Abstract
Mitochondria are the oxygen consuming power plants of the cells. They provide a critical milieu for the synthesis of many essential molecules and allow for highly efficient energy production through oxidative phosphorylation. The use of oxygen is, however, a double-edged sword that on the one hand supplies ATP for cellular survival, and on the other leads to the formation of damaging reactive oxygen species. Different quality control pathways maintain mitochondria function including mitochondrial DNA replication and repair, fusion-fission dynamics, free radical scavenging and mitophagy. Further, failure of these pathways may lead to human disease. We review these pathways and propose a strategy towards a treatment for these often untreatable disorders.
Keywords: Mitochondria, DNA repair, mitophagy, ROS, disease
Mitochondrial genetics
Mitochondria are a dynamic network of organelles constantly adapting their morphology and function to accommodate the needs of the cell. They are composed of an outer membrane, an intermembrane space, a highly folded inner membrane (the cristae) and a matrix space. Due to the prokaryotic origin of this organelle, the inner mitochondria membrane contains a specialized phospholipid, cardiolipin, that is also found in bacteria. More importantly the mitochondria contain their own DNA. The human mitochondrial genome, mitochondrial DNA (mtDNA), is a small circular ~16.6 kilobase molecule, which resides inside the matrix space associated with the inner membrane of the mitochondria [1]. MtDNA in humans encodes thirteen polypeptides, twenty-two tRNAs and two ribosomal genes that are essential for oxidative phosphorylation, the metabolic process by which cells convert energy stored in a range of different substrates to ATP, which is the energetic currency of the organism. All of the remaining mitochondrial proteins, including gene products necessary for mtDNA replication, transcription and DNA repair, are derived from nuclear genes and are imported into the mitochondria, typically, but not exclusively, via a mitochondrial targeting sequence [2]. In addition to the role of mitochondria in ATP production, this organelle is also central in apoptosis, heme and steroid synthesis, Ca2+ regulation, adaptive thermogenesis and other processes. Proper mitochondrial function is therefore critical for organismal health.
An understanding of mtDNA inheritance and maintenance patterns is essential for comprehending mitochondrial dysfunction in disease. MtDNA is packaged into protein-DNA structures called nucleoids containing one or more mtDNA genomes within a single nucleoid. Additionally, there are a few to several thousand copies of mtDNA per cell varying with cell type [3]. Cells can simultaneously carry a mixture of normal and mutated mitochondrial genomes, a condition known as heteroplasmy. Mutant mtDNA can be propagated along with normal mtDNA, when there is no selection pressure against the mutant genome, thereby contributing to the high sequence evolution of mtDNA [4]. When a cell divides and the nucleoids are segregated between the two daughter cells, the proportion of mutant to normal mtDNA can shift [5]. This has important ramifications for mitochondrial disease since the relative proportion of mutant mtDNA molecules must reach a certain threshold before a disease phenotype is observed.
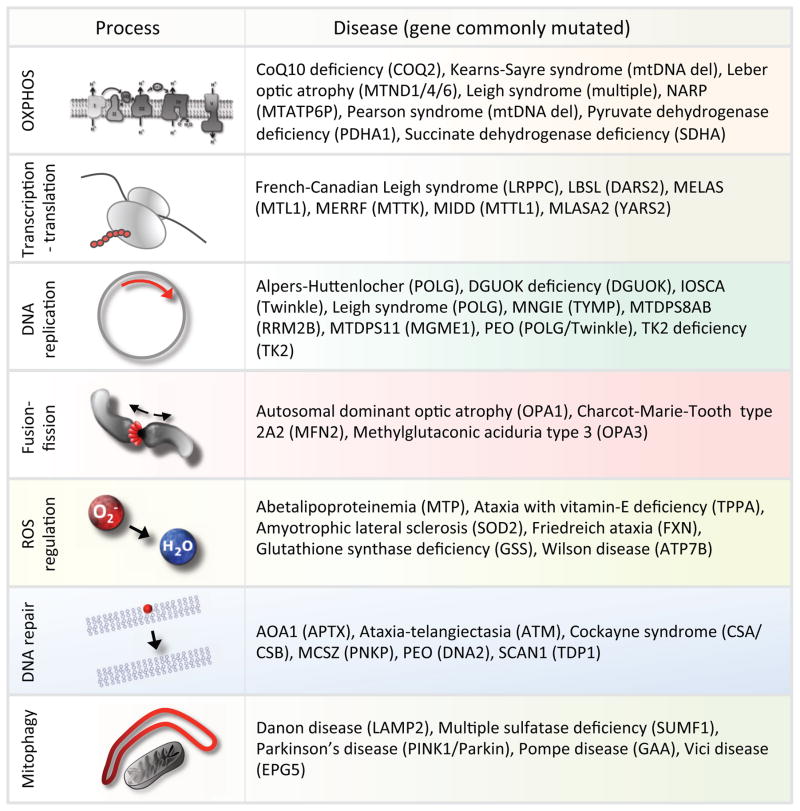
Bona fide primary mitochondrial diseases represent a heterogeneous group of disorders most often involving multiple organ systems leading to progressive degeneration and in many cases early death. Since the combined prevalence is estimated to be around 1:5000, a mitochondrial etiology should be considered when encountering any patient, particularly children, with multisystem pathology in tissues such as the central nervous system, heart, skeletal muscles, liver and in rarer cases kidney [6]. The pathogenic mutation can be located either within the mitochondrial or nuclear genome and, as in the case of mutations in Twinkle or POLG, can give rise to a great diversity of clinical syndromes (Figure 1).
Figure 1.
Examples of mitochondrial pathways that are associated with mitochondrial diseases.
