Abstract
Alzheimer’s disease (AD) is a senile dementia with increased incidence in older subjects (age>65 years). One of the earliest markers of AD is oxidative DNA damage. Recently it has been reported that preclinical AD patient brains show elevated levels of oxidative damage in both nuclear and mitochondrial nucleic acids. Moreover, different oxidative lesions in mitochondrial DNA are between 5–10-fold higher than in nuclear DNA in both control and AD postmortem brains. We previously showed that there is a significant loss of base excision repair (BER) components in whole tissue extracts of AD and mild cognitive impairment subjects relative to matched control subjects. However, comprehensive analysis of specific steps in base excision repair levels in mitochondrial extracts of AD patient brains is not available. In this study we mainly investigated various components of BER in mitochondrial extracts of AD and matched control postmortem brain samples. We found that the 5-hydroxyuracil (5OHU) incision and ligase activities are significantly lower in AD brains whereas the uracil incision, abasic site cleavage and dNTP incorporation activities are normal in these samples.
Keywords: BER, Base excision repair, 5OHU, 5hydroxyuracil, ligation, AD, Alzheimer’s disease, mitochondria
1. Introduction
AD is a progressive neurodegenerative disorder leading to dementia, a common term used to describe memory and cognitive impairment (Perez, Rivera et al. 1975). At the molecular level, AD pathology is marked by the extracellular amyloid beta plaque formation and/or intracellular tau tangle formation (Ikeda, Allsop et al. 1989; Masters and Beyreuther 1995). Previous reports have also associated mitochondrial dysfunction with AD (Trimmer, Swerdlow et al. 2000; Liang, Reiman et al. 2008). A consistent defect in AD has been a deficiency in cytochrome c oxidase, an enzyme essential for OXPHOS, which was initially reported in AD platelets (Bosetti, Brizzi et al. 2002) and later in postmortem brain tissue from patients with AD (Maurer, Zierz et al. 2000). Loss of mitochondrial genomic integrity is also implicated in AD pathology (Coskun, Wyrembak et al. 2012). The mitochondrial inner membrane anchors mitochondrial DNA (mtDNA) which is the site for oxidative phosphorylation (OXPHOS) that generates ATP, however reactive oxygen species (ROS) are also produced as a byproduct. The close proximity of mtDNA to the ROS source makes mtDNA more vulnerable to oxidative damage and damaged mtDNA, which codes for OXPHOS protein components, can cause mitochondrial dysfunction (Onyango, Khan et al. 2006; Yang, Weissman et al. 2008). MtDNA deletions and elevated levels of specific oxidative lesions have been reported to be present in postmortem brain samples (Aliyev, Chen et al. 2005; Melberg, Nennesmo et al. 2005; Wang, Xiong et al. 2005; Wang, Markesbery et al. 2006; Krishnan, Ratnaike et al. 2012; Phillips, Simpkins et al. 2013). Additionally, degraded mtDNA and proteins were found in the secondary lysozomes and swollen mitochondria with damaged cristae are found in the neurons of AD brains (Baloyannis, Costa et al. 2004; Baloyannis 2006; Reddy 2009). Together these data and many others (Coskun, Wyrembak et al. 2012) indicate that mitochondrial dysfunction and genomic instability may be a main contributor to AD pathology.
The importance of base excision repair (BER) proteins in normal brain function, including memory retention and cognition, are not well understood at this time, nor is the relative importance of nuclear versus mtDNA BER repair. Notably, DNA repair processes in mitochondria are not as comprehensive as they are in the nucleus (Croteau, Stierum et al. 1999), but BER is well conserved in this compartment (Bohr and Dianov 1999; Dianov, Souza-Pinto et al. 2001). Previously, we showed that the BER pathway is defective in AD postmortem brain whole tissue lysates (Weissman, Jo et al. 2007), however it is not known whether there are deficits in specific steps of base excision repair in mitochondria of AD brain samples. BER consists of four major steps. DNA glycosylases recognize and remove specific damaged bases leaving an abasic site. AP endonucleases process abasic sites generating a single stranded gap in the DNA. The gap is filled in by a DNA polymerase, and then ligase seals the nick to complete the DNA repair process(Wilson and Bohr 2007). Interestingly, we recently showed that loss of NEIL1, a versatile DNA glycosylase, in mice led to defective spatial memory retention during Morris watermaze tests (Canugovi, Yoon et al. 2012). Further, loss of NEIL1 delayed motor function recovery post ischemic reperfusion injury. These mice showed decreased incision activity in both whole tissue and mitochondrial extracts for 5OHU, one of the major substrates for NEIL1. These results suggest that loss of DNA repair may also contribute to brain dysfunction and pathology.
In this study, we examined the relative activity levels of BER components in the inferior parietal (IPL) region of postmortem AD and age-matched control brains. IPL is one of the most affected areas impacted by AD pathology and the relative ratio of oxidatively damaged bases per number of normal bases is high in this region (Gabbita, Lovell et al. 1998). Here, we studied the activities of several enzymes of the BER pathway including: DNA glycosylases, abasic site cleavage (AP endonuclease), gap filling (polymerase) and nick-sealing ligase. We found a deficiency in 5OHU incision and ligase activity in AD mitochondrial samples but no change in the uracil incision, abasic site cleavage or gap filling polymerase steps.
2. Materials and methods
2.1 Brain samples and source
The samples used in this study were obtained from Harvard Brain Tissue Resource Center (Belmont, MA). All samples belong either to Braak stage 5 or 6. The samples were isolated and stored within approximately 24 hours after death (Table 1). The samples were derived from part of inferior parietal region of the brains. The sample size is n=6 for both control and AD brains. Details of the patients are listed in Table 1.
