Significance
In Alzheimer’s disease, the microtubule-associated protein Tau is invariably found in a hyperphosphorylated and aggregated form. Whether (hyper)phosphorylation can drive aggregation is less clear, and no precise phosphorylation pattern leading to aggregation has been described. Combining in vitro phosphorylation assays with purified kinases and a rat brain extract with the analytical power of NMR spectroscopy, we unravel here the phosphorylation pattern of Tau that drives its aggregation. The results point to the importance of this posttranslational modification in Tau's aggregation and suggest orienting therapeutic strategies toward phosphorylated Tau.
Keywords: Tau, phosphorylation, aggregation, Alzheimer’s disease, NMR
Abstract
Determining the functional relationship between Tau phosphorylation and aggregation has proven a challenge owing to the multiple potential phosphorylation sites and their clustering in the Tau sequence. We use here in vitro kinase assays combined with NMR spectroscopy as an analytical tool to generate well-characterized phosphorylated Tau samples and show that the combined phosphorylation at the Ser202/Thr205/Ser208 sites, together with absence of phosphorylation at the Ser262 site, yields a Tau sample that readily forms fibers, as observed by thioflavin T fluorescence and electron microscopy. On the basis of conformational analysis of synthetic phosphorylated peptides, we show that aggregation of the samples correlates with destabilization of the turn-like structure defined by phosphorylation of Ser202/Thr205.
The brains of patients with Alzheimer’s disease (AD) are characterized at the molecular level by two different lesions: extraneuronal amyloid plaques and intraneuronal tangles (1). Whereas the former are composed of the Aβ40/Aβ42 peptide (2), the tangles contain fibrillary structures mainly composed of the microtubule-associated Tau protein (3–5). Early on, it was found that the Tau molecules in these fibrillary structures are phosphorylated at distinct sites and to elevated levels, of which neither is found in a healthy brain (6). These observations led to the term of hyperphosphorylated Tau, with a characteristic gel shift in SDS/PAGE (7, 8). The latter shift can, however, also be caused by a single phosphorylation event (9), so a precise definition of hyperphosphorylation in terms of sites and stoichiometry is still lacking. Phosphorylation of Tau gained direct clinical importance when the traditional assay to determine postmortem the progression of AD (based on Gallyas silver staining of the amyloid structures in the brain) was replaced by an immunochemistry assay with the AT8 antibody (10). The epitope of this antibody was mapped for more than 20 y to a doubly phosphorylated peptide of Tau centered at positions Ser202/Thr205 (following the numbering of the longest 2N4R adult isoform, Tau441) (11–13). A combined NMR and molecular dynamics study showed that this phosphopeptide adopts a particular turn-like structure in solution (14). Only very recently has the crystal structure of its fragment antigen body (Fab) with different phosphorylated peptides suggested that a third phosphorylation event at position Ser208 might lead to an epitope with an even better affinity (15). However, whether phosphorylation is a driver of aggregation remains unclear, and even more so the precise definition of the phosphopattern or phosphopatterns that can cause the aggregation of Tau.
Although earlier work with recombinant Tau phosphorylated by a rat brain extract suggested this posttranslational modification (PTM) is sufficient to promote fiber formation (16), the Tau aggregation inhibitor methylene blue was discovered in an assay independent of this PTM (17, 18). Its clinical failure underscores that further knowledge of the aggregation process is still required. More recently, significant efforts to discover Tau aggregation inhibitors have relied on the discovery that poly-anions such as heparin promote the assembly of nonphosphorylated recombinant Tau (19, 20), leading to fibrillary structures with the same morphology as the paired helical filaments (PHFs) observed in AD brains (21). While seeding aggregation with AD brain-derived PHFs has been reported in vitro for recombinant Tau (22, 23), as well as in vivo (24–26), it still does not answer whether phosphorylation can drive the initial nucleation of fibers.
We show here that disrupting the turn-like structure of the pSer202/pThr205-centered Tau peptide (14) by the substitution of Gly207 to a Val induces fiber formation for a fragment of Tau after phosphorylation by activated ERK2 kinase. This mutation never having been described for patients, we then use the kinase activity of rat brain extract to phosphorylate not only the Ser202/Thr205 sites but equally the Ser208 site in the native sequence. Because this kinase activity also phosphorylates Ser262, which was reported to interfere with Tau aggregation (27), we mutate this Ser to an Ala and show that the resulting phosphorylation pattern induces aggregation of a full-length Tau sample without the addition of any exogenous poly-anion. Our present work hence confirms that phosphorylation can induce Tau aggregation, defines the precise pattern of phosphorylation that leads to aggregation, and links it to disruption of the turn-like structure induced by phosphorylation of Ser202/Thr205. It thereby provides a framework to study potential aggregation inhibitors in the context of a well-defined phosphorylated Tau species.
Results
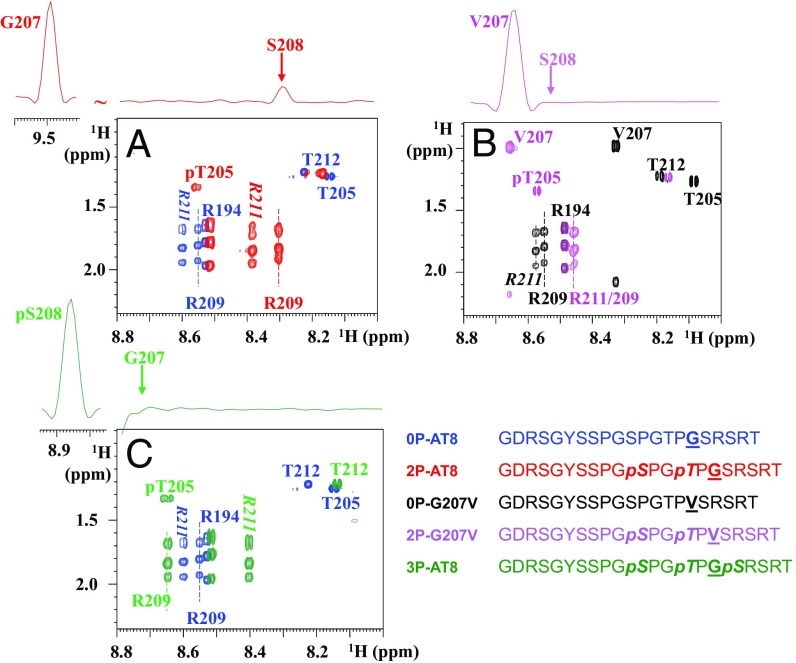
The Gly207Val Substitution Breaks the 2P-AT8 Turn-Like Structure in a Peptide Model.