A number of well-defined syndromes are caused by maternally inherited mutations/deletions in mtDNA. Leigh syndrome is one of the most severe mitochondrial syndromes and can be caused by mutations in a number of mitochondrial as well as nuclear encoded genes. The patients usually die in infancy or early childhood due to rapidly progressing neurodegeneration [7]. Pearson syndrome is caused by inherited deletions in the mitochondrial DNA [8]. This early childhood disease is characterized by lactic acidosis as well as pancreatic insufficiency and anemia, two clinical characteristics that are rare in other mitochondrial diseases. Interestingly, Pearson syndrome patients that survive through childhood may later develop Kearns-Sayre syndrome, a disease characterized by ophthalmoplegia (loss of eye movement), retinal degeneration and cardiomyopathy [9]. Other notable diseases caused by inherited mtDNA mutations are myoclonic epilepsy with ragged red fibres (MERRF) and myopathy, encephalopathy, lactic acidosis and stroke like episodes (MELAS). MERRF and MELAS are most commonly caused by mutations in the tRNA genes MTTK and MTTL1 respectively and there is often substantial overlap in the clinical presentation of the two syndromes [10]. [11]. Indeed, identical mutations in the mitochondrial genome can lead to both MERRF and MELAS as well as maternally inherited diabetes and deafness (MIDD). In addition, mitochondrial DNA mutations can lead to Leber’s optic atrophy, neuropathy, ataxia, retinitis pigmentosum (NARP) as well as a number of other defined diseases. It is thus clear that an intact mitochondrial genome is essential for organismal fitness. We will in this review attempt to explain the pathogenesis of mitochondrial disease based on the failure of a number of maintenance pathways that preserve the integrity of this crucial organelle.
Mitochondrial maintenance pathways
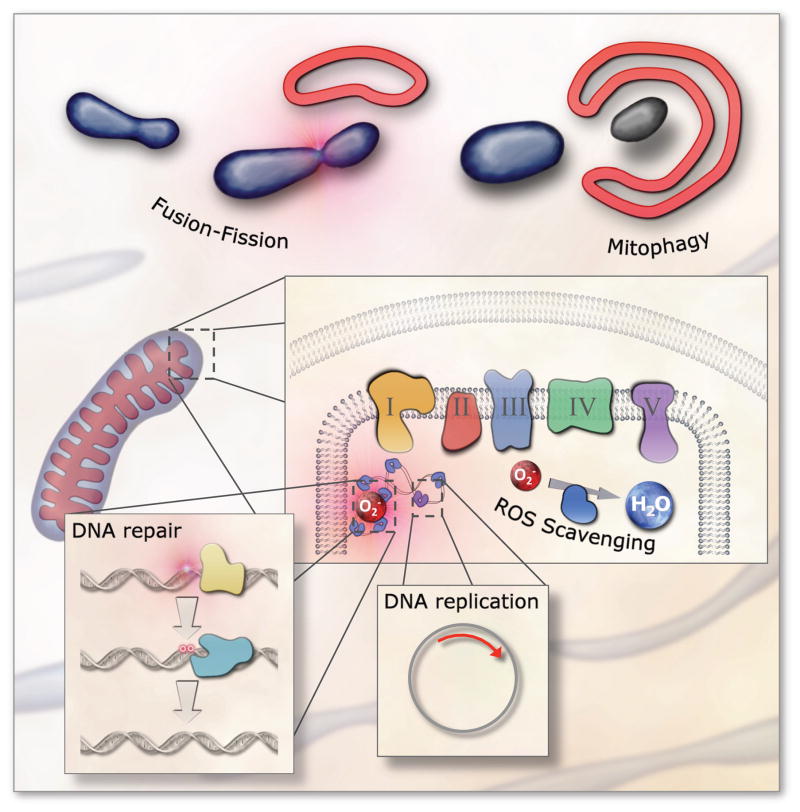
To preserve mtDNA integrity and mitochondrial function several conserved pathways have evolved that are required for life (Figure 2). These include: (i) faithful DNA replication, transcription and translation, (ii) processes that maintain redox homeostasis and prevent oxidative stress, (iii) systems that regulate proper mitochondrial fusion and fission, (D) pathways that remove and repair mtDNA modifications, and (iv) responses that eliminate excessively damaged mitochondria from the cell (i.e. mitochondrial autophagy/mitophagy). The importance of these pathways is underscored by the findings that defects in any of them can lead to human pathology. Notably, these pathways are highly regulated in particular by reactive oxygen species (ROS) that can signal fragmentation of the mitochondrial network [12], activation of anti-oxidant defenses [13,14], recruitment of DNA repair enzymes [15] and induction of mitophagy [16].
Figure 2. Mitochondrial maintenance pathways.
Several pathways have evolved that maintain mitochondrial integrity. These include faithful DNA replication; efficient DNA repair; enzymes and molecules that remove reactive oxygen species (ROS); mitochondrial morphology regulation through fission and fusion; and whole organelle removal through mitophagy.
In addition to the above pathways, several clinically important processes have been discovered that preserve mitochondrial function, such as mitochondrial proteostasis [17], lipid membrane upkeep [18] and others. Mitochondrial proteostasis may be important in the context of aging since activation of the mitochondrial unfolded protein response (mtUPR) leads to lifespan extension in lower organisms and possibly mammals [19]. The mtUPR is thought to be initiated by the accumulation of misfolded proteins in the mitochondria leading to a signaling cascade involving at least two central players, activating transcription factor associated with stress 1 (ATFS1) and the kinase GCN-2. GCN-2 inhibits cytosolic protein translation thereby decreasing the import of new protein to the mitochondria [20]. ATFS1 that is normally localized to the mitochondria, relocalizes to the nucleus upon mitochondrial proteotoxic stress and activates transcription of proteins, such as the heat shock proteins (HSPs), which attempt to decrease the proteotoxic stress. HSPs form a central node in the mtUPR response and, interestingly, HSP60 mutations leads to mitochondrial disease in humans characterized by neurodegeneration [21]. Although the role of the mtUPR in organismal health of lower organisms is well established, the function of this pathway in mammals is still being uncovered. We refer readers to a recent review on the mtUPR for additional reading [19].