Table 1.
Patient sample information obtained from Harvard brain bank
| Sample | PMI | Background | Age | Braak stage | Sex | Area | Source |
|---|---|---|---|---|---|---|---|
| AN14331 | 17.18 | AD | 82 | 5 | F | inferior perietal | Harvard brain bank |
| AN14184 | 15.42 | AD | 78 | 5 | M | inferior perietal | Harvard brain bank |
| AN06848 | 22.84 | AD | 60 | 6 | F | inferior perietal | Harvard brain bank |
| AN12633 | 21.08 | AD | 77 | 5 | F | inferior perietal | Harvard brain bank |
| AN08341 | 20.5 | AD | 74 | 5 | M | inferior perietal | Harvard brain bank |
| AN10763 | 13.5 | AD | 62 | 6 | M | inferior perietal | Harvard brain bank |
| AN11017 | 23.12 | CONTROL | 67 | NA | M | inferior perietal | Harvard brain bank |
| AN01077 | 19.38 | CONTROL | 58 | NA | F | inferior perietal | Harvard brain bank |
| AN12916 | 25.43 | CONTROL | 77 | NA | M | inferior perietal | Harvard brain bank |
| AN00349 | 17.58 | CONTROL | 79 | NA | F | inferior perietal | Harvard brain bank |
| AN18592 | 24.42 | CONTROL | 82 | NA | F | inferior perietal | Harvard brain bank |
| AN10212 | 20.53 | CONTROL | 74 | NA | M | inferior perietal | Harvard brain bank |
2.2 Oligonucleotide substrates
All oligonucleotides were ordered from IDT (San Diego, CA). The oligonucleotide sequence and location of the specific lesions can be found in Table 2.
Table 2.
Names and sequences of oligonucleotides used in the study
| Name | Sequence |
|---|---|
| 5OHU-in bubble-51mer | 5′-GCTTAGCTTGGAATCGTATC-ATGTA5ACTCG-TGTGCCGTGTAGACCGTGCC-3′ |
| 3′-CGAATCGAACCTTAGCATAG–GCACCCGACAA-ACACGGCACATCTGGCACGG-5′ | |
| Nick-1-91mer | 5′-TAATTAATGCTTGTAGGACATAATAATAACAATTGAATGTCTG-OH/P-CACA … |
| 3′-ATTAATTACGAACATCCTGTATTATTATTGTTAACTTACAGA(C)GTGT … | |
| … GCCACTTTCCACACAGACATCATAACAAAAAATTTCCACCAAAC-3′ | |
| … CGGTGAAAGGTGTGTCTGTAGTATTGTTTTTTAAAGGTGGTTTG-5′ | |
| Nick-2-91mer | 5′-TAATTAATGCTTGTAGGACATAATAATAACAATTGAATGTCTG-OH/OH-CACA … |
| 3′-ATTAATTACGAACATCCTGTATTATTATTGTTAACTTACAGA(C)GTGT … | |
| … GCCACTTTCCACACAGACATCATAACAAAAAATTTCCACCAAAC-3′ | |
| … CGGTGAAAGGTGTGTCTGTAGTATTGTTTTTTAAAGGTGGTTTG-5′ | |
| U-91mer | 5′-TAATTAATGCTTGTAGGACATAATAATAACAATTGAATGTCT(U)CACA … |
| 3′-ATTAATTACGAACATCCTGTATTATTATTGTTAACTTACAGA(C)GTGT … | |
| … GCCACTTTCCACACAGACATCATAACAAAAAATTTCCACCAAAC-3′ | |
| … CGGTGAAAGGTGTGTCTGTAGTATTGTTTTTTAAAGGTGGTTTG-5′ | |
| THF-91mer | 5′-TAATTAATGCTTGTAGGACATAATAATAACAATTGAATGTCT(F)CACA … |
| 3′-ATTAATTACGAACATCCTGTATTATTATTGTTAACTTACAGA(C)GTGT … | |
| … GCCACTTTCCACACAGACATCATAACAAAAAATTTCCACCAAAC-3′ | |
| … CGGTGAAAGGTGTGTCTGTAGTATTGTTTTTTAAAGGTGGTTTG-5′ | |
| GAP-91mer | 5′-TAATTAATGCTTGTAGGACATAATAATAACAATTGAATGTCTGCACA … |
| 3′-ATTAATTACGAACATCCTGTATTATTATTGTTAACTTACAGA( )GTGT … | |
| … GCCACTTTCCACACAGACATCATAACAAAAAATTTCCACCAAAC-3′ | |
| … CGGTGAAAGGTGTGTCTGTAGTATTGTTTTTTAAAGGTGGTTTG-5′ |
5OHU contains a 5 hydroxyuracil represented by ‘5’ in a bubble shown by dashes; nick-1: contains a ligase specific nick with a 3′-OH and a 5′-P groups on the DNA ends within the nick; nick-2: contains 3′ and 5′-OH groups on the DNA ends within the nick, this was made by polymerase incorporation of a nucleotide with in a gap substrate in the last row; U = contains a uracil; THF = contains a tetrahydrofuran abasic site analog; GAP =containing a single nucleotide gap.
