Significance
DNA alkylation represents a major form of DNA damage that is of high clinical and human health relevance; however, the molecular mechanisms of transcriptional lesion recognition, stalling, and bypass remain unknown. Herein, we carried out a comprehensive investigation to compare the effects of three regioisomeric EtdT lesions on transcription. Intriguingly, we found that the location of alkyl group dictates transcriptional stalling profile and lesion bypass routes, and we identified a novel minor-groove–sensing motif, termed Pro-Gate, which plays an important role in detecting the minor-groove lesion. This work provides important mechanistic insights into DNA alkylation-induced transcriptional stalling and mutagenesis. Our study also provides knowledge about cancer etiology and for the future design of effective cancer chemotherapeutic agents.
Keywords: transcription, DNA alkylation, transcriptional lesion bypass, transcriptional mutagenesis, RNA polymerase II
Abstract
Alkylated DNA lesions, induced by both exogenous chemical agents and endogenous metabolites, interfere with the efficiency and accuracy of DNA replication and transcription. However, the molecular mechanisms of DNA alkylation-induced transcriptional stalling and mutagenesis remain unknown. In this study, we systematically investigated how RNA polymerase II (pol II) recognizes and bypasses regioisomeric O2-, N3-, and O4-ethylthymidine (O2-, N3-, and O4-EtdT) lesions. We observed distinct pol II stalling profiles for the three regioisomeric EtdT lesions. Intriguingly, pol II stalling at O2-EtdT and N3-EtdT sites is exacerbated by TFIIS-stimulated proofreading activity. Assessment for the impact of the EtdT lesions on individual fidelity checkpoints provided further mechanistic insights, where the transcriptional lesion bypass routes for the three EtdT lesions are controlled by distinct fidelity checkpoints. The error-free transcriptional lesion bypass route is strongly favored for the minor-groove O2-EtdT lesion. In contrast, a dominant error-prone route stemming from GMP misincorporation was observed for the major-groove O4-EtdT lesion. For the N3-EtdT lesion that disrupts base pairing, multiple transcriptional lesion bypass routes were found. Importantly, the results from the present in vitro transcriptional studies are well correlated with in vivo transcriptional mutagenesis analysis. Finally, we identified a minor-groove–sensing motif from pol II (termed Pro-Gate loop). The Pro-Gate loop faces toward the minor groove of RNA:DNA hybrid and is involved in modulating the translocation of minor-groove alkylated DNA template after nucleotide incorporation opposite the lesion. Taken together, this work provides important mechanistic insights into transcriptional stalling, lesion bypass, and mutagenesis of alkylated DNA lesions.
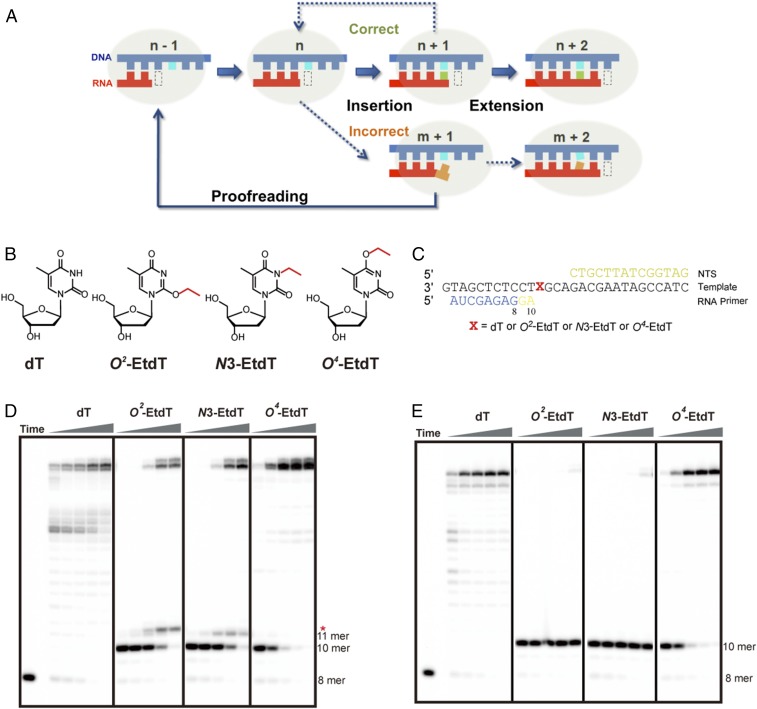
Transcription is the first key step in gene expression, where RNA polymerase II (pol II) is the central enzyme responsible for accurate pre-mRNA and noncoding RNA synthesis in eukaryotic cells. During pol II-mediated transcription elongation, there are at least three transcription fidelity checkpoints to ensure high accuracy in transcription. These fidelity checkpoints include an insertion step for specific nucleotide selection and incorporation, an extension step for preferential extension from a matched over mismatched 3′-RNA terminus, and a proofreading step for preferential removal of misincorporated nucleotides from the 3′-RNA terminus (1) (Fig. 1A).
Fig. 1.
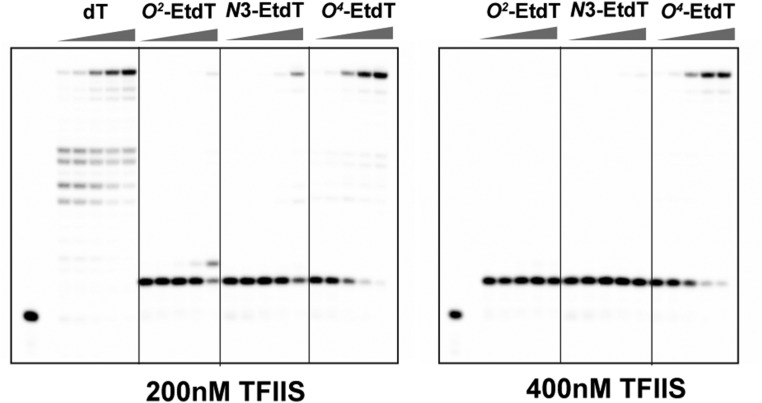
RNA pol II transcriptional elongation in the damaged template containing an alkylated thymine base. (A) A schematic diagram showing the transcription elongation process, and the three steps of transcription checkpoint control, that is, insertion, extension, and proofreading. (B) Alkylation of thymine at different positions. (C) Scaffolds used in transcription elongation experiments. The position of damaged thymine base is marked as X. (D and E) Gel analysis of RNA pol II transcriptional elongation in the absence (D) and presence (E) of TFIIS. The concentration of NTP was 1 mM; the time points were 15 s, 1 min, 5 min, 20 min, and 1 h, respectively. The concentration of TFIIS is 1 µM.
The integrity of genomic DNA constantly faces numerous attacks from both endogenous and environmental agents, some of which cause significant structural and chemical alterations to DNA (1–11). These DNA lesions, when situated in actively transcribed genomic regions, can significantly perturb pol II-catalyzed transcriptional elongation (1, 5, 6, 10–16). RNA pol II may bypass these lesions, which are sometimes accompanied with nucleotide misincorporation into the nascent transcripts, termed transcriptional mutagenesis, or stall at these DNA lesions, which may initiate a specialized DNA repair pathway that preferentially repairs damaged DNA on transcribed template strand, a process known as transcription-coupled repair (TCR) (4–6, 13, 17, 18).
Understanding how pol II recognizes DNA lesions constitutes a crucial step toward elucidating downstream pathways and consequences of transcriptional mutagenesis or TCR. Previous studies have revealed the structural basis for a few types of DNA lesions on transcription efficiency, such as those induced by reactive oxygen species (19, 20), UV light (21, 22), and platinum-based drugs (23, 24). While these studies provided important insights into the recognition of certain classes of DNA damage by pol II, little is known about the mechanisms of transcriptional lesion recognition and processing of alkylated DNA lesions (25).
Alkylation represents a major form of DNA damage that has significant implications in public health and has important clinical relevance (26). For example, tobacco-specific nitrosamines, after metabolic activation, can lead to DNA alkylation (27). It was reported that smokers’ tissue contains much higher levels of ethylated DNA lesions than nonsmokers (28–35), and the respective levels of N3-, O2-, and O4-EtdT in leukocyte DNA of smokers are ∼224-, ∼10-, ∼48-fold higher than those of nonsmokers (32). On the other hand, DNA alkylating agents (e.g., temozolomide and streptozotocin) are among the most widely used chemotherapeutic agents for cancer treatment (26).
DNA alkylation is unique in that the size of the alkyl groups is diverse, and so are the alkylation sites in DNA. In this vein, the size of alkyl group conjugated to DNA can vary significantly by different alkylating agents. While conjugation of bulky alkyl groups with DNA generally causes severe stalling of pol II-mediated transcription, covalent attachment of small alkyl group with DNA can exert very different effects on transcription (36–44). Additionally, alkyl groups can react with many different sites in DNA, including N3 and N7 of adenine, O6 and N7 of guanine, N3, O2, and N4 of cytosine, N3, O2, and O4 of thymine, as well as the phosphate backbone (45, 46). Intriguingly, the effects of alkylated DNA lesions on transcription appears to be influenced by the locations of the alkyl group on the nucleobase. Indeed, our previous studies using double-stranded plasmids harboring site-specifically inserted and structurally defined ethylated thymidine lesions revealed that the transcriptional bypass efficiency and mutation frequency are modulated by the ethyl group being conjugated with the O2, N3, or O4 position of thymidine (41, 42). However, the molecular mechanisms of transcriptional lesion bypass (TLB) and mutagenesis of these DNA lesions remain elusive.
Here, we systematically investigated how pol II recognizes and bypasses three regioisomeric ethylated thymidine lesions (O2-, N3-, or O4-EtdT) (Fig. 1B) and how these lesions affect pol II transcription fidelity checkpoints. This systematic investigation on three regioisomeric EtdT lesions provided important mechanistic insights into the recognition of small alkylated DNA lesions by RNA pol II and revealed different roles of fidelity checkpoints during TLB, which may confer distinct biological consequences. Our study also provides knowledge about cancer etiology and for the future design of effective cancer chemotherapeutic agents.
Results
Distinct Effects of O2-, N3-, and O4-EtdT on Transcription Elongation Mediated by RNA Pol II.
