Significance
Mitochondria generate the cellular fuel ATP to sustain complex life. Production of ATP depends on the oxidation of energy-rich compounds to produce a chemical potential difference for hydrogen ions, the proton motive force (pmf), across the inner mitochondrial membrane (IMM). Disruption of the IMM, dissipation of the pmf, and cell death occur if the concentration of calcium ions inside mitochondria is sufficiently elevated to open a pore in the IMM. The identity of the pore is disputed. One proposal is that the pore is in the enzyme that makes ATP. Here, we show that proteins in the enzyme’s peripheral stalk are not involved in the formation or regulation of the pore.
Keywords: human mitochondria, ATP synthase, permeability transition pore, ATP5F1 subunit b, ATP5O oligomycin sensitivity conferral protein
Abstract
The opening of a nonspecific channel, known as the permeability transition pore (PTP), in the inner membranes of mitochondria can be triggered by calcium ions, leading to swelling of the organelle, disruption of the inner membrane and ATP synthesis, and cell death. Pore opening can be inhibited by cyclosporin A mediated via cyclophilin D. It has been proposed that the pore is associated with the dimeric ATP synthase and the oligomycin sensitivity conferral protein (OSCP), a component of the enzyme’s peripheral stalk, provides the site at which cyclophilin D interacts. Subunit b contributes a central α-helical structure to the peripheral stalk, extending from near the top of the enzyme’s catalytic domain and crossing the membrane domain of the enzyme via two α-helices. We investigated the possible involvement of the subunit b and the OSCP in the PTP by generating clonal cells, HAP1-Δb and HAP1-ΔOSCP, lacking the membrane domain of subunit b or the OSCP, respectively, in which the corresponding genes, ATP5F1 and ATP5O, had been disrupted. Both cell lines preserve the characteristic properties of the PTP; therefore, the membrane domain of subunit b does not contribute to the PTP, and the OSCP does not provide the site of interaction with cyclophilin D. The membrane subunits ATP6, ATP8, and subunit c have been eliminated previously from possible participation in the PTP; thus, the only subunits of ATP synthase that could participate in pore formation are e, f, g, diabetes-associated protein in insulin-sensitive tissues (DAPIT), and the 6.8-kDa proteolipid.
In 1976, Hunter et al. (1) demonstrated that bovine heart mitochondria respond to the elevation of the concentration of exogenous Ca2+ ions to high levels by opening a nonspecific channel, now known as the mitochondrial permeability transition pore (PTP). Consequently, the mitochondria take up water, their cristae become swollen, and their membranes are disrupted. Since then, these observations have been replicated in mitochondria in situ in many cell types, and other effectors of PTP opening besides an elevated Ca2+ ion concentration have been identified, including phosphate, adenine nucleotide depletion, and thiol oxidants (2).
Today, it is well established that opening of the PTP disrupts ion homeostasis and ATP synthesis, and the mitochondrial membranes lose their integrity, leading to cell death (3). The PTP in isolated mitochondria can be opened experimentally by the introduction of thapsigargin (4), an inhibitor of the Ca2+-ATPase in the sarcoplasmic and endoplasmic reticula, at high nonspecific concentrations. In cultured human cells, the PTP can be opened by providing a route for ingress of exogenous Ca2+ ions by permeabilizing the plasma membrane either with ionophores, such as ferutinin (5), or with the mild detergent digitonin (6). The cytoplasmic Ca2+ ions are taken up into the mitochondrial matrix by the Ca2+ uniporter (7, 8), a component of the inner membrane, and when the total concentration of Ca2+ ions in the mitochondrial matrix is sufficiently elevated, the PTP opens. A characteristic feature of the PTP is that its opening can be inhibited by cyclosporin A (CsA) (9) via the binding of the drug to cyclophilin D (10–13). Cyclophilin D is a prolyl cis-trans isomerase found in the mitochondrial matrix, and it is thought to interact with and modulate the PTP rather than being an integral component (14).
PTP opening and its associated effects have been linked to various human diseases, including cardiac ischemia, neurodegeneration, cancer, and muscle dystrophy, and thus knowledge of the proteins forming the PTP has considerable medical relevance (15). Several possible protein constituents of the PTP have been proposed, including the ADP/ATP translocase, which is the predominant transport protein in the inner membranes of mitochondria, and the voltage dependent anion channel found in the outer membrane of the organelle, but neither of these has withstood scrutiny (16, 17). An alternative idea, that another component of the inner mitochondrial membrane, the AAA-protease SPG7, participates in formation of the PTP has been disputed (18, 19); (AAA is ATPase associated with diverse cellular activities, and SPG7 is a paraplegin matrix AAA-peptidase subunit). Another proposition, which we investigated in the present story, is that the PTP is associated with the dimeric ATP synthase complex (20), another abundant constituent of the inner mitochondrial membrane.
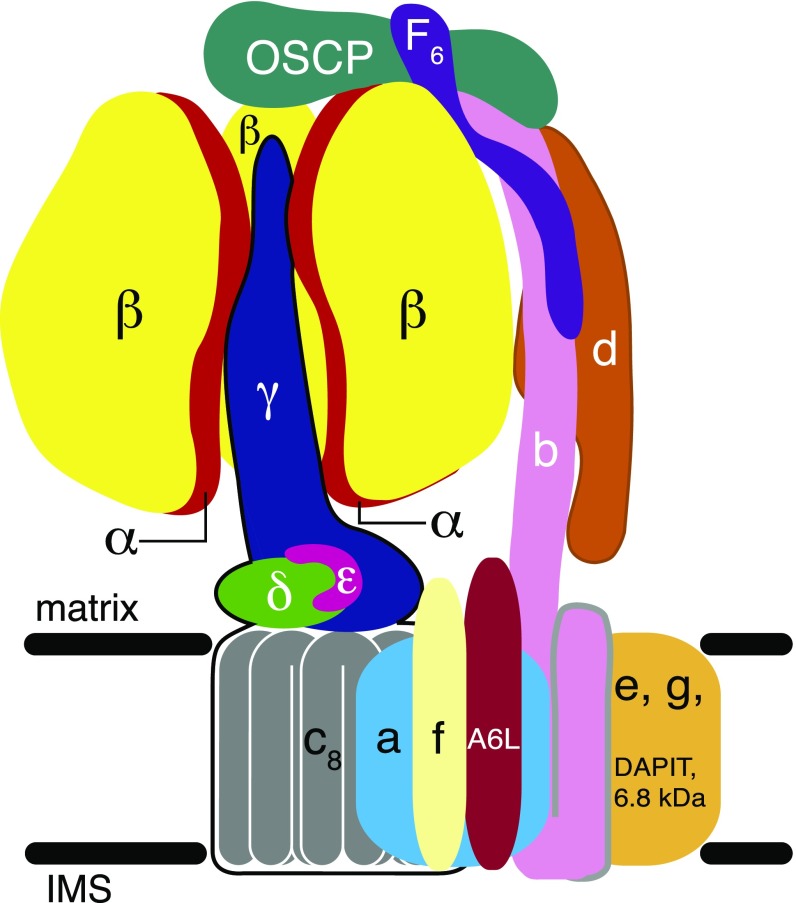
Each monomer of the dimeric mammalian complex is an assembly of 28 proteins of 18 different types organized into two domains (Fig. 1). The F1-catalytic domain sits above the membrane domain, and the two domains are linked by the central stalk (subunits γ, δ, and ε) and the peripheral stalk [subunits b, d, and F6 and oligomycin sensitivity conferral protein (OSCP)] (21). The OSCP has been proposed to provide the site of PTP–cyclophilin D interaction (20).