A combined NMR and molecular dynamics study on a doubly phosphorylated Tau peptide centered on the AT8 epitope (2P-AT8) showed that as a result of phosphorylation of Ser202/Thr205, a hydrogen bond forms over the ring of the Pro206 between the pThr205 phosphate side-chain and the amide proton of Gly207 (14) (SI Appendix, Fig. S1). As a result, the amide proton of Gly207 experiences a downfield chemical shift from 8.4 to 9.5 ppm (14) (SI Appendix, Figs. S1 and S2A). Although Arg209 and Arg211 further stabilize this helical turn motif, molecular dynamics simulations showed that conformational heterogeneity is still possible for this site (14). Phosphorylation of threonine in two identical Thr-Pro-Gly motifs of the initially intrinsically disordered 4E-BP2 protein also induces similar turns and leads to the folding of 4E-BP2 into a stable four-stranded β-sheet (28) (SI Appendix, Fig. S1). In 4E-BP2, these two phosphorylation-induced turn motifs are completely stable in the resulting β-sheet, as witnessed by their glycine amide protons shifting to ∼11 ppm after phosphorylation (28). When valines were substituted for the glycines in both 4E-BP2 Thr-Pro-Gly motifs, they were found to disrupt hairpin formation through steric hindrance, and even prevent folding of 4E-BP2 after phosphorylation (28).
In light of these observations, we first substituted the Gly207 by a Val in the 2P-AT8 peptide model to see whether the helical turn in Tau would be disrupted by this mutation, and compared the NMR spectra of both peptides (Fig. 1 A and B and SI Appendix, Figs. S2 and S3). The amide proton of Val207 in the 2P-G207V peptide shifts a mere 0.3 ppm on phosphorylation of Ser202/Thr205 (SI Appendix, Fig. S3A), far from the 0.9-ppm shift experienced by the Gly207 in the native Tau sequence (SI Appendix, Fig. S2A). The absence of the turn in the 2P-G207V peptide is evident by the absence of an NOE contact between the amide protons of Val207 and Ser208 (Fig. 1B and SI Appendix, Fig. S3B). Whereas the Arg209 amide proton shifts 0.3 ppm upfield on phosphorylation of Thr205 in the 2P-AT8 peptide (SI Appendix, Fig. S2A), phosphorylation of Ser202/Thr205 in this Gly207Val-modified peptide causes only a minor 0.1-ppm shift of the Arg209 amide proton (SI Appendix, Fig. S3A). The Gly-to-Val substitution at position 207 hence destabilizes the turn induced by phosphorylation of Ser202/Thr205 and decouples effectively the down- and upstream residues of Pro206.
Fig. 1.
Breaking the turn-like structure by the Gly207Val mutation or by phosphorylation of Ser208. Zooms of the homonuclear TOCSY spectra of the nonmodified and phosphorylated peptides centered on the Thr205 and Arg209/211 residues. (Upper) The 1D traces are extracted from the corresponding NOESY spectra on the phosphorylated peptides. (A) Phosphorylation of the WT peptide at Ser202/Thr205, as in the 2P-AT8 peptide, leads to an important shift for the Arg209 amide proton and is accompanied by an NOE contact between the amide protons of Gly207 and Ser208, indicative of the phosphorylation-induced turn (14). (B) When Gly207 is substituted for a Val, the absence of an NOE contact in the 2P-G207V peptide between the amide protons of Val207 and Ser208 and the reduced shift for Arg209 indicate a destabilization of the turn-like structure. (C) Additional phosphorylation of Ser208 in the 3P-AT8 peptide has a similar effect as the Gly-to-Val substitution at position 207.
The Gly207Val Mutation in a Tau Fragment Promotes Phosphorylation-Induced Aggregation.
To ascertain that the same substitution also destabilizes the turn-like conformation in a larger Tau construct, we extended the functional TauF4 fragment (Tau 208–324) (29, 30) by 16 residues so as to start at the same Gly192 residue as the above-described AT8 peptide models (SI Appendix, Fig. S4). Incubation with the recombinant activated ERK2 kinase led to the p-TauF8 sample, incorporating phosphates at many sites, similar to what we observed previously for full-length Tau (31) (SI Appendix, Fig. S5). NMR assignment and quantification of p-TauF8 showed in particular that both Ser202 and Thr205 were completely phosphorylated, with concomitant induction of the turn-like structure, as validated by the downfield shift of the Gly207 amide proton, as seen in the peptide model (SI Appendix, Fig. S5). Mutating Gly207 to a Val in the same TauF8 fragment and subsequent phosphorylation by the activated ERK2 kinase led to a comparable phosphorylation pattern, as assessed by NMR (SI Appendix, Fig. S6). However, Val207 in the spectrum of the phosphorylated TauF8-G207V fragment does not shift significantly, but only to a position equivalent to that in the peptide model. We equally observed the absence of a significant chemical shift change for Arg209 in the p-TauF8-G207V spectrum (SI Appendix, Fig. S6). The effective decoupling of the residues N and C terminal to the central Pro206 due to a Val at position 207 hence is also observed in the Erk-phosphorylated TauF8 fragment.
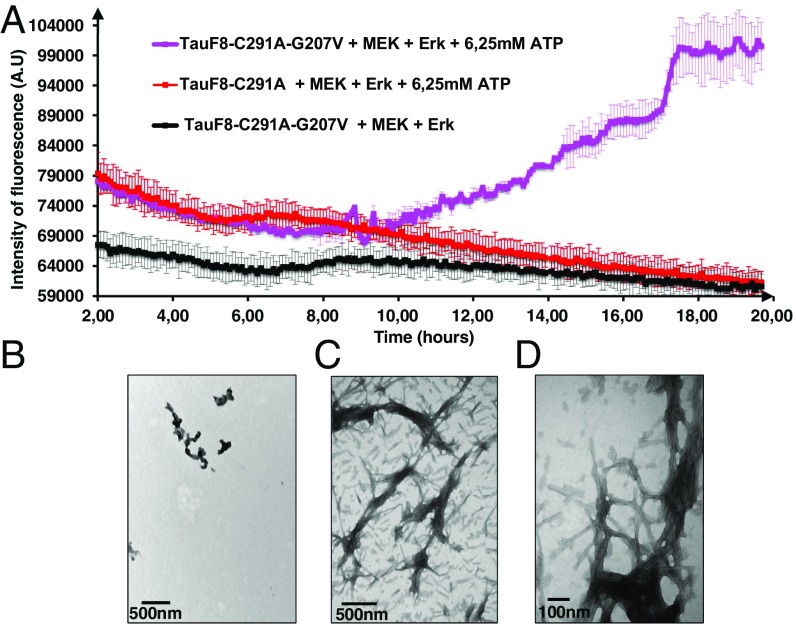
During the 20-h incubation to phosphorylate the TauF8-G207V fragment with the activated ERK2 kinase, a sizeable amount of aggregated material visible to the naked eye appeared in the reaction mixture. To test whether disrupting the turn-like structure leads to amyloid-like structures, we set up a thioflavin T (ThT) assay (32) during the phosphorylation reaction. TauF8 and TauF8-G207V fragments were incubated with the recombinant MEK3 and ERK2 kinases, and aggregation on phosphorylation was monitored in situ by ThT fluorescence emission at 490 nm. An increase of ThT signal was observed after a lag time of 10 h only for the TauF8-G207V fragment (Fig. 2A), confirming aggregation takes place during the overnight TauF8-G207V phosphorylation process. Electron microscopy confirmed the presence of many bona fide fibrils for the phosphorylated TauF8-G207V fragment (Figs. 2 B and C). In agreement with our previous results on full-length Tau (31), no ThT response could be detected during the phosphorylation of the TauF8 sample, and further analysis by electron microscopy of this sample showed very few structures after 20 h of incubation (Fig. 2D). When combined with ERK2 catalyzed phosphorylation, the turn-like disrupting G207V mutation in TauF8 hence leads to fast aggregation that already occurs during the phosphorylation reaction.