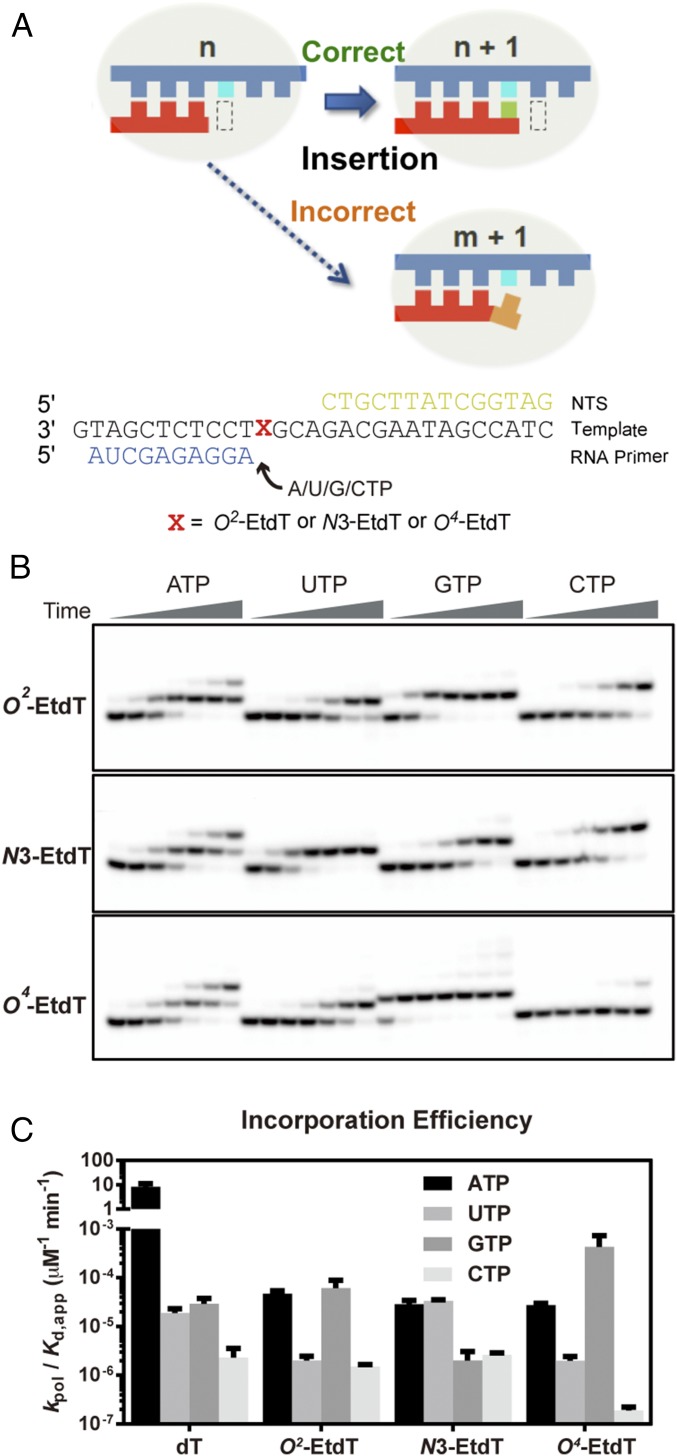
To test the impact of the three regioisomeric O2-, N3-, and O4-EtdT lesions on RNA pol II-catalyzed transcription elongation, we assembled recombinant pol II elongation complex and performed in vitro transcription assays using the DNA scaffolds containing a downstream site-specific EtdT lesion (47–49) (O2-, N3-, and O4-EtdT) (Fig. 1C and Figs. S1–S3).
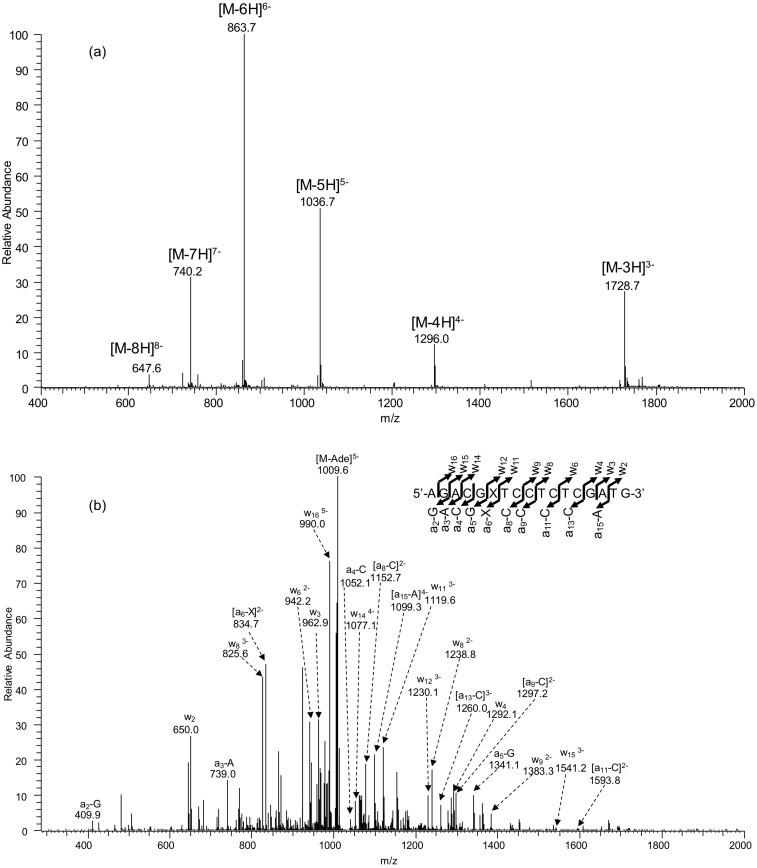
Fig. S1.
ESI-MS and MS/MS characterizations of d(AGACGXTCCTCTCGATG), X = O2-EtdT. (A) Negative-ion ESI-MS; (B) the product-ion spectrum of the [M-5H]5− ion (m/z 1,036.7).
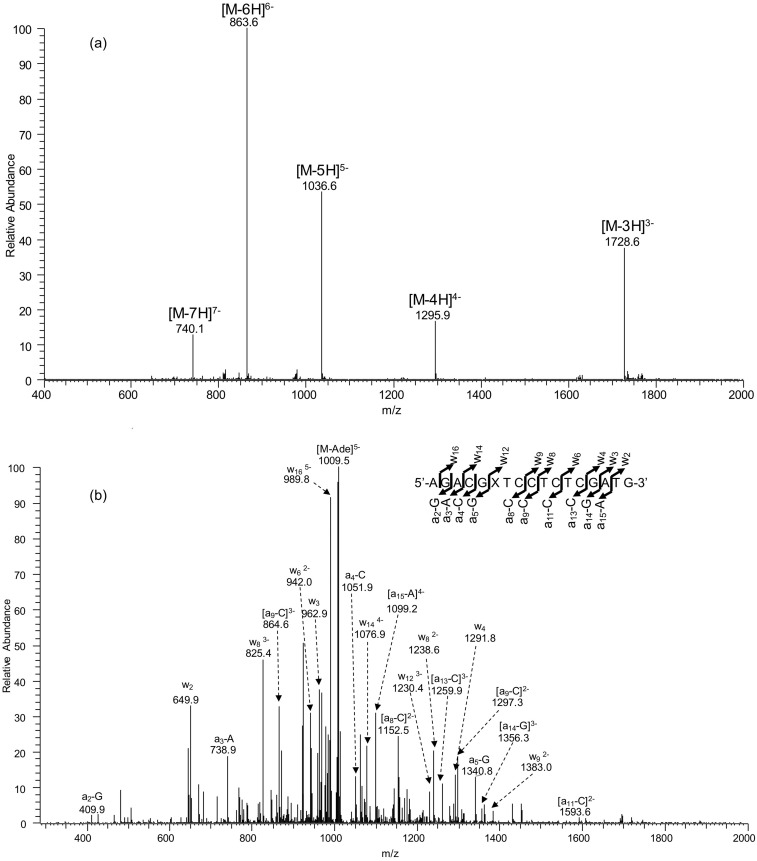
Fig. S3.
ESI-MS and MS/MS characterizations of d(AGACGXTCCTCTCGATG), X = O4-EtdT. (A) Negative-ion ESI-MS; (B) the product-ion spectrum of the [M-5H]5− ion (m/z 1,036.6).
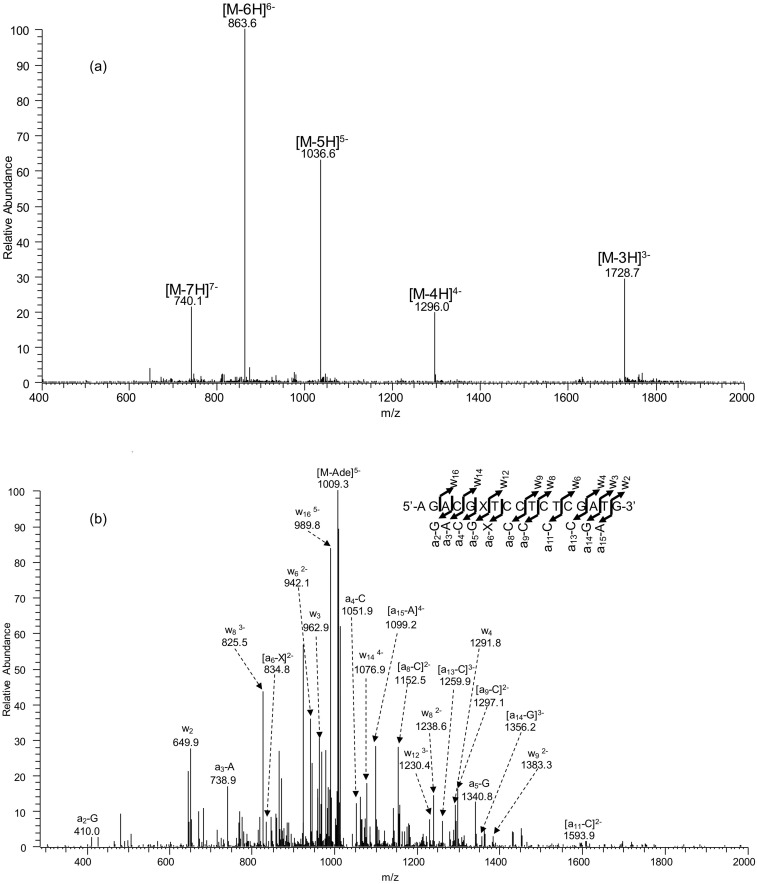
Fig. S2.
ESI-MS and MS/MS characterizations of d(AGACGXTCCTCTCGATG), X = N3-EtdT. (A) Negative-ion ESI-MS; (B) the product-ion spectrum of the [M-5H]5− ion (m/z 1,036.6).
