Significance
Neurotransmitter release occurs upon fusion of synaptic vesicles with the plasma membrane, and it is orchestrated by synaptic proteins, including SNAREs, synaptotagmin, complexin, and other factors. The system is exquisitely fine-tuned to increase the probability of membrane fusion by orders of magnitude upon Ca2+ binding to a Ca2+ sensor, such as synaptotagmin. Although crystal structures are available for some of the key complexes of soluble fragments of synaptic proteins, and the process has been reconstituted, the association of these complexes with the synaptic and plasma membranes remains unclear. We visualized functionally active synaptic proteins reconstituted into proteoliposomes and their interactions in a native membrane environment by electron cryotomography with a Volta phase plate for improved resolvability.
Keywords: synaptic vesicle fusion, neurotransmitter release, Volta phase plate, electron cryotomography, Munc13
Abstract
Neurotransmitter release is orchestrated by synaptic proteins, such as SNAREs, synaptotagmin, and complexin, but the molecular mechanisms remain unclear. We visualized functionally active synaptic proteins reconstituted into proteoliposomes and their interactions in a native membrane environment by electron cryotomography with a Volta phase plate for improved resolvability. The images revealed individual synaptic proteins and synaptic protein complex densities at prefusion contact sites between membranes. We observed distinct morphologies of individual synaptic proteins and their complexes. The minimal system, consisting of neuronal SNAREs and synaptotagmin-1, produced point and long-contact prefusion states. Morphologies and populations of these states changed as the regulatory factors complexin and Munc13 were added. Complexin increased the membrane separation, along with a higher propensity of point contacts. Further inclusion of the priming factor Munc13 exclusively restricted prefusion states to point contacts, all of which efficiently fused upon Ca2+ triggering. We conclude that synaptic proteins have evolved to limit possible contact site assemblies and morphologies to those that promote fast Ca2+-triggered release.
The process of vesicle trafficking is central for transporting materials both inside and outside of cells and is essential for maintaining cellular homeostasis (1, 2). Synaptic vesicle fusion releases neurotransmitter molecules into the synaptic cleft upon an action potential—a fast (<5 ms) and highly regulated process. The neuronal SNARE proteins synaptobrevin-2/VAMP2 and syntaxin-1A are anchored in the synaptic vesicle and plasma membranes, respectively, whereas SNAP-25 is peripherally associated with the plasma membrane. The N- to C-terminal zippering of the trans SNARE complex between synaptic and plasma membranes provides the force for membrane juxtaposition and fusion (3), but SNAREs alone are not Ca2+-sensitive. Synaptotagmins are membrane-tethered proteins containing two Ca2+-binding C2 domains, and a subset of the 16 isoforms of mammalian synaptotagmins act as Ca2+ sensors for neurotransmitter release: e.g., synaptotagmin-1 (Syt1) is the Ca2+ sensor for evoked synchronous neurotransmitter release (4). The system is exquisitely fine-tuned to increase the probability of fusion between synaptic vesicles and the plasma membrane by orders of magnitude upon Ca2+ binding to Syt1. Synaptotagmins simultaneously interact with anionic phospholipid membranes (5) and the neuronal SNARE complex (6, 7). The SNARE complex also interacts with complexin (Cpx), a small soluble protein that both activates evoked release and regulates spontaneous release (8, 9). Moreover, Cpx forms a tripartite complex with the SNARE complex and Syt1 (10). This tripartite complex is part of the prefusion, “primed” state of the synaptic vesicle fusion machinery. Both Munc13 and Munc18 are important for priming, and, at the molecular level, they facilitate proper SNARE complex assembly (11, 12).
Although high-resolution structures are available for some of the key complexes of soluble fragments of synaptic proteins (6, 10, 13, 14), and the process has been reconstituted (11, 15, 16), the association of these complexes with the synaptic and plasma membranes remains unclear. Previous cryo-EM projection studies revealed only membrane shapes but no protein densities (16–18). All synaptic proteins are relatively small (ranging from 45 to 200 kDa) and form heterogeneous and dynamic complexes, posing challenges for imaging by conventional electron tomography. Moreover, the complex nature of cellular synapses consisting of dynamic and dense networks of protein–protein and protein–membrane interactions makes it difficult to study the synaptic fusion protein machinery in vivo. Therefore, we used a bottom-up approach where we reconstituted synaptic proteins into liposomes in a stepwise fashion: Synaptobrevin-2 and Syt1 were reconstituted into liposomes with a defined synthetic lipid composition that mimics synaptic vesicles (referred to as “SV vesicles”), and syntaxin-1A and SNAP-25A were reconstituted into liposomes with a lipid composition that mimics the plasma membrane obtained from lipid brain extracts (referred to as “PM vesicles”) (19).
To visualize the morphology of membrane-associated synaptic proteins and contact sites between membranes of these functionally active proteoliposomes, we used electron cryotomography (cryo-ET), which allows the study of membranes (20) and membrane proteins under close to native conditions (21). However, low-dose imaging of radiation-sensitive biological samples leads to poor contrast (that is, poor signal-to-noise ratio) of the images. The conventional method of artificially defocusing improves the phase contrast of the images, but at the expense of inducing contrast transfer function-related artifacts that have to be dealt with during image processing. This limitation becomes even more important for imaging of the relatively small synaptic proteins. Introduction of defocus fundamentally limits the achievable resolution of EM images unless accurately corrected, but, in tomography, the contrast transfer function (CTF) correction is technically challenging due to the tilt-induced complexity in the sample (22). Consequently, alignments become very sensitive to defocus artifacts exaggerating resolution loss in the original image when reconstructed in 3D, more so in small particles due to poor alignment features. To overcome the limitations of conventional cryo-ET, we used phase contrast imaging with the Volta phase plate (VPP), allowing in-focus imaging of the samples with significantly higher contrast (23, 24).
Results
Volta Phase Plate Electron Cryomicroscopy of Functional Synaptic Proteoliposomes.
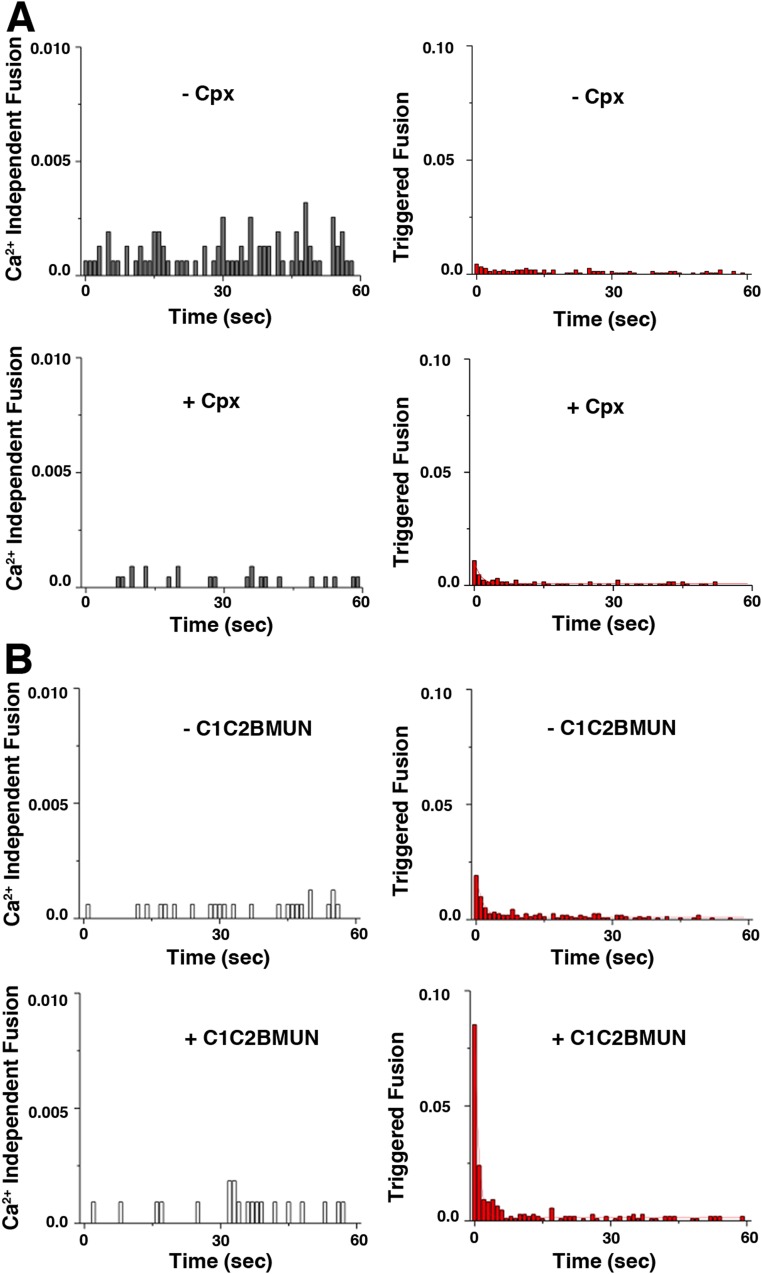
When mixed together, and Ca2+ is added, SV and PM vesicles fuse as shown by a single vesicle fusion assay as previously reported (12, 16) (Fig. S1): i.e., the SV and PM vesicles represent a minimal system for Ca2+-triggered fusion. Inclusion of additional synaptic proteins, in particular, Cpx and Munc13, greatly increases the efficiency, synchrony, and sensitivity of Ca2+-triggered fusion (Fig. S1).