Fig. 1.
Organization of subunits in one of the monomers of the dimeric ATP synthase complex in mammalian mitochondria. Black horizontal lines represent the limits of the inner membrane between the matrix and the intermembrane space (IMS). The F1 catalytic domain (subunit composition α3β3γδε) is above the membrane; one of the α subunits (red) has been removed to expose the γ subunit (dark blue), lying approximately along the central axis of the spherical α3β3 domain. The γ, δ, and ε subunits are bound to the c8-ring (gray), and together these subunits constitute the rotor. Rotation is generated by the translocation of protons through the interface between the c8-ring and ATP6 (or subunit a; light blue). The peripheral stalk (subunits OSCP, b, d, and F6) is on the right; b has two N-terminal transmembrane α-helices. The membrane domain also contains subunits ATP8 (or A6L), e, f, g, DAPIT and a 6.8-kDa proteolipid (6.8 kDa, or 6.8PL), each with a predicted transmembrane α-helix. The C-terminal region of ATP8, extends into the peripheral stalk; subunits ATP8 and b help keep subunit a in contact with the rotating c8-ring. Subunits e, f, g, DAPIT and 6.8PL are “supernumerary,” with no known roles in the generation or hydrolysis of ATP. In the dimeric complex, subunits e and g likely form the interface between monomers.
If the PTP is associated with the ATP synthase complex, then it likely will involve one or more of the membrane subunits of the enzyme. One specific proposal, that the PTP is provided by a ring of eight c subunits in the membrane sector of the enzyme’s rotor (22, 23), has been disproved in a clonal cell line in which the three genes encoding subunit c have been disrupted (24). Although these cells are incapable of making subunit c, the characteristic properties of the PTP persist (24). Another idea, that two other membrane components of the ATP synthase, subunits a (or ATP6) and A6L (or ATP8), might participate in PTP formation has been disproved as well. In human ρo cells, which lack the mitochondrial genome and thus are devoid of both subunits, the PTP persists (24, 25).
Here, we tested the possible participation of subunit b in the PTP. This subunit has two transmembrane α-helices that help hold subunit a against the c-ring (Fig. 1) (26–28). The remainder of the protein is folded into a single α-helix 150 Å long, extending away from the inner membrane toward the α3β3 domain and providing the core of the peripheral stalk (29, 30). The associated d and F6 subunits are largely α-helical as well, and form a bundle of parallel α-helices with subunit b (26–30). We disrupted the corresponding gene, ATP5F1, in a near haploid cell line, and studied whether removal of subunit b affects the functioning of the PTP. In addition, we investigated whether the OSCP provides the site of binding for cyclophilin D. The OSCP is located at the upper end of the peripheral stalk (Fig. 1) and has two domains. The N-terminal α-helical domain is joined to the α3β3 domain via interactions with the N-terminal regions of the three α subunits, and the C-terminal domain connects the OSCP with the C-terminal region of subunit b (26–28, 30). In the experiments described below, we disrupted ATP5O, the gene encoding the OSCP, and examined the effect of removing the OSCP on the susceptibility to inhibition of PTP opening by CsA mediated via cyclophilin D.
Results
Human Cells Devoid of Subunit b and the OSCP.
HAP1 cells have a haploid karyotype, but a fragment of chromosome 15 is located in chromosome 19, and there is a reciprocal translocation between chromosomes 9 and 22 (31, 32). Neither of these features affects ATP5F1 and ATP5O encoding subunit b and the OSCP, respectively, because ATP5F1 is on chromosome 1 and AT5PO is on chromosome 21. Pairs of guide RNA molecules characteristic of exon I and intron A in ATP5F1 and ATP5O genes were selected (SI Appendix, Fig. S1 and Table S1). Each pair was introduced independently into HAP1-WT cells, and clones arising from single cells, identified as expressing Cas9, were screened for the absence of either subunit b or the OSCP. In this way, HAP1-Δb and HAP1-ΔOSCP cells, lacking subunit b and the OSCP, respectively, were identified (Fig. 2). Analysis of the DNA sequences in the regions of the human genome where ATP5F1 is found in HAP1-Δb and ATP5O is found in HAP1-ΔOSCP (SI Appendix, Table S2 and Fig. S1) showed deletion of 62 and 214 bp, respectively. In addition, a single base had been inserted at the deletion site in ATP5O. Each deletion had arisen from two gRNAs and nonhomologous end-joining of the deleted genomic DNA. Human cells encode a precursor of subunit b where the mature protein is preceded by a mitochondrial import sequence of 42 residues (33). The deletion in ATP5F1 removed the translational initiator codon of the precursor, 20 bases upstream, codons 2–13 plus the first base of codon 14, and extended 2 bases into intron A. The OSCP has an N-terminal mitochondrial import sequence of 23 amino acids (34), and the deletion in ATP5O also removed the translational initiator codon plus 88 bases upstream and codons 2–12, and extended 90 bases into intron A.
Fig. 2.