As shown in Fig. 1D, pol II elongation complex was significantly stalled immediately before the O2-EtdT or N3-EtdT site, whereas no stalling was observed at the same position for nondamaged template (dT). Accumulation of 10-mer transcript for the O2-EtdT– and N3-EtdT–harboring templates indicated inefficient nucleotide insertion opposite the lesion sites. Interestingly, after prolonged incubation, pol II was able to move forward by one nucleotide to a second stalling site (11-mer) immediately after the nucleotide incorporation opposite the O2-EtdT or N3-EtdT, suggesting that the extension past the damaged site was also substantially hampered. No further significant stalling was observed once pol II escaped these two pausing/stalling sites. Strikingly, the second pol II stalling band for the O2-EtdT template (11-mer transcript, marked by a red asterisk in Fig. 1D) exhibited different mobility on the gel from that observed for the N3-EtdT template, suggesting that pol II preferentially inserts different nucleotides opposite O2-EtdT and N3-EtdT. In stark contrast to the two strong consecutive stallings of pol II observed for N3-EtdT and O2-EtdT, we only found a single pausing at the 10-mer position for O4-EtdT. In addition, the bypass across O4-EtdT site was highly efficient relative to the other two alkylated thymidine derivatives. Taken together, our transcription runoff assay showed that the same size of alkyl group conjugated at the three different positions of dT can exert markedly different effects on transcriptional pausing. While N3-EtdT and O2-EtdT cause two strong consecutive pol II stallings (with a different migration in the second stalling), minimal stalling of RNA pol II elongation was observed for O4-EtdT.
TFIIS Prevents Transcriptional Bypass of O2-EtdT and N3-EtdT, but Not O4-EtdT.
Transcription factor TFIIS can stimulate transcript cleavage and reactivate backtracked pol II, and therefore it promotes pol II to bypass a variety of DNA lesions and transcription barriers such as pausing sequences and nucleosomes (50–53). Thus, we also examined whether TFIIS could facilitate transcriptional bypass of the three EtdT lesions. Intriguingly, in the presence of TFIIS, we observed prolonged pol II stalling immediately before pol II reaches the O2-EtdT and N3-EtdT sites (10-mer position, Fig. 1E). In sharp contrast to the expected roles of TFIIS in promoting transcriptional bypass, we observed that TFIIS pronouncedly prevents the bypass of O2-EtdT and N3-EtdT, where, in the presence of TFIIS, no full-length products emanating from transcriptional bypass of the two lesions were observed (Fig. 1E, O2-EtdT and N3-EtdT panels, and Fig. S4). Notably, the concentrations of TFIIS we used for in vitro TFIIS-stimulated transcript cleavage assays (200, 400, and 1,000 nM) are comparable to its estimated cellular concentration (∼250–400 nM; Materials and Methods). This result suggests that the TFIIS-stimulated proofreading activity of pol II (backtrack and transcript cleavage) is much more efficient than the pol II-mediated forward transcriptional bypass of the O2-EtdT and N3-EtdT lesions (i.e., insertion and extension). Consequently, the majority of pol II was stalled at the sites of the two lesions. In contrast to the findings made for the O2-EtdT and N3-EtdT lesions, we observed no apparent effect of TFIIS on the pol II-mediated bypass of O4-EtdT. As a positive control, we found that TFIIS promotes pol II-mediated bypass of the pausing sequences on the nondamaged template (Fig. 1E, dT panel). Taken together, we found that the stalling of pol II at the O2-EtdT and N3-EtdT sites is exacerbated by TFIIS-stimulated proofreading activity, whereas transcription bypass across O4-EtdT site is relatively efficient and not affected by the presence of TFIIS.
Fig. S4.
RNA pol II transcriptional elongation in the presence of TFIIS. The concentration of TFIIS is 200 and 400 nM, respectively. The time points were 15 s, 1 min, 5 min, 20 min, and 1 h, respectively.
Impact of EtdT Lesions on the Three Transcriptional Fidelity Checkpoint Steps of RNA Pol II.
To further investigate whether the transcriptional fidelity of pol II is altered during lesion bypass, we carried out a systematic investigation on the impacts of the EtdT lesions on the three transcriptional fidelity checkpoint steps: insertion, extension, and proofreading.
For the first fidelity checkpoint step (Fig. 2A, insertion), we observed distinct preferences in nucleotide incorporation opposite the three EtdT lesions (Fig. 2B). As shown in Fig. 2B, with the use of 1 mM nucleoside triphosphates, we observed that AMP could be incorporated opposite all three EtdT lesions. We also detected significant nucleotide misincorporation, to an extent that is comparable to or higher than the canonical AMP incorporation. Intriguingly, GMP was preferentially inserted opposite O2- and O4-EtdT, whereas UMP was the most favorable nucleotide inserted opposite N3-EtdT (Fig. 2B). To assess quantitatively the nucleotide discrimination in the insertion step, we performed pre–steady-state kinetic experiments under single-turnover conditions to measure the rate constant (kpol) (Fig. S5) and specificity constant (kpol/Kd) for nucleotide insertion opposite all three EtdT lesions or undamaged dT (Fig. 2C, Figs. S5 and S6, and Tables S1 and S2).
Fig. 2.
Nucleotide incorporation opposite the EtdT lesions. (A) A scheme illustrating the first fidelity checkpoint step (insertion) and the scaffold used in this assay. (B) Representative images of gels for monitoring single-nucleotide addition opposite the three ethylated thymine nucleosides. The concentration of NTP was 1 mM; the time points were 1 min, 5 min, 20 min, 1 h, 3 h, 8 h, and 1 d, respectively. (C) Kinetic analysis of single-nucleotide incorporation opposite the ethylated thymidines in comparison with the undamaged thymidine.
Fig. S5.
Kinetic analysis of kpol for the incorporation and extension steps for the three regioisomeric EtdT lesions.
Fig. S6.
Representative kinetic fitting curves for nucleotide incorporation opposite to the N3-EtdT lesion (Upper) and extension past the N3-EtdT lesion (Lower).
Table S1.
Kinetic data of lesion bypass by RNA pol II: Incorporation
| Lesion | Incorporation | |||
| NTP | kpol, min−1 | Kd, µM | kpol/Kd, min−1⋅µM−1 | |
| O2-EtdT | ATP | 0.34 ± 0.03 | 7,100 ± 800 | (4.7 ± 0.7) × 10−5 |
| UTP | 0.0095 ± 0.0010 | 4,700 ± 800 | (2.0 ± 0.4) × 10−6 | |
| GTP | 0.24 ± 0.06 | 3,900 ± 1,500 | (6.1 ± 2.7) × 10−5 | |
| CTP | 0.0087 ± 0.0005 | 6,000 ± 500 | (1.5 ± 0.2) × 10−6 | |
| N3-EtdT | ATP | 0.11 ± 0.01 | 3,800 ± 600 | (2.9 ± 0.5) × 10−5 |
| UTP | 0.096 ± 0.002 | 2,900 ± 100 | (3.4 ± 0.2) × 10−5 | |
| GTP | 0.017 ± 0.005 | 8,300 ± 3,500 | (2.0 ± 1.1) × 10−6 | |
| CTP | 0.018 ± 0.001 | 7,100 ± 600 | (2.6 ± 0.3) × 10−6 | |
| O4-EtdT | ATP | 0.12 ± 0.01 | 4,500 ± 400 | (2.7 ± 0.3) × 10−5 |
| UTP | 0.017 ± 0.002 | 2,600 ± 400 | (2.0 ± 0.4) × 10−6 | |
| GTP | 3.1 ± 1.2 | 7,100 ± 4,000 | (4.3 ± 3.0) × 10−4 | |
| CTP | (4.9 ± 0.4) × 10−4 | 2,600 ± 400 | (1.9 ± 0.3) × 10−7 | |
Table S2.
Kinetic data of lesion bypass by RNA pol II: Extension
| Lesion | Extension | |||
| RNA primer | kpol, min−1 | Kd, µM | kpol/Kd, min−1⋅µM−1 | |
| O2-EtdT | 11A | 4.1 ± 0.1 | 1,600 ± 100 | 0.0026 ± 0.0001 |
| 11U | 7.1 ± 0.8 | 4,600 ± 700 | 0.0015 ± 0.0003 | |
| 11G | 0.0014 ± 0.0001 | 700 ± 70 | (1.9 ± 0.2) × 10−6 | |
| 11C | 27 ± 4 | 10,000 ± 2,000 | 0.0026 ± 0.0006 | |
| N3-EtdT | 11A | 0.13 ± 0.01 | 1,600 ± 200 | (8.1 ± 0.8) × 10−5 |
| 11U | 1.2 ± 0.1 | 4,600 ± 700 | (2.7 ± 0.5) × 10−4 | |
| 11G | 0.0042 ± 0.0001 | 800 ± 80 | (5.0 ± 0.5) × 10−6 | |
| 11C | 2.9 ± 0.7 | 3,200 ± 1,100 | (9.1 ± 3.8) × 10−4 | |
| O4-EtdT | 11A | 12 ± 1 | 4,300 ± 700 | 0.0027 ± 0.0005 |
| 11U | 34 ± 1 | 470 ± 30 | 0.072 ± 0.005 | |
| 11G | 87 ± 8 | 38 ± 20 | 2.3 ± 1.2 | |
| 11C | 0.95 ± 0.16 | 4,800 ± 1,200 | (2.0 ± 0.6) × 10−4 | |
These kinetic results, as summarized in Fig. 2C, led to several important observations. First, the rate constants for the incorporations of the correct AMP opposite all three EtdT lesions were greatly diminished (by ∼105-fold) relative to that for the control undamaged template. Second, we observed unique patterns in nucleotide selection for the three EtdT lesions, indicating that these regioisomeric EtdT lesions elicit distinct impacts on substrate selection in the first checkpoint step (insertion). For the minor-groove O2-EtdT lesion, AMP and GMP incorporations are strongly favored over CMP and UMP incorporations (Fig. 2C, O2-EtdT panel). On the other hand, AMP and UMP are preferentially inserted opposite N3-EtdT over GMP and CMP (Fig. 2C, N3-EtdT panel). For the major-groove O4-EtdT lesion, incorporation of GMP was highly efficient, that is, at a rate that is ∼10-fold higher than that of AMP incorporation (Fig. 2C, O4-EtdT panel). As a result of this high frequency of GMP misincorporation, O4-EtdT was the easiest among the three EtdT lesions to be bypassed by pol II. These results indicate the functional importance of regioisomeric alkylation in DNA, where alkylation at different positions of thymine base exerts pronouncedly distinct effects on substrate nucleotide selectivity in the insertion step, thereby differentially modulating transcriptional fidelity in the first step of fidelity checkpoint.