Fig. 2.
Destabilizing the turn by the G207V mutation combined with ERK2-mediated phosphorylation leads to aggregation of the TauF8 fragment. (A) Aggregation of TauF8 (without external inducers) during the phosphorylation reaction, as followed by ThT emission at 490 nm. An increase in ThT emission is observed only for the G207V mutant. Error bars correspond to three independent aggregation experiments, with new batches of TauF8, TauF8-G207V, MEK, and ERK2 kinases. (B–D) TEM at the end point of the phosphorylation reactions shows a large amount of fibrils after phosphorylation of the TauF8-G207V mutant (B and C), but only a smattering of fibril-like structures for the wt TauF8 (D).
Triple Phosphorylation at the AT8 Epitope Equally Breaks the Turn and Promotes Tau Aggregation.
Because no G207 Tau mutation has ever been described, we wondered whether a PTM might destabilize the turn-like structure in a similar manner. An obvious candidate is the phosphorylation of the nearby Ser208, recently added to the AT8 epitope on top of the phosphorylated Ser202/Thr205 residues (15). The crystal structure of a triply phosphorylated Tau peptide in complex with the AT8 Fab shows the peptide in an extended conformation compatible with a model whereby it would interfere with the turn-like structure in a similar way as the G207V mutation (SI Appendix, Fig. S7). To characterize the potential structural changes induced by the simultaneous phosphorylation of Ser208 and Ser202/Thr205, we synthesized the triply phosphorylated peptide (3P-AT8) and analyzed its conformation by NMR spectroscopy (Fig. 1C and SI Appendix, Fig. S8). The most obvious change induced by the additional phosphorylation of Ser208 is the amide proton shift of Gly207 that returns from 9.5 ppm in the doubly phosphorylated peptide (2P-AT8; SI Appendix, Fig. S2) to 8.7 ppm when Ser208 is additionally phosphorylated in the 3P-AT8 peptide (SI Appendix, Fig. S8). Further evidence for disruption of the turn-like structure in this 3P-AT8 peptide comes from the absence of a Gly207-pSer208 HN-HN NOE contact (Fig. 1C). Additional phosphorylation at position 208 hence prevents the turn-like structure in a manner similar to the Gly-to-Val mutation at position 207.
We have previously confirmed that ERK2, as a proline-directed kinase, does not phosphorylate the Ser208 residue (31). However, the kinase activity of a rat brain extract (RBE) in the presence of serine/threonine phosphatase inhibitor okadaic acid can lead to phosphorylation of the Ser208 site, but also phosphorylates Ser262 (31) As phosphorylation of Ser262 has been described as inhibitory for Tau aggregation (27), we mutated Ser262 to alanine in the TauF8 fragment and phosphorylated the resulting TauF8-S262A sample by the RBE. After only 5 h of incubation with RBE, we observed a quasi-total loss of the soluble fraction and the formation of a pellet composed of aggregates and fibers (SI Appendix, Fig. S9).
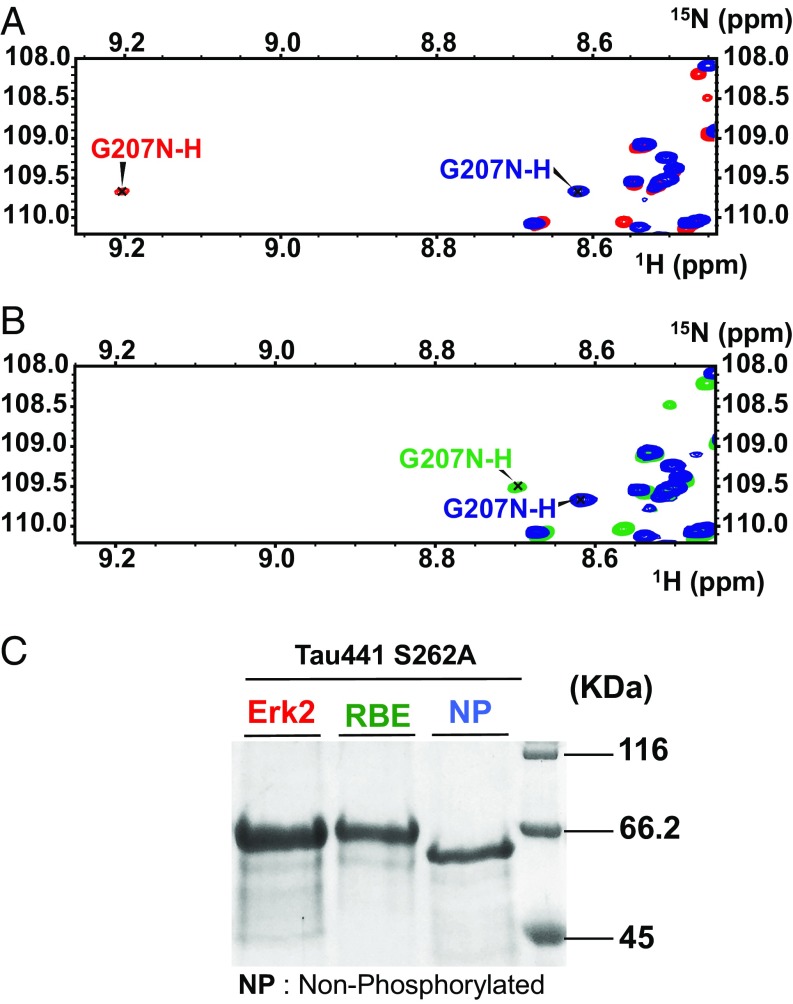
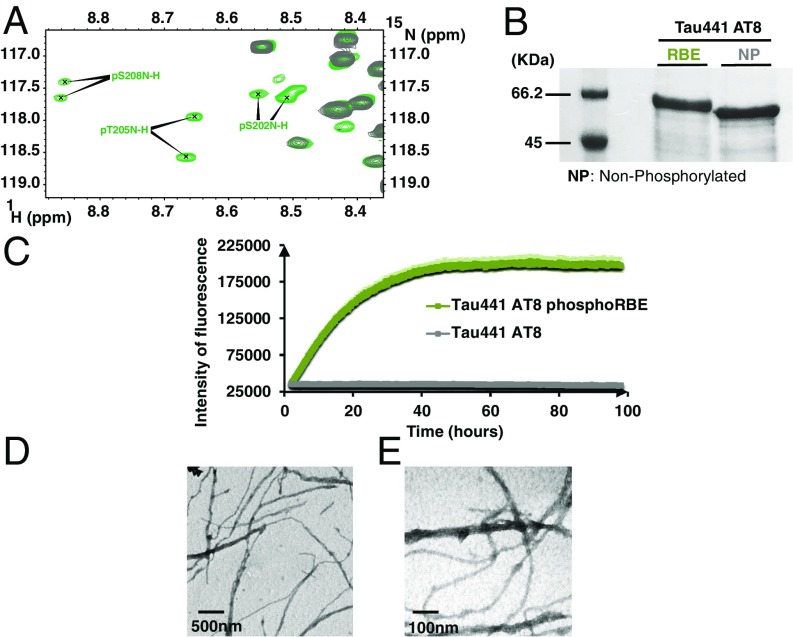
