Abstract
Mechanically interlocked molecules that change their conformation in response to stimuli have been developed by synthetic chemists as building blocks for molecular machines. Here we describe a natural product, the lasso peptide benenodin-1, which exhibits conformational switching between two distinct threaded conformers upon actuation by heat. We have determined the structures of both conformers and have characterized the kinetics and energetics of the conformational switch. Single amino acid substitutions to benenodin-1 generate peptides that are biased to a single conformer, showing that the switching behavior is potentially an evolvable trait in these peptides. Lasso peptides such as benenodin-1 can be recognized and cleaved by enzymes called lasso peptide isopeptidases. We show that only the native conformer of benenodin-1 is cleaved by its cognate isopeptidase. Thus thermally-induced conformational switching of benenodin-1 may also be relevant to the biological function of these molecules.
Graphical abstract
Authors are required to submit a graphic entry for the Table of Contents (TOC) that, in conjunction with the manuscript title, should give the reader a representative idea of one of the following: A key structure, reaction, equation, concept, or theorem, etc., that is discussed in the manuscript. Consult the journal’s Instructions for Authors for TOC graphic specifications.

INTRODUCTION
Lasso peptides, one of the many families of ribosomally synthesized and post-translationally modified peptides (RiPPs),1 are naturally-occurring [1]rotaxanes: threaded molecules with one free end and one linked to the ring.2–3 The threaded slipknot structure is achieved via installation of an isopeptide bond between the N-terminal amine and either a glutamic acid or aspartic acid sidechain. The rings of lasso peptides range from 7 aa to 9 aa, or from 23 to 29 backbone atoms.3 Large amino acid sidechains on the thread, referred to as steric locks, often serve as the stopper moieties in lasso peptides. A subset of these peptides can unthread upon heating,4 technically making these compounds pseudo-[1]rotaxanes. In all examples studied thus far, the thermal unthreading of lasso peptides is presumed to be irreversible.
With the advent of genome mining tools for lasso peptides,5–6 alpha-proteobacteria have been a rich source for new examples of these natural products.6–7 In addition to genes for the precursor protein and the two maturation enzymes,8–10 lasso peptide gene clusters from alpha-proteobacteria include genes for an isopeptidase enzyme, a putative TonB-dependent transporter, and several putative regulatory proteins.6 The lasso peptide isopeptidase cleaves the sole isopeptide bond within the lasso peptide, rendering the peptide substrate linear.11 The isopeptidase only cleaves threaded substrates, accommodating the “loop” region of the rotaxane thread in a cavity within the enzyme active site.12–13
Here we describe a new lasso peptide from the alpha-proteobacterium Asticcacaulis benevestitus. This cold-tolerant organism, originally isolated from a soil sample near the town of Vorkuta, Russia at the northern end of the Ural Mountains14 (Figure 1A). We originally identified the cluster using our precursor-directed approach to lasso peptide genome mining,6 but it was also identified using publicly-available genome mining tools such as antiSMASH15 and BAGEL.16 The gene cluster is also identified by more recent global surveys of RiPP biosynthetic gene clusters.17–18 Because the lasso peptide is derived from A. benevestitus, we have named it benenodin-1. Like all other characterized alpha-proteobacterial lasso peptides, the benenodin-1 gene cluster encodes an isopeptidase which linearizes the threaded lasso peptide substrate.11–13, 19 Remarkably, benenodin-1 exhibits switching between two distinct [1]rotaxane conformers upon heating, and thus is a natural example of a switchable mechanically-interlocked molecule (MIM).20–21 Synthetic examples of [1]rotaxanes, several of which exhibit switching, have been reported,22–28 but this property has not been observed previously in any lasso peptide. In addition, we have generated two variants of benenodin-1 with single amino acid substitutions, each of which is biased toward one of the two conformers upon heating. The benenodin-1 isopeptidase is only able to recognize and cleave one of the two conformers, providing new insights into isopeptidase substrate recognition.
Figure 1.
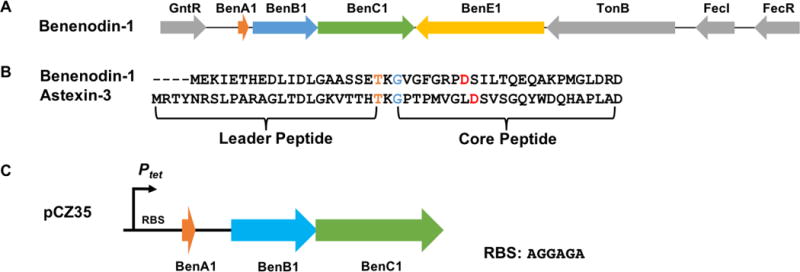
Benenodin-1 gene cluster and sequence. A: The gene cluster for lasso peptide benenodin-1 showing the highly conserved architecture typical of lasso peptides produced in alpha-proteobacteria. The A, B, and C genes are required for lasso peptide biosynthesis while the E gene encodes lasso peptide isopeptidase. B: Sequence of benenodin-1 aligned with astexin-3. The conserved threonine in the leader peptide is colored orange, the isopeptide-bonded glycine is colored blue, and the isopeptide-bonded aspartic acid is colored red. C: Engineered gene cluster for the heterologous expression of benenodin-1 in E. coli.