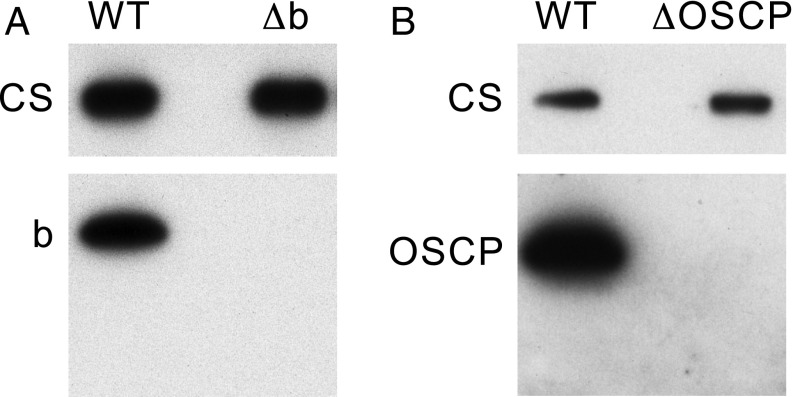
Expression of the b and OSCP subunits of human ATP synthase in HAP1-WT and mutated clonal cells. Mitoplasts from HAP1-WT cells (A) and from HAP1-Δb and HAP1-ΔOSCP cells (B), extracted with dodecylmaltoside, fractionated by SDS/PAGE, and subjected to Western blotting with antibodies against the corresponding subunits b and OSCP. Citrate synthase (CS) served as a loading control.
Characteristics of HAP1-Δb and HAP1-ΔOSCP Cells.
The HAP1-Δb and HAP1-ΔOSCP cells grew more slowly than the HAP1-WT cells (SI Appendix, Fig. S2A), and the copy numbers of mitochondrial DNA were reduced by 8% in the former and by 30% in the latter (SI Appendix, Fig. S2B). Relative to HAP1-WT cells, the levels of complexes I, III, and IV, but not of complex II, were reduced in both derivative cell lines (SI Appendix, Fig. S2C), and thus they have a lower respiratory capacity (SI Appendix, Fig. S2 D and E).
Pore Opening in HAP1-Δb and HAP1-ΔOSCP Cells.
Under the optimum conditions established previously (24), PTP opening in intact HAP1-WT cells was demonstrated in the presence of both thapsigargin and the calcium ionophore ferutinin, and was prevented by the addition of CsA. Similar results were obtained with HAP1-Δb and HAP1-ΔOSCP cells (Fig. 3). Other experiments on PTP opening were conducted with HAP1-WT, HAP1-Δb, and HAP1-ΔOSCP cells in which their plasma membranes had been permeabilized with digitonin. In one set of experiments, the responses of the cells to successive pulses of Ca2+ were monitored with Calcium green-5N in the absence and presence of CsA (Fig. 4 and SI Appendix, Tables S3–S5). On average, the ratios of the number of calcium pulses required to induce the PTP in the presence and absence of CsA were similar: 2.63 ± 0.48 in HAP1-WT cells (n = 8), 2.48 ± 0.42 in HAP1-Δb cells (n = 4), and 2.22 ± 0.36 in HAP1-ΔOSCP cells (n = 6). Thus, in response to pulses of exogenous Ca2+, there was no significant difference in PTP opening in the presence and in the absence of either subunit b or the OSCP. As expected, in HAP1-WT and HAP1-Δb cells, inhibition of the mitochondrial calcium uniporter with Ru360 immediately after a single calcium injection prevented any further uptake of Ca2+ by mitochondria (SI Appendix, Fig. S3).
Fig. 3.
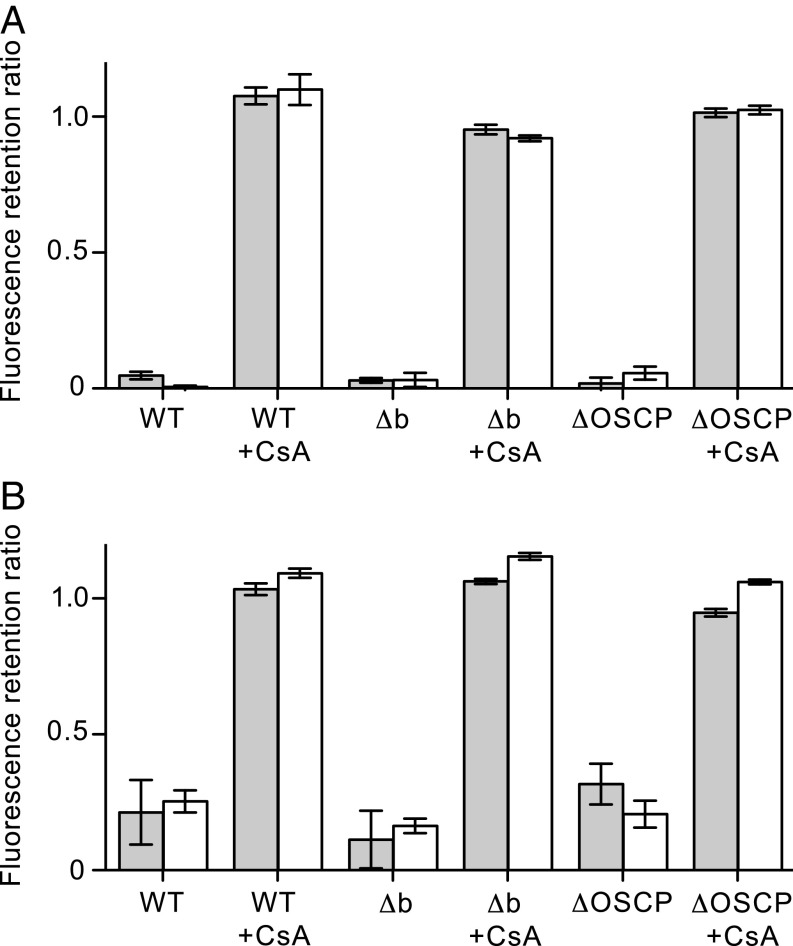
Opening of the PTP in HAP1 cells. PTP opening induced with 40 μM thapsigargin (A) and with 25 μM ferutinin (B). HAP1-WT, HAP1-Δb, and HAP1-ΔOSCP cells were stained with calcein and tetramethylrhodamine methyl ester (TMRM) and then incubated for 1 h in the presence of either thapsigargin or ferutinin. Duplicate samples were incubated first in the presence of 5 μM CsA and then treated with either thapsigargin or ferutinin. Gray and white columns correspond to the retention ratios for calcein and TMRM, respectively, compared with cells treated with the vehicle dimethyl sulfoxide only. The data are mean ± SD (n = 4).
Fig. 4.