Fragments of Tau devoid of their N or C termini tend to aggregate faster than full-length Tau (33, 34), leading us to consider the effect of the phosphorylation of Ser208 in the context of Tau441. However, when previously using the RBE kinase activity with Tau441 as a substrate, we did not obtain significant amounts of protein aggregates (31). With hindsight, and suspecting the inhibitory role of Ser262 phosphorylation not only in a heparin-derived assay (27) but equally when phosphorylation is the driver, we produced both WT and the S262A mutant of Tau441 (Tau441-S262A). On independent incubation with the activated ERK2 kinase or with the RBE, none of the full-length Tau samples aggregated to a significant extent during the phosphorylation reaction. Closer NMR analysis of the resulting phosphorylation patterns showed that Thr205 is fully phosphorylated both by the activated ERK2 kinase (SI Appendix, Fig. S10) and the kinase activity present in RBE (SI Appendix, Fig. S11), but Ser208 is exclusively phosphorylated by kinases of the RBE (SI Appendix, Fig. S11). The similar gel shift of both phosphorylated samples in an SDS/PAGE analysis (Fig. 3C) shows the superior analytical power of NMR spectroscopy in deciphering complex PTM patterns. Similar to the peptide models, the downfield shift of the Gly207 amide proton resonance observed in the ERK2-phosphorylated sample (Fig. 3A) is absent after phosphorylation with RBE (Fig. 3B). On the basis of a 3D NMR experiment on a RBE phosphorylated 15N,13C -labeled sample, we identified its resonance for Gly207 in the crowded region where other glycines are found, with an amide proton chemical shift identical to that found in the 3P-AT8 peptide (Fig. 3B).
Fig. 3.
Phosphorylation of Ser208 by RBE, but not ERK2, kinase activity promotes aggregation of Tau441-S262A. (A and B) Glycine region from the 1H-15N HSQC spectra of Tau441-S262A (blue), Tau441-S262A phosphorylated by ERK2 (red), and Tau441-S262A phosphorylated by RBE (green). The downfield shift for the G207 amide proton resonance observed in the ERK2-phosphorylated sample is absent after phosphorylation with RBE. (C) SDS/PAGE analysis of Tau441-S262A before and after phosphorylation by ERK2 and RBE. By comparison with the native protein (lane NP), a similar gel shift is observed for the two phosphorylated samples. The pS208 and pS356 sites phosphorylated solely by the RBE cannot be distinguished by SDS/PAGE.
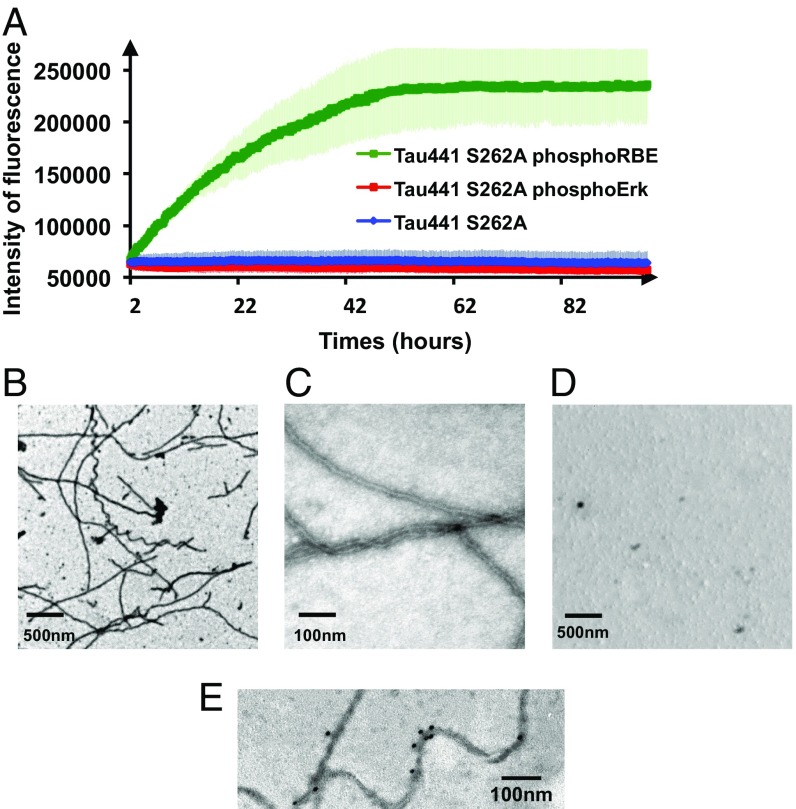
Setting up a ThT aggregation assay (32) with the phosphorylated samples in the absence of any other inducer, we obtained a sizeable fluorescence signal for RBE phosphorylated Tau441-S262A protein (Fig. 4A). The ERK2-phosphorylated Tau441-S262A sample (without phosphorylated Ser208, and hence with the Ser202/Thr205 phosphorylation-induced helical turn) gave no ThT signal in the same conditions (Fig. 4A). In agreement with this result, the electron microscopy images showed plenty of fibers for the RBE phosphorylated sample (Fig. 4 B and C), whereas very few fibers were detected in the phospho-ERK sample (Fig. 4D). Further morphological characterization of the resulting fibers was performed by immunohistochemistry and NMR spectroscopy. As for AD patient brain–derived fibers, our synthetic phospho-Tau fibers were stained by the AT8 antibody (Fig. 4E). The flexible nature of the N and C termini of Tau in the resulting fibers was further confirmed by the presence of NMR signal after aggregation for residues in these regions (35, 36), whereas the rigid core was further confirmed by disappearance of the signals of the core region and notably the PHF6 peptide (SI Appendix, Fig. S12).
Fig. 4.