RESULTS AND DISCUSSION
Heterologous expression of benenodin-1 lasso peptide
Based on its sequence, benenodin-1 has an 8 aa ring with an isopeptide bond between Gly-1 and Asp-8 (25 backbone atoms, Figure 1B). The isopeptide-bonded ring of benenodin-1 is one amino acid shorter than the previously characterized astexin-3 (Figure 1B).11 We grew A. benevestitus, extracted the cell lysate and supernatant, and subjected the extracts to MALDI-MS, but did not find any masses consistent with benenodin-1. We6 and others7, 29–30 have demonstrated that alpha-proteobacterial lasso peptides are often produced at mg/L yields via heterologous expression in E. coli. We constructed a plasmid for the heterologous production of benenodin-1 (pCZ35, see Supporting Information for details) in E. coli (Figure 1C). The native benenodin-1 cluster was cloned into the pASK75 expression vector containing a strong tet promoter and a ribosome binding site upstream. The natural stem-loop sequence between benenodin-1 lasso peptide precursor and the maturation enzymes was kept intact. We expressed the construct in M9 media, and the cell lysate was analyzed by mass spectrometry and HPLC. For cells harboring pCZ35, both MALDI-MS and HPLC data revealed the dominant product was a truncation of benenodin-1 lacking its 5 C-terminal aa (benenodin-1 ΔC5, retention time 14.7 min) (Figure S1). Smaller amounts of a ΔC4 truncation (retention time 14.4 min) were observed in HPLC, while a small signal for the full-length peptide was observed by mass spectrometry.
NMR structure of benenodin-1 ΔC5
Based on HPLC analysis, benenodin-1 ΔC5 was the major product of heterologous expression, so we chose this variant for structural studies. A sample of benenodin-1 ΔC5 (2.5 mg/mL) was prepared in 95% H2O/D2O, and COSY, TOCSY, NOESY, 13C-HSQC and 15N-HSQC spectra were acquired. TOCSY and NOESY spectra (Figure S2) were used for proton assignment (Table S1) and generating distance constraints. Simulated annealing of the structure was carried out using CYANA 2.1, and followed by an energy minimization step in explicit water using GROMACS (see supplemental methods for additional details on NMR and structure calculations). NOEs were observed between Gln-15 and each of the 8 aa in the ring (Figure 2A, Table S2). Similarly, Glu-14 exhibited NOEs to 6 of the 8 ring amino acids (Figure 2A. Table S2). These data strongly implicate Glu-14 and Gln-15 as the steric lock residues bracketing the ring. This is in contrast to most other lasso peptides which have bulkier aromatic or large positively-charged amino acids as steric lock residues.2–3, 31–32 The 20 lowest energy structures of benenodin-1 ΔC5 are shown in Figure 2. Overall, benenodin-1 ΔC5 has an 8 aa ring with 6 aa in the threaded loop and 5 aa in the tail.
Figure 2.
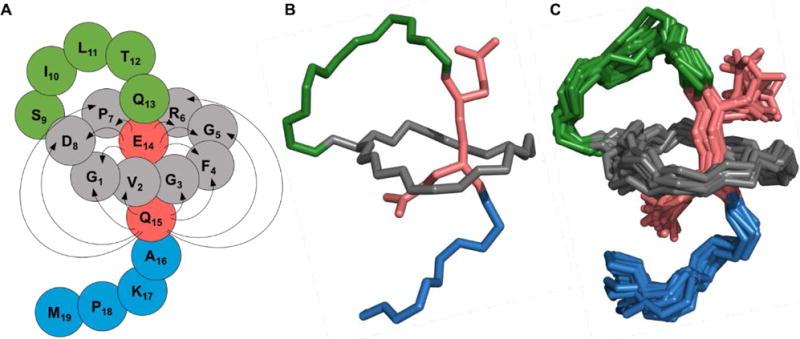
NMR structure of benenodin-1 ΔC5. A: Cartoon of the structure with grey residues representing the ring of the peptide, lime green residues in the loop, and blue residues in the tail. The steric lock residues Glu-14 and Gln-15 are shown in red. Arrows show NOEs observed between ring residues and steric lock residues. B: The lowest energy conformer of benenodin-1 showing the backbone of the peptide and the sidechains of the steric lock residues. Coloring is the same as in the cartoon in part A. C: As in part B, but showing the 20 lowest energy structures.