Fig. S1.
Single vesicle fusion experiments. Ca2+-triggered fusion experiments using the same SV and PM vesicles preparations as used for the cryo-ET experiments shown in Figs. 2–4. Fusion between single vesicles was observed by using a content-mixing assay as previously described (12, 19). (A) Fusion experiments without and with 2 μM Cpx. (B) Fusion experiments in the presence of 2 μM Cpx and without and with 0.5 μM C1C2BMUN fragment, as described in ref. 12. Note that the experiments shown in B included 0.5% DAG, resulting in a slightly increased Ca2+-triggered amplitude (compare B, Top Right vs. A, Bottom Right). After SV-PM vesicle association, vesicle pairs either undergo Ca2+-independent fusion or remain associated until fusion is triggered by 500 μM Ca2+ addition. The histograms were normalized with respect to the corresponding number of analyzed SV-PM vesicle pairs (Table S6).
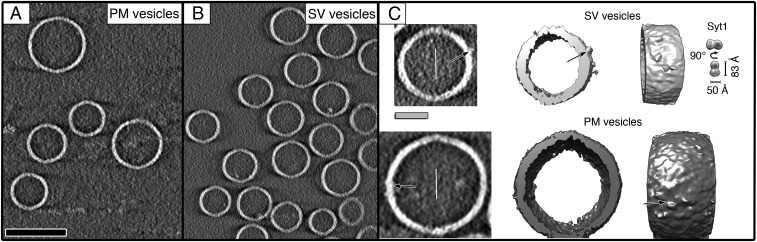
Cryo-ET images of individual PM and SV vesicles under optimized conditions (Methods) showed evenly distributed and well-separated vesicles that do not form any contacts or fusion interfaces (Fig. 1 A and B). VPP greatly improved the resolvability and reliability of imaging of membranes and of individual synaptic proteins (Fig. 1C and Fig. S2). The vesicles had an effective bilayer thickness of ∼40 Å (Fig. S3 A–C). The diameter distributions of the PM and SV vesicles had pronounced maxima at ∼90 and 55 nm, respectively (Fig. S3D). Some densities protruded from the surface of SV vesicles that likely correspond to Syt1 C2A-C2B domains, based on the shape and volume of these densities (Fig. 1C, Top and Fig. S2C). Smaller densities that protruded from PM vesicles likely correspond to complexes between syntaxin-1A and SNAP-25A (Fig. 1C, Bottom). The differences in vesicle diameters and the protruding densities made it possible to uniquely identify the particular type of vesicle in mixtures of PM and SV vesicles.
Fig. 1.
Cryo-ET of individual PM and SV vesicles. (A and B) Gray scale tomographic slice views of individual PM and SV vesicles that were separately frozen on EM grids, respectively. SV vesicles include reconstituted Syt1 and synaptobrevin-2 whereas PM vesicles include syntaxin-1A and SNAP-25A. (C) Representative subtomograms (tilt axes along the solid white lines) of SV (light gray, Top) and PM vesicles (dark gray, Bottom). Protruding densities (arrows) from both vesicles are seen both in 2D-slice (Left) and 3D isosurface views (Middle and Right). These densities likely correspond to Syt1 in SV vesicles and syntaxin-1A/SNAP-25A complexes in PM vesicles. Tomogram 2D slice thickness: 1 pixel = 3.42 Å. (Scale bars: black, 100 nm; gray, 25 nm.) Thin white lines, tilt axes.
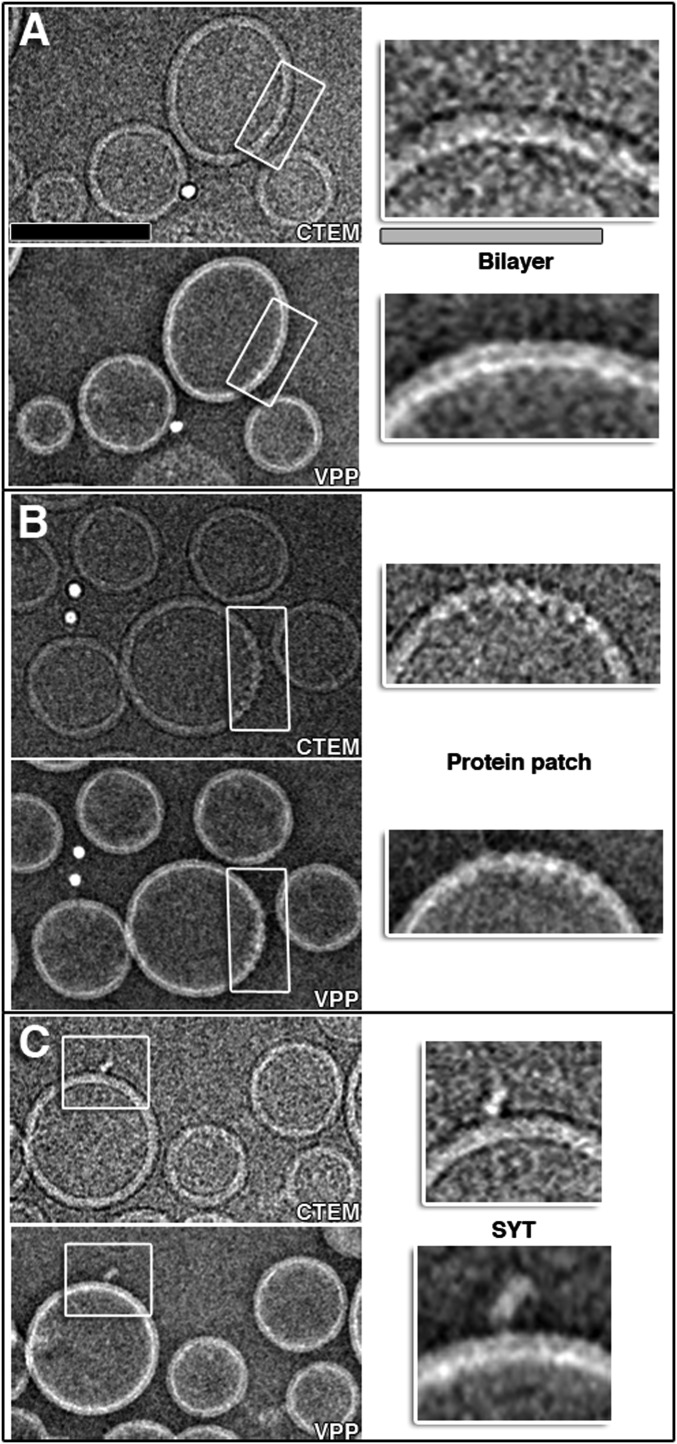
Fig. S2.