Additional phosphorylation of Ser208 by RBE promotes aggregation of Tau441-Ser262A (A) Aggregation of Tau441-S262A (blue), Tau441-S262A phosphorylated by ERK2 (red), and Tau441-S262A phosphorylated by RBE (green) followed by ThT emission at 490 nm. An increase in ThT emission at 490 nm is observed only for the 3P-AT8 Tau441-S262A protein. Error bars correspond to three independent aggregation experiments, with new batches of proteins and different RBEs. (B–D) TEM images at the end point of the aggregation assay of Tau441-S262A phosphorylated by RBE (B and C) or by Erk (D) confirm the results obtained in the aggregation assay. Large amounts of fibrils are observed only for Tau441-S262A phosphorylated by RBE. (E) Immunogold electron microscopy of the fibers obtained with Tau441-S262A phosphorylated by RBE. AT8 indeed stains the fibers in a similar manner as AD brain-derived fibers.
Confirmation of the inhibitory role of Ser262 phosphorylation came from the fluorescence curve of RBE phosphorylated Tau441. The phosphorylation patterns of the Tau441 and Tau441-S262A samples after incubation with RBE were almost identical, except for Ser262, which was only phosphorylated to 70% in the wt Tau441 sample (SI Appendix, Fig. S13). Concomitantly, we did observe a weaker ThT signal than for Tau441-S262A (SI Appendix, Fig. S14), roughly corresponding to the 30% fraction of Tau441 that is not phosphorylated at the Ser262 position.
Triple Phosphorylation at the AT8 Epitope Is Sufficient to Promote Tau Aggregation.
RBE kinases phosphorylate many residues outside the abovementioned 3P-AT8 motif (SI Appendix, Fig. S13). To investigate whether these additional sites also contribute to the aggregation process, we made a mutant whereby all previously identified phosphorylation sites by the RBE kinase activity except for Ser202/Thr205/Ser208 were mutated into alanine residues (SI Appendix, Fig. S15). Phosphorylation of this Tau441 AT8 protein by RBE leads to a simpler spectrum than for the phosphorylated Tau441-Ser262A protein, albeit with several resonances for each phospho-site because of incomplete phosphorylation (37) (Fig. 5A). Even with fewer phosphorylation sites, a gel shift on phosphorylation is still observable on SDS/PAGE (Fig. 5B). The ThT assay with the RBE phosphorylated Tau441 AT8 protein (Fig. 5C) reached similar levels as the RBE-phosphorylated Tau441-Ser262A sample (Fig. 4A), and electron microscopy again confirmed the presence of many fibers (Fig. 5 D and E). The sole 3P-AT8 motif hence is capable of driving the aggregation of Tau.
Fig. 5.
The 3P-AT8 motif alone is sufficient to drive aggregation. (A) Region of the 1H-15N HSQC spectrum of RBE phosphorylated Tau441-AT8. Because of incomplete phosphorylation, two resonances for each phospho-site are identified. (B) SDS/PAGE analysis of Tau441-AT8 before and after phosphorylation by RBE. Whereas a gel shift is still visible on phosphorylation, it is less pronounced in this mutant, where most phosphorylation sites have been removed. (C) Aggregation of RBE phosphorylated Tau441-AT8 as followed by ThT emission at 490 nm confirms phosphorylation of the Ser202/Thr205/Ser208 epitope is sufficient to trigger aggregation. (D and E) TEM images at the end point of the aggregation reveal a network of fibers when RBE-phosphorylated Tau-AT8 is allowed to aggregate for 4 d. Error bars correspond to two independent experiments with the same Tau441-AT8 batch, but with different RBEs.
Discussion
Phosphorylation was identified early on as a defining PTM of those Tau molecules that make up the intraneuronal tangles that characterize AD patient brains (6). In analogy with the situation of unmodified Tau, however, where in vitro production of fibrillary structures with full-length Tau proved difficult (38), obtaining AD-like fibers with phosphorylated Tau has not been an easy task. One study phosphorylated Tau with a RBE to 12–15 phosphates per Tau molecule and obtained fibers with the macroscopic aspect of those isolated from AD patient brains (16). However, no precise definition of the phosphorylation pattern generated by the RBE was determined, and attempts to repeat this experiment did not yield any appreciable amount of fibers (31, 39).
The analytical challenge of determining a precise phosphorylation pattern of Tau is significant, mostly because of its large number of potential phosphorylation sites and their clustering in regions such as the proline-rich region or the extreme C terminus. Recent progress in mass spectrometry has improved the coverage of Tau’s possible PTMs (40, 41), but antibodies remain the workhorse in a clinical setting. Notably, Braak staging, whereby the cognitive decline of deceased patients is correlated with the extent of the lesions in their brain, has been standardized on the basis of immunochemistry with the AT8 antibody (10). The recent finding that the optimal epitope of this clinical antibody is not limited to the double phosphorylation at Ser202/Thr205 of Tau but also can include the modification of the Ser208 residue was quite unexpected (15). Whereas the former 2P-AT8 epitope is characterized by a turn-like structure (14), we show here that this structure can be disrupted either by mutating the Gly207 to Val or by phosphorylation of the Ser208 (Fig. 1). Our finding that the resulting Tau species with phosphorylation at Ser202/Th205, but with a disrupted turn-like structure, forms abundant fibers detectable by thioflavin fluorescence or electron microscopy (Figs. 2 and 4) suggests the initial turn-like structure induced by the phosphorylation of only Ser202 and Thr205 is protective against aggregation. A protective role for the phosphorylation of Tau at the Thr205 position was equally proposed for the synaptic pool of Tau, not in the aggregation process but, rather, in the attenuation of the Aβ oligomer-induced toxicity (42). Importantly, here, our resulting fibers show several characteristics of the natural fibers that fill the neurons of patients with AD, such as robust AT8 staining (Fig. 4), a core region composed of the microtubule-binding repeats of Tau characterized by disappearing NMR signals, as well as a fuzzy coat around this core with residual NMR intensity (43) (SI Appendix, Fig. S12). Some morphological differences exist between the twisted fibrils observed with RBE-phosphorylated Tau441-S262A (Fig. 4) and the straight and less well-defined fibers obtained after aggregation of Tau441-AT8 (Fig. 5). These transmission electron microscopy (TEM) observations suggest a link between the exact phosphorylation patterns and the morphology of fibrils that might be relevant for the different tauopathies. Phosphorylation at Ser208 might be catalyzed by Casein kinase 1 (44), and its identification as a potential site for O-GlcNacylation (45) points to the important role of this residue. The exact mechanism of the aggregation process is not clear at this moment, but is subject to intense study by the combined approach of synthetic phosphorylated peptides, in vitro kinase assays, and NMR spectroscopy we have used here to define the aggregation-inducing phosphorylation pattern of Tau.