Thermally induced conformational switching of benenodin-1 ΔC5
Based on its NMR structure, we expected benenodin-1 ΔC5 to be thermolabile due to its small steric lock residues, Glu-14 and Gln-15. To examine its thermal stability, benenodin-1 ΔC5 was incubated at 95 °C for up to 18 h, and then analyzed via HPLC and MALDI-MS at different times. A second peak with the same mass but a different retention time (14.3 min) was observed. In contrast to previous studies of lasso peptide unthreading, the area of the second peak plateaued after an incubation time of 2 hours (Figure 3A). This peak was collected and treated with carboxypeptidase B and Y, which reports on the threaded state of the peptide.4, 7 No C-terminal degradation products were observed (Figure S3), suggesting that the peptide in this new peak is still threaded. The peptide in the 14.3 min retention time peak was isolated and subjected to heat treatment at 95 °C. The same ratio of the two peaks was achieved upon heating for 2 h or more (Figure 3B). These data suggest that, upon heating, benenodin-1 ΔC5 achieves an equilibrium between two threaded conformers: conformer 1, the native state, and conformer 2, a putative partially unthreaded state. In this way, benenodin-1 functions as a thermally actuated rotaxane switch.
Figure 3.
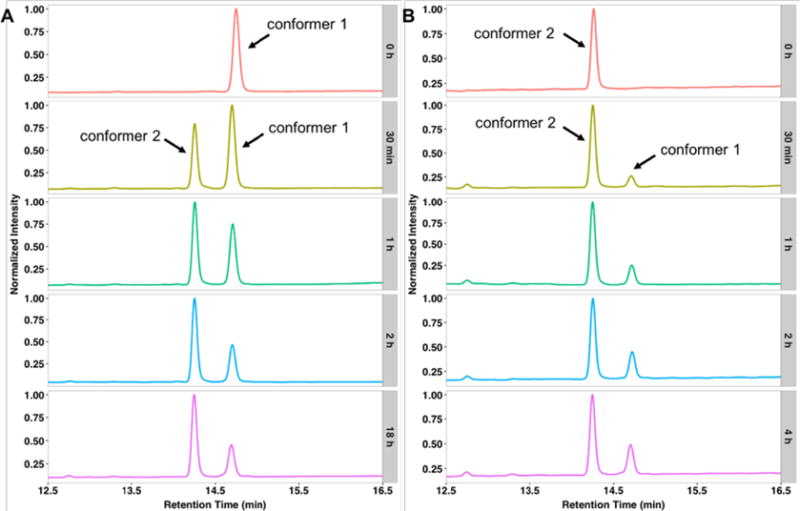
Benenodin-1 establishes an equilibrium between two conformers upon heating. A: Chromatograms of native benenodin-1 ΔC5 heated at 95 °C for the indicated times (grey boxes). About 60% of the native peptide (conformer 1, retention time 14.7 min) is converted into a new species (conformer 2, retention time 14.3 min). B: Heating of conformer 2 of benenodin-1 at 95 °C leads to the same equilibrium ratio of the two conformers.
To gain further insight into this conformational switch, we solved the structure of conformer 2 of benenodin-1 ΔC5 (Table S1). In stark contrast to the NOESY spectrum of the native conformer 1 structure, conformer 2 has only a single NOE between Glu-14 and the ring amino acids (Figure S4, Table S2). The number of NOEs between Gln-15 and the ring is also reduced in conformer 2 (Table S2). Instead, the ring is now in close proximity to Lys-17, as evidenced by NOEs between this residue and 6 of the 8 ring residues (Figure 4A). The energy minimized structure of conformer 2 (Figure 4B) shows that the Ala-16 sidechain is nearly completely buried within the ring of the peptide, and that Lys-17 is functioning as a secondary steric lock/stopper that prevents further unthreading of the peptide. Whereas conformer 1 has loop length of 6 aa and a tail of 5 aa, conformer 2 has a loop of 8 aa (including Ala-16) and a tail of only 3 aa (Figure 4C).
Figure 4.