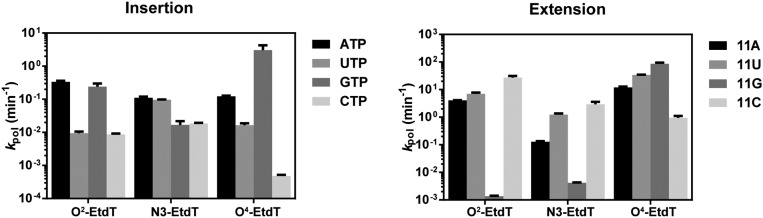
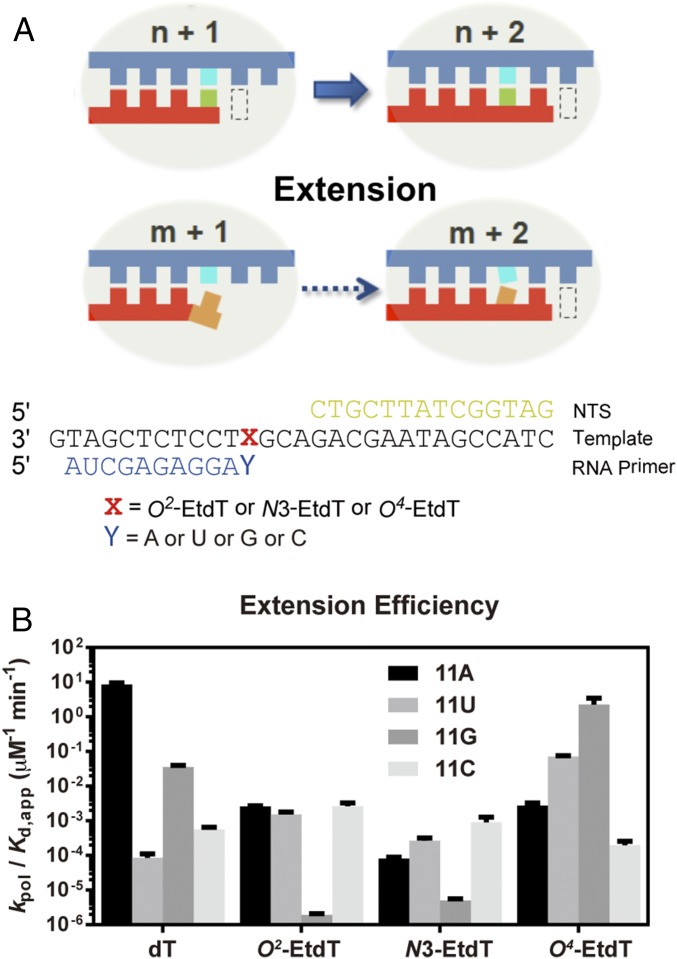
Next, we investigated the impacts of these EtdT lesions on the second step of fidelity checkpoint (Fig. 3A, extension). We found that, for all three EtdT lesions, the efficiency of extension from the matched primer 11A was markedly reduced (by ∼104- or 105-fold) relative to that for the undamaged dT template (Fig. 3B, 11A columns). Furthermore, we observed that the presence of DNA lesions significantly changes the efficiency of extension from mismatched primers. For the undamaged template (dT), apart from the efficient extension from the matched primer 11A, we observed that extension from the mismatched 11G primer was also relatively efficient (∼10−1 µM−1⋅min−1, only ∼100-fold less than extension from 11A), whereas extension from the mismatched 11U or 11C primer was extremely inefficient (105-fold and 104-fold less than extension from 11A, respectively) (Fig. 3B, dT panel). In stark contrast, we found that the presence of O2-EtdT or N3-EtdT greatly disfavored the extension from the mismatched 11G (∼10−6 µM−1⋅min−1, five orders of magnitude lower than that of 11G extension for the control dT template), whereas these DNA lesions modestly favored 11U and 11C extension (Fig. 3B, O2-EtdT and N3-EtdT panels). Strikingly, we found that the extension after GMP incorporation was the most efficient for the O4-EtdT–containing template, which is ∼1,000- and 30-fold more efficient than extension from 11A and 11U, respectively (Fig. 3B, O4-EtdT panel). Taken together, the three regioisomeric EtdT lesions significantly alter the extension selectivity in a position-specific manner, thereby influencing differently transcriptional fidelity in the second fidelity checkpoint step.
Fig. 3.
Analysis of the subsequent extension step after the nucleotide addition opposite the alkylated thymine site. (A) Scaffold used in this assay. (B) Kinetic analysis of the subsequent extension after the ethylated thymine in comparison with the undamaged thymine base.
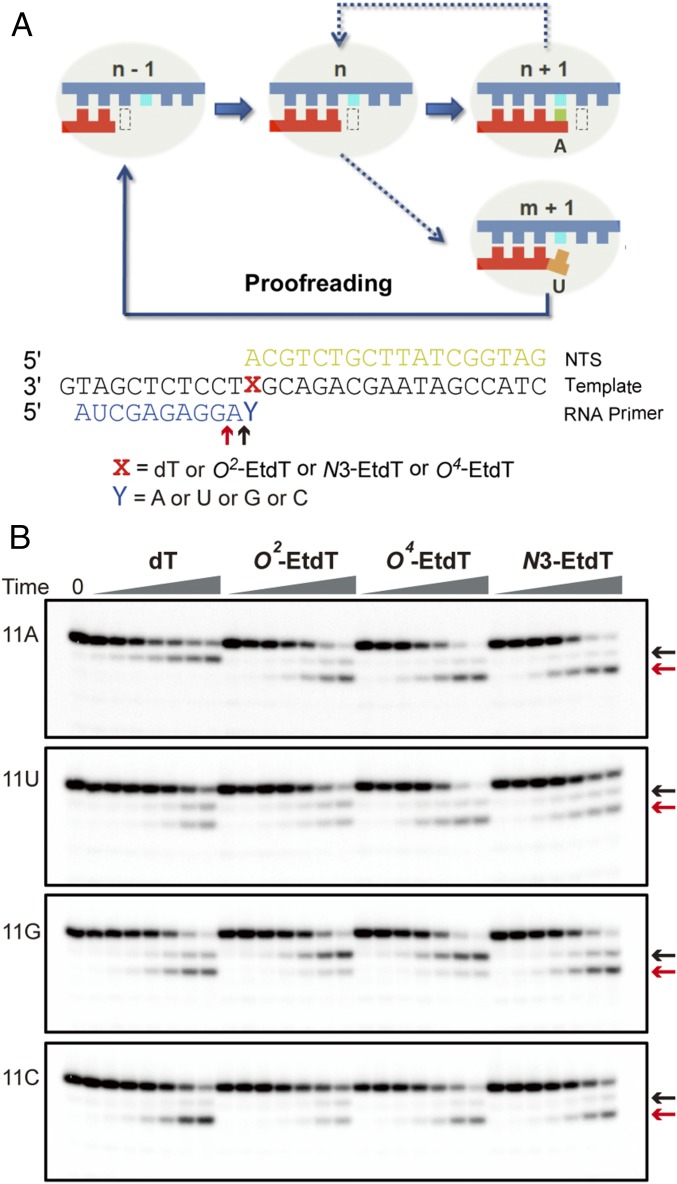
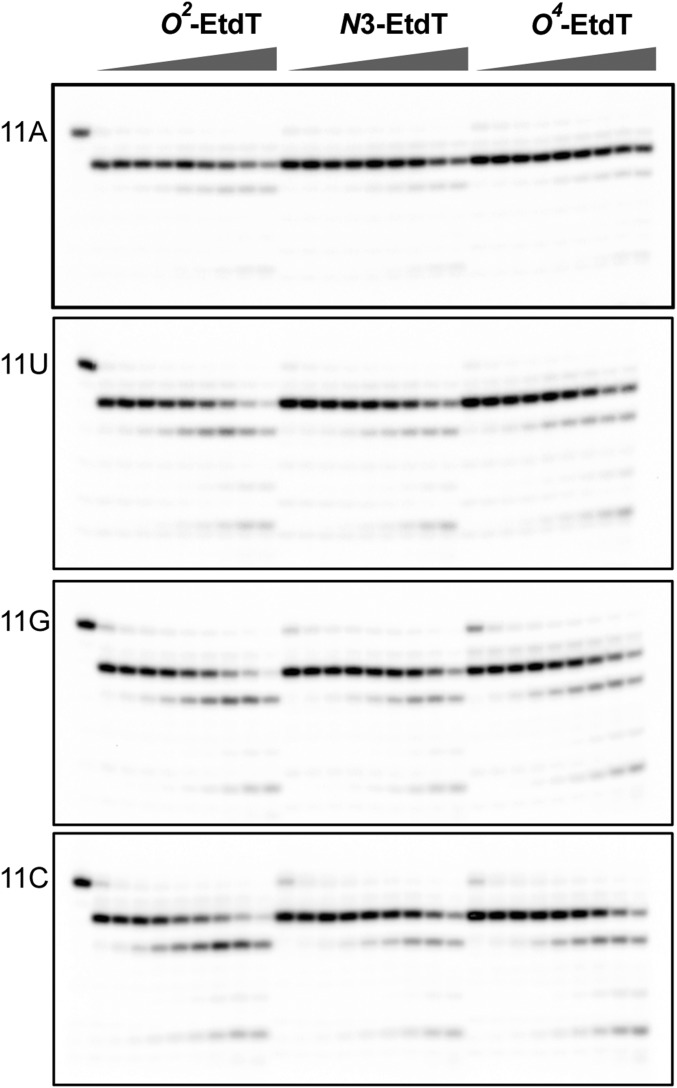
We then assessed the third transcription fidelity checkpoint step (Fig. 4A, proofreading). To this end, we first performed intrinsic transcript cleavage assay, which not only allowed us to measure the transcript cleavage rates but also revealed pol II translocation states based on the transcript cleavage pattern (Fig. 4B). It turned out that the presence of EtdT lesions significantly changed the transcript cleavage pattern of pol II complex containing matched 11A. For the matched 11A/dT scaffold, we only observed a single dominant transcript cleavage product (Fig. 4B, n-1, black arrows) corresponding to the cleavage from pol II at pretranslocation state (the 3′-terminal rA still occupies the insertion site and forms Watson–Crick base pairing with dT in the template) (Fig. 4B, 11A and dT panel). In contrast, we observed a dominant n-2 cleavage product (Fig. 4B, red arrows) and a minor n-1 cleavage product (Fig. 4B, black arrows) from all three 11A/Et-dT lesion scaffolds, indicating that the presence of the EtdT lesions promotes pol II backtracking, where pol II translocates backward from the pretranslocation to backtracked state and 3′-terminal rA is extruded toward the secondary channel (Fig. 4B, 11A panel). In addition, consistent with the previous literature finding that 3′-mismatched RNA or damaged DNA scaffolds promote pol II backtracking (51), we observed two cleaved transcripts (n-1 and n-2) for all other mismatched or damaged scaffolds (Fig. 4B, 11C, 11G, and 11U panels). Notably, the formation of n-2 cleavage product was the major event observed for all of the mismatched or damaged templates, except for those carrying 11G:O2-EtdT or 11G:O4-EtdT (Fig. 4B, 11G). The cleavage products of these two scaffolds mainly arise from pretranslocation cleavage, indicating that their base parings at the 3′-terminus were more stable than other damaged scaffolds. We also performed TFIIS-stimulated cleavage assay and found that, in the presence of TFIIS, pol II can efficiently cleave the transcripts at the damage sites (Fig. S7). Collectively, these results suggest that pol II, in the presence of TFIIS, can efficiently cleave transcripts for the damage-containing scaffolds, but the translocation patterns are altered and depend on the positions on the thymine base where the alkyl groups are situated.