2.3 Tissue fractionation and lysate preparation
Whole tissue lysates were prepared as described previously (Gabbita, Lovell et al. 1998). Mitochondrial fractionation was performed using the mitochondria isolation kit (Thermo scientific/Pierce, Logan, UT) as per the manufacturer’s suggestions. The isolated mitochondrial lysates were prepared by resuspending mitochondria (1 mg/ml) in buffer A (20 mM HEPES (pH 7.0), 150 mM KCl, 2 mM EGTA, 1% (w/v) CHAPSO (Sigma, St Louis, MO), 1X protease inhibitor mixture (Roche Diagnostics Corporation, Indianapolis, IN and incubating at 4° C for 1 h with end-over-end rotation. The lysates were centrifuged at 37, 000 g for 1 h, and the supernatants were collected. The samples were flash frozen in liquid nitrogen and stored at −80° C. Protein concentration was determined using the BCA protein assay kit (ThermoScientific, Rockford, IL).
2.4 Western blotting
Whole tissue lysates or mitochondrial lysates were used to assay for the purity of mitochondrial lysates. About 40 μg of lysates were separated by SDS PAGE on 4–15% gradient pre-cast gels (Biorad, Hercules, CA). The separated samples were then blotted on to a PVDF membrane at 200 volts for 2 hr on ice. The blots were blocked with 5% milk and incubated with the respective primary antibodies followed by secondary antibodies and washed with TBST (Tris buffer saline with 0.1 % Tween20) washes. Lamin B1 (ab 16048), VDAC (ab 14734), NEIL1 (ab 21337) DNA ligase III (ab36499) and beta Actin (ab8227) antibodies were purchased from ABcam (Cambridge, MA ).
2.5 Incision assays
Incision assays were performed as described in Weissmann et al. (Weissman, Jo et al. 2007). Briefly, Incision reactions contained 100 fmol of 32P-labelled duplex oligonucleotide with a specific modification. The reactions were incubated with 4 μg of mitochondrial lysates or 12 μg of whole tissue lysates respectively at 32° C. The reactions were terminated by the addition of 5 mgs/ml proteinase K and 10% SDS and incubated at 37° C for 30 min followed by the addition of formamide containing buffer and continued incubation at 95° C for 5 min. These samples were resolved on a denaturing 15% poly acrylamide gel containing 7M urea. The gels were visualized by a Phosphorimager (Molecular dynamics, GE Healthcare, Ramsey, MN). The images were analyzed using ImageQuant 5.2 software (GE Healthcare, Pittsburgh, PA). The percent incision was calculated by dividing the product band intensity by the total intensity in the lane. All samples were also normalized by the background band near product in the untreated samples in each assay. The incubation times varied for each type of activity assay. While assaying for uracil or 5OHU incision, we incubated the reactions overnight whereas the incubation time was only 5 min when assaying for abasic site analog tetrahydrofuran (THF) cleavage in the same lysates.
2.6 Incorporation assay
Single nucleotide gap filling reaction was conducted using an unlabeled DNA oligo substrate with a single gap in the middle of the substrate (Table 2). Samples were diluted in 10 mM Tris-HCl (pH 7.4) containing 100 mM KCl to achieve similar protein concentrations in each of the independent samples. Reactions (10 μl) contained 50 mM Tris–HCl (pH 7.4), 50 mM KCl, 1 mM DTT, 5 mM MgCl2, 5% glycerol, 5 mM dCTP (Roche Applied Sciences, Indianapolis, IN), 1 pmol of duplex gap oligonucleotide 4 mCi of 32P-dCTP (GE Healthcare, Pittsburgh, PA ) and 4 μg protein. Reactions were incubated at 37° C for 3 h and terminated by the addition of 1 μl of 0.5 M EDTA and formamide dye followed by the heating at 95° C for 5 min.
2.7 Ligation assay
Ligation was measured using two substrates. First with a single nick in a radio-labeled 91-mer oligonucleotide containing 3′-OH and 5′-P substrate and second with a gap containing substrate that was pretreated with pol beta ( courtesy of Dr. Wilson DM III Lab, NIA). Reactions contained 50 mM Tris–HCl (pH 7.4), 50 mM KCl, 1 mM DTT, 5 mM MgCl2, 5% glycerol, 10 mM ATP, 100 fmol of oligonucleotide substrate and 4 μg of mitochondrial lysate or 12 μg of whole tissue lysate. 12 μg Hela cell lysate has been used for positive control and no protein was added in the negative control. Reactions were incubated at 37° C for 3 h and terminated by the addition of formamide dye followed by the heating at 95° C for 5 min.
2.8. In vitro plasmid ligation assay
Brain tissue extracts were prepared as described (Weissman, Jo et al. 2007) and stored at −80°C, samples along with positive control (AG11359 cells) were treated with ice cold 3M KCl to get a final salt concentration of 0.5 M and incubated on a rotor for 30 min at 4° C. The extracts were centrifuged at 27,000 X g and dialyzed for 2 h at 4° C with dialysis buffer (25 mM HEPES, pH 7.5, 100 mM KCl, 1 mM EDTA, 10% glycerol, 0.2 mM PMSF and 0.5 mM DTT). Protein concentration of dialyzed extracts was estimated from the supernatant fractions isolated after centrifugation at 10,000 X g, 5 min. 4° C. In vitro plasmid ligation assays were performed as mentioned elsewhere (Wang, Perrault et al. 2003; Shamanna, Hoque et al. 2011) using 2.5 μg of extract and 10 ng of pUC18 plasmid DNA linearized with BamHI restriction enzyme (New England Biologicals, Ipswich, MA). Briefly, ligation was carried out for 2 h in a 20 μl reaction buffer (20 mM HEPES [pH 7.5], 10 mM MgCl2, 80 mM KCl, 1 mM dithiothreitol [DTT], 1 mM ATP, and 50 μM dATP, dGTP, dCTP, and dTTP) at 25° C. Reactions were terminated by adding 50 mM EDTA and 2% SDS, and deproteinated with proteinase K (Invitrogen, Grand Island, NY). End products of the ligation reaction were resolved on 0.7% agarose gels and visualized by imaging the SYBR gold stained gels using Molecular Imager Gel Doc XR system (Bio-Rad, Hercules, CA). DNA bands were quantified with ImageJ 1.45r software (NIH).