Our results indicate “hyperphosphorylation” as a driving force for Tau aggregation and describe this widely used expression in terms of sites and occupancy. Indeed, Tau phosphorylation at the three positions, Ser202/Thr205/Ser208, while not at Ser262, is sufficient to induce aggregation without the addition of any exogenous aggregation inducer. The former three sites correspond to the AT8 epitope, as recently redefined on the basis of the AT8 Fab crystal structure in complex with the triply phosphorylated peptide (15), and hence suggest the antibody, clinically used to stage the disease progression (10), indeed immunostains a Tau species that can aggregate without any other factors. Whereas the quest for aggregation inhibitors has been mainly performed with heparin as an external aggregation inducer (46–49), our present description of a phosphorylation-driven Tau aggregation assay opens up new avenues and should allow retesting of the previously defined classes of molecules or the discovery of potentially more specific molecules that counteract the aggregation of phosphorylated Tau.
Materials and Methods
Peptide Synthesis and Purification.
Peptides were synthesized manually, using standard Fmoc-SPPS protocols, in a fritted syringe on a 0.1-mmol scale, using Rink amide resin (0.4–0.7 mmol⋅g−1). Coupling was carried out using the requisite Fmoc protected amino acids (10 eq.), O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU; 9.5 eq.), and N,N-diisopropylethylamine (DIPEA; 20 eq.) with shaking for 1 h. Phosphorylated amino acids were double coupled (2 × 3 h), using protected phospho-amino acids (5 eq.), N-[(dimethylamino)-1H-1,2,3-triazolo-[4,5-b]pyridin-1-ylmethylene]-N-methylmethanaminium hexafluorophosphate N-oxide (HATU) (4.9 eq.), and DIPEA (15 eq.).
Peptide Purification/Analysis.
RP-HPLC was performed using a Waters 1525 binary pump system with a Waters 2487 dual-wavelength absorbance detector and UV detection at 220 nm. Solvent A was 0.1% TFA in water, and solvent B was 0.1% TFA in CH3CN. Analytical RP-HPLC was performed using a Higgins Analytical Proto200 C18 3 μM column (4.6 × 100 mm), at a flow rate of 1.0 mL⋅min−1 with a gradient of 5–20% B in 10 min. Preparative RP-HPLC was performed using an Xbridge C18 5 μM (19 × 50 mm) column with a flow rate of 10 mL⋅min−1, using the appropriate gradient. MALDI-TOF mass spectrometry analyses were performed in the ion-positive reflector mode on an ABI Voyager DE-Pro MALDI-TOF mass spectrometer (Applied Biosystems), using as matrix a saturated solution of α-cyano-4-hydroxycinnamic acid in CH3CN:H2O:CF3COOH (50:50:0.1).
Production of 15N- and 13C-Labeled Tau441 and TauF8 Fragment in Escherichia coli.
The Tau441 (residues 1–441 of human MAPT) and TauF8 fragments (residues 192–324) in their WT or mutant forms (G207V or S262A) were produced in the Escherichia coli BL21 DE3 strain carrying the pET15b recombinant plasmid (Novagen). Cells were grown at 37 °C in M9 minimal medium containing 2 g/L 13C6-glucose, 1 g/L 15N-ammonium chloride, 0.5 g/L 15N,13C-Isogro (Sigma), 1 mM MgSO4, 100 μM CaCl2, MEM vitamins mixture (Sigma), and ampicillin (100 mg/L). When the OD at 600 nm reached 0.8, the induction phase was initiated by the addition of 0.5 mM IPTG and continued for 3 h at 37 °C. Cells were harvested by centrifugation at 4,000 × g for 25 min, and the pellet was resuspended in 50 mM NaH2PO4/Na2HPO4 (pH 6.2), 2.5 mM EDTA, 2 mM DTT, and 0.5% triton ×100 complemented with a protease mixture inhibitor (Complete, Roche). The lysate was obtained by homogenization of this suspension with a high-pressure homogenizer (Emulsiflex, Avestin), followed by centrifugation at 30,000 × g for 30 min. The soluble extract was incubated at 75 °C for 15 min. The soluble proteins were isolated by centrifugation at 30,000 × g for 30 min and purified by cation exchange chromatography (HiTrap SP HP 1 mL; GE Healthcare). Lyophilized proteins were kept at −20 °C until further use.
In Vitro Phosphorylation of Tau and TauF8 Fragments by ERK2.
Next, 100 μM 15N,13C Tau or TauF8 (WT or mutant forms) was mixed with 1 μM ERK2 and about 0.1 μM MEK R4F (MEK3), the constitutively active mutated form of MEK3. The mixture was incubated at 37 °C overnight in 400 μL phosphorylation buffer (50 mM Hepes KOH at pH 8.0, 12.5 mM MgCl2, 50 mM NaCl, 2 mM EDTA, 1 mM DTT, 1 mM EGTA, 12.5 mM ATP). Enzymatic reaction was stopped by heating the reaction mixture at 75 °C for 15 min, followed by centrifugation at 16,000 × g for 20 min. Then, the supernatant was buffer-exchanged in 50 mM ammonium bicarbonate before lyophilization. Before further analyses, a gel shift of protein band on SDS/PAGE (T = 10% for p-Tau441 and T = 12% for p-TauF8) allowed a qualitative control of protein phosphorylation. A control experiment was performed under the same conditions in which ERK2 was omitted. Each set of phosphorylation experiments was independently repeated three times with new batches of Tau proteins and kinases.
In Vitro Phosphorylation of Tau and TauF8 Fragments by RBE.
RBE was prepared from adult rat Sprague-Dawley rats by homogenizing a brain (about 2 g) in 5 mL homogenizing buffer [10 mM Tris at pH 7.4, 5 mM EGTA, 2 mM DTT, 1 μM okadaic acid (Sigma)] supplemented with 20 μg/mL leupeptin and 40 mM Pefabloc (Sigma-Aldrich). Ultracentrifugation was performed at 100,000 × g at 4 °C for 1 h. The supernatant was directly used for its kinase activity. Total protein concentration was estimated at 11 mg/mL by BCA colorimetric assay (Pierce). The 15N,13C Tau and TauF8 fragments, in their WT or mutant forms, were dissolved, respectively, at 10 and 25 μM in 2.5 mL phosphorylation buffer (40 mM Hepes at pH 7.3, 2 mM MgCl2, 5 mM EGTA, 2 mM DTT, 2 mM ATP, and 1 μM okadaic acid) complemented with a protease inhibitor mixture (Complete, Roche). The phosphorylation reaction was performed at 37 °C for 24 h with 500 μL brain extract. Enzymatic reaction was stopped by heating the mixture at 75 °C for 15 min and centrifugation at 16,000 × g for 20 min. Then, the supernatant was buffer-exchanged in 50 mM ammonium bicarbonate before lyophilization. Qualitative control of protein phosphorylation was performed by SDS/PAGE before NMR analyses and aggregation assays. Each set of phosphorylation experiment was independently repeated three times with new batches of Tau proteins and fresh RBE.
NMR Spectroscopy.