Mitochondrial DNA replication and transcription
The mtDNA replication machinery constitutes a clinically important group of proteins because a large number of diseases are associated with mutations in genes involved in this process. The core components and minimal set of proteins needed for mtDNA replication consists of DNA polymerase γ (POLG), the only DNA polymerase present in mitochondria; Twinkle, a DNA helicase; and mitochondrial single stranded binding protein (mtSSB). A growing number of proteins are associated with mitochondrial replication, and we will refer the reader to recent reviews for a comprehensive list [22,23]. Despite decades of research into the process of mtDNA replication, the exact mechanism remains unclear. At least three models are currently under consideration including (i) standard leading and lagging strand replication as seen in the nucleus, (ii) a strand displacement mode where the leading strand synthesizes a large fragment of DNA before lagging strand synthesis is initiated, and (iii) a model whereby the lagging strand is continuously or intermittently hybridized with complementary RNA (termed RITOLs) perhaps to protect the exposed strand [22]. Regardless of which model may be correct, faithful and efficient mtDNA replication is central for mtDNA stability, and numerous mitochondrial diseases are associated with mtDNA replication failure. Indeed, POLG mutations can lead to Leigh syndrome and early death as well as Alpers-Huttenlocher syndrome that usually manifests at around 2–4 years of age, with progressive neurological decline in combination with seizures and liver disease [7]. Infantile onset spinocerebellar ataxia (IOSCA) patients typically debuts in early childhood with progressive ataxia and muscle weakness leading to loss of ambulation by the teenage years [24]. Early adult onset is usually seen in mitochondrial neurogastrointestinal encephalopathy (MNGIE) where nerves in the digestive system deteriorate leading to eating difficulties and weight loss, while the central nervous system becomes affected at later stages in the disease progression [25]. The mildest cases of Twinkle and POLG mutations can lead to adult onset progressive external ophthalmoplegia, symptoms that are often also observed in the more severe syndromes [26].
Mitochondrial RNA is transcribed as long transcripts containing multiple genes without introns (ie. polycistronic), a remnant of the bacterial ancestry of this organelle. Transcription is initiated from a specialized structure in the genome called the displacement loop (D-loop). In this region the DNA forms a loop and an RNA molecule is hybridized to the light strand. Three transcripts are believed to be generated from three promotors in the D-loop region of the mitochondrial genome, two on the heavy strand (HSP1 and HSP2) and one on the light strand (LSP). The essential transcription apparatus consists of the mitochondrial RNA polymerase, mitochondrial transcription factor B2 (TFB2M) and mitochondrial transcription factor A (TFAM) [27,28]. It has, however, been proposed that TFAM may not be essential for transcription but could play a role as a transcriptional regulator [29]. Regardless, mitochondrial transcription is essential for replication, at least in part because of the need for RNA primers in replication initiation. Indeed, loss of mitochondrial transcriptional regulation can lead to severe infantile neurodegeneration [30]. Posttranscriptional splicing and translation of the mitochondrial RNA occur within the mitochondrial matrix [31]. The exact mechanisms remain largely unknown, although a significant amount of diseases appear associated with these processes. For further detail we will refer the reader to a recent review on this topic [31].
Fission and Fusion
Mitochondria are dynamic in part because they are constantly undergoing fission and fusion reactions. Fission is the process by which one mitochondrial organelle divides and becomes two while fusion is the opposite [32]. Even though the process is still being elucidated, some mechanistic insight into these complicated pathways has been achieved. Both processes are regulated by a number of guanosine triphosphatases (GTPase) that are conserved from yeast to mammals. Fission is believed to be initiated by the stabilization of the protein dynamin-related protein 1 (Drp1) on the outer membrane [33]. Drp1 forms a spiral around the outer membrane that creates a curvature in the membrane and eventually constricts both the inner and outer membrane to allow fission to occur. Fusion is mediated through proteins such as Mitofusin-1 (Mfn1) and Mfn2 that facilitate the fusion of the outer membrane [34]. Upon fusion of the outer membrane, proteins such as Optic Atrophy-1 (OPA1) and OPA3 are required for proper fusion of the inner membrane [35]. Interestingly, the endoplasmic reticulum (ER) may play a specific role in mitochondrial fission. Physical and functional interactions between the ER and the mitochondria have been known to occur for decades at so called ER-mitochondria encounter structures (ERMES) [36,37]. During fission ER appears to associate with and wrap around the constricting neck between the mitochondria that are being separated [38]. The role of the ER in this process is not well understood, but could entail local alterations in calcium levels [39], regulation of the lipid composition in the outer mitochondrial membrane [40], changes in MFN1/2 activity [41] and others. In addition, mitochondria may associate with other endomembranes such as the golgi apparatus [42] and, at least in yeast, the lysosomal compartment that can supply phospholipids for mitochondrial membrane synthesis [43]. Interestingly, the mitochondrial-lysosomal interaction may also occur in mammals where budding of vesicles from the mitochondria may deliver cargo to the lysosomes for degradation [44].
Although fission may logically be necessary to increase the number of mitochondria for cell division, there is no a priori reason for why fusion should occur. One intriguing possibility is that fusion may facilitate the exchange of mtDNA to complement deficits in mitochondria carrying mutated genomes. This inter-organelle complementation was elegantly shown by Gilkerson in a study using two cell lines containing different mtDNA deletions [45]. When the cells were fused, both nucleoid populations were visible and transcomplemented one another thus allowing mitochondrial protein synthesis [45]. Importantly, this implies that each mitochondrion has the intrinsic ability to exchange mtDNA with other mitochondria. From a maintenance perspective increasing mitochondrial fusion will allow for transfer of nucleoids and the preservation of mitochondrial function but may thereby simultaneously hamper the cellular detection of damaged mitochondria. Accordingly, mitochondrial fission predominates during oocyte development leading to fragmentation of the mitochondrial network and perhaps allowing for the discrimination and removal of organelles containing mutated DNA [46]. Notably, increasing evidence suggests that degradation of mitochondria is regulated by fission of damaged mitochondria and subsequent degradation of the damaged part. Accordingly, fission and fusion are intimately linked with how nucleoids are distributed within a cell and with mitochondrial quality control [47]. Fission and fusion may be particularly important during cell division where non-random segregation of mitochondria can facilitate the retention or removal of bad mitochondria from the mother cell. This has been demonstrated in yeast where the mother cell retains mitochondria of lower quality while the daughter cell receives the functionally best mitochondria [48]. Whether this occurs in mammals remains to be elucidated but the mechanism could represent a fascinating way of selecting the fittest mitochondria for transmission during for example germ cell development. Intriguingly, increased fusion and decreased fission were observed in the accelerated aging disorders Cockayne syndrome, xeroderma pigmentosum group A and ataxia-telangiectasia, highlighting the possibility that alterations in this pathway may be involved in the aging process [49]. This is supported by recent data suggesting that senescent cells show increased fusion through downregulation of the fission proteins Fission-1 (Fis1) and Drp1 [50]. Loss of fission could thus contribute to an age-associated accumulation of mtDNA damage.