Fig. 4.
Backtrack and proofreading of RNA pol II after nucleotide insertion opposite the ethylated thymidine lesions. (A) Scaffold used in the cleavage assay. (B) Results of RNA pol II intrinsic cleavage. The black arrow refers to the pretranslocation cleavage product; the red arrow refers to the backtrack cleavage product. The time points were 5 min, 20 min, 60 min, 3 h, 8 h, 24 h, and 48 h, respectively.
Fig. S7.
TFIIS-stimulated RNA pol II backtrack and transcript cleavage. Time points were 10 s, 30 s, 1 min, 2 min, 4 min, 8 min, 15 min, 30 min, and 60 min, respectively.
Distinct Transcriptional Bypass Routes of O2-, N3-, and O4-EtdT Lesions.
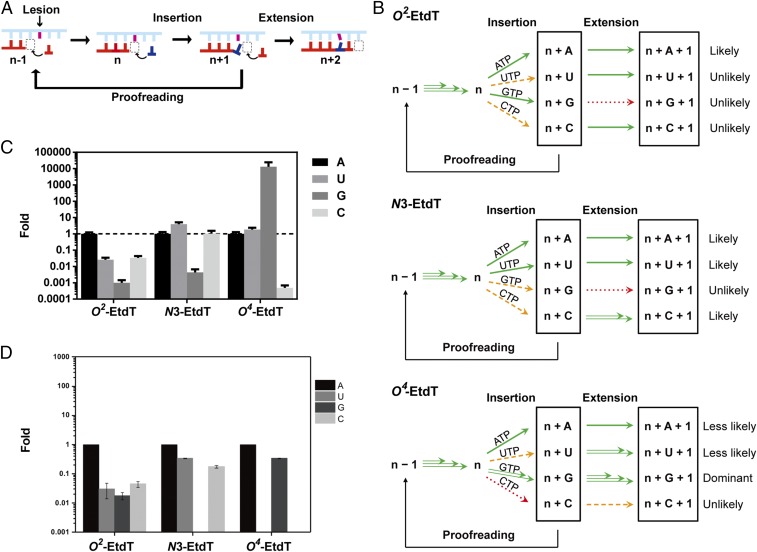
By integrating the kinetic data of the three transcriptional fidelity checkpoints for the EtdT lesions, an overall transcriptional bypass scheme can be derived, as summarized in Fig. 5. Alkylation at different positions confers distinct routes for TLB (Fig. 5B). For the DNA template containing a minor-groove O2-EtdT lesion, both ATP and GTP were favored in the insertion step. However, the subsequent extension after GMP incorporation was extremely disfavored. As a result, AMP incorporation and subsequent extension constituted the major TLB pathway, which essentially maintained the transcriptional fidelity, albeit with compromised bypass efficiency. The lesion bypass routes for the N3-EtdT template, however, were very different. In the nucleotide addition step, AMP and UMP could be more readily incorporated than CMP and GMP; nevertheless, the subsequent extension after CMP incorporation was highly efficient. Consequently, all three TLB routes (from AMP, UMP, and CMP) are likely to occur for N3-EtdT, where only the GMP pathway is blocked in both the insertion and extension steps. In this scenario, transcriptional fidelity was compromised with potential A→U or A→C mutations (Fig. 5B). Strikingly, a distinct and dominant TLB route was observed for the major-groove O4-EtdT lesion, where GMP was preferentially inserted in the addition step with over 10-fold higher efficiency than the matched AMP (Fig. 2). Furthermore, the extension after GMP incorporation was remarkably efficient, which is ∼1,000- and 100-fold more efficient than the extension after AMP and UMP insertions, respectively (Fig. 3). In addition, the efficiency in extension after GMP insertion is similar to what was observed for the matched nucleotide extension for the undamaged template (Fig. 3). Hence, the GMP pathway predominates the lesion bypass route for O4-EtdT, and the transcriptional fidelity is almost abolished with a high frequency of A-to-G mutation.
Fig. 5.
Alkylation at different positions at the hydrogen bonding face of thymidine leads to distinct bypass preferences by RNA pol II. (A) Lesion bypass was controlled by different fidelity checkpoints. (B) A summary of transcription lesion bypass for the three regioisomeric EtdT lesions. Green, orange, and red arrows designate ribonucleotide incorporations that are efficient, difficult, and extremely difficult, respectively. This scheme was depicted based on the relative kinetic values of each transcriptional bypass step. (C) A comprehensive analysis of relative bypass efficiency of RNA pol II for the three regioisomeric EtdT lesions. (D) The distributions of nucleotides inserted opposite the regioisomeric EtdT lesions based on transcription assays conducted in XPA-deficient human skin fibroblasts, where the data represent the mean and SE of results from three independent transfection experiments (the panel was plotted based on data reported in ref. 41).
To quantify the alteration in fidelity for TLB, we introduced an index, termed TLB efficiency (Materials and Methods for more details), to gauge the overall lesion bypass efficiency by combining the rates for all three fidelity checkpoints (i.e., insertion, extension, and proofreading). Considering that the proofreading rates for all three lesions are similar (Fig. 4 and Fig. S7), the TLB value is thus mainly controlled by the insertion and extension steps, and it reflects the likelihood of a particular translesion bypass route or stalling. By normalizing the TLB value with “error-free” TLB value for each template (e.g., the AMP route; see Materials and Methods for more details), we can directly compare the different TLB routes for the three lesion-bearing DNA templates, as summarized in Fig. 5C. Therefore, for each lesion, we can quantitatively evaluate the likelihood of particular TLB routes (Fig. 5C). A value above 1 indicates that this specific route is more efficient than the error-free TLB route, whereas a value below 1 suggests that this specific route is disfavored. As shown in Fig. 5C, error-free route (AMP route) is strongly favored for transcriptional bypass of the minor-groove O2-EtdT lesion, whereas a dominant error-prone route (GMP route) is preferred for transcriptional bypass of the major-groove O4-EtdT lesion. For the N3-EtdT lesion that disrupts base pairing, multiple transcriptional bypass routes are possible, which could be error free (AMP route) or error prone (UMP and CMP routes).
Importantly, the aforementioned in vitro transcription results are well correlated with what we observed previously from cell-based transcription studies (41). We found that, in nucleotide-excision repair-deficient human skin fibroblasts, O4-EtdT induces exclusively A→G mutation and N3-EtdT triggers A→C and A→U mutations, whereas the nucleotide incorporation opposite O2-EtdT is promiscuous (41) (Fig. 5D). Hence, the results obtained from the present in vitro biochemical study are remarkably similar to those from cell-based study, especially in the context that the in vitro assay was conducted with purified RNA pol II in the presence of one nucleotide at a time, whereas the cellular transcription reaction is mediated by the entire human transcription machinery in the mutual presence of all four nucleotides.
Structural Insights into Lesion Bypass and Stalling of Major- and Minor-Groove Lesions.
The above results showed that the pol II-mediated bypass routes for major-groove and minor-groove alkylation lesions are distinct; while GMP insertion is permitted for both O2-EtdT and O4-EtdT in the insertion step (Fig. 2C), the subsequent extension after the GMP insertion is extremely inefficient for O2-EtdT, but strongly favored for O4-EtdT (Fig. 3B). To further understand how pol II distinguishes the GMP route for O2-EtdT and O4-EtdT lesions during the insertion and extension steps, we modeled, based on previous structural studies (54, 55), the active site of the pol II elongation complex when it encounters the O2-EtdT and O4-EtdT lesions, followed by energy minimization (Materials and Methods for details) (Fig. 6). Molecular-modeling results indicate that guanine can form wobble base pairs with both O2-EtdT and O4-EtdT in different orientations (Fig. 6B). These hydrogen bonds may stabilize the rG:O2-EtdT or rG:O4-EtdT pair, which also finds its support from the intrinsic cleavage patterns as shown in Fig. 4B. Thus, these wobble base pairs at the +1 site of pol II active center could favor GMP binding and addition in the first insertion step (Fig. 2). Note that because the efficiency of GMP addition is several orders of magnitude lower than that of canonical ATP incorporation for the undamaged dT template (trigger loop-dependent nucleotide addition, in which the trigger loop adopts an active, closed conformation to catalyze nucleotide addition) (56), we speculate that nucleotide addition opposite the lesion likely occurs in a trigger loop-independent manner, in which the trigger loop remains in inactive, open conformations (1, 57).
Fig. 6.
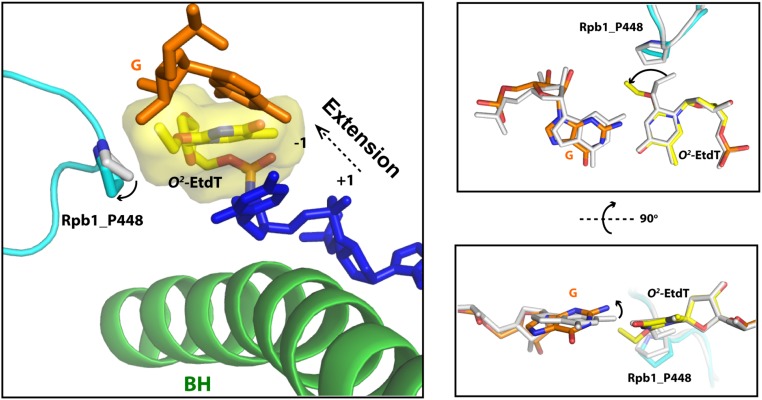
Transcriptional bypass of O4- and O2-EtdT has distinct structural effects. (A) During the incorporation of GMP, the guanine base can pair with both O4- and O2-EtdT. (B) In the extension step, the alkylation in the major groove (O4-EtdT) has limited disruption during translocation, whereas the minor-groove alkylation (O2-EtdT) has strong steric clash with the P448 residue, and further altered this minor-groove–interacting loop in Rpb1.