2.9. Statistical analysis
The results are reported as mean± standard error. Each assay was performed at least twice (on six independent biological samples per set). The differences among human control and AD samples were analyzed by the Student’s t-test and a p<0.05 was considered statistically significant.
3. Results
3.1. AD patient brain samples have decreased 5OHU incision activity in both whole tissue and mitochondrial
Previously it was reported that there was a statistically significant elevation of, 5-hydroxyuracil, 5-hydroxycytosine, 8-hydroxyguanine and 8-hydroxyadenine in AD brain compared with control subjects (Gabbita, Lovell et al. 1998). While our lab showed that there is a significantly lower level of 8oxoG incision activity in AD postmortem brain samples (Weissman, Jo et al. 2007) compared to controls, no reports exist of the incision capacities of 5OHU in AD brain samples. Loss of NEIL1 in mice led to a memory retention deficit and reduced 5OHU incision activity in both whole tissue and mitochondrial extracts(Canugovi, Yoon et al. 2012). This suggests that accumulation of 5OHU repair may be important. Therefore we studied the levels of 5OHU incision activity from AD and control brain samples (n=6 of each) using biochemical assays described in the Methods section. We found that the 5OHU incision is significantly reduced in AD whole brain tissue lysates when compared to control samples (p<0.01) (Fig. 1A) (Table 3).
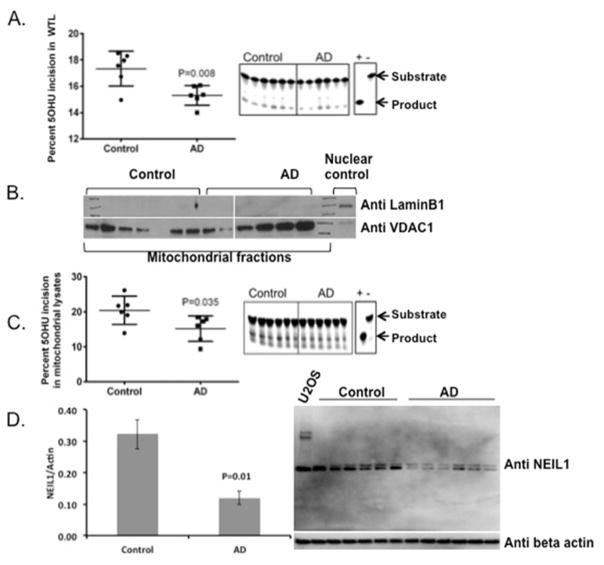
Figure 1.
5OHU incision activity in AD and control extracts. A) Whole tissue lysates from control and AD postmortem brains were assessed for 5OHU incision using 5OHU in a bubble (Table 2). The data are represented by a mean ± standard error, n=6. B) Mitochondrial purity was tested by western analysis of nuclear and mitochondrial markers. C) Mitochondrial lysates from control and AD postmortem brains were assessed for 5OHU incision activity using the same substrate as in panel A containing 5OHU in the bubble. The data are represented by a mean ± standard error, n=6. D) Western analysis of NEIL1 protein in the whole tissue lysates of AD and control samples.
Table 3.
Summary of DNA repair tests conducted in this study
| Sample | Repair step | Substrate | DNA repair activity§ |
|---|---|---|---|
| Brain nuclear lysates | DNA Glycosylase | 5-hydroxyuracil in a bubble DNA | Yes* |
| Ligase | Nick-1 | No (P=0.06) | |
| Linearized plasmid | Yes* | ||
| Brain mitochondrial lysates | DNA Glycosylase | 5-hydroxyuracil | Yes* |
| Uracil | No | ||
| AP endonuclease | Abasic site | No | |
| Polymerase | Gap | No (p=0.056) | |
| Ligase | Nick-1 | Yes* | |
| Nick-2 | Yes* |
In the column#4 titled DNA repair activity, the symbol § indicates whether there is a significant decrease in repair activity in the AD samples relative to control samples. In each row within this column, * symbol indicates that the p-value is <0.05. in cases of no difference but with a trend we indicated the p-values. Nick-1 is a heteroduplex with a 5′-P and 3′OH containing nick, where as Nick-2 is a heteroduplex with a nick containing 3′- and a 5′-OH groups. Nick to is generated by gap filling activity by a polymerase on a gap substrate.