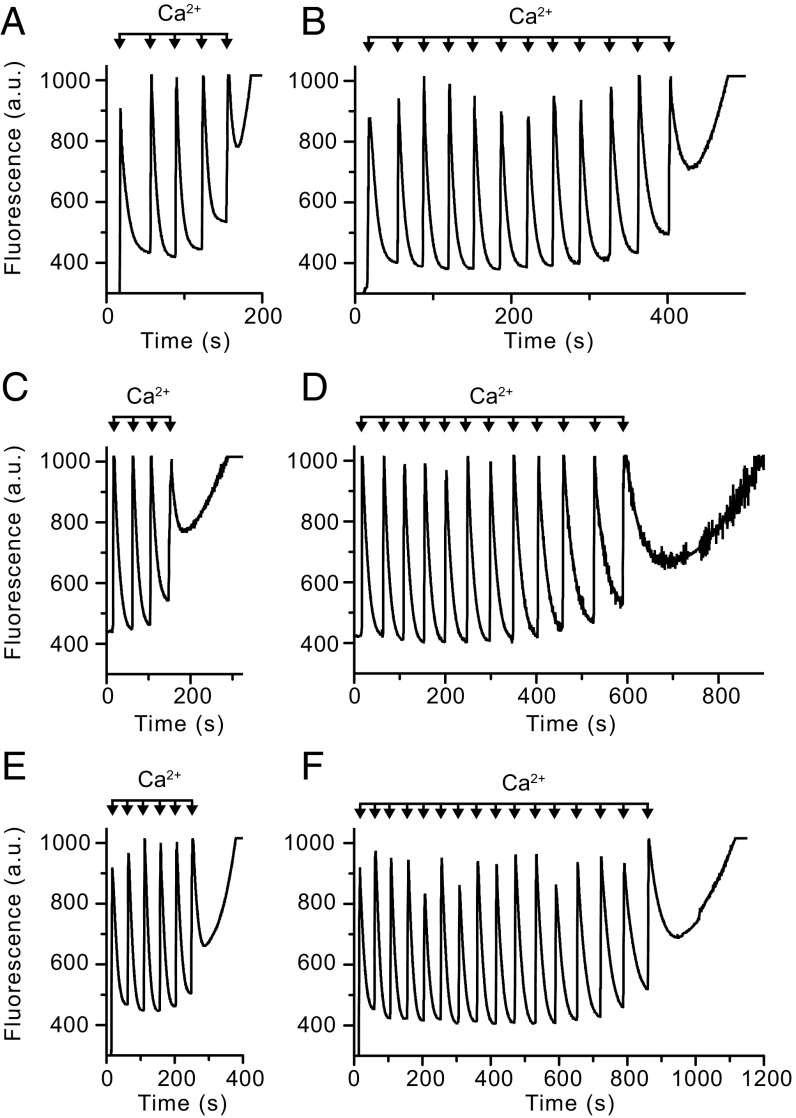
Calcium-induced opening of the PTP in permeabilized HAP1 cells. (A and B) HAP1-WT cells. (C and D) HAP1-Δb cells. (E and F) HAP1-ΔOSCP cells. The calcium retention capacity of mitochondria in digitonin-permeabilized cells (20 × 106 cells/mL) was examined in response to pulses of 10 μM CaCl2. Extramitochondrial Ca2+ was measured with Calcium green-5N fluorescence (a.u., arbitrary unit). Shown is the calcium retention capacity in the absence (A, C, and E) and presence (B, D, and F) of CsA (1 μM).
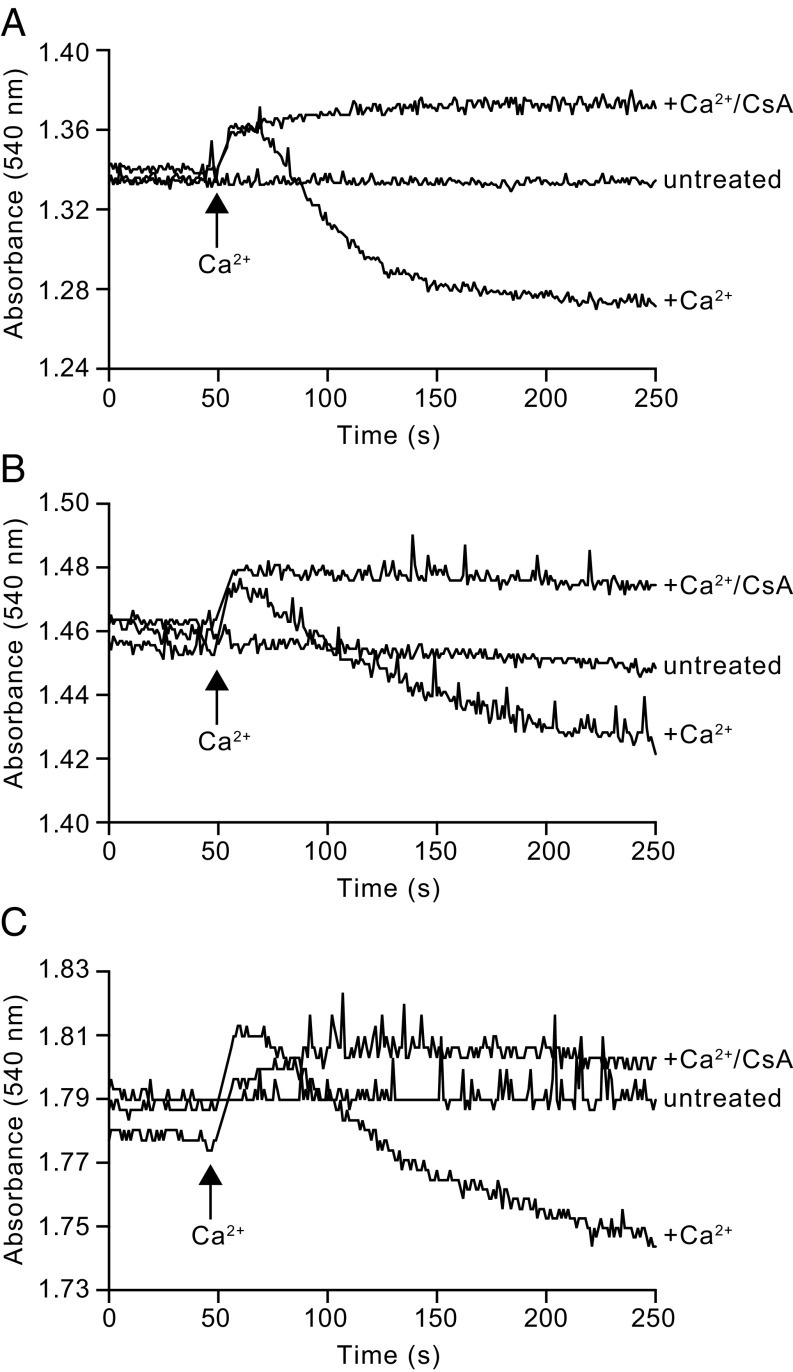
In a second set of experiments with HAP1-WT, HAP1-Δb, and HAP1-ΔOSCP cells, with their plasma membranes permeabilized with digitonin, the decrease in absorbance at 540 nm following the addition of exogenous Ca2+, was consistent with the opening of the PTP and the swelling of the mitochondria in all three cell types. In each case, in the presence of CsA, the addition of exogenous Ca2+ was not accompanied by a decrease in absorption at 540 nm (Fig. 5).