VPP cryo-EM. (A–C, Top Left) Conventional cryo-TEM images of a mixture of SV + PM vesicles collected at ∼6.5 μm defocus. (Lower Left) The TEM images of the same mixture of SV + PM vesicles that were collected with VPP at 0 μm defocus. Increased resolvability of the images at focus allowed improved interpretation of the images, as is shown in the close-up views of the areas indicated by gray boxes in the Right Top and Bottom panels showing vesicle bilayers, protein patches, and protruding membrane proteins, such as Syt1. (Scale bars: black, 100 nm; gray, 25 nm.) All other tomograms throughout this work were collected with VPP. Tomogram 2D slice thickness: 1 pixel = 3.42 Å.
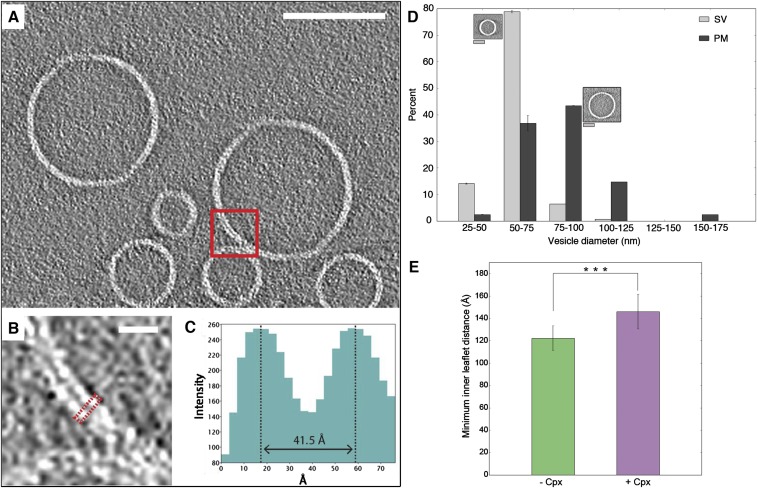
Fig. S3.
Vesicle and membrane analysis. (A–C) Bilayer thickness measurement. (A) The lipid bilayer thickness measurement was performed for the selected region (red box) using a VPP tomographic 2D slice of a mixture of SV + PM vesicles. (Scale bar: 100 nm.) (B) Close-up view of the selected region. (Scale bar: 10 nm.) (C) Line plot of the density across the bilayer (red dotted box in B). The bilayer thickness was estimated from the peak-to-peak distance. Tomogram 2D slice thickness: 1 pixel = 3.42 Å. (D) Bar chart of vesicle diameter distributions of individual SV (light gray) vesicles and PM (dark gray) vesicles. The majority of the individual SV vesicles have a diameter of 55 nm whereas the majority of the individual PM vesicles have a diameter of 90 nm. Representative images of SV and PM vesicles shown next to the two top bars. (Gray scale bar: 25 nm.) (E) Inner leaflet distance with and without Cpx. The bar graph shows the average and SD for the minimum distance between inner leaflets at contact sites between vesicles in the absence and presence of Cpx. Means and SDs are shown (146 ± 16 Å in the presence of Cpx and 122 ± 11 Å in the absence of Cpx). Statistical significance was assessed by Student’s t test (***P < 0.001).
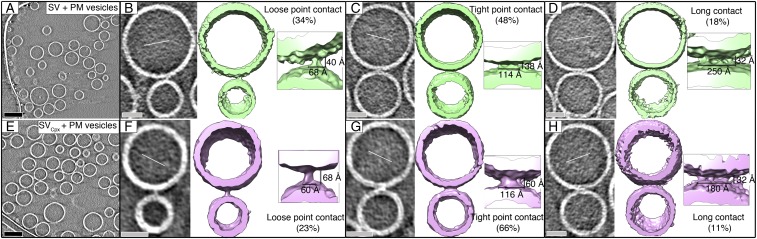
We trapped prefusion protein–membrane contact sites by incubating a mixture of PM and SV vesicles in the absence of Ca2+ for 1 min before freezing. Cryo-ET of the mixture revealed connected densities between many pairs of docked vesicles. These contact sites are strong densities as seen in 2D slice views (gray left subpanels in Fig. 2 B–D, Fig. S4, and Movie S1) and in isosurface 3D maps (Fig. 2 B–D, green surface subpanels). The isosurface contour level of the 3D maps was adjusted such that the membrane or membrane-associated proteins, such as Syt1, were visible and matched the expected size.
Fig. 2.
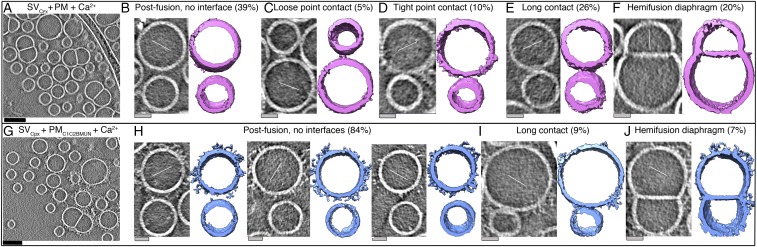
SV + PM vesicle contact site morphologies with and without Cpx. (A) Tomographic 2D-slice view of a mixture of SV + PM vesicles in the absence of Cpx. (B–D) Representative morphologies of contact sites of a mixture of SV + PM vesicles: loose, tight, or long contacts (Methods for definition). Subtomograms of vesicles with contact sites are shown in 2D-slice view (gray, Left, tilt axes along the solid white lines) and corresponding isosurface representations [green, Right, insets show close-up views (4×) of the contact sites]. (E) Tomographic 2D-slice view of a mixture of SVCpx + PM vesicles (i.e., SV vesicles were first incubated with 2 μM Cpx, and Cpx continued to be present during mixing with PM vesicles). (F–H), Representative morphologies of contact sites of a mixture of SVCpx + PM vesicles shown in tomographic 2D-slice view (gray, Left, tilt axes along the solid white lines) and isosurface representation [pink, Right, insets show close-up views (4×) of the contact sites]. Additional views of the isosurface representations are shown in Fig. S5 E–J, more representative gray scale tomographic 2D-slice views are shown in Fig. S4, and videos of representative tomograms are available in Movies S2 and S3. Percentages were calculated with respect to the total number of observed interfaces (Table S1). Tomogram 2D slice thickness: 1 pixel = 3.42 Å. (Scale bars: black, 100 nm; gray, 25 nm.) Thin white lines, tilt axes.
Fig. S4.