Recently, there have been reports suggesting the possible presence of elevated levels of specific oxidative damage in mitochondrial DNA of preclinical AD patient brains (Wang, Markesbery et al. 2006; Lovell, Soman et al. 2011). Therefore, we evaluated 5OHU incision activity in mitochondrial extracts isolated from AD postmortem brain samples. First, we tested the purity of the mitochondrial lysates by western analysis of nuclear and mitochondrial specific markers such as Lamin B1 and VDAC proteins, respectively (Fig. 1B). The mitochondrial extracts show no detectable levels of nuclear contamination. We then tested the incision activities in these extracts. The 5OHU incision capacities were found to be significantly lower in AD mitochondrial lysates compared to controls (p= 0.039) (Fig. 1C) (Table 3). One of the major DNA glycosylases that remove 5OHU lesions from DNA is NEIL1. Therefore we measured the levels of NEIL1 in the whole brain extracts of AD and control samples. We found significantly lower levels of NEIL1 protein in the AD whole brain extracts (Fig. 1D). Together these data suggest that there is a significantly lower amount of 5OHU incision activity, likely due to the loss of NEIL1, in the AD post mortem brain samples both in whole brain and mitochondrial lysates.
3.2. AD patient brain samples have decreased ligase activity
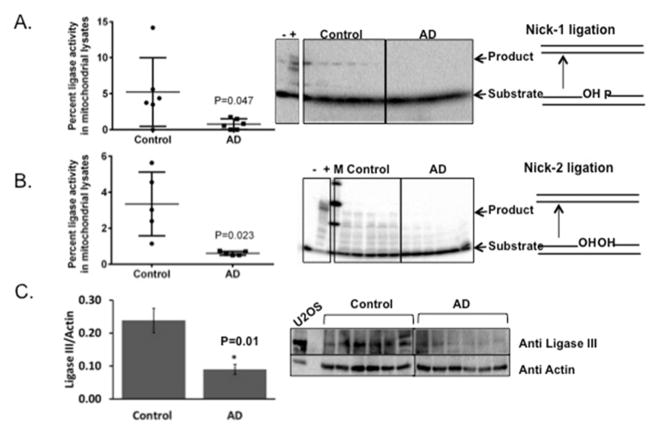
Ligase activity is the crucial final step in the completion of the BER process. Loss of ligase activity can lead to nicks in DNA that can be detrimental to the cell and the importance of ligase is exemplified by the fact that in a mouse model without mitochondrial ligase activity, the phenotype is embryonically lethal at day 8.5 (Puebla-Osorio, Lacey et al. 2006). We therefore studied the ligase activity in mitochondrial lysates using two substrates. The first substrate contained a nick with 3′-OH and 5′-P groups representing “clean” DNA ends and the second substrate contained a nick with 3′- and 5′-OH ends (Table 3). While the ligation assay with the former substrate measures ligase activity, the latter measures ligase activity in concert with polynucleotide kinase. In both of these reactions, we found minimal to no activity in AD patient samples whereas the control brain samples showed activity (fig. 2A and B). It is possible that the very low level of activity present in AD may not have been detected in our assay. However there is a striking significant difference in the activity levels of AD mitochondrial samples when compared to control samples p=0.047 and p=0.023, respectively in Fig. 2A and 2B. Further, Ligase III levels normalized to actin levels show a significant decrease (p=0.01) in AD brain samples relative to control samples supporting the loss of ligase activity.
Figure 2.
Ligase activity in AD and control brain mitochondrial extracts. A) Ligation in mitochondrial lysates of control and AD postmortem brains was studied using a nick substrate with a 5′-P and a 3′-OH in an oligomeric DNA hetro-duplex. The data are represented by a mean ± standard error, n=6. B) Ligation in mitochondrial lysates of control and AD postmortem brains was studied using a gap substrate pretreated with purified polymerase beta and cold dCTP. The data are represented by a mean ± standard error, n=5 for control and n=6 for AD samples. Hela cell lysate has been used for positive control represented by a ‘+’ sign and no protein was included in negative control represented by ‘−’sign. C) A representative western blot of Ligase III levels in both control and AD whole tissue lysates are shown. The graph indicates the quantitation of Ligase III levels shown in the blot normalized to the actin levels.
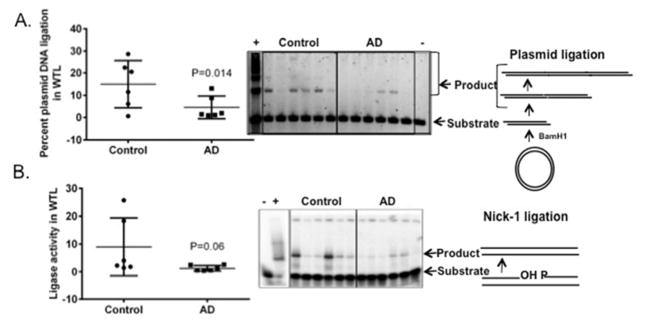
We then tested the ligase activity in whole tissue lysates using a DNA heteroduplex substrate containing a nick with 3′-OH and 5′-P ends and an independent assay employing linearized plasmid DNA substrates (Table 3). In vitro end joining assays, with a variety of DNA substrates, have been used to demonstrate the importance of several players in the DNA repair pathways (Pastwa, Somiari et al. 2009). Based on previous reports that indicate dominant ligase activity in in vitro end joining assays (Paul, Wang et al. 2013), we used a linearized plasmid DNA substrate with cohesive ends to check the activity of ligases in AD brain extracts. In our assays, we found a significant decrease in plasmid ligation in the whole tissue lysates of AD samples relative to control samples (p= 0.014) (Fig. 3A). The nick ligation of the radiolabeled heteroduplex was not significantly different (Fig. 3B), however the AD samples showed a tendency toward lower levels of ligase activity (p=0.06). Together we found that there is lower nick ligation activity in mitochondrial lysates and plasmid ligation activity in the AD whole tissue lysates.
Figure 3.