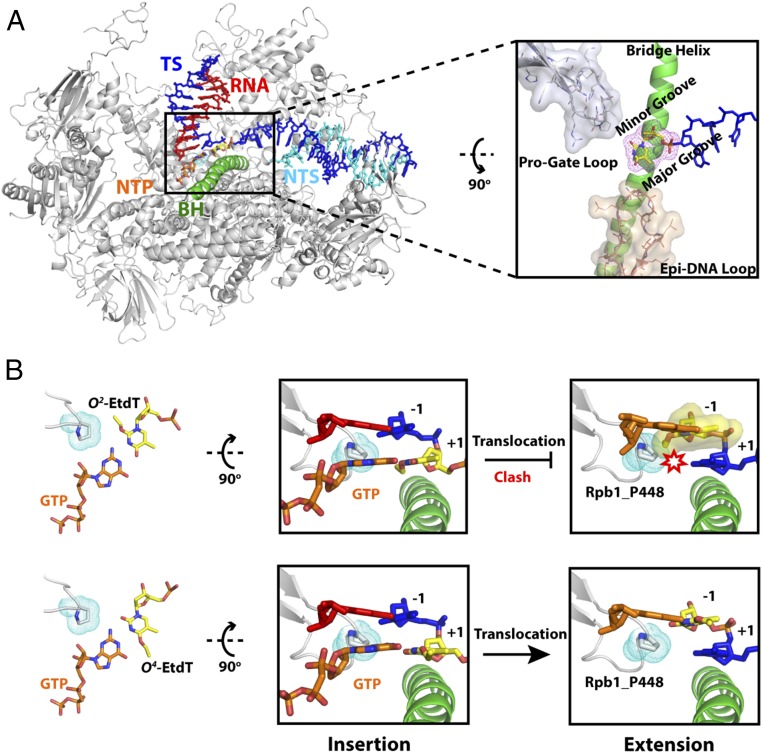
To proceed with extension after GMP incorporation, the newly formed rG:O4-EtdT or rG:O2-EtdT base pair needs to translocate into the −1 site so as to facilitate the subsequent nucleotide addition. Interestingly, during translocation, the ethyl group in the major and minor grooves of DNA could experience drastically different environments (Fig. 6). Upon translocation of the O4-EtdT from the +1 to −1 site, the ethyl group, which is located in the major groove of DNA, is well tolerated in the active site of pol II without any steric clash. Hence, subsequent extension from the rG:O4-EtdT pair is highly efficient (Fig. 3B). In contrast, the ethyl group of the O2-EtdT lesion, which points to the minor groove, encounters a severe steric clash with P448 in Rpb1 when there is no local rearrangement. P448 is located in a motif consisting of a connection loop between the two β-sheets (β14 and β15) in the active-site domain. Because the P448-containing loop (Rpb1 440–460) faces toward the minor groove of RNA:DNA hybrid and acts as a “steric gate” to prevent or slow down the translocation of nucleotide with alkylation in the minor groove, we termed it the “Pro-Gate” loop. This clash may severely hinder the translocation of the minor-groove O2-EtdT lesion, which may significantly compromise the extension efficiency (Figs. 3A and 6B). Notably, our energy minimization results showed that P448 is displaced from the original position to minimize the steric clash, suggesting a potential mechanism for tolerating small minor-groove DNA alkylation and a slow transcriptional bypass (Fig. S8), whereas the presence of bulky minor-groove DNA alkylation could lead to a strong pol II arrest.
Fig. S8.
Energy minimization to minimize the steric clash between Rpb1 P448 from the Pro-Gate loop and O2-EtdT lesion. The color code is the same as Fig. 6. The energy-minimized Pro-Gate loop, O2-EtdT lesion, and GTP are highlighted in cyan, yellow, and orange, respectively.
Discussion
Our previous plasmid-based cellular transcription assays revealed that the three regioisomeric EtdT lesions exhibited different transcriptional bypass efficiencies and mutation frequencies (41), although the molecular mechanisms underlying the differences were unclear. In this study, we systematically investigated the mechanisms contributing to the regiospecific effects of DNA alkylation on pol II-mediated transcription, elucidating important relationships between DNA lesion structure and transcriptional stalling, lesion bypass, and fidelity. The addition of an ethyl group to different positions (O2, N3, and O4) of thymine elicited distinct transcriptional responses of RNA pol II. Here, we revealed how RNA pol II handles regioisomeric DNA alkylation and how such alkylation affects transcriptional fidelity. A comparison of TLB efficiencies of the three lesions showed that O2- and N3-alkylation greatly blocked pol II elongation, whereas O4-alkylation exerted limited blockage effect. The major pausing of O2- and N3-alkylation occurred in both the insertion and the subsequent extension steps at the damaged site. For O4-alkylation, only weak pausing at the insertion step was found, and no obvious pausing was observed at the extension step (Fig. 1D).
Further mechanistic insights came from our study about the impact of these regioisomeric EtdT lesions on the individual pol II fidelity checkpoints (Fig. 5). Strikingly, although GMP was selected for both O2-EtdT and O4-EtdT in the first fidelity checkpoint, the eventual transcriptional readout was different. For O4-alkylation, a high frequency of A→G mutation was observed, whereas transcriptional fidelity was maintained for the O2-alkylation template. The opposite consequences were actually elicited by the second fidelity checkpoint (extension step). For the O4-EtdT template, the extension step was highly efficient owing to pol II’s tolerance of alkylation in the DNA major groove. On the contrary, the extension after GMP incorporation opposite O2-EtdT was extremely difficult, which is potentially attributed to the steric blockage of P488 residue situated in the DNA minor groove during translocation.
Our systematic investigations of alkylation at different positions of thymine base suggested a potentially unique recognition motif of RNA pol II that may serve as a gatekeeper for monitoring the structural integrity of DNA minor groove. Our previous studies unveiled a couple of important recognition motifs in RNA pol II that target the DNA major-groove modifications or bulky, helix-distorting lesions during translocation (10, 11, 58, 59). Furthermore, we recently identified two important residues (Rpb 1 R1386/H1387) in switch 1 region that are important for the early detection of minor-groove DNA binder Py-Im molecules even before it reaches the downstream bubble edge (59). Here, we found that the Pro-Gate loop containing a highly conserved Pro (i.e., P448 in Rpb 1) residue might play an additional inspective role during translocation to sense the environment of the minor groove after nucleotide incorporation. This postinsertion minor-groove sensor could serve as another checkpoint for minor-groove modifications following the nucleotide insertion step. We proposed that Pro-Gate loop acts as a steric gate to prevent or slow down the translocation of O2-EtdT lesion after GMP incorporation (GMP route) but permit other extension routes that bypass O2-EtdT lesions (AMP, UMP, or CMP routes). Based on our intrinsic cleavage assay, we found that GMP is the only nucleotide that can form stable hydrogen bonds with O2-EtdT in pretranslocation state and thus observed a dominant n-1 cleavage product (Fig. 4B, in 11G:O2-EtdT panel), whereas other nucleotides (AMP, UMP, or CMP) fail to stably interact with O2-EtdT. We speculate that the wobble base pairing between guanine and O2-EtdT may restrain the flexibility of the thymine base, leading to the direct clash between the ethyl group and P448, whereas the pairing of O2-EtdT with other nucleobases may be flexible enough to allow for local rearrangement of the lesion, thereby avoiding such severe clash. Therefore, we observed extremely slow extension once GMP is incorporated opposite O2-EtdT, which arises from the blockage of translocation mediated by the Pro-Gate loop and is aggravated by wobble base pairing. Future studies are warranted for understanding the biological roles of P448 in transcribing undamaged template, in modulating the transcriptional bypass and stalling of other minor-groove lesions (e.g., N2-alkylated dG derivatives) (48), and in stimulating TCR of these lesions. Taken together, our structural analysis provides mechanistic insights into why the extension step blocked the GMP pathway during the bypass of O2-EtdT, even though this pathway is favorable in the initial insertion step.
The distinct profiles of transcriptional stalling and lesion bypass routes may also confer very different biological consequences. For O2-EtdT, even though the transcription progression is initially stalled, a subsequent error-free TLB is favored and the transcriptional fidelity is essentially maintained. Therefore, O2-EtdT may not severely perturb the accuracy in transmission of genetic information during transcription. On the other hand, transcription elongation through N3-EtdT is slow and mutagenic, which can compromise the fidelity of genetic information flow. It is also interesting to note that the pol II transcriptional bypass of N3-EtdT lesion was greatly compromised in the presence of TFIIS in vitro. This persistent TFIIS-induced transcriptional stalling/arrest may provide enough time for the arrested pol II complex to recruit CSB and other repair factors to initiate TC-NER. Indeed, our previous studies showed that N3-alkylated thymidine derivatives could be substrates for TC-NER (41, 42). It would be interesting for future studies to investigate the potential roles of TFIIS in the TC-NER of N3-EtdT lesion. For O4-EtdT, TLB is relatively efficient and highly mutagenic. Clearly, from our comprehensive analysis of transcriptional effect of alkylation at different positions in thymidine, distinct downstream cellular response could be envisaged. Herein, studies of these regioisomeric alkylated DNA lesions not only elucidated the biochemical basis of transcriptional mutagenesis but also offered fundamental insights into the biological consequences of regioisomeric alkylated thymidine lesions.
Another interesting observation from our study is that the presence of TFIIS causes an elevated transcriptional stalling at N3-EtdT and O2-EtdT lesions, which is in sharp contrast to the canonical well-established roles of TFIIS in promoting transcriptional bypass of a variety of DNA lesions and transcription barriers such as pausing sequences and nucleosomes (50–53). Mechanistically, TFIIS stimulates the cleavage of backtracked transcript (from the inactive form of pol II) and therefore facilitates the recovery of the backtracked pol II (inactive form) back into the active form of pol II (posttranslocation state) to allow new nucleotide addition. Why does the presence of TFIIS apparently lead to different outcomes (bypass/stalling) for different transcriptional barriers? We think that there is a key difference between the cases of natural pause sites/nucleosomes and O2-, N3-EtdT covalent DNA lesions that contributes to the different outcomes of pol II bypass in the presence of TFIIS. For the noncovalent transcription barriers, such as natural pause sites or nucleosomes, while these barriers cause a large portion of pol II to backtrack due to steric hindrance, these barriers can “breathe” (such as transient disruption of local histone–DNA contacts for nucleosome barriers) to transiently relieve the steric hindrance and allow pol II to proceed (for new nucleotide addition). In this scenario, the recovery of pol II from the inactive backtracked state is still the rate-limiting step (i.e., the rate of nucleotide incorporation is still much faster than TFIIS-stimulated cleavage rate). As a result, TFIIS promotes the overall pol II’s bypass efficiency of these barriers. This is also true in the case of certain types of covalent DNA lesions as long as the recovery of pol II from the inactive backtracked state is still the rate-limiting step. In sharp contrast, for O2- and N3-EtdT lesions, the forward translocation and nucleotide incorporation step opposite to these lesions is significantly slowed down (by 105-fold) and becomes the rate-limiting step (i.e., much slower than the backtracking/cleavage rate). In this scenario, the recovery of pol II from the inactive backtracked state is no longer the rate-limiting step. Because TFIIS stimulates the transcript cleavage and drives the equilibrium further favored for the backtracking/cleavage route over the transcriptional bypass route. As a result, we observed futile cycles of backtracking/TFIIS-stimulated cleavage route that lead to persistent pol II stalling at these DNA lesions.