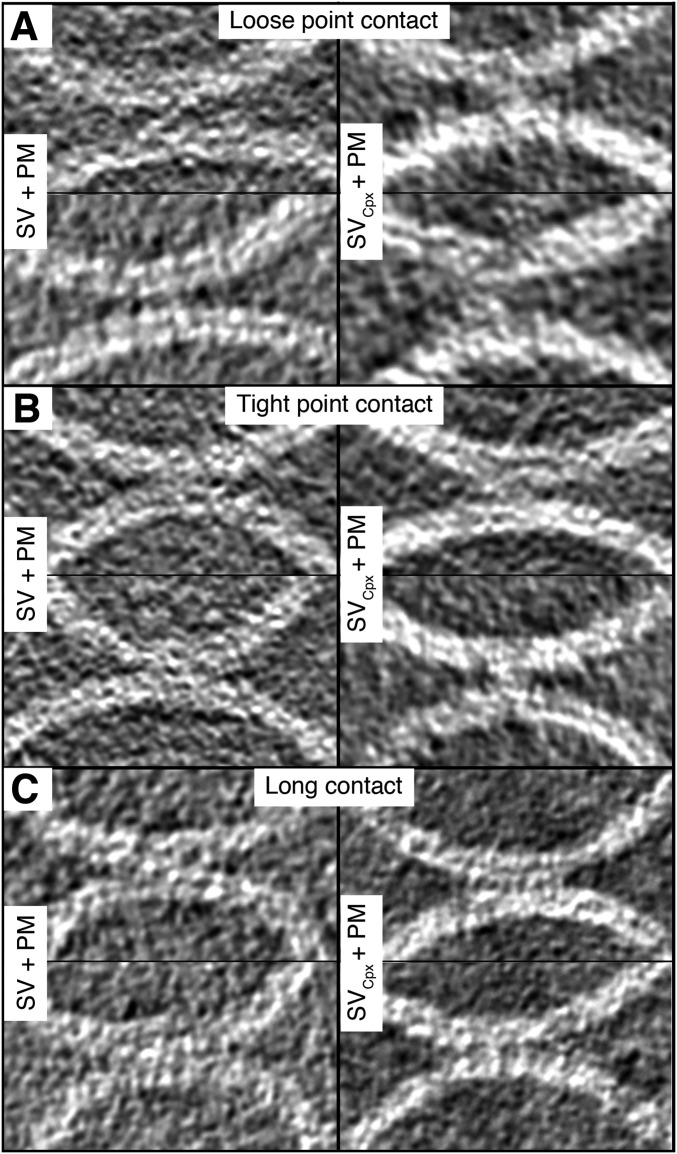
Contact sites of SV + PM vesicles with and without Cpx. Representative gray scale tomographic 2D-slice views of contact sites, in addition to those shown in Fig. 2. Contact sites with and without Cpx are shown in the right and left panels, respectively. (A) Loose point contact, (B) tight point contact, and (C) long contact. Tomogram 2D slice thickness: 1 pixel = 3.42 Å.
An analysis of tomograms with several hundred contact sites of the SV + PM vesicle mixtures revealed a limited number of distinct contact site morphologies. We characterized them based on the contact site length (Methods). The majority of the observed contact sites are tight point contacts (site length ∼115 to 160 Å, 48%), along with smaller populations of loose point contacts (site length ∼50 to 80 Å, 34%) and long point contacts (site length ≥180 Å, 18%) (Fig. 2 B–D, Table S1, and Methods). Volumetric occupancy analysis of representative contact sites suggests that the density of a tight-contact site can accommodate approximately two SNARE–Syt1 complexes (Fig. S5 A and B and Tables S2 and S3).
Table S1.
| Vesicles | No. of loose point contacts (with no C1C2BMUN at or near site) | No. of tight point contacts (with no C1C2BMUN at or near site) | No. of long contacts | No. of cases with C1C2BMUN near contact site | No. of cases with C1C2BMUN at contact site | No. of hemifusion diaphragms | Total no. of analyzed contact sites | Total no. of vesicles without contacts |
| SV + PM | 59 | 84 | 32 | N/A | N/A | 0 | 175 | 183 |
| SVCpx + PM | 74 | 218 | 37 | N/A | N/A | 0 | 329 | 287 |
| SVCpx + PMC1C2BMUN | 50 | 94 | 0 | 179 | 113 | 0 | 436 | 338 |
| SVCpx + PM + Ca2+ | 18 | 35 | 83 | N/A | N/A | 66 | 202 | 136 |
| SVCpx + PMC1C2BMUN + Ca2+ | 0 | 0 | 25 | 0 | 0 | 21 | 46 | 237 |
N/A, not applicable.
Fig. S5.
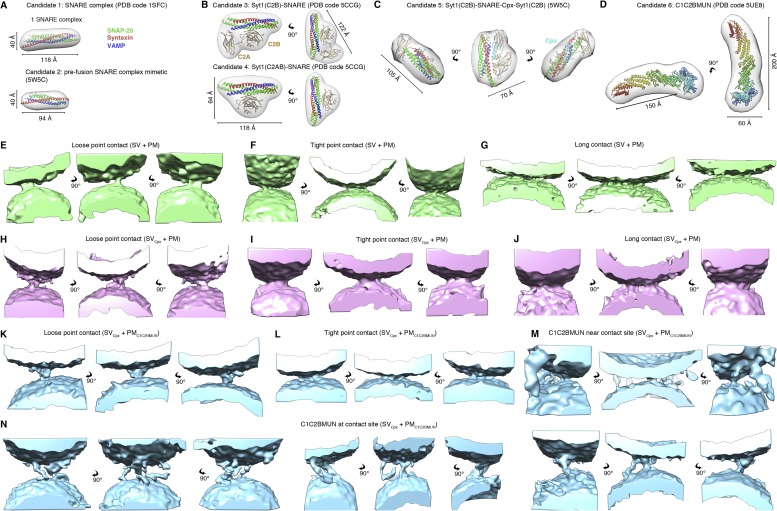
Contact site candidates and morphologies. (A–D) Contact site candidates consisting of synaptic proteins and their complexes with known structures. Molecular surface representation (gray) of crystal structures were filtered to 45 Å and superimposed on schematic representations of the crystal structures. (E–N) Additional views of the isosurface representation of subtomograms of the representative contact sites shown in Figs. 2 and 3. In N, three examples of contact sites with C1C2BMUN at the sites are shown in three orthogonal views.
Table S2.
Volumes of synaptic proteins with known structure
| Candidate no. | Synaptic protein(s) | Volume (V × 103 Å3) |
| 1 | SNARE complex (PDB ID code 1SFC) (13) | 145 |
| 2 | SNARE complex, prefusion mimetic (PDB ID code 5W5C) (10) | 97 |
| 3 | Syt1(C2A-C2B)-SNARE complex (PBD ID code 5CCG) (6) | 302 |
| 4 | Syt1(C2B)-SNARE complex (PDB ID code 5CCG) (6) | 218 |
| 5 | SNARE-Cpx-Syt1 (C2AB) complex (PDB ID code 5W5C) (10) | 227 |
| 6 | C1C2BMUN fragment (PDB ID code 5UE8) (25) / subtomogram average of the C1C2BMUN fragment | 347/461 |
Table S3.
Candidates for SV + PM contact sites
| Type | Measured CS volume/103 Å3 (Fig. 2B–D) | Possible candidates based on volume and composition | Maximum no. |
| Loose point contact | 92 | 2 | 1 |
| Tight point contact | 295 | 3,4 | 2 |
| Long contact | 877 | 3,4 | 6 |
Effect of Complexin-1 on Membrane Separation and Contact Site Morphologies.
We next determined the effect of Cpx on the contact site morphologies. We incubated the SV vesicles for 30 min with 2 μM full-length Cpx (referred to as SVCpx vesicles) before mixing with PM vesicles. As previously reported (12), inclusion of Cpx increased the amplitude of Ca2+-triggered fusion between SVCpx and PM vesicles using the single vesicle fusion assay (Fig. S1). Inclusion of Cpx also increased the number of contact sites, thereby increasing the apparent number of vesicles in the EM tomograms in the absence of Ca2+ (Fig. 2E). At the morphological level, similar classes of contact sites were observed as without Cpx (Fig. 2 B–D and F–H and Figs. S4 and S5 E–J). However, the population of tight point contact sites increased from 48 to 66% whereas the populations of loose and long contact sites decreased. Additionally, the mean membrane separation increased by ∼24 Å for loose and tight point contact sites (Fig. S3E). Volumetric occupancy analysis of representative contact sites suggests that the density of a tight-contact site can accommodate approximately two tripartite complexes consisting of Syt1, SNAREs, and Cpx (Fig. S5 C and H–J and Table S4).
Table S4.
Candidates for SVCpx + PM contact sites
| Type | Measured CS volume/103 Å3 (Fig. 2F–H) | Possible candidates based on volume and composition | Maximum no. |
| Loose point contact | 460 | 3,4,5 | 1 |
| Tight point contact | 899 | 3,4,5 | 2 |
| Long contact | 850 | 3,4,5 | 6 |
Imaging of Flexible Munc13 Bound to Membranes.