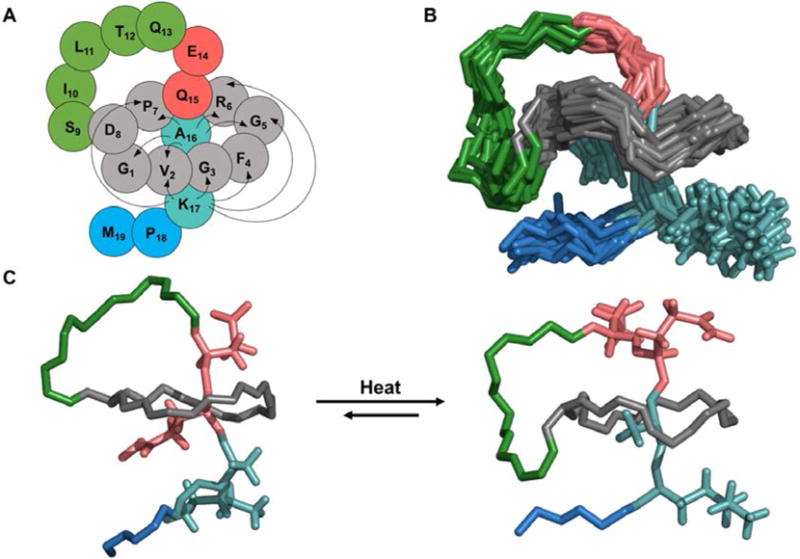
Conformer 2 of benenodin-1 ΔC5 is partially unthreaded. A: Cartoon representation of the benenodin-1 ΔC5 conformer 2 structure showing NOEs between the ring residues (grey) and the new steric lock residues (Ala-16 and Lys-17, turquoise). B: 20 lowest energy structures of benenodin-1 ΔC5 conformer 2 with the same coloring as part A and showing the sidechains of Glu-14, Gln-15, Ala-16, and Lys-17. C: Comparison of the lowest energy structures of conformer 1 (left) and conformer 2 (right) of benenodin-1 ΔC5.
Kinetics and energetics of conformational switching
To probe the kinetics and energetics of the switch between the two benenodin-1 conformers, we carried out a series of heating experiments at temperatures ranging from 35–95 °C (Figure 5). The kinetics of the conformational switch from 55–95 °C fit well to a simple two state model, and the resulting rate constants fit well to an Arrhenius model (Figure S5), allowing for an estimation of the barrier height between the two states. We did not include the data for 35 °C and 45 °C in these fits because equilibrium was not reached over the course of a week-long experiment. For the conversion of conformer 1 to conformer 2, the activation energy was estimated to be 27 kcal/mol while the reverse activation energy was estimated as 23 kcal/mol. Assuming no hysteresis in the conformational switch, this implies that conformers 1 and 2 have comparable stability, differing by only about 4 kcal/mol (Figure 5). The equilibrium ratio [conformer 2]/[conformer 1] also varies with temperature such that ~63% of the peptide switches to conformer 2 at 95 °C, but only ~45% of the peptide is switched to conformer 2 at 55 °C (Table S3). The equilibrium data fit reasonably well to a van’t Hoff model (Figure S5), which can be extrapolated to estimate the ratio of the peptide in each conformer at the temperature at which A. benevestitus lives. At equilibrium at the average summer temperature of Vorkuta, Russia (13 °C), ~35% of the peptide will exist in conformer 2. However, based on extrapolations of our kinetic data, it would take months for this equilibrium to be achieved. These fits also explain why we did not observe any benenodin-1 conformer 2 in our expressions of the peptide, which were carried out at 20 °C. At this temperature, only 0.2% of the peptide is expected to exist in conformer 2 (Eq. 2, Supplementary Methods).
Figure 5.
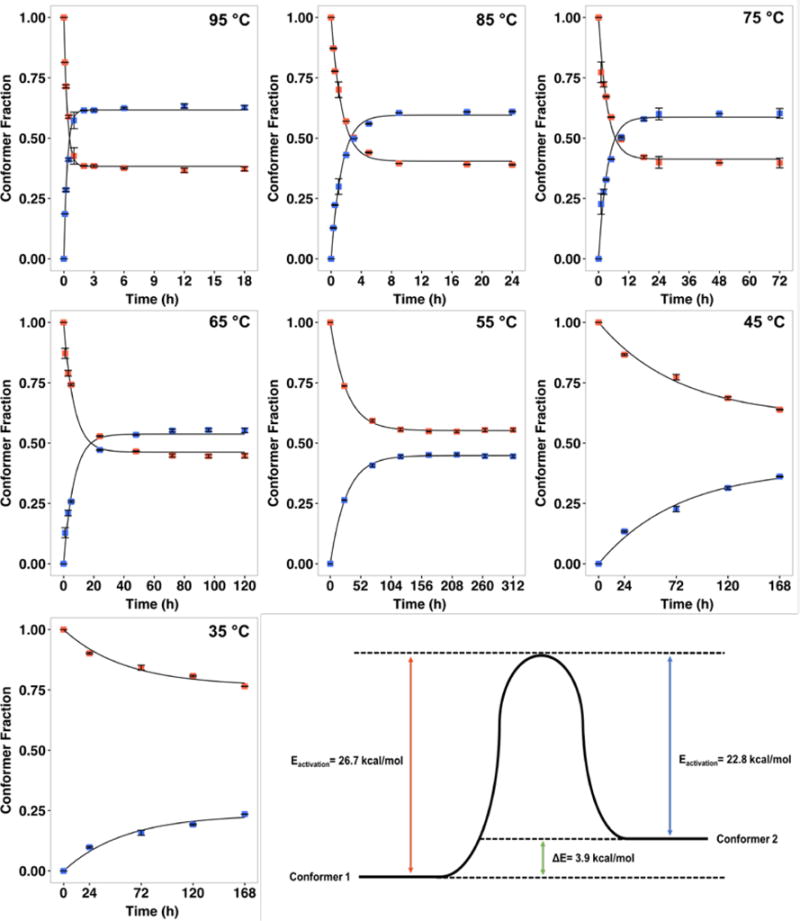
Kinetics and energetics of conformational switching in benenodin-1. Native benenodin-1 ΔC5 (conformer 1) was heated at each of the indicated temperatures and the extent of switching was determined using HPLC. Red points represent conformer 1, blue points are conformer 2, and the solid lines represent fits to a two-state model (see Supporting methods for details). Error bars represent the standard deviation of two measurments, goodness of fit R2 is greater than 0.97 for all fits. These data were used to calculate rate constants of switching and equilibrium ratios of the two conformers (Table S3). The temperature dependence of the rate constants was fit to an Arrhenius model in order to estimate the activation energy of switching, giving an estimate of the energy difference between the two conformers (lower right panel).