The importance of fusion-fission in human health is highlighted by a number of diseases caused by mutations in proteins involved in these processes. This is best exemplified by mutations in OPA1 that leads to optic atrophy often in combination with hearing loss and ophthalmoplegia [51]. Loss of MFN2, on the other hand, leads to Charcot-Marie-Tooth type 2A, a disease characterized by neuropathy and associated muscle weakness as well as in some cases hearing loss, optic atrophy and pyramidal signs [52,53]. It is thus clear that fusion-fission plays a critical role in maintaining a healthy pool of mitochondria.
Reactive Chemical Species Homeostasis
Mitochondria are the major source of intracellular ROS, creating superoxide that can be dismutated into hydrogen peroxide. Besides the formation of ROS via respiration, intracellular accumulation of metals such as iron significantly increase the level of reactive chemical species through Fenton chemistry where a hydroxyl radical is produced from the reaction of reduced iron with hydrogen peroxide. This is particularly pertinent in iron-overload disorders such as Friedreich ataxia and hemochromatosis where Fenton chemistry leads to increased ROS generation [54,55]. Although ROS have indispensable roles in cellular signaling, in the immune response, and may be involved in pro-longevity signaling in C. elegans [56], they maintain the potential to damage all known macromolecules, including nucleic acids, proteins and lipids. Such damage may contribute to the etiology of human disease and likely promotes at least some of the functional deterioration associated with aging (the so-called “free radical theory of aging”) [57].
To avoid oxidative stress, cells maintain a balance between the production of ROS and the ability to detoxify the reactive intermediates. Mitochondrial ROS production is regulated by uncoupling proteins (UCPs) that lower the membrane potential of the mitochondria allowing electrons to flow through the electron transport chain with less resistance and thereby decreases the likelihood of spurious reduction of molecular oxygen to superoxide [13]. Notably, loss of UCPs has been linked to neurodegeneration in animal models and decreased levels of UCP2 were found in several neurodegenerative accelerated aging disorders [58]. However, once free radicals have been produced the antioxidants supply adds an additional level of protection. The primary cellular antioxidant enzymes are superoxide dismutases (SODs) that convert superoxide to hydrogen peroxide; catalase that converts hydrogen peroxide to water; glutathione peroxidase, and the peroxiredoxins that neutralizes hydrogen peroxide by disulfide bond formation. In addition, non-enzymatic factors such as glutathione, vitamin C, and vitamin E function as scavengers of various oxidants (Table 1).
Table 1.
A list of common antioxidants and their reactions.
| Reducing agent | Reaction | |
|---|---|---|
| superoxide dimutase |
|
|
| Catalase | 2H2O2 → 2H2O + O2 | |
| Peroxiredoxins | 2RSH + H2O2 → RSSR + 2H2O | |
| Glutathione peroxidase/Glutathione | 2GSH + H2O2 → GSSG + 2H2O | |
| Vitamin E/C | Vitred + RO2 → VitOx + RH |
A number of diseases are associated with loss of ROS homeostasis, including primary vitamin E deficiency (AVED), glutathione synthase deficiency, abetalipoproteinemia, Friedreich ataxia, Wilson’s disease and possibly amyotrophic lateral sclerosis (ALS). AVED is caused by defects in TTPA, the enzyme responsible for the uptake of the essential antioxidant, vitamin E, from the gut [59]. Similarly, in abetalipoproteinemia defective uptake of lipid soluble vitamins leads to loss of vitamin E and A [60]. Patients suffering from AVED and abetalipoproteinemia display cerebellar degeneration as well as retinal pathology with a subset of patients showing cardiomyopathy. Due to the concomitant loss of vitamin A, abetalipoproteinemia patients also display night blindness that can progress to complete vision loss. Mitochondrial defects have long been associated with vitamin E deficiency [61] and, although not formally investigated, presumably also occur in abetalipoproteinemia. Friedreich ataxia is caused by a trinucleotide repeat expansion in the protein FXN leading to defects in iron sulfur cluster and heme biosynthesis, and loss of FXN leads to loss of iron transporting enzymes [55]. This is interpreted by the cell as a loss of iron and, as an adaptive response, the cells increase the iron uptake leading to intracellular iron accumulation and ROS generation through Fenton chemistry. Like AVED and abetalipoproteinemia, Friedreich ataxia is characterized by cerebellar ataxia and degeneration as well as neuropathy, myopathy and in some cases cardiomyopathy. Wilson disease is caused by mutations in the copper transporter ATP7B and leads to copper overload and ROS generation. Patients suffering from this disorder develop Parkinsonian symptoms as well as hepatic disease, perhaps connecting Wilson disease to the mitophagy pathway (see below) [62]. In addition, patients develop anemia, a feature also observed in glutathione synthase deficient patients [63]. In summary, the close clinical similarities between these ROS related diseases and known mitochondrial diseases suggest that mitochondrial defects may be central in the pathogenesis of these disorders.
MtDNA Repair
DNA repair mechanisms have evolved to faithfully restore damaged genetic material back to its original, undamaged state [64]. DNA damage can arise via a wide range of events, including spontaneous decay of the nucleic acid structure, reactions with intracellular chemical species, such as ROS, and interactions with chemical and physical environmental agents. Each of these events can lead to a range of DNA alterations such as complete loss of a base (known as an apurinic or apyrimidinic (AP) site), cleavage of the phosphodiester backbone to create a discontinuous strand break, or more complex lesions such as intra- or inter-strand crosslinks. If unrepaired, DNA damage can block transcription or replication as well as lead to mutagenesis. In either case, cellular dysfunction can arise, resulting in cell death or transformation, outcomes that likely contribute to neurodegeneration, cancer, and aging [65].