Ligase activity in AD and control brain whole tissue lysates (WTL). A) Right panel is an agarose gel showing plasmid ligation activity. The control and AD post mortem brain WTLs were tested for their ability to ligate BamH1 cut plasmid (sticky end ligation). Positive control is represented by AG11359 and negative control is represented by the EDTA. Left panel is the quantification of the percent plasmid ligation activity represented in the gel. Data are mean ± standard deviation, n=6. B) Ligation in WTLs of control and AD postmortem brains using the same substrate as in 2A. The data are represented by a mean ± standard error, n=6.
3.3. AD mitochondrial lysates have normal uracil incision and AP endonuclease activity
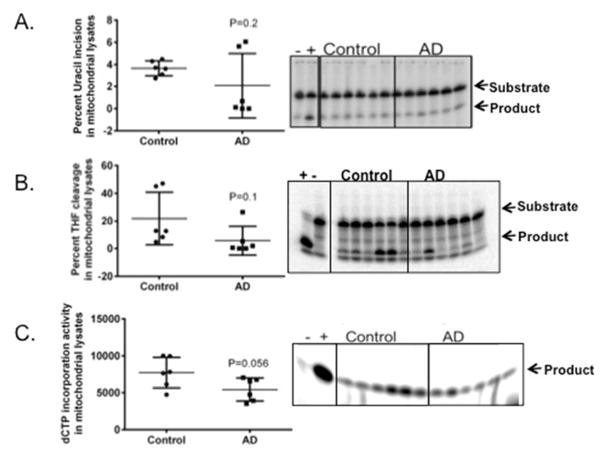
Previous data from our lab indicates that there is a significant decrease in uracil incision but not in the abasic site cleavage activity in AD postmortem brain whole tissue lysates (Weissman, Jo et al. 2007). Mitochondrial uracil incision and abasic site cleavage were not tested previously. Therefore, we tested the uracil and abasic site cleavage activities using radiolabeled oligomeric DNA substrates containing a site specific uracil or THF adduct, respectively. We found no significant difference in uracil incision activity (p=0.2) (Fig. 4A) or in abasic site cleavage activity (p=0.1) (Fig. 4B) (Table 3) in the mitochondrial lysates from AD patient brain samples.
Figure 4.
Uracil incision, abasic site cleavage and dNTP incorporation activities in AD and control postmortem brain mitochondrial lysates. A) Uracil incision in mitochondrial extracts. B) Abasic site (THF) cleavage in mitochondrial extracts. C) dCTP incorporation activity in mitochondrial lysates. Data are represented by the mean± standard error of six samples for both control and AD, n=6.
3.4. AD mitochondrial lysates show a strong trend towards decreased polymerases activity
Our lab previously published that the polymerase beta gap filling activity is significantly lower in AD postmortem brain samples (Weissman, Jo et al. 2007). However no reports exist of the comparative polymerase activity in mitochondrial polymerase function from AD postmortem brains. In pursuit of profiling the strand break repair and BER levels in AD patient brain samples, we tested the polymerase activity in mitochondrial lysates. Our results indicate that the mitochondrial fractions do not show significant differences in the polymerase gap filling activity in AD mitochondrial samples when compared to controls (Fig. 4C)(Table 3). However there was a trend of p=0.056 toward lower polymerase gap filling activity in AD samples.
4. Discussion
The primary goal of this study was to determine whether there is a loss of mitochondrial DNA repair in AD postmortem brain samples. To address this question, we tested multiple enzymatic steps in the BER pathway that are important in DNA damage recognition, removal or repair of oxidative DNA damage including base modifications, abasic sites as well as single strand breaks or nicks in the DNA backbone. Isolating mitochondria from small amounts of flash frozen brain samples is technically challenging. However our mitochondrial samples showed no nuclear contamination based on the western analysis showed in Fig. 1B. Amongst the DNA glycosylase activities tested 5OHU incision activity was found to be significantly lower in both whole tissue extracts as well as mitochondrial extracts of AD samples compared to controls (Fig. 1 A and C). 5OHU is a substrate for DNA glycosylases such as NEIL1 and NTH1 that remove oxidatively damaged pyrimidine bases from the DNA. Therefore we looked for the levels of NEIL1 protein using western analysis in the whole tissue samples. We found significantly less protein in AD brains (Fig. 1D). Hegde et al. reported that NEIL1 initiates repair of oxidized base damage is specifically inhibited by copper and iron (Hegde, Hegde et al. 2010). Based on this, we initially hypothesized that the AD brains might have elevated iron deposition that may inhibit NEIL1 activity without loss of protein quantity. However we found less NEIL1 protein in AD samples, which likely directly contributes to the lower 5OHU incision activity. Although there is a significant difference in uracil incision in the whole tissue lysates tested previously by our group (Weissman, Jo et al. 2007), the mitochondrial uracil incisions tested in this study were not significantly different in AD when compared to control brain samples which may be attributed to the high variance within the group of samples tested in AD mitochondrial lysates compared to the controls (p=0.2) (Fig. 4A). The AP endonuclease activity is also intact in the mitochondrial lysates (Fig. 4B) similar to the whole tissue samples tested in the earlier report (Weissman, Jo et al. 2007). We also tested the mitochondrial polymerase nucleotide incorporation activity, which strongly tended (p=0.056) towards loss of function in AD samples (Fig. 4C). However it was not found to be significant as was seen in the whole tissue samples reported by our lab in the past (Weissman, Jo et al. 2007).