Conversion of benenodin-1 conformers into [2]rotaxanes
It has been demonstrated previously that the lasso peptide microcin J25 can be converted into a [2]rotaxane via protease cleavage of its loop region.33–34 Microcin J25 has large steric lock residues (Phe and Tyr), so its thread is locked firmly in place within the ring. We were interested in whether similar [2]rotaxanes could be generated from the two conformers of benenodin-1 given its small steric lock residues. To this end, we generated a T12R variant of benenodin-1, installing a trypsin cleavage site within the loop of the peptide (Figure S6A). This variant was successfully produced by the benenodin-1 maturation machinery, though at ~10% of the yield of the wild-type peptide. After heating the peptide at 95 °C for 3h, it exhibited switching similar to the wild-type peptide (Figure S6B). We subjected the mixture of conformer 1 and conformer 2 of this variant to trypsin cleavage for 16 h at 37 °C to generate potential [2]rotaxanes. Under these conditions, conformer 2 was cleaved completely while only ~1/4 of the conformer 1 peptide was cleaved (Figure S6B). This finding is consistent with our structures for the two conformers. In conformer 2, the Arg-12 residue is more accessible to the protease than in conformer 1 since the Arg-12 is moved away from the ring in conformer 2 (Figure S6A). LC-MS analysis of these cleaved peptides shows that they are indeed [2]rotaxanes with the threaded structure remaining intact (Figure S6C).
Mutagenesis of steric lock residues in benenodin-1
Alanine scanning mutagenesis is commonly employed in studies of lasso peptide stability,4, 35 as it can confirm proposed roles of steric lock residues. We generated the K17A, Q15A, and E14A variants of benenodin-1 ΔC5 and studied their thermostability at 95 °C. After heating the K17A variant (retention time 15.6 min) for 3 h at 95 °C, nearly all of the material shifted to a new peak with retention time 13.5 min (Figure S7). This large change in retention time suggested that the peptide may have fully unthreaded. To confirm this, the material at 13.5 min was collected and subjected to carboxypeptidase treatment. After carboxypeptidase treatment, a new peak at 9.5 min appeared, and MALDI-MS analysis revealed a m/z of 873.39, corresponding to removal of 10 residues from the C-terminal tail of benenodin-1 ΔC5. Collectively, these data provide a strong argument that the Lys-17 residue is indeed serving as a secondary steric lock residue in conformer 2 as its removal allows for further irreversible unthreading of the lasso.
Based on both HPLC and the carboxypeptidase assay (Figure S8), the Q15A construct remained in a single conformation after heating for 3 h at 95 °C and thus did not display the thermally induced switching of the wild-type peptide. This indicates that the Q15A variant is biased toward conformer 1. The E14A variant had unusual behavior on the HPLC, running as a broad peak with a retention time of 17.5 min (Figure S9A), suggesting that this peptide may form a non-specific aggregate at the concentrations used for HPLC analysis. Heat treatment of the E14A variant resulted in a new, sharp peak at 14.4 min, back in the range of other benenodin variants (Figure S9C). We propose that this new peak corresponds to a conformer 2 state. Carboxypeptidase treatment of the material at 17.5 min resulted in the cleavage of a single amino acid from the C-terminus, generating benenodin-1 E14A ΔC6 (Figure S9B). In contrast, carboxypeptidase did not cleave the material eluting at 14.4 min (Figure S9D), providing further support for the proposal that this peak corresponds to a peptide resembling conformer 2.