To counteract the destructive effects of DNA damage, five primary corrective systems exist in the nucleus, exhibiting complementary, yet in some instances overlapping, substrate specificity: direct reversal, mismatch repair, base excision repair, nucleotide excision repair, and DNA double stranded break repair. The presence or absence of these DNA repair pathways within the mitochondria is still a topic of debate and it is therefore of importance to understand the function of the nuclear DNA repair pathways. 06-methylguanine-DNA-methyltransferase [66] carries out direct reversal, transferring certain methyl group adducts from DNA to an active site cysteine residue [66]. DNA mismatch repair copes with errors that arise during DNA replication, such as mismatched nucleotides or short hairpin or loop structures [67]. Base excision repair is the predominant pathway for coping with non-bulky base modifications, namely oxidative DNA base lesions, as well as AP sites and different chemical forms of single-strand breaks [68]. Nucleotide excision repair handles bulky base modifications or other helix-distorting alterations to DNA [69]. Mismatch, base excision and nucleotide excision repair all involve a series of enzymatic steps that aim to excise the damage or substrate lesion, restore the original nucleotide content via a re-synthesis step, and seal the final, remaining nick via ligation. Double stranded break repair has two major branches: homologous recombination and non-homologous end-joining [70]. The former, which is unquestionably the most complex of the pathways, resolves DNA double-strand breaks or collapsed replication forks through a process that entails the faithful exchange of genetic information from an undamaged sister chromatid or a homologous chromosome as the template. Non-homologous end-joining involves specific processing steps that re-connect two DNA ends via ligation, yet often results in error-prone resolution of double-strand breaks.
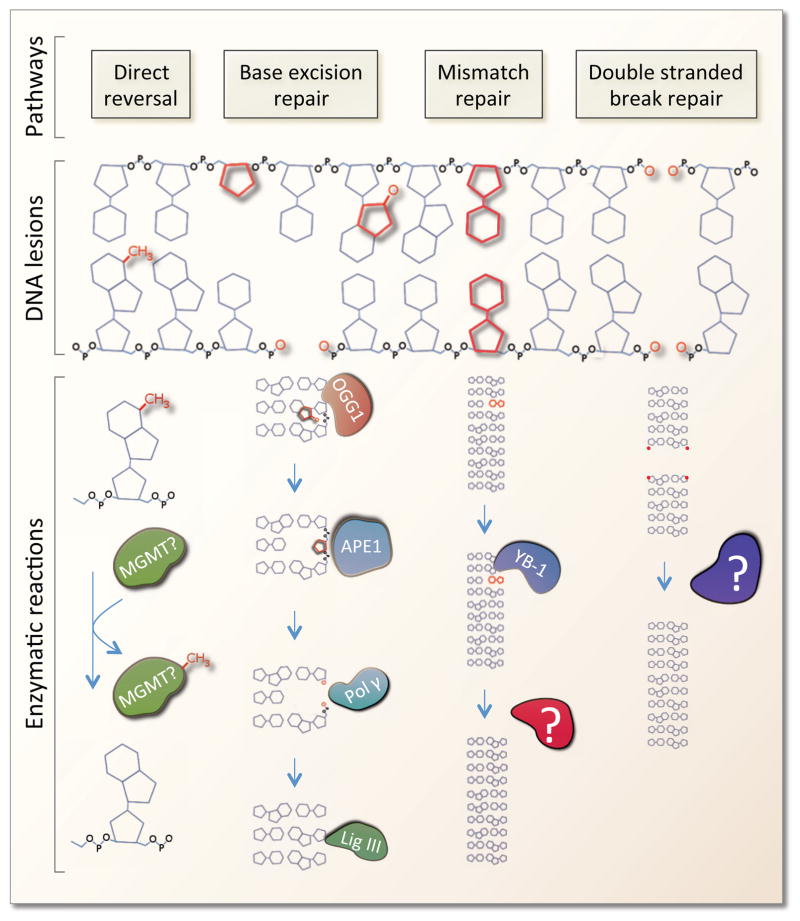
As eluded to earlier, the collection of DNA repair proteins stationed in the mitochondria is derived from the nuclear genome. Significantly, the proximity of mtDNA to the respiratory chain and oxidative phosphorylation results in an increased likelihood of ROS-induced mtDNA damage. Within mitochondria, the predominant DNA repair mechanism is base excision repair [71], which is the primary system for coping with oxidative DNA damage (Figure 3). Importantly, POLG is believed to be the only DNA polymerase in the mitochondria and thereby participates in base excision repair as well as replication. In addition, POLG contains a specialized exonuclease domain that acts as a proof reading mechanism allowing higher fidelity replication, perhaps minimizing the need for mismatch DNA repair. Indeed, the nuclear form of mismatch repair appears to be absent in mitochondria, but Y-box binding protein 1 (YB-1) may facilitate a unique mitochondrial mismatch repair process in human cells [72]. Repair of more complex lesions to the DNA such as UV-induced dimerization of the bases or interstrand DNA crosslinks does not appear to occur in mitochondria [73]. Further, repair of DNA double stranded breaks is absent or much less efficient in the mitochondria as compared to the nucleus. However, mitochondrial extracts can rejoin blunt-ended and cohesive linearized plasmid DNA [74] and, at least a few key double stranded break repair proteins, such as Rad51 and Mre11, may be present in mitochondria [75,76].
Figure 3. Mitochondrial DNA repair pathways.