Fig. 5.
Swelling of mitochondria in permeabilized HAP1 cells associated with opening of the PTP. Swelling of 30 × 106 digitonin-permeabilized cells/mL was induced by the addition of 200 μM CaCl2 and monitored by the decrease in absorbance at 540 nm measured in the presence or absence of 1 μM CsA. (A) HAP1-WT cells. (B) HAP1-Δb cells. (C) HAP1-ΔOSCP cells.
Vestigial ATP Synthase Complexes in HAP1-Δb and HAP1-ΔOSCP Cells.
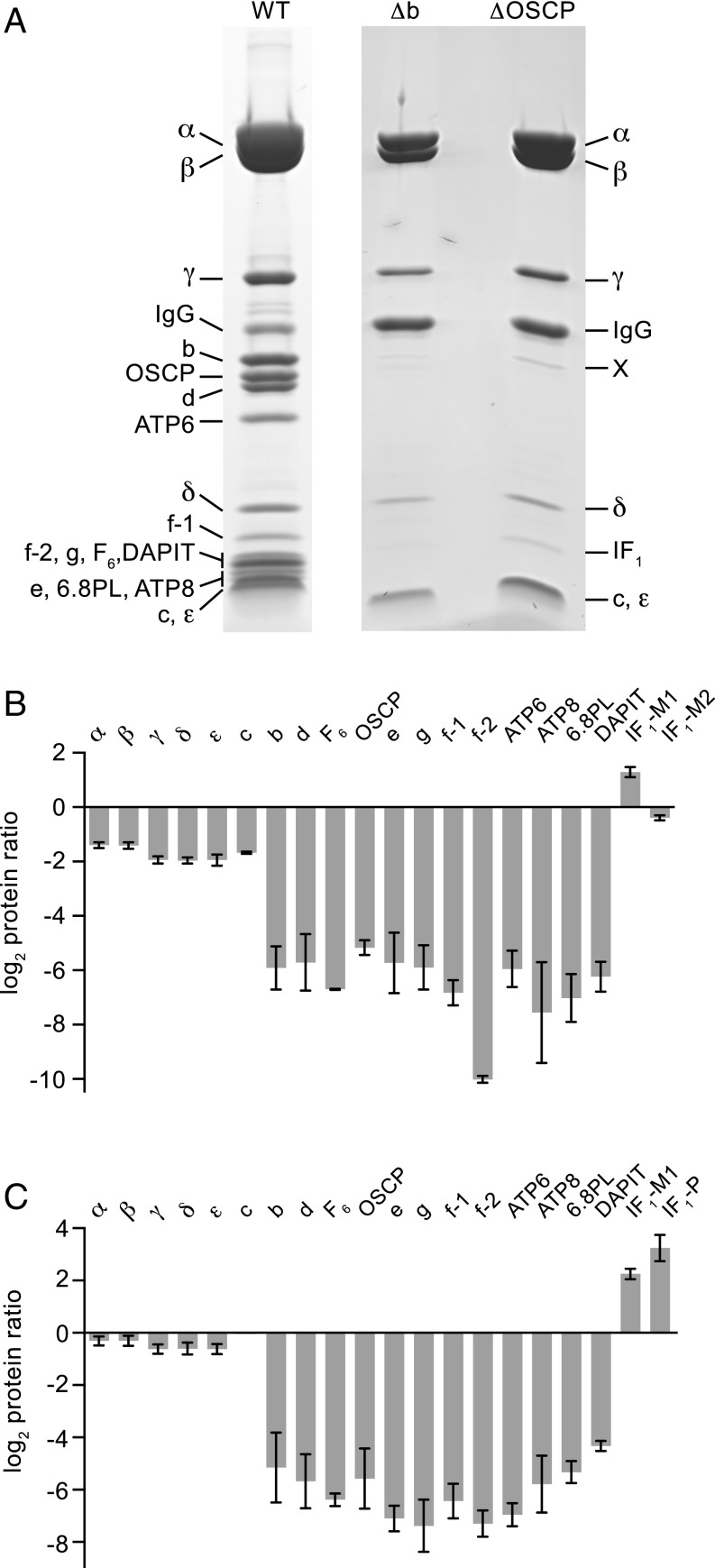
Despite the significant effect of the removal of either subunit b or the OSCP on cellular respiration, the mitochondria of both HAP1-Δb and HAP1-ΔOSCP cells still retain an assembled vestigial ATP synthase complex. Analysis of this complex by SDS/PAGE and MS of the bands revealed a complete complement of the subunits that form the F1-catalytic domain (subunits α, β γ, δ, and ε) plus subunit c; these are the components of the F1-c8 ring subcomplex (Fig. 6A). In HAP1-ΔOSCP cells, an elevated level of one mature form of IF1, IF1-M1, was associated with the complex. Examination of the vestigial complexes by quantitative MS confirmed their subunit compositions (Fig. 6 B and C and SI Appendix, Fig. S4 and Datasets S1 and S2). Also detected were elevated levels of mature IF1 in the complexes from both HAP1-Δb and HAP1-ΔOSCP cells, and of the import precursor of IF1, IF1-P, in the complex from HAP1-ΔOSCP cells. There was a small amount of OSCP in the complex from HAP1-Δb cells, but peripheral stalk subunits d and F6; supernumerary subunits e, f, g, DAPIT and 6.8PL; and the mitochondrial encoded subunits ATP6 and ATP8 were not detected at significant levels. Relative to the levels of intact ATP synthase present in HAP1-WT cells, the level of the vestigial complex was reduced to ≈30% in HAP1-Δb cells and 65% in HAP1-ΔOSCP cells. The quantitative analysis of samples of mitoplasts from HAP1-Δb and HAP1-ΔOSCP cells (SI Appendix, Figs. S4 and S5 and Datasets S3 and S4) confirmed the findings with the purified vestigial complexes, but significant (albeit low) levels of peripheral stalk subunits (F6 and OSCP in HAP1-Δb cells and F6 in HAP1-ΔOSCP cells) remained in the mitoplasts from both cells as well. In addition, supernumerary subunits (e, f, g, DAPIT and 6.8PL) and ATP8 were present at low levels in HAP1-ΔOSCP cells.
Fig. 6.