Substrate recognition by benenodin-1 isopeptidase
As discussed above, a hallmark of alpha-proteobacterial lasso peptide gene clusters is an isopeptidase (Figure 1) that specifically cleaves the threaded [1]rotaxane form of the lasso peptide into a linear product. Based on our recently published structure of the astexin-2/astexin-3 isopeptidase AtxE2,13 we generated a homology model of the benenodin-1 isopeptidase BenE using I-TASSER36–37 (Figure S10). Overall, the predicted structure of BenE is very similar to that of AtxE2 with a slight narrowing of the substrate cleft, consistent with the smaller ring size of the benenodin-1 substrate relative to astexin-3. We were interested in whether the two different conformers of benenodin-1 ΔC5 could be cleaved by the BenE enzyme associated with the benenodin-1 gene cluster. Reactions were set up under conditions that led to complete cleavage of the lasso peptide astexin-3 by its cognate isopeptidase11–12 with the benenodin-1 substrate present at 100 μM and the BenE enzyme at 100 nM. After a 16 h reaction at 20 °C, all of benenodin-1 conformer 1 was hydrolyzed, as evidenced by a change in retention time on HPLC, and confirmation by MALDI. However, when an equilibrium mixture of conformers 1 and 2 was treated under the same conditions, conformer 2 was not cleaved (Figure 6A, Figure S11). This finding is consistent with our previous mutagenesis and structural studies on the astexin-3 isopeptidase.12–13 In this work we showed that the loop region of the lasso peptide was most important for its recognition by the isopeptidase. Since conformer 2 has a larger loop than the native form of the peptide (conformer 1), we propose that conformer 2 is no longer able to be accommodated within the active site of the enzyme.
Figure 6.
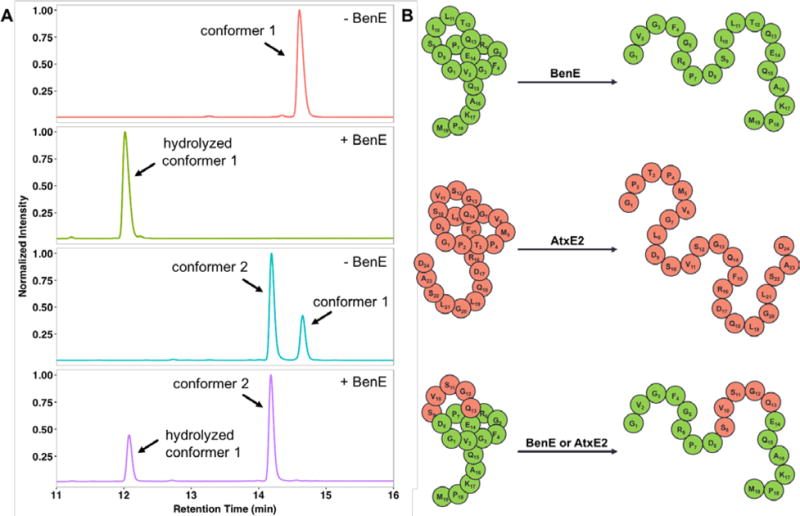
Substrate specificity of benenodin-1 isopeptidase BenE. A: BenE can only hydrolyze conformer 1 of benenodin-1. Benenodin-1 ΔC5, conformer 1 (red trace) is completely hydrolyzed by BenE (green trace). In a mixture of the two conformers generated by heating (turquoise trace), only conformer 1 is hydrolyzed (purple trace). B: A chimeric substrate comprised of the loop residues of astexin-3 (red) grafted onto benenodin-1 (green) is successfully hydrolyzed by both the benenodin-1 isopeptidase BenE and the astexin-3 isopeptidase AtxE2.
To probe the importance of the loop size further, we generated a chimeric lasso peptide in which the first five loop residues of astexin-3 (SVSGQ, Figure 1B) replaced the same 5 loop residues of benenodin-1 (SILTQ). This “loop swap” only requires 3 relatively conservative amino acid substitutions (Figure 6B), and was correctly processed into a lasso peptide by the benenodin maturation machinery. We also attempted to produce the opposite “loop swap” chimera in which the benenodin-1 loop replaced the astexin-3 loop. Unfortunately, the astexin-3 maturation machinery did not process this substrate into a lasso peptide. While the loop size of astexin-3 and benenodin-1 is identical, it should be noted that astexin-3 has a ring of 9 aa while benenodin’s ring is only 8 aa. The benenodin-1/astexin-3 chimera was successfully hydrolyzed by both the astexin-3 isopeptidase AtxE211–12 and the benenodin-1 isopeptidase BenE (Figure 6B, Figure S12). The chimeric peptide is actually a better substrate for AtxE2 than it is for BenE, despite the fact that all of the chimera except for the loop is derived from benenodin-1. In control experiments (Figure S13), we found that AtxE2 did not cleave wild-type benenodin-1 ΔC5 (conformer 1) and that BenE did not cleave astexin-3. These data further support the idea that the loop segment of lasso peptides serves as the primary recognition element for the isopeptidase. In the case of AtxE2, the chimeric substrate has the “wrong” ring size (i.e. one aa smaller than its native substrate, Figure S14), but the enzyme is still able to cleave the chimera due to the shape complementarity of the loop.