A number of different DNA lesions have been shown to be reparable in the mitochondria. Direct reversal of spurious methylation has been shown to occur, however, the enzyme(s) involved is not currently known. Base excision repair is the major DNA repair pathway in mitochondria and most proteins involved in this pathway have been shown to be present in mitochondria. Base excision repair consists of multiple steps. The lesion is initially identified by a number of different glycosylases (eg. OGG1, NEIL1 etc.) that remove the damaged base leaving an abasic site. The abasic site is then recognized by APE1 that removes the ribose allowing for final repair by DNA polymerase γ and ligase III. Mismatch repair (MMR) has been shown in human mitochondria where Y-box-binding protein 1 (YB-1) is proposed to be a key player. The downstream enzymes in mitochondrial MMR are, however, still not known. DNA double stranded break repair (DSBR) may occur in the mitochondria by blunt end ligation, although the enzymes involved in this pathway are not known.
From a clinical perspective DNA repair is important. Recently, mutations in two mitochondrial DNA nucleases, DNA2 and MGME1, have been shown to cause ophthalmoplegia, myopathy and loss of mitochondrial DNA [77,78]. Although it remains speculative as to whether these enzymes participate in DNA repair, the catalytic function of these nucleases could be compatible with base excision repair. In addition, several primary DNA repair disorders have been shown to have alterations in mitochondrial function. These include spinocerebellar ataxia with axonal neuropathy 1 (SCAN1), caused by defects in the base excision repair-related enzyme TDP1 [79]; ataxia with oculomotor apraxia 1 (AOA1), caused by defects in another base excision repair-related enzyme APTX [80]; Cockayne syndrome (CS), caused by defects in the nucleotide excision repair proteins CSA/CSB [81]; ataxia-telangiectasia (AT), caused by defects in the double stranded break repair kinase ATM [82]; and xeroderma pigmentosum group A, caused by defects in the nucleotide excision repair protein XPA [83]. Although, the phenotype is quite varied among these diseases, they all display progressive cerebellar pathology. SCAN1 and AOA1 are clinically very similar with cerebellar degeneration and peripheral neuropathy, and both are caused by defects in base excision repair-related proteins, ATPX and TDP1, that have been found to be present in mitochondria [84,85]. AOA1 patients can, however, present indistinguishable from AT patients that likewise show progressive cerebellar neurodegeneration as a prominent feature. In addition to the intracranial aspects of AT, these patients develop telangiectasia and are cancer prone, likely due to the role of ATM in DNA double stranded break repair. CS and XPA patients also display severe cerebellar neurodegeneration, but additionally show hypersensitivity to sunlight caused by defective repair of UV-induced DNA lesions. Interestingly, although ATM, CSA and CSB have all been found in mitochondria [86–88], nucleotide excision repair and classic double stranded break repair are not thought to occur in this organelle. Indeed, XPA, a necessary protein for nucleotide excision repair, does not appear to be present in mitochondria, and recent findings suggest that the mitochondrial dysfunction observed in these disorders may be secondary to activation of a nuclear DNA damage response [49]. This outcome appears to involve the activation of the DNA damage response enzyme poly-ADP-ribose-polymerase 1, leading to increased ATP consumption, depletion of NAD+ and SIRT1 attenuation [49]. Neurodegeneration is prevalent in mitochondrial disorders perhaps because the brain has a higher energy demand than other tissues. It is therefore possible that the neurodegeneration observed in the DNA repair disorders may not be caused by a defect in ATP supply, but by an increase in ATP demand leading to secondary energetic failure. Accordingly, increased energy expenditure and progressive fat loss is common in these diseases [89]. Regardless of whether the mitochondrial involvement in these disorders is primary or secondary, the outcome is mitochondrial dysfunction. DNA repair may thus be quite important when considering patients with mitochondrial defects.
Autophagy and Mitophagy
Autophagy is an evolutionarily conserved fundamental intracellular degradative pathway that operates as a homeostatic mechanism in all eukaryotic cells [90]. It is a tightly-regulated process that plays a normal part in cell growth and development, maintaining a balance between the synthesis, degradation, and subsequent recycling of cellular products. There are several forms of autophagy, each of which involves delivering intracellular cargo to lysosomes for degradation. Macroautophagy (autophagy hereafter), entails the production of vesicles called autophagosomes that capture and deliver cytoplasmic material to lysosomes. The autophagy-related genes (the atg genes) are conserved from yeast to mammals and regulate the cannibalism of intracellular content such as proteins, and organelles. Failure of autophagy is thought to lead to the accumulation of cell damage, and defects in the process are linked to liver disease, neurodegeneration, Crohn’s disease, aging, cancer, and metabolic syndrome [91].
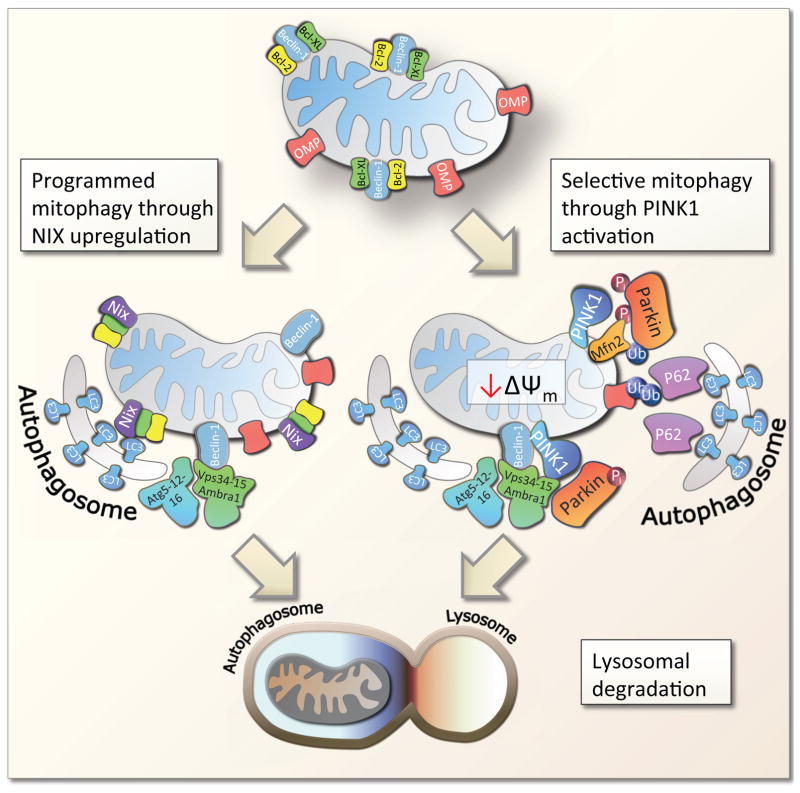
Interestingly, a specialized sub-pathway of autophagy, conserved from yeast to mammals, has evolved to specifically deal with the degradation of dysfunctional mitochondria (Figure 4). The pathway, termed mitophagy, utilizes many components of normal autophagy, yet the initiation that leads to the selective degradation of mitochondria is specific to this cellular process. Notably, mitophagy can be initiated in several different ways. Programmed mitophagy occurs during erythroblast maturation to rid mature red blood cells of mitochondria. This process is believed to be initiated through increased expression of the promitophagic factor NIX (BNIP3L) [92]. NIX can directly interact with LC3 (the mammalian homologue of yeast atg-8) that acts as a scaffolding protein coating the double lipid membrane of the autophagosome. In addition, upregulation of NIX may lead to the de-repression of Beclin-1, a key autophagic regulator. Beclin-1 stimulates autophagy likely through both initiation as well as elongation of the autophagosome [93].