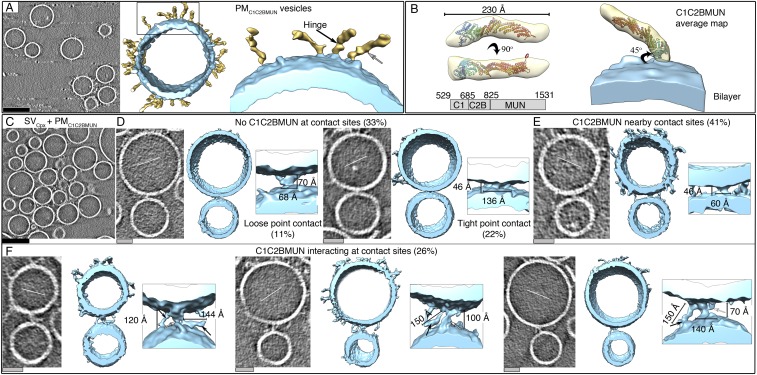
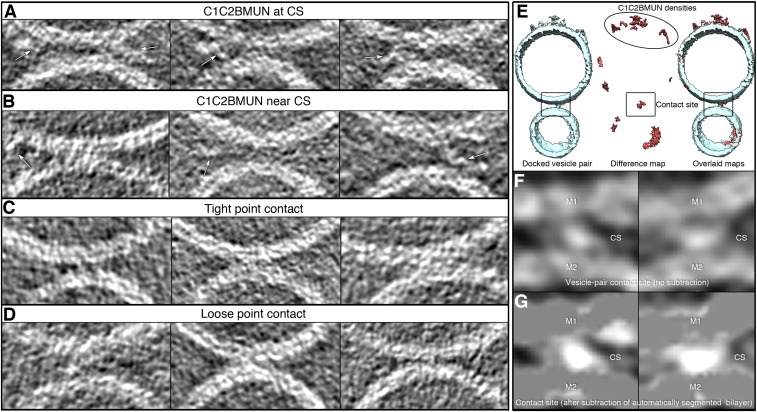
Next, we preincubated PM vesicles with 0.5 μM C1C2BMUN fragment of Munc13 for 30 min (referred to as PMC1C2BMUN vesicles) and also included 0.5% diacylglycerol (DAG) in the preparation of the PM vesicles as previously described (12). The cryo-ET tomograms of the PMC1C2BMUN vesicles alone revealed several flexible filamentous densities protruding from them, roughly corresponding to the dimensions of the C1C2BMUN fragment based on its crystal structure (25) (Fig. 3A). The C1C2BMUN fragments appear to be loosely or tightly attached to the membrane at varying angles. The loosely bound C1C2BMUN fragments were in contact with the membrane whereas the tightly bound fragments had a pronounced density at the base (Fig. 3A). The loosely bound C1C2BMUN fragments might involve binding to DAG in the membrane because the C1 domain of Munc13 binds to DAG (26, 27), and both DAG and PIP2 were included in the preparation of the PM vesicles (12). The tighter bound C1C2BMUN fragments may additionally involve an interaction with the reconstituted complex of syntaxin-1A and SNAP-25A in the PM vesicles.
Fig. 3.
SV + PM vesicle contact site morphologies in the presence of Cpx and C1C2BMUN. (A) Tomographic 2D-slice view of individual PMC1C2BMUN vesicles (i.e., PM vesicles were incubated with the C1C2BMUN fragment of Munc13). Multiple flexible densities (gold) protrude from the vesicles that correspond to C1C2BMUN fragments. The black arrow points to the hinge of a bent filament. The grey arrow shows a straight filament protruding at an ∼45 degree angle from the membrane. (B) Subtomogram average of the C1C2BMUN fragment (Left, two orthogonal isosurface views) and superposition with the automatically docked crystal structure of the C1C2BMUN fragment (N- to C-terminal, blue-to-red rainbow-colored ribbon representation, PDB ID code 5UE8) (25) and crystal structures of the MUN domain (red ribbon representation, PDB ID codes 5UF7 and 4Y21). Bottom, domain structure of the C1C2BMUN fragment. (Right) A composite model consisting of a subtomogram of the membrane and the subtomogram average of the C1C2BMUN fragment in an orientation as seen for a C1C2BMUN fragment in A (gray arrow in A). (C) Gray scale tomographic 2D-slice views of mixtures of SVCpx + PMC1C2BMUN vesicles (i.e., SV vesicles were preincubated with 2 μM Cpx, PM vesicles were preincubated with 0.5 μM C1C2BMUN fragment, and both Cpx and C1C2BMUN were present during mixing of the two classes of vesicles). (D–F) Corresponding vesicle contact site morphologies. Subtomograms of vesicles with contact sites are shown in tomographic 2D-slice view (gray, Left, tilt axes along the solid white lines) and corresponding isosurface representations [blue, Right, insets show close-up views (4×) of the contact sites]. Black arrows in F point to C1C2BMUN fragments, and white arrows point to other densities that may correspond to complexes of SNAREs, Syt1, and Cpx. Additional views of the isosurface representations are shown in Fig. S5 K–N, more representative gray scale tomographic 2D-slice views are shown in Fig. S7 A–D, and a video of a representative tomogram is available in Movie S4. Percentages are calculated with respect to the total number of observed interfaces (Table S1). Tomogram 2D slice thickness: 1 pixel = 3.42 Å . (Scale bars: black, 100 nm; gray, 25 nm.) Thin white lines, tilt axes.
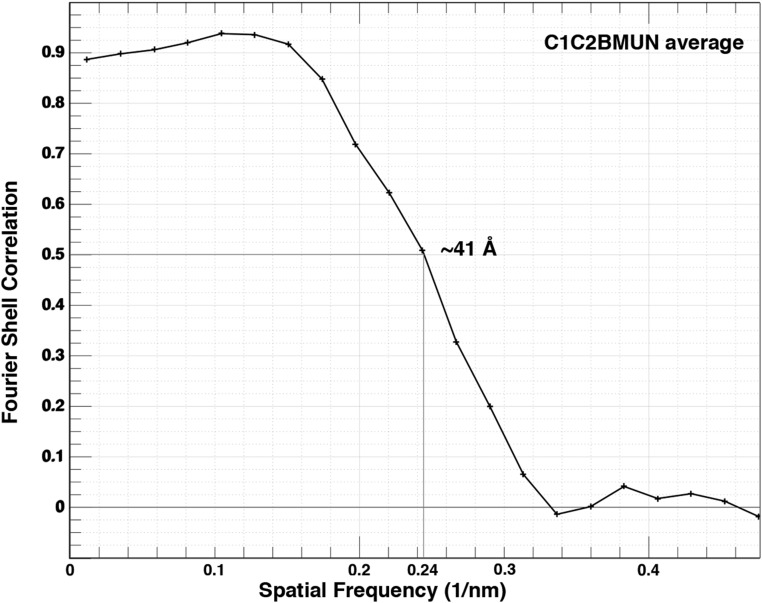
The C1C2BMUN fragments exhibited flexibility near the membrane-attached base and in the middle of the fragment (Fig. 3A). Nevertheless, we managed to generate a low-resolution (∼41 Å) (Fig. S6) subtomogram average from particles that projected away from the membrane plane at an ∼45° angle (Fig. 3B). The averaged C1C2BMUN EM density represents the arrangement of the C1C2BMUN fragment in one conformation with respect to the membrane (Fig. 3B). The subtomogram average fits the crystal structure of the C1C2BMUN fragment reasonably well (Fig. 3B), where the C1 and C2B domains at the base of the filament interact with the membrane, whereas the MUN domain projects away. The extra densities in the EM map are likely due to 32 N-terminal and 38 C-terminal residues of the C1C2BMUN fragment that are missing in the crystal structure.
Fig. S6.
Fourier shell correlation analysis. Fourier shell correlation (FSC) analysis for the subtomogram average of the C1C2BMUN fragment at ∼41-Å resolution.
Effect of Munc13 on Contact Site Morphologies.