Alkylating agents are ubiquitously present in the environment and they can also be produced by endogenous metabolism (26). In addition, alkylating agents are among the most frequently prescribed cancer chemotherapeutic agents (26). Hence, alkylation constitutes one of most common forms of DNA damage. The results from the present study suggest that the impact of DNA alkylation on transcriptional efficiency and fidelity could be significantly modulated by the positions in DNA to which the alkyl groups are conjugated. This work reveals that minor-groove DNA alkylation is more effective at blocking pol II transcription than its major-groove DNA alkylation counterpart. Future strategies for the rational design of effective alkylating agents can be envisaged, that is, through modulating the dominant conjugation position and the size of the alkyl group. In addition, altering the level or activity of TFIIS could be another layer to modulate the efficacy and toxicity of DNA alkylating agents. Interestingly, it was reported that modulation of TFIIS level could be a potential strategy for anticancer therapy (60). Thus, our work provides important knowledge for the risk assessment of human exposure to DNA alkylating agents and for guiding the development of cancer therapeutic agents that can maximize their toxicity to tumor cells while minimizing the induction of transcriptional mutagenesis.
Materials and Methods
Preparation of Damaged DNA Templates.
Oligodeoxyribonucleotides (ODNs) harboring a site-specifically inserted O2-, N3-, and O4-EtdT were synthesized following previously published procedures (49). The identities and purities of the lesion-containing ODNs were confirmed by electrospray ionization–mass spectrometry (ESI-MS) and tandem-MS (MS/MS) analyses (Figs. S1–S3). The synthesized lesion-containing ODNs were subsequently ligated to provide the templates for in vitro transcription studies.
Transcriptional Elongation.
The Saccharomyces cerevisiae RNA pol II was purified as previously described with IgG affinity column followed by HiTrap Heparin and Mono Q columns for further purification (56, 58). The RNA pol II elongation complexes for in vitro transcription assays were assembled using established methods (57–59, 61–63). Briefly, an aliquot of 5′-32P–labeled RNA (10 µM) was annealed with a 1.5-fold amount of template DNA (15 µM) and twofold amount of nontemplate DNA (20 µM) from 65 °C to room temperature over a 2-h period to form the RNA/DNA scaffold (final stock concentration: 1 µM, defined by RNA concentration) in an elongation buffer, which contained 20 mM Tris⋅HCl (pH 7.5), 40 mM KCl, and 5 mM MgCl2. An aliquot of the annealed RNA/DNA scaffold (50 nM) was then incubated with a fourfold amount of pol II (200 nM) at room temperature for 10 min to ensure the formation of pol II elongation complex for all RNA/DNA scaffold. The in vitro transcription reaction was initiated by mixing the pol II elongation complex with an equal volume of various twofold concentration of NTP solution. The final reaction mixture contained 25 nM scaffold, 100 nM pol II, 5 mM DTT, 5 mM MgCl2, 40 mM KCl, 20 mM Tris⋅HCl (pH 7.5), and NTP. The transcription reactions were quenched at various time points by addition of 1 vol of 0.5 M EDTA (pH 8.0). Samples were then mixed with 1 vol of denaturing PAGE running buffer (100% formamide supplemented with 0.25% bromophenol blue and xylene cyanol) and heated to 95 °C for 5 min to denature the scaffolds. The quenched products were then resolved on 16% denaturing urea/TBE polyacrylamide gels and visualized using a storage phosphor screen and quantitated by Molecular Imager PharosFX Plus system (Bio-Rad).
Single-Turnover Nucleotide Incorporation Transcription Assays.
The assay was carried out as previously described (61–63). Briefly, nucleotide incorporation assays were conducted by preincubating 50 nM scaffold with 200 nM pol II for 10 min in the elongation buffer at 22 °C. The preincubated enzyme:scaffold complex was then mixed with an equal volume of solution containing 40 mM KCl, 20 mM Tris⋅HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, and twofold concentrations of various nucleotides. The final reaction mixture contained 25 nM scaffold, 100 nM pol II, 5 mM MgCl2, and various nucleotide concentrations in the elongation buffer. The transcription reactions were quenched at various time points by addition of 1 vol of 0.5 M EDTA (pH 8.0). Reactions requiring time points shorter than 5 s were quenched using a RQF-3 Rapid Quench Flow (KinTek Corporation) as described (61). Samples were then mixed with 1 vol of denaturing PAGE running buffer (100% formamide supplemented with 0.25% bromophenol blue and xylene cyanol) and heated to 95 °C for 5 min to denature the scaffolds. The quenched products were then resolved on 16% denaturing urea/TBE polyacrylamide gels and visualized using a storage phosphor screen and quantitated by Molecular Imager PharosFX Plus system (Bio-Rad).
Nonlinear-regression data fitting was performed using Prism 6. The time dependence of product formation was fit to a one-phase association equation to determine the observed rate (kobs). The substrate concentration dependence was fit to a hyperbolic equation to obtain values for the maximum rate of NTP incorporation (kpol) and apparent Kd (Kd,app) governing NTP binding, as previously described (64). The specificity constant was determined by kpol/Kd,app.
Intrinsic and TFIIS-Stimulated Transcript Cleavage Assays.
The elongation complex was assembled as described above in 20 mM Tris⋅HCl (pH 7.5) without Mg2+, and intrinsic cleavage was initiated by the addition of Mg2+. The mixture for the final intrinsic cleavage reaction contained 20 mM Tris⋅HCl (pH 9.0), 100 nM pol II, 25 nM scaffold, and 50 mM MgCl2. The reaction was quenched by addition of an equal volume of 0.5 mM EDTA at various time points and analyzed by denaturing PAGE.
Recombinant TFIIS was purified as described (61–63). The elongation complex was assembled as described above in a 20 mM Tris⋅HCl (pH 7.5) buffer without Mg2+. The solution was then mixed with an equal volume of solution containing TFIIS (from 200 nM to 1 µM as indicated in each figure) and MgCl2 in elongation buffer. The final reaction solution contained 100 nM pol II, 25 nM scaffold, 1 μM TFIIS, and 5 mM MgCl2. The reactions were quenched at various time points by addition of an equal volume of 0.5 M EDTA (pH 8.0). The quenched products were again analyzed by 16% denaturing urea/TBE PAGE and visualized using a storage phosphor screen and Pharos FX imager (Bio-Rad).
Analysis of TLB Efficiency.
Transcription bypass efficiency is controlled by the three fidelity checkpoints, that is, nucleotide addition, subsequent extension, and proofreading, where the nucleotide addition and subsequent extension determine the forward transcription elongation behavior, whereas the proofreading induces pol II backtrack (Fig. 5A). Thus, a simplified equation can be generated to describe the efficiency of transcriptional elongation:
where K1, K2, and K−1 designate the efficiencies of nucleotide addition, subsequent extension, and proofreading activity, respectively. Hence, comparing with the bypass after the correct AMP insertion, the relative or normalized TLB efficiency can be described as the following:
where A represents the TLB route after AMP insertion opposite the lesion site and x represents TLB route(s) after the insertion of other nucleotides opposite the lesion site. Considering the proofreading activities of TLB routes were similar for all three EtdT lesions studied here (note: this may not always be the case), the normalized TLB efficiency approximates to the following simplified equation:
Here, the specificity constants (kpol/Kd) can be used to represent the K values in the aforementioned equations. Thus, the normalized TLB efficiency can be calculated as described below:
Estimation of Cellular Concentration of TFIIS.
We estimate cellular concentration TFIIS based on recent global analysis of protein expression and MS-based proteomic data of TFIIS abundance. It was reported that there are 6,260 (65) or 7,254 TFIIS molecules per cell (64), which corresponds to 1.04 × 10−20 [6,260/(6.022 × 1023)] or 1.20 × 10−20 [7,254/(6.022 × 1023)] mol of TFIIS per cell, given that the average volume of a Saccharomyces cerevisiae cell is around ∼30–40 µm3 (or 3–4 × 10−14 L) (66). Therefore, the cellular concentration of TFIIS is estimated to be ≈250 nM [1.04 × 10−20 mol/(4 × 10−14 L)] to ∼400 nM [1.20 × 10−20 mol/(3 × 10−14 L)], which is comparable to the concentrations of TFIIS we used for in vitro TFIIS-stimulated transcript cleavage assays (i.e., 200, 400, and 1,000 nM).
Molecular Modeling and Energy Minimization Methods.
To determine the partial charges of damaged nucleosides (O2-EtdT and O4-EtdT) during GMP incorporation and the subsequent translocation, we performed geometry optimization and electrostatic potential calculation for each molecule. Geometry optimizations were performed using Gaussian 03 with the Hartree–Fock method and the 6-31G* basis set. Electrostatic potential calculations were conducted using the same method and basis set. The partial charges were then calculated by RESP method using resp module in AmberTools 13, where partial charges were determined by a two-stage RESP procedure (67). In the first stage, partial charges of base atoms and the phosphate group were fitted to the electrostatic potential while maintaining the partial charges of sugar groups assigned by AMBER 99SB-ILDN (68). The bonded and Lennard–Jones parameters were taken from general Amber force field (69).
Four structural models were prepared: GTP insertion stages in both the O2-dT and O4-dT templates and the corresponding extension stages thereafter. Each model was solvated in a dodecahedron box with ∼103,000 TIP3P (70) water molecules. For the insertion stages, 408 Na+ and 335 Cl− were added.
For the extension stages, 404 Na+ and 335 Cl− were added. Apart from the damaged sites, AMBER 99SB-ILDN (68) force field was used to describe bonded and nonbonded parameters of amino acid residues and nucleotides. Energy minimization for each model was then performed using the steepest descent method. GROMACS 4.5 (71) simulation package was used for energy minimization. Notably, for O2-dT template with GTP insertion, the first energy minimization was performed by restraining the position of O2-dT, followed by another minimization without any restraint.
Acknowledgments
We acknowledge Dr. Xuhui Huang for his insightful discussion of molecular modeling and simulation. This work was supported by National Institutes of Health Grants R01 GM102362 (to D.W.) and R01 ES025121 (to Y.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708748114/-/DCSupplemental.
