryonic stem cells derived from this strain (14, 15). The C57BL/6NJ strain has been established, starting from C57BL/6J mice (derived from the Jackson Laboratories in 1951) at NIH. Now, 66 y later, compared with the reference C57BL/6J, the strain C57BL/6NJ is still closely related, but no longer identical. A comparison between C57BL/6J and C57BL/6NJ was performed in the past (16). Using our tool, only 17 transcripts were shown to contain an SG variation, some of which might, however, be important (Table 2); for example, the gene Crb1, which appears to have a 14% shorter protein and is cause for retinal degeneration in this strain (17). Also in these mice, only a few MUT changes have been described; for example, the Cyfip2S968F mutation, which we find in our database with a PROVEAN score −5.251, and which leads to an unstable protein, the CyfipM1N allele (18), which was linked to a reduced acute and sensitized response to cocaine and methamphetamine (18). The point mutation in the Adamts12 gene (Adamts12C1518F with PROVEAN score of −7.131) might lead to a specific cancer phenotype in these mice, as the knockout allele of this gene leads to increased tumor angiogenesis and invasion (19). An increased susceptibility for development of colon cancer in these C57BL/6NJ mice compared with C57BL/6J has been described (20). Finally, several genes of the UDP glucuronosyltransferase 1 family, responsible for the glucuronidation of hydrophobic substrates, are mutated in this strain: Ugt1a1, Ugt1a6a, Ugt1a7c, Ugt1a5, Ugt1a9, Ugt1a2, and Ugt1a10. All these mutations are exactly the same missense mutation (R > S; PROVEAN score, −4.545 to −4.791), as these different genes all share the affected exon (Fig. 2).
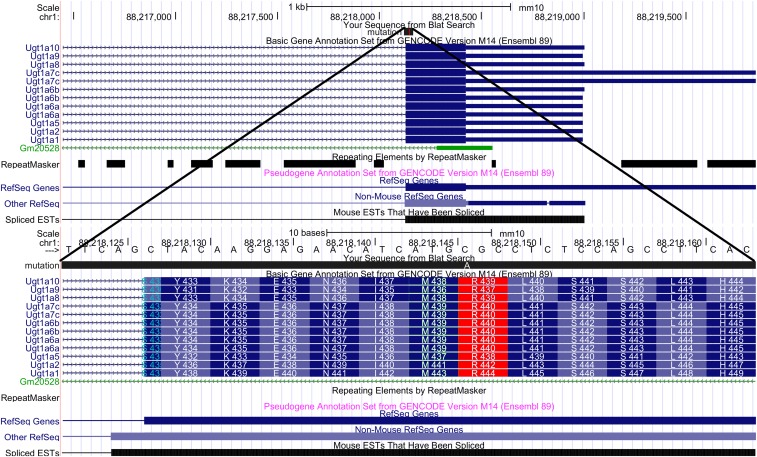
Fig. 2.
Overview of the Ugt1a genomic locus on mouse chromosome 1, and the sequence variation in the exon shared by multiple Ugt1a genes. The genes that were found mutated in the C57BL/6NJ strain, Ugt1a1, Ugt1a6a, Ugt1a7c, Ugt1a5, Ugt1a9, Ugt1a2, and Ugt1a10, all make use of a common exon. In this exon, the affected codon leading to the mutation (R > S) is found, and is enlarged in the figure and labeled in red. For each gene, this relevant sequence change leads to a specific mutation; for example, for Ugt1a10 and Ugt1a10R439S.
Because of the big size of their oocytes and zygotes, FVB/NJ mice have been used as the preferential strain for transgenic overexpression by injection of DNA in zygote pronuclei. Therefore, many biological systems have been studied in these mice. We found that 242 transcripts of these mice have a SG, that is, a nonsense mutation, compared with the reference genome, comprising a long list of very important genes, such as Adamts4. This gene encodes a protein of 648 aa, but in FVB/NJ, only 27 aa. Adamts4 knockout mice are resistant to high-fat-diet–induced atherosclerosis (21), a trait also described in FVB/NJ mice (22). Among the 2,100 MUT variations, many interesting sequence variations with impressively low PROVEAN score are found; for example, in the gene coding for the important chemokine receptor CCR5, Ccr5, a P185L variation, is found, leading to a PROVEAN score of −9.103. As CCR5 knockout mice were found to be resistant to acetaminophen (23, 24), this mutant version found in FVB/NJ might explain their resistance to this inducer of hepatitis (25).
Mouse strains that are, from an evolutionary point of view, very distant from C57BL/6J, for example, SPRET/EiJ and CAST/EiJ, display many thousands of potentially important sequence variations. CAST/EiJ, a strain generated from the Mus musculus castaneus subspecies, shows 634 SG, 317 SL, and 6,001 MUT transcripts. These mice, for example, carry an exceptional SL mutation in the Ahr gene, leading to a 43-aa-longer protein, but also they have a MUT in Brca1 (Brca1N623S) with PROVEAN score of −3.369 and 10 sequence variations (with PROVEAN scores of −2.5 or less) in the Brca2 gene, the most severe one (PROVEAN score −12.166) being a single-aa-deletion Brca2L1495del. Their Nlrp3 gene (coding for a major inflammasome protein) has a single MUT leading to Nlrp3P214A (PROVEAN score of −7.09). In fact, these mice have severe MUT versions in most of their Nlrp genes. By studying the variant alleles of these mice, their value as a reservoir of interesting alleles becomes apparent.