We next studied the effect of both C1C2BMUN and Cpx on vesicle contact site morphologies and performed cryo-ET of SV and PM vesicles preincubated with these synaptic proteins, respectively. Functionally, inclusion of the C1C2BMUN fragment resulted in a further large increase of the Ca2+-triggered fusion amplitude of SVCpx + PMC1C2BMUN vesicles (Fig. S1). In the absence of Ca2+, the mixture of SVCpx + PMC1C2BMUN vesicles clearly revealed the two vesicles types: PM vesicles with protruding C1C2BMUN fragment densities and SV vesicles with protruding Syt1 densities (Fig. 3C). We observed loose or tight point contact sites (33%) that have no C1C2BMUN fragments at the site (Fig. 3D, Figs. S5 K–N and S7 A–D, and Table S5). Another large population (41%) are point contact sites with nearby C1C2BMUN fragment densities (Fig. 3E and Figs. S5M and S7B). Remarkably, no long contact sites were observed: i.e., the presence of the C1C2BMUN fragment suppresses formation of these long contact sites. A smaller fraction (26%) of the contact sites also included filamentous densities interacting with the contact site densities (Fig. 3F and Figs. S5N and S7A). The filamentous densities (black arrows in close-up views in Fig. 3F) at these sites corresponded in size and shape to C1C2BMUN fragments (Fig. 3B and Fig. S5D). Additionally, there were corresponding densities at these contact sites that probably consist of complexes of SNAREs, Syt1, and Cpx (white arrows in close-up views in Fig. 3F).
Fig. S7.
Contact sites of SVCpx + PMC1C2BMUN vesicles and difference map analysis. (A–D) Representative gray scale tomographic 2D-slice views of contact sites, in addition to those shown in Fig. 3. Some of the C1C2BMUN fragments are indicated by arrows. The left, center, and right panels show three examples of the particular contact site. Tomogram 2D slice thickness: 1 pixel = 3.42 Å. (E–G) Difference map analysis. (E, Left) A docked SVCpx + PMC1C2BMUN vesicle pair (blue). (Middle) A difference map (red) obtained after bilayer subtraction. (Right) A superposition of the difference map and the segmentation of the subtomogram. All maps are shown in isosurface representation. (F and G) Gray scale tomographic 2D slices of the contact site (CS) observed in the docked SVCpx + PMC1C2BMUN vesicle pair shown in A along the XZ/XY directions (relative to the tomogram axis) before and after subtraction of the two membranes (M1 and M2), respectively. F shows a contact site with no bilayer subtraction in two orthogonal views (left and right panels). G shows the same contact site as in F after bilayer subtraction in the same orthogonal views. A video of the contact site analysis is available in Movie S1.
Table S5.
Candidates for SVCpx + PMC1C2BMUN contact sites
| Type | Measured CS volume/103 Å3 (Fig. 3D–F) | Possible candidates based on volume and composition | Maximum no. |
| Loose point contact without C1C2BMUN at or nearby the site | 454 | 3,4,5 | 1 |
| Tight point contact without C1C2BMUN at or nearby the site | 827 | 3,4,5 | 2 |
| Contact site with C1C2BMUN at the site | 1,511 | 3,4,5,6 | 1, 1–2* |
Maximum number of C1C2BMUN molecules.
Effect of Ca2+ Addition on Contact Site Morphologies.
To study the effect of Ca2+ addition on the vesicle contact site morphologies, we performed cryo-ET of SVCpx + PM and SVCpx + PMC1C2BMUN mixtures after addition of 500 μM Ca2+ for 1 min, followed by flash-freezing. Single vesicle fusion experiments with the same sets of vesicle preparations indicated Ca2+-triggered events as previously reported (12, 16) (Fig. S1). Fusion between a pair of PM and SV vesicles was expected to result in a relatively small increase in the diameter of the resulting fused vesicle, so it was difficult to extract this increase from vesicle size diameter distributions considering the observed variation in diameters (Fig. S3D). However, for both SVCpx + PM and SVCpx + PMC1C2BMUN, we also observed several large circular or elliptical vesicles (≥200 nm) (Fig. 4 A and G), which were likely caused by multiple fusion events with the same vesicle.
Fig. 4.
Contact site morphologies after Ca2+-triggered fusion. (A) Gray scale tomographic 2D-slice view of SVCpx + PM vesicles after incubation with 500 μM Ca2+ for 1 min. (B–F) Corresponding 2D-slice views (gray, Left, tilt axes along the solid white lines) and isosurface representations (dark pink, Right) of subtomograms of the vesicles. (G) Gray scale tomographic 2D-slice view of SVCpx + PMC1C2BMUN vesicles after incubation with 500 μM Ca2+ for 1 min. We also observed a substantial increase in the number of C1C2BMUN fragments that were bound to PM vesicles relative to Fig. 3C, consistent with an increase of membrane binding of the C2B domain in the presence of Ca2+ (26). (H–J) Corresponding 2D-slice views (gray, Left, tilt axes along the solid white lines) and isosurface representations (dark blue, Right) of subtomograms of the vesicles. Videos of representative tomograms are available in Movies S5 and S6. Percentages were calculated with respect to the sum of the total number of observed interfaces plus the number of vesicles without interfaces (Table S1). Tomogram 2D slice thickness: 1 pixel = 3.42 Å. (Scale bars: black, 100 nm; gray, 25 nm.) Thin white lines, tilt axes.
Analysis of the contact site morphologies revealed substantial changes in the population of the sites upon Ca2+ addition. Some point and long contacts remained even after Ca2+ addition for SVCpx + PM vesicle mixtures: i.e., without C1C2BMUN (Fig. 4 C–E). These remaining point contacts may include protein complexes that are not properly assembled and do not undergo fast Ca2+ triggered fusion. Also, several hemifusion diaphragms (20%) (Fig. 4F and Table S1) were observed along with the point and long contacts. Such hemifusion diaphragms are long-lived metastable states (16, 17) that should be considered “dead-end” states in the context of fast neurotransmitter release.
In marked contrast, when C1C2BMUN was included, all point contact sites of the SVCpx + PMC1C2BMUN vesicle mixtures disappeared after Ca2+ addition (Fig. 4 G–I), and the fraction of hemifusion diaphragms was much smaller (Fig. 4 G and J), which suggests that nearly all docked SVCpx + PMC1C2BMUN vesicles completely fused upon Ca2+ addition, except for small fractions of hemifusion diaphragms (7%) and long contacts (9%) (Fig. 4 I and J and Table S1). The lack of C1C2BMUN fragments at a considerable number of contact sites before Ca2+ addition (Fig. 3D) suggests that, in these cases, Munc13 catalyzed proper SNARE complex assembly and then dissociated from the primed prefusion complexes before Ca2+ triggering.
Discussion
The number of synaptic proteins and their assembly into higher order complexes have been the subjects of intense investigation and controversy (28–32). We visualized synaptic protein prefusion states in a native membrane environment at 41-Å resolution (Fig. S6). We built the system in a step-wise fashion to visualize individual synaptic protein interactions and their regulation. Our images show that SNAREs and Syt1 produce a distinct set of contact morphologies between membranes. Point contacts include at most two complexes whereas long contacts contain more complexes (>∼6) that are likely related to higher order assemblies (Tables S3–S5): i.e., the precise copy number of complexes involving SNAREs and Syt1 is variable. Inclusion of Cpx increased the membrane mean separation between synaptic and target membranes and also increased the occurrence of tight point contacts while reducing the occurrence of long contacts in our tomography datasets (Fig. 2 and Fig. S3E). Upon further inclusion of the C1C2BMUN fragment of Munc13, the contacts were largely restricted to point contacts only: i.e., no long contacts were present, and the large majority of vesicles completely fused upon Ca2+ addition (Fig. 4 G–I). This effect of Munc13 is probably related to its molecular function to promote proper synaptic complex assembly (12). Although hemifusion diaphragms are possible outcomes of membrane fusion with a system that consists of SNAREs only, they were suppressed as the reconstituted system was made more complete with Cpx and Munc13.