Finally, we tested the ligase activity, which had not been evaluated previously. We measured ligase activity in both mitochondrial and whole brain lysates. This will to some extent separate the activities of different ligases as the localization of different ligases is slightly different within the cellular compartments (Puebla-Osorio, Lacey et al. 2006). Ligase 3α is known to be mainly present in mitochondria due to the presence of a mitochondrial targeting sequence whereas all others are found to be present in the nucleus (Lakshmipathy and Campbell 1999). To gain additional information, we used two substrates with different end groups, either 5′-P and a 3′-OH or 5′- and 3′-OH moieties. Utilizing these substrates can separate the simple ligase activity from concerted polynucleotide kinase and ligase activities in nick processing in mitochondrial lysates (Karimi-Busheri, Lee et al. 1998). Ligase activity was detectable in all the control mitochondrial samples whereas in the AD mitochondrial lysates it was undetectable. We do not believe that the loss of activity is due to the inactivation of the samples during preparation and handling but that the baseline level of activity in our samples is not detectable due to the sensitivity of the assay we used. However, it is significantly lower in AD mitochondrial samples compared to control samples indicating defective ligase activity. Further, the same samples have shown detectable 5OHU incision, uracil incision, abasic site cleavage and nucleotide incorporation by polymerase indicating that the ligase activity maybe too limited to detect in this assay. To independently confirm that the ligase activity is lower in AD samples, we performed the same assay using the whole tissue lysates. Although we did not find a significant difference in the ligase activity in AD whole tissue lysates with the traditional nick containing hetero-duplex substrate, the significance of the data (p=0.06) was tending towards lower ligase activity in whole tissue lysates. We further tested plasmid ligation activity in the whole tissue lysates of these samples using a linearized plasmid with sticky ends. Around 90% of products from such substrates depend on simple ligase activity (Shamanna, Hoque et al. 2011). Results from our plasmid ligation assay indicate compromised ligase activity in AD brain tissue representing a probable reason for decreased DNA repair activity in the affected brains.
The postmortem intervals (PMIs) were slightly high, averaging 18.4 hours in AD patients and 21.7 hours in control brains. Therefore we plotted the PMI against each of the assays, except ligase, for both AD and control samples to see if there was any correlation in the activity with respect to the length of PMI (Supplementary fig 1). No correlation was found between length of PMI and repair activities.
Overall, this study sought to profile BER in mitochondria as well as the activities of 5OHU incision and ligation in whole tissue lysates from AD and control postmortem samples. Previous reports by Wang j et al., show that oxidatively damaged DNA lesions are much higher (5–10 folds) in mitochondrial DNA relative to nuclear DNA in both control and AD postmortem samples. In addition, many of the lesions measured were elevated in AD samples compared to control samples. Further, it is established that oxidative damage is one of the earliest sign of AD as indicated by the recent studies on pre-clinical AD samples, which also showed elevated staining of oxidative damage specific antibodies in nuclear and extra-nuclear regions possibly including mitochondrial DNA and RNA damage (Wang, Markesbery et al. 2006; Lovell, Soman et al. 2011). Accumulation of DNA damage is a function of both the rate of creation by ROS and removal by DNA repair mechanisms. This paper and prior literature (Lior, refs), make the case that decreased BER could potential contribute to the elevated levels of oxidized bases in AD patient DNA. Whether this defect in BER in mitochondria described in this study actually contributes to the disease pathology is still an open question.
Supplementary Material
Supplementary figure 1. Postmortem intervals (PMIs) were plotted against the respective repair activities to determine if a correlation existed between the enzyme activity and postmortem interval in both AD (A–B) and Control (C–D) brain mitochondrial extracts.
Acknowledgments
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging. We would like to thank Harvard brain bank for the Human postmortem brain samples used in this study. Additionally, we would like to thank Huiming Lu and Somnath Ghosh for critically reading this manuscript.
Footnotes
Disclosure Statement: Authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aliyev A, Chen SG, et al. Mitochondria DNA deletions in atherosclerotic hypoperfused brain microvessels as a primary target for the development of Alzheimer’s disease. J Neurol Sci. 2005;229–230:285–292. doi: 10.1016/j.jns.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Baloyannis SJ. Mitochondrial alterations in Alzheimer’s disease. J Alzheimers Dis. 2006;9(2):119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- Baloyannis SJ, Costa V, et al. Mitochondrial alterations in Alzheimer’s disease. Am J Alzheimers Dis Other Demen. 2004;19(2):89–93. doi: 10.1177/153331750401900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA, Dianov GL. Oxidative DNA damage processing in nuclear and mitochondrial DNA. Biochimie. 1999;81(1–2):155–160. doi: 10.1016/s0300-9084(99)80048-0. [DOI] [PubMed] [Google Scholar]
- Bosetti F, Brizzi F, et al. Cytochrome c oxidase and mitochondrial F1F0-ATPase (ATP synthase) activities in platelets and brain from patients with Alzheimer’s disease. Neurobiol Aging. 2002;23(3):371–376. doi: 10.1016/s0197-4580(01)00314-1. [DOI] [PubMed] [Google Scholar]