Since the BenE isopeptidase can discriminate between the two conformers of wild-type benenodin-1, we tested each of the alanine variants described above, both before and after heating, as substrates for BenE. As produced by the bacteria, in a putative conformer 1 state, each of the alanine variants (E14A, Q15A, and K17A) was hydrolyzed by the iso-peptidase (Figure S15). As expected, the unthreaded K17A variant, generated by heating, was not a substrate for BenE (Figure S15). We proposed above that the Q15A variant remains in a conformer 1 state after heating, and this was borne out by isopeptidase treatment experiments that showed complete hydrolysis of the heated Q15A peptide by BenE (Figure S16). Similarly, we proposed that, after heating, the E14A variant was biased toward conformer 2. In accordance with this, the heated E14A peptide was not cleaved by BenE (Figure S16). These isopeptidase assays confirm that the Q15A variant of benenodin-1 is biased toward conformer 1 upon heating while the E14A variant displays the opposite behavior and switches exclusively to conformer 2.
CONCLUSION
Here we describe the isolation of a new lasso peptide natural product from A. benevestitus that we have named benenodin-1. This name, which translates roughly to “good knot,” turned out to be particularly apt, because the peptide remains in its knotted state upon extensive heating. In fact, upon heating, the peptide establishes an equilibrium between two distinct slipknotted conformers, a property never before observed for lasso peptides. Fitting of the conformational switching data at different temperatures to an Arrhenius model allowed us to determine the energy landscape of the two conformers. This work shows that, in addition to synthetic chemists, nature has also built MIMs that can act as thermally actuated switches. Since lasso peptides are directly gene-encoded, it means that these MIMs are subject to evolutionary pressures in nature as well as being readily engineered in the laboratory. We have shown here that single amino acid substitutions in the steric lock/stopper amino acids can lead to three different outcomes upon heating: irreversible unthreading, bias toward the native conformer, and bias toward the switched conformer.
In other recent work from our laboratory, we demonstrated that the lasso peptide microcin J25 could be cleaved in its loop region, generating a [2]rotaxane, and reassembled using disulfide bonding into radial [3] and [4]catenanes.34 Due to the bulky steric lock residues of microcin J25, however, these catenanes were rigid and thus unable to exhibit any switching behavior. The benenodin-1 peptide, with its built-in switching capability and its ability to be cleaved within its loop (Figure S6), is a prime candidate for building conformationally complex peptide-based MIMs such as catenanes that can exhibit switching behavior and more sophisticated properties.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM107036
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:
The supplementary document includes detailed methods, 16 figures and 3 tables. Coordinates for both benenodin-1 conformers have been uploaded to the PDB and BMRB: conformer 1 (PDB: 5TJ1 and BMRB: 30188) and conformer 2 (PDB: 5TJ0 and BMRB: 30187).
ORCID
A. James Link: 0000-0001-5066-9691
Notes
The authors declare no competing financial interests.
References
- 1.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maksimov MO, Pan SJ, Link AJ. Nat Prod Rep. 2012;29:996–1006. doi: 10.1039/c2np20070h. [DOI] [PubMed] [Google Scholar]
- 3.Hegemann JD, Zimmermann M, Xie X, Marahiel MA. Acc Chem Res. 2015;48:1909–1919. doi: 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- 4.Allen CD, Chen MY, Trick AY, Le DT, Ferguson AL, Link AJ. ACS Chemical Biology. 2016;11:3043–3051. doi: 10.1021/acschembio.6b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maksimov MO, Link AJ. J In Microbiol Biotechnol. 2014;41:333–344. doi: 10.1007/s10295-013-1357-4. [DOI] [PubMed] [Google Scholar]
- 6.Maksimov MO, Pelczer I, Link AJ. Proc Natl Acad Sci U S A. 2012;109:15223–15228. doi: 10.1073/pnas.1208978109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegemann JD, Zimmermann M, Zhu SZ, Klug D, Marahiel MA. Biopolymers. 2013;100:527–542. doi: 10.1002/bip.22326. [DOI] [PubMed] [Google Scholar]