Effects of the separate deletion of subunit b and the OSCP in HAP1 cells on the human ATP synthase complex. (A) Impact of removal of subunit b and the OSCP on the subunit compositions of the vestigial ATP synthase complexes. The complexes were purified from mitoplasts derived from HAP1-WT, HAP1-Δb, and HAP1-ΔOSCP cells and fractionated by SDS/PAGE. Proteins were stained with Coomassie blue dye and identified by mass spectrometry analysis of tryptic digests of gel bands. In the ΔOSCP track, band X contained peptides from α and γ subunits, and from TMED9, PRDX3 and Rab-7a. (B and C) Relative abundances of subunits of ATP synthase and of forms of the ATPase inhibitor protein, IF1 (24). The intact ATP synthase and the vestigial complexes were purified from 1:1 mixtures of SILAC-labeled cells with HAP1-WT and HAP1-Δb cells (B) and HAP1-WT and HAP1-ΔOSCP cells (C). Tryptic peptides were analyzed by quantitative mass spectrometry. The experiment was performed twice with reciprocal protein labeling. The bars represent median values of both relative abundance ratios determined for proteins identified in the complementary SILAC labeling experiments. Error bars show the range of the two values. In B, for subunit b, peptide data assigned to the truncated form detected in HAP1-Δb cells were excluded from the calculation of the abundance ratio. In C, the OSCP protein ratio for the control light/ΔOSCP heavy mixture was calculated with all available peptide values (n = 174), rather than by the standard procedure, which limits the calculation to four values assigned for peptide pairs with an identified control sequence and an isotopic cluster for the ΔOSCP partner. The relative abundance of subunit c in HAP1-ΔOSCP samples was unchanged, with error bars smaller than the abscissa line. The histograms are derived from SI Appendix, Fig. S4 and Datasets S1 and S2.
The most surprising aspect of these quantitative MS analyses is that they provide evidence for the presence at low levels (0.6%, based on peptide intensities) in the HAP1-Δb cells of five tryptic peptides representing residues 74–89, 90–97, 98–112, 122–129, and 130–141 of the membrane extrinsic region of the mature subunit b. These peptides are derived from a region of the SDS/PAGE gel corresponding to a truncated subunit b (apparent molecular weight 17.5 kDa); however, there was no evidence of peptides originating from the N-terminal region of the b protein. To understand the origin of these peptides, RNA transcripts covering the region that could code for these peptides in HAP1-Δb cells were amplified by PCR and sequenced. These analyses showed that transcription had occurred in HAP1-Δb cells by use of an alternative splice site in intron A (SI Appendix, Fig. S6), allowing translation to begin from the ATG codon encoding methionine-67 in the WT subunit b, thereby producing a truncated subunit b (residues 67–214) lacking the membrane-intrinsic region of the mature subunit b. The quantitative MS experiments showed no evidence of the production of a truncated OSCP subunit by a similar mechanism.
Oligomeric State of Vestigial ATP Synthases.
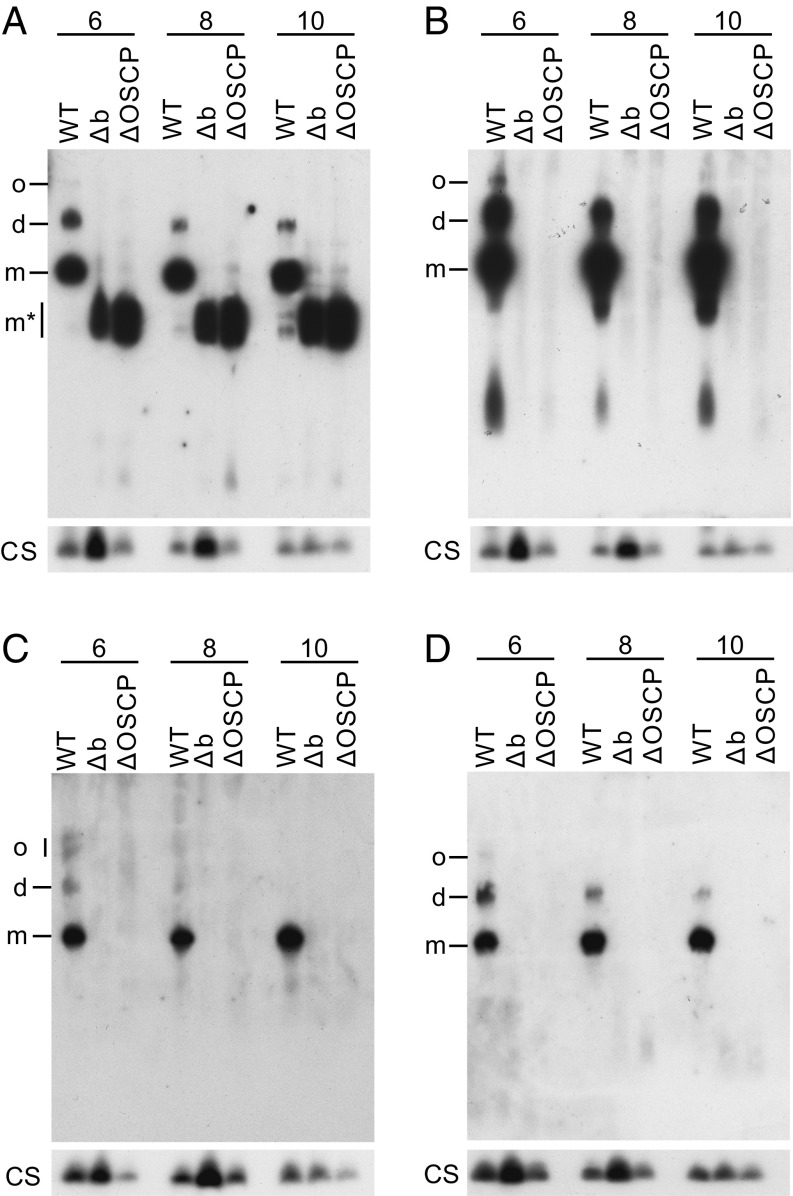
The PTP has been proposed to be associated with dimers and not monomers of the ATP synthase complex. Therefore, the oligomeric state(s) of the vestigial complexes in HAP1-Δb and HAP1-ΔOSCP cells, which are identical in their subunit compositions, were investigated by native gel electrophoresis of digitonin extracts of mitoplasts (Fig. 7). These analyses, done at several concentrations of digitonin, showed that the vestigial ATP synthase complexes from both HAP1-Δb and HAP1-ΔOSCP cells ran to a position on the gels corresponding to a monomeric F1-c8 subcomplex of ATP synthase (Fig. 7A). There was no evidence of a dimeric form of the subcomplex. Moreover, when the gels were probed with antibodies against supernumerary subunit g (Fig. 7B) and peripheral stalk subunits d and F6, there was no evidence of any separate subcomplex containing these subunits.