- Canugovi C, Yoon JS, et al. Endonuclease VIII-like 1 (NEIL1) promotes short-term spatial memory retention and protects from ischemic stroke-induced brain dysfunction and death in mice. Proc Natl Acad Sci U S A. 2012;109(37):14948–14953. doi: 10.1073/pnas.1204156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun P, Wyrembak J, et al. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim Biophys Acta. 2012;1820(5):553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, Stierum RH, et al. Mitochondrial DNA repair pathways. Mutat Res. 1999;434(3):137–148. doi: 10.1016/s0921-8777(99)00025-7. [DOI] [PubMed] [Google Scholar]
- Dianov GL, Souza-Pinto N, et al. Base excision repair in nuclear and mitochondrial DNA. Prog Nucleic Acid Res Mol Biol. 2001;68:285–297. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- Gabbita SP, Lovell MA, et al. Increased nuclear DNA oxidation in the brain in Alzheimer’s disease. J Neurochem. 1998;71(5):2034–2040. doi: 10.1046/j.1471-4159.1998.71052034.x. [DOI] [PubMed] [Google Scholar]
- Hegde ML, Hegde PM, et al. Specific Inhibition of NEIL-initiated repair of oxidized base damage in human genome by copper and iron: potential etiological linkage to neurodegenerative diseases. J Biol Chem. 2010;285(37):28812–28825. doi: 10.1074/jbc.M110.126664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Allsop D, et al. Morphology and distribution of plaque and related deposits in the brains of Alzheimer’s disease and control cases. An immunohistochemical study using amyloid beta-protein antibody. Lab Invest. 1989;60(1):113–122. [PubMed] [Google Scholar]
- Karimi-Busheri F, Lee J, et al. Repair of DNA strand gaps and nicks containing 3′-phosphate and 5′-hydroxyl termini by purified mammalian enzymes. Nucleic Acids Res. 1998;26(19):4395–4400. doi: 10.1093/nar/26.19.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan KJ, Ratnaike TE, et al. Mitochondrial DNA deletions cause the biochemical defect observed in Alzheimer’s disease. Neurobiol Aging. 2012;33(9):2210–2214. doi: 10.1016/j.neurobiolaging.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol. 1999;19(5):3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105(11):4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell MA, Soman S, et al. Oxidatively modified nucleic acids in preclinical Alzheimer’s disease (PCAD) brain. Mech Ageing Dev. 2011;132(8–9):443–448. doi: 10.1016/j.mad.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CL, Beyreuther K. Molecular neuropathology of Alzheimer’s disease. Arzneimittelforschung. 1995;45(3A):410–412. [PubMed] [Google Scholar]
- Maurer I, Zierz S, et al. A selective defect of cytochrome c oxidase is present in brain of Alzheimer disease patients. Neurobiol Aging. 2000;21(3):455–462. doi: 10.1016/s0197-4580(00)00112-3. [DOI] [PubMed] [Google Scholar]
- Melberg A, Nennesmo I, et al. Alzheimer pathology associated with POLG1 mutation, multiple mtDNA deletions, and APOE4/4: premature ageing or just coincidence? Acta Neuropathol. 2005;110(3):315–316. doi: 10.1007/s00401-005-1047-z. [DOI] [PubMed] [Google Scholar]
- Onyango I, Khan S, et al. Mitochondrial genomic contribution to mitochondrial dysfunction in Alzheimer’s disease. J Alzheimers Dis. 2006;9(2):183–193. doi: 10.3233/jad-2006-9210. [DOI] [PubMed] [Google Scholar]
- Pastwa E, Somiari RI, et al. In vitro non-homologous DNA end joining assays--the 20th anniversary. Int J Biochem Cell Biol. 2009;41(6):1254–1260. doi: 10.1016/j.biocel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Wang M, et al. DNA ligases I and III cooperate in alternative non-homologous end-joining in vertebrates. PLoS One. 2013;8(3):e59505. doi: 10.1371/journal.pone.0059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez FI, V, Rivera M, et al. Analysis of intellectual and cognitive performance in patients with multi-infarct dementia, vertebrobasilar insufficiency with dementia, and Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 1975;38(6):533–540. doi: 10.1136/jnnp.38.6.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips NR, Simpkins JW, et al. Mitochondrial DNA deletions in Alzheimer’s brains: A review. Alzheimers Dement. 2013 doi: 10.1016/j.jalz.2013.04.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla-Osorio N, Lacey DB, et al. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol Cell Biol. 2006;26(10):3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH. Amyloid beta, mitochondrial structural and functional dynamics in Alzheimer’s disease. Exp Neurol. 2009;218(2):286–292. doi: 10.1016/j.expneurol.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanna RA, Hoque M, et al. The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining. Mol Cell Biol. 2011;31(23):4832–4843. doi: 10.1128/MCB.05849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer PA, Swerdlow RH, et al. Abnormal mitochondrial morphology in sporadic Parkinson’s and Alzheimer’s disease cybrid cell lines. Exp Neurol. 2000;162(1):37–50. doi: 10.1006/exnr.2000.7333. [DOI] [PubMed] [Google Scholar]
- Wang H, Perrault AR, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31(18):5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Markesbery WR, et al. Increased oxidative damage in nuclear and mitochondrial DNA in mild cognitive impairment. J Neurochem. 2006;96(3):825–832. doi: 10.1111/j.1471-4159.2005.03615.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Xiong S, et al. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer’s disease. J Neurochem. 2005;93(4):953–962. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- Weissman L, Jo DG, et al. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35(16):5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6(4):544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Yang JL, Weissman L, et al. Mitochondrial DNA damage and repair in neurodegenerative disorders. DNA Repair (Amst) 2008;7(7):1110–1120. doi: 10.1016/j.dnarep.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Postmortem intervals (PMIs) were plotted against the respective repair activities to determine if a correlation existed between the enzyme activity and postmortem interval in both AD (A–B) and Control (C–D) brain mitochondrial extracts.