Figure 4. The mechanism of mitophagy.
Mammalian mitophagy is believed to occur through at least two pathways, programmed mitophagy and selective mitophagy, although significant cross talk between these has been found. Programmed mitophagy was discovered in maturing red blood cells through upregulation of the protein NIX that may facilitate the dissociation of the anti-mitophagic proteins Bcl-2 and Bcl-XL from the pro-mitophagic protein Beclin-1. This potentially derepresses Beclin-1 allowing recruitment of the Vps34-15-AMBRA complex that in turn will associate with the autophagosome elongation machinery atg5-12/atg16 leading to the formation of an autophagosome. NIX coats the mitochondria and associates directly with LC3 which binds to the growing autophagosome. Mitophagy is completed by the fusion of the mitophagosome with a lysosome. In selective mitophagy the initiating event is believed to be mitochondrial inner membrane depolarization leading to PINK1 accumulation at the outer membrane. PINK1 phosphorylates Parkin, Mfn2 and other proteins leading to the activation of Parkin. Parkin is a E3-ubiquitin ligase that upon activation ubiquitinates outer mitochondrial membrane proteins (OMPs), possibly VDAC, are ubiquitinated. Vps34-15-AMBRA complex and the Atg5-12/atg16 complex is recruited by activated PINK1/Parkin. Outer membrane ubiquitination leads to the recruitment of p62 which will associate with LC3 on the growing autophagosome. When the autophagosome is completed, fusion with a lysosome will lead to complete degradation of the mitochondria.
In selective mitophagy, mitochondrial membrane depolarization is believed to lead to the accumulation and activation of the kinase PINK1, an enzyme mutated in familial Parkinson’s disease, at the outer membrane [94]. Here PINK1 phosphorylates Parkin, an E-3 ubiquitin ligase, as well as ubiquitin, leading to the activation of Parkin, ubiquitination of outer mitochondrial membrane proteins and recruitment of downstream factors such as p62 (SQSTM1) and Beclin-1 [16,95]. p62 can directly interact with LC3, and p62 accumulation leads to degradation of depolarized mitochondria. Notably, NIX is also recruited upon membrane depolarization perhaps upstream of Parkin and can thereby directly stimulate mitophagy, possibly acting as a secondary mitophagy switch [96]. More recently, PINK1 and Parkin activation was shown to lead to ubiquitination of the fusion protein Mfn2 connecting mitophagy with the fusion-fission machinery [97]. Indeed, Mfn2 ubiquitination may lead to degradation of this protein and inhibition of mitochondrial fusion. This leads to fragmentation of mitochondria perhaps allowing for identification of dysfunctional mitochondria that can be degraded by mitophagy. Interestingly, mitophagic defects, mitochondrial membrane hyperpolarization, PINK1 degradation and increased mitochondrial fusion have been found in some accelerated aging models [49]. Thus, mounting evidence suggests that selective mitophagy is important for the maintenance of mitochondrial health and perturbations in this pathway will lead to disease.
As suggested by the discussion above, mitophagy is a highly complex pathway involving numerous proteins and at least two organelles: the mitochondria and the lysosome. Defects in proteins associated with either of these structures as well as proteins involved in general autophagy can therefore lead to mitophagic dysfunction. Currently, Vici syndrome, caused by mutations in EPG5, is the only known human disease with a defect in general autophagy [98]. Patients suffering from Vici syndrome show immune deficiencies, neurological structural deficits, cardiomyopathy and skeletal myopathy. Although the specific molecular role of EPG5 is under investigation, it is believed to participate in the autophagosome-lysosome fusion process [98]. Interestingly, EPG5 knockout mice develop ALS-like phenotypes with specific degeneration of motor neurons [99]. Regarding mitophagy, two landmark papers showed that PINK1 and Parkin, two proteins mutated in familial Parkinson’s disease, are involved in the selective degradation of damaged mitochondria [16,100]. Loss of these proteins may contribute to the accumulation of damaged mitochondria and death of dopaminergic neurons in the substantia nigra in the mesencephalon. Further support of a mitochondrial etiology in Parkinson’s disease comes from the early observations that exposure to various mitochondrial toxins leads to Parkinson’s disease in humans and rodents [101]. Interestingly, Parkinsonism is relatively rare in primary mitochondrial diseases indicating that mitochondrial dysfunction does not automatically lead to dopaminergic neuronal death. Conversely, Parkinson’s disease is not characterized by the severe neurodegeneration that commonly debuts in early adult hood in primary mitochondrial diseases. This may indicate that alternative mitophagy pathways may compensate for defects in PINK1 or Parkin [102] or that mitophagy plays a relatively minor role in overall mitochondrial maintenance. Recent findings of defective mitophagy in neurodegenerative accelerated aging disorders do, however, support a significant role of this pathway in overall mitochondrial maintenance [49].