NMR of peptides was performed on 800- or 900-MHz Bruker spectrometers equipped with a triple resonance cryogenic probe head. Homonuclear nuclear Overhauser (NOESY) and total correlation (TOCSY) spectra were acquired at 293 K with standard pulse programs on 2-mM peptide samples in a phosphate buffer (50 mM phosphate buffer at pH 6.4, 25 mM NaCl, 2.5 mM EDTA, and 5% D2O). 1H,15N heteronuclear single quantum coherence (HSQC) spectra were acquired at natural abundance, with 128 scans per increment and 2,048 × 256 complex points in the direct and indirect dimensions, respectively. For protein NMR experiments, Tau proteins were dissolved at a concentration of 200–300 μM in NMR sample buffer (50 mM phosphate buffer at pH 6.4, 25 mM NaCl, 2.5 mM EDTA, 1 mM DTT, and 10% D2O). To estimate phosphorylation levels at each site, the ratio of the peak integral corresponding to phosphorylated forms over the sum of peak integrals corresponding to nonphosphorylated and phosphorylated forms were calculated.
In Vitro Aggregation Assay and TEM.
Tau aggregation assays were performed at 10 μM Tau441 WT or S262A phosphorylated by RBE in aggregation buffer (100 mM Mes buffer at pH 6.9, 2 mM EGTA, 1 mM MgCl2, 0.33 mM DTT, and 50 μM ThT). Lyophilized proteins were resuspended in the aggregation buffer at the desired concentration, and 50,μM ThT was added. Aggregation kinetics were followed by ThT emission at 490 nm, using a plate reader (PHERAStar, BMG Labtech). In parallel, aggregation experiments were performed under the same conditions in 1.5-mL tubes. Mixtures were incubated for 5 d at 37 °C without agitation. At the end of incubation, 10 μL samples from 1.5-mL tubes and plates were withdrawn and applied on 400-mesh hexagonal formvar-coated grids for 90 s. The sample-loaded grids were washed three times with ultrapure water and drained. The grids were next negatively stained with 2% uranyl-acetate solution for 90 s and washed two times with ultrapure water. TEM was performed with a HITACHI H7500 microscope at 80 kV.
Immunogold TEM.
For immunogold labeling of the fibrils, 10 μL of the aggregation sample was adsorbed on carbon-coated EM grids (Electron Microscopy Science) for 20 min. Excess fibrils were removed by placing the EM grids three times on drops of fresh PBS buffer without drying. After blocking in 1% (wt/vol) BSA in PBS for 60 min, grids were incubated with primary anti-Tau antibody (Human PHF-TAU clone AT8) diluted 1:100 in 1% BSA for 1 h. After further washing the grids five times in PBS, they were incubated with the gold-labeled secondary antibody [6 nm-Glod donkey IGG anti-mouseIgG] diluted 1:20 in 1% BSA for 60 min, followed by five washes in PBS. Before staining, the sample was washed 10 times in water to avoid precipitation of uranyl-acetate in PBS. The sample was negatively stained by incubation in 2% (wt/vol) uranyl-acetate in water for 8 min, followed by four washing/drying steps in water. TEM was performed with a HITACHI H7500 microscope at 80 kV.
Supplementary Material
Acknowledgments
The NMR facilities were funded by the Région Hauts-de-France, CNRS, Pasteur Institute of Lille, European Community [Fonds européen de développement régional (FEDER)], French Research Ministry, and the University of Lille. We acknowledge support from the TGE RMN THC (Très Grands Equipements Résonance Magnétique Nucléaire Très Hauts Champs, FR-3050, France), FRABio (Fédération de Recherche “Biochimie Structurale & Fonctionnelle des Assemblages Biomoléculaires,” FR 3688, France), and from MetaToul (Toulouse metabolomics & fluxomics facilities, www.metatoul.fr), which is part of the French National Infrastructure for Metabolomics and Fluxomics MetaboHUB-AR-11-INBS-0010 (www.metabohub.fr). Our research is supported by grants from the LabEx (Laboratory of Excellence) DISTALZ (Development of Innovative Strategies for a Transdisciplinary approach to Alzheimer's disease).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708448114/-/DCSupplemental.
References
- 1.Goedert M, Spillantini MG. A century of Alzheimer’s disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 2.Glenner GG, Wong CW. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 3.Kosik KS, Joachim CL, Selkoe DJ. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc Natl Acad Sci USA. 1986;83:4044–4048. doi: 10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundke-Iqbal I, et al. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- 5.Wood JG, Mirra SS, Pollock NJ, Binder LI. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau (tau) Proc Natl Acad Sci USA. 1986;83:4040–4043. doi: 10.1073/pnas.83.11.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundke-Iqbal I, et al. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci USA. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindwall G, Cole RD. The purification of tau protein and the occurrence of two phosphorylation states of tau in brain. J Biol Chem. 1984;259:12241–12245. [PubMed] [Google Scholar]
- 8.Baudier J, Cole RD. Phosphorylation of tau proteins to a state like that in Alzheimer’s brain is catalyzed by a calcium/calmodulin-dependent kinase and modulated by phospholipids. J Biol Chem. 1987;262:17577–17583. [PubMed] [Google Scholar]
- 9.Godemann R, Biernat J, Mandelkow E, Mandelkow EM. Phosphorylation of tau protein by recombinant GSK-3beta: Pronounced phosphorylation at select Ser/Thr-Pro motifs but no phosphorylation at Ser262 in the repeat domain. FEBS Lett. 1999;454:157–164. doi: 10.1016/s0014-5793(99)00741-3. [DOI] [PubMed] [Google Scholar]
- 10.Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006;112:389–404. doi: 10.1007/s00401-006-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biernat J, et al. The switch of tau protein to an Alzheimer-like state includes the phosphorylation of two serine-proline motifs upstream of the microtubule binding region. EMBO J. 1992;11:1593–1597. doi: 10.1002/j.1460-2075.1992.tb05204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goedert M, Jakes R, Vanmechelen E. Monoclonal antibody AT8 recognises tau protein phosphorylated at both serine 202 and threonine 205. Neurosci Lett. 1995;189:167–169. doi: 10.1016/0304-3940(95)11484-e. [DOI] [PubMed] [Google Scholar]
- 13.Porzig R, Singer D, Hoffmann R. Epitope mapping of mAbs AT8 and Tau5 directed against hyperphosphorylated regions of the human tau protein. Biochem Biophys Res Commun. 2007;358:644–649. doi: 10.1016/j.bbrc.2007.04.187. [DOI] [PubMed] [Google Scholar]
- 14.Gandhi NS, et al. A phosphorylation-induced turn defines the Alzheimer’s disease AT8 antibody epitope on the tau protein. Angew Chem Int Ed Engl. 2015;54:6819–6823. doi: 10.1002/anie.201501898. [DOI] [PubMed] [Google Scholar]