Taken together, the occurrence of point contacts between membranes consisting of approximately two synaptic complexes increases in our reconstituted system as Cpx and Munc13 are added. It is therefore tempting to speculate that the synaptic fusion machinery has evolved to prefer point contacts rather than more extensive contacts. We note that each synaptic complex composed of SNAREs, Syt1, and Cpx has two states: locked and Ca2+-triggered (10). Only the Ca2+-triggered state promotes fusion whereas the locked state resists fusion by keeping membranes apart. We note that inclusion of Cpx increases the membrane separation (Fig. S3E), supporting the notion that a locked complex lowers the probability of fusion, as also suggested by the dominant negative phenotype of certain mutations of the C2B domain of Syt1 (10). Suppose that there are n synaptic complexes in a contact site; then the probability that all complexes are triggered upon Ca2+ addition is pn, where p is the probability that an individual synaptic complex switches to the Ca2+-triggered state. If a locked complex is strongly inhibitory (i.e., fusion cannot occur if there remains at least one locked complex in a particular contact site upon Ca2+ addition), then a small contact site is more likely to fuse than a large one. Alternatively, if the locked and triggered complexes are energetically balanced (i.e., a simple majority of triggered complexes is sufficient for fusion, and the triggered complexes have to be spatially adjacent to each other in the contact site to promote fusion), again, a smaller contact site would have a higher probability of Ca2+-triggered fusion. Of course, further studies are required that will include additional factors, such as Munc18. Nevertheless, these simple models could explain why a parsimonious system consisting of the smallest required number of synaptic complexes would have been evolutionarily favored for fast Ca2+-triggered neurotransmitter release.
Methods
Vesicle Sample Preparation and Fusion Assay.
SV and PM vesicles were prepared and fusion experiments (Fig. S1 and Table S6) were performed using the protocols published in refs. 12 and 19.
Table S6.
Data summary table for the fusion experiments shown in Fig. S1
| Vesicles | Ca2+-independent fusion events | Ca2+-triggered fusion events | No. of analyzed SV-PM vesicle pairs | Repeats (n) |
| SV + PM | 84 | 100 | 1,562 | 3 |
| SVCpx + PM | 22 | 76 | 1,279 | 4 |
| SVCpx + PM* | 24 | 154 | 1,624 | 3 |
| SVCpx + PMC1C2BMUN* | 20 | 227 | 1,079 | 7 |
With DAG.
Cryo-ET Sample Preparation and Data Acquisition.
The PM/SV vesicle samples were mixed for 1 min at 37 °C before plunge-freezing under liquid-N2 conditions. For experiments with Cpx, SV vesicles were preincubated with 2 μM full-length Cpx for 30 min before mixing with PM vesicles while maintaining the same Cpx concentration. For experiments that also included the C1C2BMUN fragment, 0.5% DAG was added to the PM vesicles, and PM vesicles were preincubated with 0.5 μM C1C2BMUN for 30 min before mixing with SVCpx vesicles that had been preincubated with Cpx while maintaining the same Cpx and C1C2BMUN concentrations. For experiments that imaged the vesicles after Ca2+-triggered fusion, 500 μM Ca2+ was added to the corresponding vesicle mixtures for 1 min and then frozen.
Gold fiducial markers (10 nm, EM BSA gold tracer) were added to the samples before freezing for tilt-series alignment. For freezing, aliquots of 3 μL of the various vesicle samples were applied to glow-discharged 200-mesh Lacey Carbon copper grids (Plano GmbH), which were then vitrified in liquid ethane using a Leica EM GP automatic plunger (Leica Microsystems Inc.). Sample blotting during freezing was done using ashless Whatman filter paper (grade 40 to 44; GE Healthcare).
We optimized freezing conditions to reduce the tendency of vesicles to associate with the hydrophobic edges of the grid bars and to ensure spreading of the vesicles in the holes away from the edges. We tried Quantifoil copper grids (Quantifoil Inc.) of varying hole sizes (hole sizes 0.5 to 2 μm) and with Lacey Carbon copper grids (Plano GmbH). We found that glow discharged (27 s) Lacey grids, with their varying sizes and shapes of the holes, produced the best freezing results with our samples. Before use, the grids were cleaned by organic solvent (ethyl acetate) overnight, followed by twice washing in deionized water and drying overnight.
Tilt-series for cryo-ET were acquired on a Titan Krios field emission gun (FEG) electron microscope (FEI) operated at an acceleration voltage of 300 kV. The microscope was equipped with a Gatan postcolumn energy filter and an FEI Volta phase plate. The images were recorded on a direct detector camera (K2 summit; Gatan) operated in counting mode. The tilt-series were collected using SerialEM (33, 34) using the Volta phase plate protocol at 0 µm defocus (35). All tilt series were acquired at a magnification of 42,000× at a sample spacing of 3.42 Å, tilt range of ±64°, tilt increment of 2°, and total dose of ∼86 e−/Å2.
Image Processing.
Tilt series were aligned using the gold fiducial markers, and tomograms were reconstructed using the IMOD package (36). All unbinned tomograms (pixel size, 3.42 Å) were analyzed at a ∼45-Å resolution (low-pass filtered), at which all of the features were clearly distinguishable against the background with minimal noise. A total of ∼15 tomograms were collected for each condition (SV/PM/PMC1C2BMUN only; SV + PM; SVCpx + PM; SVCpx + PMC1C2BMUN), with ∼100 vesicle pairs observed per tomogram.
From each of the SV + PM conditions, ∼300 of the best contact sites were identified such that the contact site was formed between larger PM and smaller SV vesicles; protruding Syt1 densities could be seen from smaller SV vesicles. In the SVCpx + PMC1C2BMUN tomograms, PM vesicles additionally showed filamentous C1C2BMUN proteins protruding from the surface. All contact sites between docked vesicles were identified such that no missing wedge was seen along the contact sites (perpendicular to the tilt axis). The subtomograms of docked vesicle pairs were extracted and analyzed using Chimera (37). Map segmentation and visualization were performed using the scientific visualization and analysis tool of AMIRA (ZIB, Zuse Institute Berlin; FEI) and Chimera (37).
Vesicle Diameter Measurement.
The diameters of the SV and PM vesicles were determined using the 3DMOD module of IMOD, with adjustable sliders allowing diameter measurement of spherical objects such as vesicles. The vesicle diameters were measured in the nonmissing wedge direction with an approximate diameter measurement error of ±10 nm. A regular histogram of 25-nm bin size was plotted with an error estimation based on misclassification likelihood, based on the ±10-nm measurement error, into nearby bins using ipython’s matplotlib module (DOI 10.5281/zenodo.15423) (Fig. S3D).
Membrane Thickness.
To measure the membrane thickness, a 4× magnified 2D tomographic slice (pixel size 3.42 Å) showing a lipid bilayer was acquired from a 4× binned tomogram (voxel size, 13.7 Å) by using the 3dmod module in the IMOD package (36). A line profile of the slice was then used to obtain the distance between peak-to-peak densities at the original pixel size of 3.42 Å using the Lineplot tool in DigitalMicrograph (Gatan Inc.) (Fig. S3 A–C).
Definition of Contact Site Morphologies.
The observed contact sites for SV + PM and SVCpx + PM vesicle mixtures fell into three discrete classes that we characterized by contact site length (that is, the maximum length of the density of the contact site parallel to the membranes): “loose” point contact (∼50 to 80 Å), “tight” point contact (∼115 to 160 Å), and long contact sites (≥180 Å). We chose the site length rather than the site height to characterize the contact site morphologies because the site length could be efficiently identified in the tomograms.
Measurement of Membrane Separation at Contact Sites.