- 8.Duquesne S, Destoumieux-Garzón D, Zirah S, Goulard C, Peduzzi J, Rebuffat S. Chem Biol. 2007;14:793–803. doi: 10.1016/j.chembiol.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Pan SJ, Rajniak J, Cheung WL, Link AJ. Chembiochem. 2012;13:367–370. doi: 10.1002/cbic.201100596. [DOI] [PubMed] [Google Scholar]
- 10.Yan KP, Li YY, Zirah S, Goulard C, Knappe TA, Marahiel MA, Rebuffat S. Chembiochem. 2012;13:1046–1052. doi: 10.1002/cbic.201200016. [DOI] [PubMed] [Google Scholar]
- 11.Maksimov MO, Link AJ. J Am Chem Soc. 2013;135:12038–12047. doi: 10.1021/ja4054256. [DOI] [PubMed] [Google Scholar]
- 12.Maksimov MO, Koos JD, Zong C, Lisko B, Link AJ. J Biol Chem. 2015;290:30806–30812. doi: 10.1074/jbc.M115.694083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chekan JR, Koos JD, Zong C, Maksimov MO, Link AJ, Nair SK. J Am Chem Soc. 2016;138:16452–16458. doi: 10.1021/jacs.6b10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasilyeva LV, Omelchenko MV, Berestovskaya YY, Lysenko AM, Abraham WR, Dedysh SN, Zavarzin GA. International Journal of Systematic and Evolutionary Microbiology. 2006;56:2083–2088. doi: 10.1099/ijs.0.64122-0. [DOI] [PubMed] [Google Scholar]
- 15.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Jong A, van Hijum S, Bijlsma JJE, Kok J, Kuipers OP. Nucleic Acids Res. 2006;34:W273–W279. doi: 10.1093/nar/gkl237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skinnider MA, Johnston CW, Edgar RE, Dejong CA, Merwin NJ, Rees PN, Magarvey NA. Proc Natl Acad Sci U S A. 2016;113:E6343–E6351. doi: 10.1073/pnas.1609014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tietz JI, Schwalen CJ, Patel PS, Maxson T, Blair PM, Tai HC, Zakai UI, Mitchell DA. Nature Chemical Biology. 2017;13:470–478. doi: 10.1038/nchembio.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fage CD, Hegemann JD, Nebel AJ, Steinbach RM, Zhu SZ, Linne U, Harms K, Bange G, Marahiel MA. Angew Chem-Int Edit. 2016;55:12717–12721. doi: 10.1002/anie.201605232. [DOI] [PubMed] [Google Scholar]
- 20.Yang WL, Li YJ, Liu HBA, Chi LF, Li YL. Small. 2012;8:504–516. doi: 10.1002/smll.201101738. [DOI] [PubMed] [Google Scholar]
- 21.Erbas-Cakmak S, Leigh DA, McTernan CT, Nussbaumer AL. Chem Rev. 2015;115:10081–10206. doi: 10.1021/acs.chemrev.5b00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Flood AH, Stoddart JF. J Am Chem Soc. 2004;126:9150–9151. doi: 10.1021/ja048164t. [DOI] [PubMed] [Google Scholar]
- 23.Inoue Y, Kuad P, Okumura Y, Takashima Y, Yamaguchi H, Harada A. J Am Chem Soc. 2007;129:6396–6397. doi: 10.1021/ja071717q. [DOI] [PubMed] [Google Scholar]
- 24.Clavel C, Romuald C, Brabet E, Coutrot F. Chemistry-a European Journal. 2013;19:2982–2989. doi: 10.1002/chem.201203597. [DOI] [PubMed] [Google Scholar]
- 25.Clavel C, Fournel-Marotte K, Coutrot F. Molecules. 2013;18:11553–11575. doi: 10.3390/molecules180911553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Li X, Agren H, Qu DH. Organic Letters. 2014;16:4940–4943. doi: 10.1021/ol502466x. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Sun J, Liu Z, Nassar MS, Botros YY, Stoddart JF. Chemical Science. 2017;8:2562–2568. doi: 10.1039/c6sc05035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito F, Bode JW. Chemical Science. 2017;8:2878–2884. doi: 10.1039/c7sc00021a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegemann JD, Zimmermann M, Zhu SZ, Steuber H, Harms K, Xie XL, Marahiel MA. Angew Chem-Int Edit. 2014;53:2230–2234. doi: 10.1002/anie.201309267. [DOI] [PubMed] [Google Scholar]
- 30.Hegemann JD, Zimmermann M, Xie XL, Marahiel MA. J Am Chem Soc. 2013;135:210–222. doi: 10.1021/ja308173b. [DOI] [PubMed] [Google Scholar]
- 31.Wilson KA, Kalkum M, Ottesen J, Yuzenkova J, Chait BT, Landick R, Muir T, Severinov K, Darst SA. J Am Chem Soc. 2003;125:12475–12483. doi: 10.1021/ja036756q. [DOI] [PubMed] [Google Scholar]
- 32.Knappe TA, Linne U, Zirah S, Rebuffat S, Xie XL, Marahiel MA. J Am Chem Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 33.Rosengren KJ, Blond A, Afonso C, Tabet JC, Rebuffat S, Craik DJ. Biochemistry. 2004;43:4696–4702. doi: 10.1021/bi0361261. [DOI] [PubMed] [Google Scholar]
- 34.Allen CD, Link AJ. J Am Chem Soc. 2016;138:14214–14217. doi: 10.1021/jacs.6b09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knappe TA, Linne U, Robbel L, Marahiel MA. Chem Biol. 2009;16:1290–1298. doi: 10.1016/j.chembiol.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y. BMC Bioinformatics. 2008;9 [Google Scholar]
- 37.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