- 15.Malia TJ, et al. Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8. Proteins. 2016;84:427–434. doi: 10.1002/prot.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso A, Zaidi T, Novak M, Grundke-Iqbal I, Iqbal K. Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc Natl Acad Sci USA. 2001;98:6923–6928. doi: 10.1073/pnas.121119298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wischik CM, Edwards PC, Lai RY, Roth M, Harrington CR. Selective inhibition of Alzheimer disease-like tau aggregation by phenothiazines. Proc Natl Acad Sci USA. 1996;93:11213–11218. doi: 10.1073/pnas.93.20.11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wischik CM, Harrington CR, Storey JMD. Tau-aggregation inhibitor therapy for Alzheimer’s disease. Biochem Pharmacol. 2014;88:529–539. doi: 10.1016/j.bcp.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Goedert M, et al. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature. 1996;383:550–553. doi: 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- 20.Pérez M, Valpuesta JM, Medina M, Montejo de Garcini E, Avila J. Polymerization of tau into filaments in the presence of heparin: The minimal sequence required for tau-tau interaction. J Neurochem. 1996;67:1183–1190. doi: 10.1046/j.1471-4159.1996.67031183.x. [DOI] [PubMed] [Google Scholar]
- 21.Kidd M. Paired helical filaments in electron microscopy of Alzheimer’s disease. Nature. 1963;197:192–193. doi: 10.1038/197192b0. [DOI] [PubMed] [Google Scholar]
- 22.Morozova OA, March ZM, Robinson AS, Colby DW. Conformational features of tau fibrils from Alzheimer’s disease brain are faithfully propagated by unmodified recombinant protein. Biochemistry. 2013;52:6960–6967. doi: 10.1021/bi400866w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer V, Dinkel PD, Rickman Hager E, Margittai M. Amplification of Tau fibrils from minute quantities of seeds. Biochemistry. 2014;53:5804–5809. doi: 10.1021/bi501050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clavaguera F, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci USA. 2013;110:9535–9540. doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcon B, et al. Conformation determines the seeding potencies of native and recombinant Tau aggregates. J Biol Chem. 2015;290:1049–1065. doi: 10.1074/jbc.M114.589309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo JL, et al. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J Exp Med. 2016;213:2635–2654. doi: 10.1084/jem.20160833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow EM. Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry. 1999;38:3549–3558. doi: 10.1021/bi981874p. [DOI] [PubMed] [Google Scholar]
- 28.Bah A, et al. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 29.Fauquant C, et al. Systematic identification of tubulin-interacting fragments of the microtubule-associated protein Tau leads to a highly efficient promoter of microtubule assembly. J Biol Chem. 2011;286:33358–33368. doi: 10.1074/jbc.M111.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gigant B, et al. Mechanism of Tau-promoted microtubule assembly as probed by NMR spectroscopy. J Am Chem Soc. 2014;136:12615–12623. doi: 10.1021/ja504864m. [DOI] [PubMed] [Google Scholar]
- 31.Qi H, et al. Characterization of neuronal Tau protein as a target of extracellular signal-regulated kinase. J Biol Chem. 2016;291:7742–7753. doi: 10.1074/jbc.M115.700914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedhoff P, Schneider A, Mandelkow EM, Mandelkow E. Rapid assembly of Alzheimer-like paired helical filaments from microtubule-associated protein tau monitored by fluorescence in solution. Biochemistry. 1998;37:10223–10230. doi: 10.1021/bi980537d. [DOI] [PubMed] [Google Scholar]
- 33.Abraha A, et al. C-terminal inhibition of tau assembly in vitro and in Alzheimer’s disease. J Cell Sci. 2000;113:3737–3745. doi: 10.1242/jcs.113.21.3737. [DOI] [PubMed] [Google Scholar]
- 34.Gamblin TC, Berry RW, Binder LI. Tau polymerization: Role of the amino terminus. Biochemistry. 2003;42:2252–2257. doi: 10.1021/bi0272510. [DOI] [PubMed] [Google Scholar]
- 35.Sillen A, et al. Regions of tau implicated in the paired helical fragment core as defined by NMR. ChemBioChem. 2005;6:1849–1856. doi: 10.1002/cbic.200400452. [DOI] [PubMed] [Google Scholar]
- 36.Sillen A, et al. High-resolution magic angle spinning NMR of the neuronal tau protein integrated in Alzheimer’s-like paired helical fragments. J Am Chem Soc. 2005;127:10138–10139. doi: 10.1021/ja0516211. [DOI] [PubMed] [Google Scholar]
- 37.Theillet F-X, et al. Cell signaling, post-translational protein modifications and NMR spectroscopy. J Biomol NMR. 2012;54:217–236. doi: 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wille H, Drewes G, Biernat J, Mandelkow EM, Mandelkow E. Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J Cell Biol. 1992;118:573–584. doi: 10.1083/jcb.118.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tepper K, et al. Oligomer formation of tau protein hyperphosphorylated in cells. J Biol Chem. 2014;289:34389–34407. doi: 10.1074/jbc.M114.611368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris M, et al. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat Neurosci. 2015;18:1183–1189. doi: 10.1038/nn.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mair W, et al. FLEXITau: Quantifying post-translational modifications of Tau protein in vitro and in human disease. Anal Chem. 2016;88:3704–3714. doi: 10.1021/acs.analchem.5b04509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ittner A, et al. Site-specific phosphorylation of tau inhibits amyloid-β toxicity in Alzheimer’s mice. Science. 2016;354:904–908. doi: 10.1126/science.aah6205. [DOI] [PubMed] [Google Scholar]
- 43.Crowther T, Goedert M, Wischik CM. The repeat region of microtubule-associated protein tau forms part of the core of the paired helical filament of Alzheimer’s disease. Ann Med. 1989;21:127–132. doi: 10.3109/07853898909149199. [DOI] [PubMed] [Google Scholar]
- 44.Hanger DP, Anderton BH, Noble W. Tau phosphorylation: The therapeutic challenge for neurodegenerative disease. Trends Mol Med. 2009;15:112–119. doi: 10.1016/j.molmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Smet-Nocca C, et al. Identification of O-GlcNAc sites within peptides of the Tau protein and their impact on phosphorylation. Mol Biosyst. 2011;7:1420–1429. doi: 10.1039/c0mb00337a. [DOI] [PubMed] [Google Scholar]
- 46.Taniguchi S, et al. Inhibition of heparin-induced tau filament formation by phenothiazines, polyphenols, and porphyrins. J Biol Chem. 2005;280:7614–7623. doi: 10.1074/jbc.M408714200. [DOI] [PubMed] [Google Scholar]
- 47.Bulic B, et al. Development of tau aggregation inhibitors for Alzheimer’s disease. Angew Chem Int Ed Engl. 2009;48:1740–1752. doi: 10.1002/anie.200802621. [DOI] [PubMed] [Google Scholar]
- 48.Brunden KR, et al. Tau-directed drug discovery for Alzheimer’s disease and related tauopathies: A focus on tau assembly inhibitors. Exp Neurol. 2010;223:304–310. doi: 10.1016/j.expneurol.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daccache A, et al. Oleuropein and derivatives from olives as Tau aggregation inhibitors. Neurochem Int. 2011;58:700–707. doi: 10.1016/j.neuint.2011.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.