Membrane separation at a contact site between two vesicles was estimated by measuring the minimum distance between the inner leaflet density edges at the contact site rather than the separation between outer leaflets because the presence of density at the contact site between the membranes makes the distance estimation difficult. Specifically, the distances were measured from the tomograms using Chimera (37) at the contact sites by calculating the minimum distance between two pseudo-atoms placed at the two inner leaflet membrane edges at the contact site. The outer leaflet membrane distances shown in Figs. 2 and 3 were estimated by subtracting twice the bilayer thickness (i.e., 80 Å) from the measured inner-leaflet minimum distance. The distribution of inner-leaflet minimum distances shifted to larger values upon inclusion of Cpx (Fig. S3E).
Subtomogram Averaging of the C1C2BMUN Fragment.
Subtomograms of the C1C2BMUN fragment of Munc13 were extracted from tomograms of PMC1C2BMUN vesicles (i.e., preincubated with the C1C2BMUN fragment). Fully automated particle marking using the program PEET (38) was not possible due to heterogeneity in size and shape of the filamentous C1C2BMUN particles protruding from the membrane at varying angles. Therefore, subtomograms of decorated vesicles were extracted using Chimera to identify relatively straight particles at a ∼45° angle with respect to the bilayer, which were then marked in PEET for alignment. For initial alignment, a preliminary average from ∼5 to 10 particles was generated using PEET. The reconstruction averaging process was stopped when the addition of more particles led to a resolution drop due to heterogeneity. The final average (Fig. 3B) was generated from the 40 best particles out of an initial 80 at a resolution of 41 Å at Fourier shell correlation (FSC) = 0.5 using PEET (Fig. S6).
Volumetric Occupancy Analysis of Contact Sites.
We estimated possible candidates of synaptic proteins or their complexes of known structure for the observed contact sites by calculating their volumes at the resolution level of our tomograms (∼45 Å). The possible candidates included neuronal SNAREs, Syt1, Cpx, and the C1C2BMUN fragment of Munc13 (i.e., the proteins used in our vesicle experiments) (Fig. S5 A–D). The molecular surfaces of the corresponding crystal structures were rendered and low-pass filtered to isosurface maps in chimera at a sampling of 3.42 Å at a resolution corresponding to the tomograms (∼45 Å). We calculated volumes of the low-pass filtered molecular surfaces (Table S2). The high contrast in the VPP tomograms allowed us to perform automatic density-based segmentation in AMIRA to extract only the bilayer from the docked vesicle pair. The extracted bilayer was then subtracted from the vesicle pair to calculate a difference map (Movie S1 and Fig. S7 E–G). The volume measure module in Chimera was used to estimate the volume of the observed remaining density at the contact site in the difference map (Tables S3–S5). We estimated the most likely candidate(s) by comparing the volumes of the candidates (Table S2) with the expected protein composition with the observed volumes of the contact sites (Tables S3–S5). Only those candidates were considered that included the synaptic proteins that were present in the particular vesicle mixtures. The volumetric occupancy analysis also provided the maximum number of the most likely candidate for the various sites (Tables S3–S5). We did not include shape in this analysis because the candidate complexes may not be in the same conformation as that in the crystal structures. The representative contact sites shown in Figs. 2 and 3 and Fig. S5 E–N were used for the volume calculations. The volumes were measured with maps filtered to a ∼41-Å resolution (Fig. S6). Clearly, the heterogeneity of the contact sites may result in differences for other similar contact sites.
Supplementary Material
Acknowledgments
We thank Ucheor B. Choi, Jiajie Diao, Jeremy Leitz, Jürgen Plitzko, William Weis, Minglei Zhao, and Qiangjun Zhou for discussions; Richard Pfuetzner and Austin Wang for protein expression and purification; and Wah Chiu, Htet Khant, and Caroline Fu for EM data collection during an earlier part of the project. This research was supported in part by NIH Grant R37MH63105 (to A.T.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Representative tomograms have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-8807–EMD-8811).
See Commentary on page 8920.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708492114/-/DCSupplemental.
References
- 1.Südhof TC. Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothman JE. The principle of membrane fusion in the cell (Nobel lecture) Angew Chemie Int Ed Engl. 2014;53:12676–12694. doi: 10.1002/anie.201402380. [DOI] [PubMed] [Google Scholar]
- 3.Gao Y, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernández-Chacón R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Lara Á, et al. PtdInsP2 and PtdSer cooperate to trap synaptotagmin-1 to the plasma membrane in the presence of calcium. eLife. 2016;5:1–22. doi: 10.7554/eLife.15886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, et al. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature. 2015;525:62–67. doi: 10.1038/nature14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brewer KD, et al. Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution. Nat Struct Mol Biol. 2015;22:555–564. doi: 10.1038/nsmb.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trimbuch T, Rosenmund C. Should I stop or should I go? The role of complexin in neurotransmitter release. Nat Rev Neurosci. 2016;17:118–125. doi: 10.1038/nrn.2015.16. [DOI] [PubMed] [Google Scholar]
- 9.Mohrmann R, Dhara M, Bruns D. Complexins: Small but capable. Cell Mol Life Sci. 2015;72:4221–4235. doi: 10.1007/s00018-015-1998-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Q, et al. The primed SNARE-complexin-synaptotagmin complex for neuronal exocytosis. Nature. 2017 doi: 10.1038/nature23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai Y, et al. Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron. 2017 doi: 10.1016/j.neuron.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE. 1998 doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 15.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 16.Diao J, et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. eLife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez JM, et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336:1581–1584. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bharat TA, et al. SNARE and regulatory proteins induce local membrane protrusions to prime docked vesicles for fast calcium-triggered fusion. EMBO Rep. 2014;15:308–314. doi: 10.1002/embr.201337807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai Y, et al. N-terminal domain of complexin independently activates calcium-triggered fusion. Proc Natl Acad Sci USA. 2016;113:E4698–E4707. doi: 10.1073/pnas.1604348113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dierksen K, et al. Three-dimensional structure of lipid vesicles embedded in vitreous ice and investigated by automated electron tomography. Biophys J. 1995;68:1416–1422. doi: 10.1016/S0006-3495(95)80314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dodonova SO, Appen A Von, Hagen WJH, et al. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science. 2015;349:195–198. doi: 10.1126/science.aab1121. [DOI] [PubMed] [Google Scholar]
- 22.Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. CTF determination and correction for low dose tomographic tilt series. J Struct Biol. 2009;168:378–387. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asano S, Fukuda Y, Beck F, et al. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- 24.Fukuda Y, Laugks U, Lučić V, Baumeister W, Danev R. Electron cryotomography of vitrified cells with a Volta phase plate. J Struct Biol. 2015;190:143–154. doi: 10.1016/j.jsb.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, et al. Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C 1 C 2 BMUN. eLife. 2017;6:e22567. doi: 10.7554/eLife.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee JS, et al. Beta phorbol ester- and diacylglycerol-induced augmentation of transmitter release is mediated by Munc13s and not by PKCs. Cell. 2002;108:121–133. doi: 10.1016/s0092-8674(01)00635-3. [DOI] [PubMed] [Google Scholar]
- 27.Shin O-H, et al. Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat Struct Mol Biol. 2010;17:280–288. doi: 10.1038/nsmb.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kümmel D, et al. Complexin cross-links prefusion SNAREs into a zigzag array. Nat Struct Mol Biol. 2011;18:927–933. doi: 10.1038/nsmb.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanetti MN, et al. Ring-like oligomers of synaptotagmins and related C2 domain proteins. eLife. 2016;5:e17262. doi: 10.7554/eLife.17262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi L, et al. SNARE proteins: One to fuse and three to keep the nascent fusion pore open. Science. 2012;335:1355–1359. doi: 10.1126/science.1214984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinha R, Ahmed S, Jahn R, Klingauf J. Two synaptobrevin molecules are sufficient for vesicle fusion in central nervous system synapses. Proc Natl Acad Sci USA. 2011;108:14318–14323. doi: 10.1073/pnas.1101818108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickner W. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 33.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Hagen WJH, Wan W, Briggs JAG. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J Struct Biol. 2017;197:191–198. doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci USA. 2014;111:15635–15640. doi: 10.1073/pnas.1418377111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Nicastro D, Schwartz C, Pierson J, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.