References
- 1.Xu L, et al. RNA polymerase II transcriptional fidelity control and its functional interplay with DNA modifications. Crit Rev Biochem Mol Biol. 2015;50:503–519. doi: 10.3109/10409238.2015.1087960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindsey-Boltz LA, Sancar A. RNA polymerase: The most specific damage recognition protein in cellular responses to DNA damage? Proc Natl Acad Sci USA. 2007;104:13213–13214. doi: 10.1073/pnas.0706316104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ljungman M, Lane DP. Transcription—guarding the genome by sensing DNA damage. Nat Rev Cancer. 2004;4:727–737. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- 4.Tornaletti S, Hanawalt PC. Effect of DNA lesions on transcription elongation. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 5.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: Transcription-coupled repair or transcriptional mutagenesis? Chem Rev. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 6.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 7.Belotserkovskii BP, Mirkin SM, Hanawalt PC. DNA sequences that interfere with transcription: Implications for genome function and stability. Chem Rev. 2013;113:8620–8637. doi: 10.1021/cr400078y. [DOI] [PubMed] [Google Scholar]
- 8.Brégeon D, Doetsch PW. Transcriptional mutagenesis: Causes and involvement in tumour development. Nat Rev Cancer. 2011;11:218–227. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, et al. Molecular basis of transcriptional fidelity and DNA lesion-induced transcriptional mutagenesis. DNA Repair (Amst) 2014;19:71–83. doi: 10.1016/j.dnarep.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin JH, Xu L, Wang D. RNA polymerase II acts as a selective sensor for DNA lesions and endogenous DNA modifications. Transcription. 2016;7:57–62. doi: 10.1080/21541264.2016.1168506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin JH, Xu L, Wang D. Mechanism of transcription-coupled DNA modification recognition. Cell Biosci. 2017;7:9. doi: 10.1186/s13578-016-0133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lainé JP, Egly JM. When transcription and repair meet: A complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Svejstrup JQ. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci. 2007;32:165–171. doi: 10.1016/j.tibs.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 15.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LH. DNA damage response and transcription. DNA Repair (Amst) 2011;10:743–750. doi: 10.1016/j.dnarep.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Svejstrup JQ. Rescue of arrested RNA polymerase II complexes. J Cell Sci. 2003;116:447–451. doi: 10.1242/jcs.00271. [DOI] [PubMed] [Google Scholar]
- 19.Damsma GE, Cramer P. Molecular basis of transcriptional mutagenesis at 8-oxoguanine. J Biol Chem. 2009;284:31658–31663. doi: 10.1074/jbc.M109.022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walmacq C, et al. Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions. Proc Natl Acad Sci USA. 2015;112:E410–E419. doi: 10.1073/pnas.1415186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 22.Walmacq C, et al. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol Cell. 2012;46:18–29. doi: 10.1016/j.molcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damsma GE, Alt A, Brueckner F, Carell T, Cramer P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat Struct Mol Biol. 2007;14:1127–1133. doi: 10.1038/nsmb1314. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Zhu G, Huang X, Lippard SJ. X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct. Proc Natl Acad Sci USA. 2010;107:9584–9589. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soll JM, Sobol RW, Mosammaparast N. Regulation of DNA alkylation damage repair: Lessons and therapeutic opportunities. Trends Biochem Sci. 2017;42:206–218. doi: 10.1016/j.tibs.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 28.Swenberg JA, et al. O4-ethyldeoxythymidine, but not O6-ethyldeoxyguanosine, accumulates in hepatocyte DNA of rats exposed continuously to diethylnitrosamine. Proc Natl Acad Sci USA. 1984;81:1692–1695. doi: 10.1073/pnas.81.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godschalk R, et al. Comparison of multiple DNA adduct types in tumor adjacent human lung tissue: Effect of cigarette smoking. Carcinogenesis. 2002;23:2081–2086. doi: 10.1093/carcin/23.12.2081. [DOI] [PubMed] [Google Scholar]
- 30.Den Engelse L, De Graaf A, De Brij RJ, Menkveld GJ. O2- and O4-ethylthymine and the ethylphosphotriester dTp(Et)dT are highly persistent DNA modifications in slowly dividing tissues of the ethylnitrosourea-treated rat. Carcinogenesis. 1987;8:751–757. doi: 10.1093/carcin/8.6.751. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Wang M, Villalta PW, Hecht SS. Liquid chromatography-electrospray ionization tandem mass spectrometry analysis of 7-ethylguanine in human liver DNA. Chem Res Toxicol. 2007;20:1498–1502. doi: 10.1021/tx700147f. [DOI] [PubMed] [Google Scholar]
- 32.Chen HJ, Wang YC, Lin WP. Analysis of ethylated thymidine adducts in human leukocyte DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Anal Chem. 2012;84:2521–2527. doi: 10.1021/ac203405y. [DOI] [PubMed] [Google Scholar]
- 33.Chen HJ, Lee CR. Detection and simultaneous quantification of three smoking-related ethylthymidine adducts in human salivary DNA by liquid chromatography tandem mass spectrometry. Toxicol Lett. 2014;224:101–107. doi: 10.1016/j.toxlet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Chao MR, Wang CJ, Chang LW, Hu CW. Quantitative determination of urinary N7-ethylguanine in smokers and non-smokers using an isotope dilution liquid chromatography/tandem mass spectrometry with on-line analyte enrichment. Carcinogenesis. 2006;27:146–151. doi: 10.1093/carcin/bgi177. [DOI] [PubMed] [Google Scholar]
- 35.Anna L, Kovács K, Gyorffy E, Schoket B, Nair J. Smoking-related O4-ethylthymidine formation in human lung tissue and comparisons with bulky DNA adducts. Mutagenesis. 2011;26:523–527. doi: 10.1093/mutage/ger011. [DOI] [PubMed] [Google Scholar]
- 36.Cline SD, Riggins JN, Tornaletti S, Marnett LJ, Hanawalt PC. Malondialdehyde adducts in DNA arrest transcription by T7 RNA polymerase and mammalian RNA polymerase II. Proc Natl Acad Sci USA. 2004;101:7275–7280. doi: 10.1073/pnas.0402252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dimitri A, Burns JA, Broyde S, Scicchitano DA. Transcription elongation past O6-methylguanine by human RNA polymerase II and bacteriophage T7 RNA polymerase. Nucleic Acids Res. 2008;36:6459–6471. doi: 10.1093/nar/gkn657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimitri A, et al. Transcription of DNA containing the 5-guanidino-4-nitroimidazole lesion by human RNA polymerase II and bacteriophage T7 RNA polymerase. DNA Repair (Amst) 2008;7:1276–1288. doi: 10.1016/j.dnarep.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dimitri A, Goodenough AK, Guengerich FP, Broyde S, Scicchitano DA. Transcription processing at 1,N2-ethenoguanine by human RNA polymerase II and bacteriophage T7 RNA polymerase. J Mol Biol. 2008;375:353–366. doi: 10.1016/j.jmb.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burns JA, Dreij K, Cartularo L, Scicchitano DA. O6-methylguanine induces altered proteins at the level of transcription in human cells. Nucleic Acids Res. 2010;38:8178–8187. doi: 10.1093/nar/gkq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You C, Wang P, Dai X, Wang Y. Transcriptional bypass of regioisomeric ethylated thymidine lesions by T7 RNA polymerase and human RNA polymerase II. Nucleic Acids Res. 2014;42:13706–13713. doi: 10.1093/nar/gku1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.You C, Wang J, Dai X, Wang Y. Transcriptional inhibition and mutagenesis induced by N-nitroso compound-derived carboxymethylated thymidine adducts in DNA. Nucleic Acids Res. 2015;43:1012–1018. doi: 10.1093/nar/gku1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanasova M, et al. Altered minor-groove hydrogen bonds in DNA block transcription elongation by T7 RNA polymerase. ChemBioChem. 2015;16:1212–1218. doi: 10.1002/cbic.201500077. [DOI] [PubMed] [Google Scholar]
- 44.Cheng TF, Hu X, Gnatt A, Brooks PJ. Differential blocking effects of the acetaldehyde-derived DNA lesion N2-ethyl-2′-deoxyguanosine on transcription by multisubunit and single subunit RNA polymerases. J Biol Chem. 2008;283:27820–27828. doi: 10.1074/jbc.M804086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S, Wang Y. Mass spectrometry for the assessment of the occurrence and biological consequences of DNA adducts. Chem Soc Rev. 2015;44:7829–7854. doi: 10.1039/c5cs00316d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan B, et al. The roles of DNA polymerases κ and ι in the error-free bypass of N2-carboxyalkyl-2′-deoxyguanosine lesions in mammalian cells. J Biol Chem. 2011;286:17503–17511. doi: 10.1074/jbc.M111.232835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You C, et al. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat Chem Biol. 2012;8:817–822. doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersen N, Wang P, Wang Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem Res Toxicol. 2013;26:1730–1738. doi: 10.1021/tx4002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kireeva ML, et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 51.Charlet-Berguerand N, et al. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 54.Kalnik MW, Kouchakdjian M, Li BF, Swann PF, Patel DJ. Base pair mismatches and carcinogen-modified bases in DNA: An NMR study of G.T and G.O4meT pairing in dodecanucleotide duplexes. Biochemistry. 1988;27:108–115. doi: 10.1021/bi00401a018. [DOI] [PubMed] [Google Scholar]
- 55.O’Flaherty DK, et al. Lesion orientation of O4-alkylthymidine influences replication by human DNA polymerase η. Chem Sci. 2016;7:4896–4904. doi: 10.1039/c6sc00666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu L, et al. Dissecting the chemical interactions and substrate structural signatures governing RNA polymerase II trigger loop closure by synthetic nucleic acid analogues. Nucleic Acids Res. 2014;42:5863–5870. doi: 10.1093/nar/gku238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015;523:621–625. doi: 10.1038/nature14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu L, et al. RNA polymerase II senses obstruction in the DNA minor groove via a conserved sensor motif. Proc Natl Acad Sci USA. 2016;113:12426–12431. doi: 10.1073/pnas.1612745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hubbard K, Catalano J, Puri RK, Gnatt A. Knockdown of TFIIS by RNA silencing inhibits cancer cell proliferation and induces apoptosis. BMC Cancer. 2008;8:133. doi: 10.1186/1471-2407-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kellinger MW, Ulrich S, Chong J, Kool ET, Wang D. Dissecting chemical interactions governing RNA polymerase II transcriptional fidelity. J Am Chem Soc. 2012;134:8231–8240. doi: 10.1021/ja302077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kellinger MW, Park GY, Chong J, Lippard SJ, Wang D. Effect of a monofunctional phenanthriplatin-DNA adduct on RNA polymerase II transcriptional fidelity and translesion synthesis. J Am Chem Soc. 2013;135:13054–13061. doi: 10.1021/ja405475y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 65.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 66.Ferrezuelo F, et al. The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat Commun. 2012;3:1012. doi: 10.1038/ncomms2015. [DOI] [PubMed] [Google Scholar]
- 67.Cornell WD, Cieplak P, Bayly CI, Kollman PA. Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J Am Chem Soc. 1993;115:9620–9631. [Google Scholar]
- 68.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 70.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 71.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]