Fig. 7.
Oligomeric state of ATP synthase in HAP1-WT cells and of vestigial ATP synthase complexes in HAP1-Δb and HAP1-ΔOSCP cells. Mitoplasts from the cells were extracted with the concentrations of digitonin/protein (wt/wt) indicated above each lane, and fractionated by blue native-PAGE. The ATP synthase and vestigial complexes were detected by Western blotting with antibodies against the β subunit (A), g subunit (B), d subunit (C), and F6 subunit (D). Citrate synthase (CS) served as a loading control. o, oligomers; d, dimers; m, monomers; m*, monomeric F1-c8 subcomplex.
Discussion
The experiments described above with HAP1-Δb and HAP1-ΔOSCP cells demonstrate conclusively by four independent assays of the PTP that, even when the membrane region of subunit b or the entire OSCP subunit is absent from their mitochondria, the cells retain a functional PTP. In intact HAP1-Δb and HAP1-ΔOSCP cells, the PTP opens characteristically in response to treatment with either thapsigargin or ferutinin (Fig. 3). Likewise, in permeabilized HAP1-Δb and HAP1-ΔOSCP cells, the PTP opens in response to several pulses of extramitochondrial Ca2+, and a single bolus of 200 μM calcium chloride causes the mitochondria in the permeabilized cells to swell (Figs. 4 and 5). In all of these assays, opening of the PTP could be inhibited by CsA. Therefore, the two transmembrane α-helices of subunit b are not an essential component of the PTP, because the pore remains functional in their absence. Moreover, the OSCP does not provide the site of PTP–cyclophilin D interaction, as the opening of the pore remains sensitive to inhibition by CsA in the absence of the OSCP.
The mitochondria of both HAP1-Δb and HAP1-ΔOSCP cells contain a vestigial ATPase complex with the subunit composition of the F1-domain plus subunit c, presumably representing the F1-c8 component of the intact ATP synthase. The closely related F1-c8 complex from bovine mitochondria has been isolated and characterized (35). This complex is capable of hydrolyzing ATP but is unable to carry out the synthetic reaction. However, the human vestigial complexes in HAP1-Δb and HAP1-ΔOSCP cells are associated with IF1-M1, one of two observed mature forms of IF1, where residues 1–24 have been removed from the mitochondrial import precursor, IF1-P (24). The mature inhibitor protein acts by forming a 1:1 inhibited complex with F1-ATPase (36). The relative molar quantities of IF1-M1 and the vestigial complex have not been measured in either HAP1-Δb and HAP1-ΔOSCP cells, but it is reasonable to assume that at least some of the hydrolytic activity of the vestigial complexes will be inhibited by this protein. In addition, in HAP1-ΔOSCP cells, IF1-P itself is also associated with the vestigial complex (Fig. 6). Previously, this mitochondrial import precursor protein has been observed intact in association with another related but distinct vestigial ATPase complex inside the mitochondria derived from human ρ0 cells (24, 37). Whether IF1-P follows the same pathway of entry into the mitochondria of these cells as IF1-M1, or whether indeed IF1-P is capable of inhibiting ATP hydrolysis, is unknown. In the bovine inhibited complex, the inhibitory region of mature IF1 from residues 21–50 forms an α-helix that occupies a deep groove in the catalytic interface between the αDP and βDP subunits; residues 14–18 interact with the coiled-coil region of the γ subunit via a short α-helix, residues 8–13 form an extended structure, and residues 1–7 are disordered and extend into the central aqueous cavity of F1-ATPase (38, 39). The mode of binding of IF1-P to the subcomplex is not known, but it seems possible that the presence in IF1-P of the additional 24 N-terminal residues might impede binding of the protein to the site occupied by mature IF1 in the inhibited complex.
The removal of either subunit b or the OSCP destabilizes the peripheral stalk, and none of its four constituent subunits (OSCP, b, F6, and d) or the associated ATP8 subunit, is present in the vestigial subcomplex from HAP1-ΔOSCP cells and also in HAP1-Δb cells, although traces of the OSCP were detected in the latter. In the absence of the peripheral stalk and ATP8, ATP6 no longer has any support to maintain its contact with the c8-ring (26–28). Therefore, ATP6 and the supernumerary subunits (e, f, g, DAPIT, and 6.8PL) associated with ATP6 and the membrane domain of subunit b are also absent from the subcomplex (Fig. 6).
As depicted in Fig. 1, subunit f is likely associated with subunits ATP6 and ATP8 (27), and in the dimeric complex, subunits e and g likely form the interfaces between monomers, with subunits DAPIT and 6.8PL together in a more peripheral position, relative to the dimer interface. Clearly, this dimer interface is not present in the vestigial complexes from HAP1-Δb and HAP1-ΔOSCP cells. However, subunits e, f, and g (and other supernumerary subunits) are still present in the mitoplasts of HAP1-Δb and HAP1-ΔOSCP cells, albeit at reduced levels relative to HAP1-WT cells (SI Appendix, Fig. S5). In native gels of digitonin extracts of mitoplasts (Fig. 7), although there was no evidence of a separate subcomplex, monomeric or dimeric, involving subunit g, it remains possible that such a subcomplex was present in the mitoplast membranes, and that it was disrupted by the conditions of extraction with digitonin. As noted earlier, dimers of the integral dimeric ATP synthase itself can become disrupted artifactually by this process (24, 40). Therefore, to eliminate any possibility of the participation of subunits e and g (and subunits f, DAPIT, and 6.8PL) in forming the PTP, it will be necessary to remove each of them by gene disruption experiments and to examine the consequences of doing so.
Materials and Methods
Human HAP1 and mutant cells derived from them were grown under standard conditions. Oxygen consumption rates were measured using a Seahorse XF24 analyzer (Agilent). ATP5F1 and ATP5O were disrupted in HAP1 cells by CRISPR-Cas9 technology (41). ATP synthase and its subcomplexes were purified from mitoplasts by immunocapture. Proteins were subject to stable isotope labeling in cell culture (SILAC). Labeled proteins were quantitated by MS. Opening of the PTP was assayed by four methods. In intact HAP1 cells, PTP opening was induced by thapsigargin (4) or ferutinin (5). In HAP1 cells in which the plasma membrane had been permeabilized with digitonin, PTP opening was assessed by examining the capacity of the mitochondria to retain Ca2+ introduced exogenously (42) and monitoring the swelling of mitochondria in response to a pulse of 200 μM calcium chloride (43) in the absence and presence of CsA.Full details of these processes are provided in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by Medical Research Council, United Kingdom Programme Grant MR/M009858/1 (to J.E.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711201114/-/DCSupplemental.
References
- 1.Hunter DR, Haworth RA, Southard JH. Relationship between configuration, function, and permeability in calcium-treated mitochondria. J Biol Chem. 1976;251:5069–5077. [PubMed] [Google Scholar]
- 2.Zoratti M, Szabò I. The mitochondrial permeability transition. Biochim Biophys Acta. 1995;1241:139–176. doi: 10.1016/0304-4157(95)00003-a. [DOI] [PubMed] [Google Scholar]
- 3.Kwong JQ, Molkentin JD. Physiological and pathological roles of the mitochondrial permeability transition pore in the heart. Cell Metab. 2015;21:206–214. doi: 10.1016/j.cmet.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korge P, Weiss JN. Thapsigargin directly induces the mitochondrial permeability transition. Eur J Biochem. 1999;265:273–280. doi: 10.1046/j.1432-1327.1999.00724.x. [DOI] [PubMed] [Google Scholar]
- 5.Abramov AY, Duchen MR. Actions of ionomycin, 4-BrA23187 and a novel electrogenic Ca2+ ionophore on mitochondria in intact cells. Cell Calcium. 2003;33:101–112. doi: 10.1016/s0143-4160(02)00203-8. [DOI] [PubMed] [Google Scholar]
- 6.Chauvin C, et al. Rotenone inhibits the mitochondrial permeability transition-induced cell death in U937 and KB cells. J Biol Chem. 2001;276:41394–41398. doi: 10.1074/jbc.M106417200. [DOI] [PubMed] [Google Scholar]
- 7.De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baughman JM, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crompton M, Ellinger H, Costi A. Inhibition by cyclosporin A of a Ca2+-dependent pore in heart mitochondria activated by inorganic phosphate and oxidative stress. Biochem J. 1988;255:357–360. [PMC free article] [PubMed] [Google Scholar]
- 10.Tanveer A, et al. Involvement of cyclophilin D in the activation of a mitochondrial pore by Ca2+ and oxidant stress. Eur J Biochem. 1996;238:166–172. doi: 10.1111/j.1432-1033.1996.0166q.x. [DOI] [PubMed] [Google Scholar]
- 11.Basso E, et al. Properties of the permeability transition pore in mitochondria devoid of Cyclophilin D. J Biol Chem. 2005;280:18558–18561. doi: 10.1074/jbc.C500089200. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa T, et al. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- 13.Schinzel AC, et al. Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc Natl Acad Sci USA. 2005;102:12005–12010. doi: 10.1073/pnas.0505294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elrod JW, Molkentin JD. Physiologic functions of cyclophilin D and the mitochondrial permeability transition pore. Circ J. 2013;77:1111–1122. doi: 10.1253/circj.cj-13-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasola A, Bernardi P. The mitochondrial permeability transition pore and its involvement in cell death and in disease pathogenesis. Apoptosis. 2007;12:815–833. doi: 10.1007/s10495-007-0723-y. [DOI] [PubMed] [Google Scholar]
- 16.Kokoszka JE, et al. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–465. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baines CP, Kaiser RA, Sheiko T, Craigen WJ, Molkentin JD. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat Cell Biol. 2007;9:550–555. doi: 10.1038/ncb1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanmughapriya S, et al. SPG7 is an essential and conserved component of the mitochondrial permeability transition pore. Mol Cell. 2015;60:47–62. doi: 10.1016/j.molcel.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.König T, et al. The m-AAA protease associated with neurodegeneration limits MCU activity in mitochondria. Mol Cell. 2016;64:148–162. doi: 10.1016/j.molcel.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Giorgio V, et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc Natl Acad Sci USA. 2013;110:5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 22.Bonora M, et al. Role of the c subunit of the FO ATP synthase in mitochondrial permeability transition. Cell Cycle. 2013;12:674–683. doi: 10.4161/cc.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alavian KN, et al. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc Natl Acad Sci USA. 2014;111:10580–10585. doi: 10.1073/pnas.1401591111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J, et al. Persistence of the mitochondrial permeability transition in the absence of subunit c of human ATP synthase. Proc Natl Acad Sci USA. 2017;114:3409–3414. doi: 10.1073/pnas.1702357114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masgras I, Rasola A, Bernardi P. Induction of the permeability transition pore in cells depleted of mitochondrial DNA. Biochim Biophys Acta. 2012;1817:1860–1866. doi: 10.1016/j.bbabio.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 26.Zhou A, et al. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife. 2015;4:e10180. doi: 10.7554/eLife.10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinothkumar KR, Montgomery MG, Liu S, Walker JE. Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy. Proc Natl Acad Sci USA. 2016;113:12709–12714. doi: 10.1073/pnas.1615902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn A, et al. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol Cell. 2016;63:445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickson VK, Silvester JA, Fearnley IM, Leslie AGW, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rees DM, Leslie AGW, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci USA. 2009;106:21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essletzbichler P, et al. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014;24:2059–2065. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaca Jacome AS, et al. N-terminome analysis of the human mitochondrial proteome. Proteomics. 2015;15:2519–2524. doi: 10.1002/pmic.201400617. [DOI] [PubMed] [Google Scholar]
- 34.Gevaert K, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 35.Watt IN, Montgomery MG, Runswick MJ, Leslie AGW, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Fernandez JC, Harris DA. A thermodynamic analysis of the interaction between the mitochondrial coupling adenosine triphosphatase and its naturally occurring inhibitor protein. Biochem J. 1978;176:967–975. doi: 10.1042/bj1760967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittig I, et al. Assembly and oligomerization of human ATP synthase lacking mitochondrial subunits a and A6L. Biochim Biophys Acta. 2010;1797:1004–1011. doi: 10.1016/j.bbabio.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 38.Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. How the regulatory protein, IF1, inhibits F1-ATPase from bovine mitochondria. Proc Natl Acad Sci USA. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bason JV, Montgomery MG, Leslie AGW, Walker JE. Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase. Proc Natl Acad Sci USA. 2014;111:11305–11310. doi: 10.1073/pnas.1411560111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1Fo-ATP synthase exists as a dimer: Identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy AN, Bredesen DE, Cortopassi G, Wang E, Fiskum G. Bcl-2 potentiates the maximal calcium uptake capacity of neural cell mitochondria. Proc Natl Acad Sci USA. 1996;93:9893–9898. doi: 10.1073/pnas.93.18.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke SJ, McStay GP, Halestrap AP. Sanglifehrin A acts as a potent inhibitor of the mitochondrial permeability transition and reperfusion injury of the heart by binding to cyclophilin-D at a different site from cyclosporin A. J Biol Chem. 2002;277:34793–34799. doi: 10.1074/jbc.M202191200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.