At the lysosomal side, Danon disease [103], Pompe disease [104], and multiple sulfatase deficiency [105], clinically resemble a number of mitochondrial disorders such as Barth syndrome, multiple acyl-CoA dehydrogenase deficiency and methyl-glutaconic aciduria 1 and 3. Danon and Pompe diseases are characterized by myopathy and neurodegeneration while multiple sulfatase deficiency predominantly leads to cerebral and cerebellar atrophy as well as hepatopathy. Notably, loss of these lysosomal proteins leads to a much more severe phenotype than what is seen in Parkinson’s disease. This could reflect that a complete loss of autophagy, as seen in lysosomal disorders, results in severe cellular defects, while dysfunction of an autophagic sub-pathway, such as mitophagy, may be less detrimental.
Concluding remarks
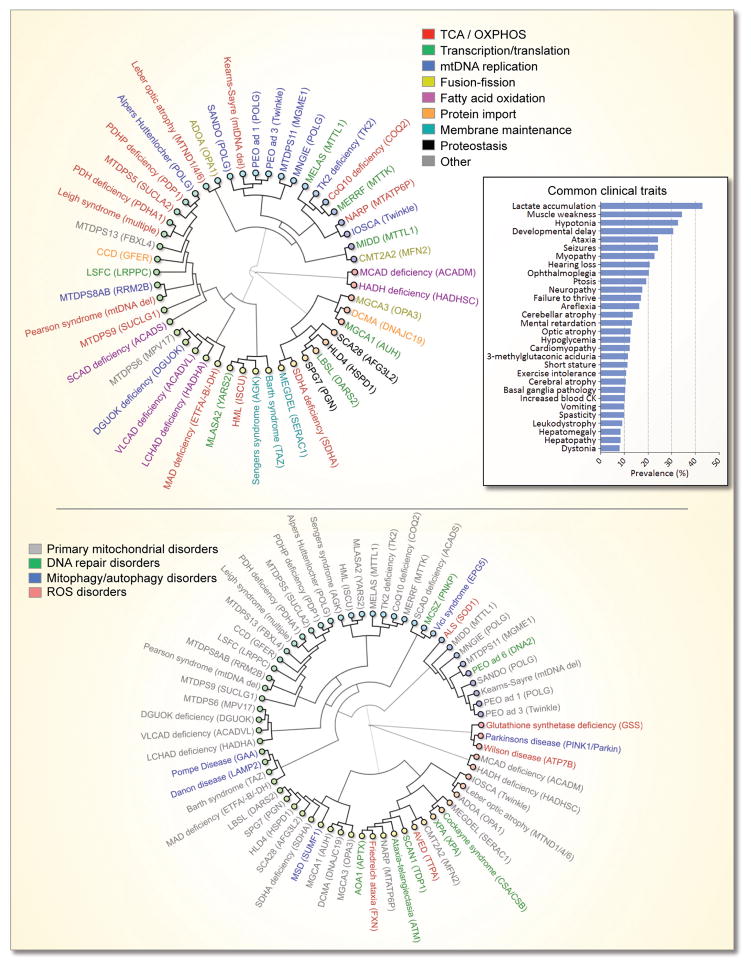
Mitochondrial diseases are extraordinarily diverse not only in their phenotypical presentation but also in their genetic background. To better understand the complexity of mitochondrial disorders, a database of these diseases, www.mitodb.com, has been created that contains the prevalence of signs and symptoms from published patient cohorts for more than 40 known mitochondrial disorders, describing more than 2,200 patients. Based on the clinical similarities between these diseases, hierarchical clustering can be performed where diseases that are clinically similar appear close to each other in a dendrogram, while diseases without overlapping features are distant (Figure 5) [106]. Although the underlying defective pathway is still being debated for a number of these disorders, it appears quite difficult to correlate defects in a specific pathway with the clinical outcome. For example, defects in mitochondrial replication can lead to a large number of different phenotypes of greatly varying severity. Indeed, even within the same defined clinical syndrome, major phenotypical variation occurs [107–109]. Other genetic factors, environmental influences, levels of heteroplasmy or perhaps simple stochastic events may be responsible for this complexity. Notably, the primary mitochondrial disorders are often phenotypically similar to a number of emerging mitochondrial diseases characterized by ROS dysregulation, DNA repair defects or mitophagic dysfunction (Figure 5). Indeed, when looking at all the mitochondrial diseases, some clinical traits, such as lactic acidosis, myopathy etc., are clearly more prevalent than others (Figure 5, insert). Based on this fact, it may still be possible to distinguish mitochondrial diseases from non-mitochondrial diseases [106]. Supporting this notion, using various bioinformatics tools developed for www.mitodb.com, mitochondrial dysfunction was found in xeroderma pigmentosum group A, a DNA repair disorder not previously shown to have mitochondrial dysfunction [49].
Figure 5. The complex clinical relationship between mitochondrial disorders.
The dendrograms show the association between diseases based on the prevalence of the clinical traits in the diseases. The closer two diseases are the more signs and symptoms are shared ei. the more similar the diseases are. Genes commonly mutated in these disorders are shown in the parentheses. The top dendrogram shows primary mitochondrial diseases and the colors represent the specific putative pathway that may be affected in that disease. The insert shows the average prevalence of commonly altered clinical traits in the mitochondrial disorders depicted in the dendrogram. The bottom dendrogram show how diseases with dysregulation of ROS, DNA repair and mitophagy associate with the primary mitochondrial disorders.
It seems clear that mitochondrial maintenance is emerging as an important factor in preserving health. Even though the mechanistic pathogenesis for many of the primary disorders are still being elucidated, the observation that a large number of etiologically very distinct diseases can lead to similar highly complex phenotypes indicate that the secondary pathogenesis could potentially be identical and that a unified mitochondrial treatment for these disorders may be possible. We propose that augmentation of mitochondrial health may be a viable pharmaceutical approach in these diseases that may show efficacy regardless of the primary etiology. Attempts at this approach will undoubtedly be difficult and are complicated by the interconnection and involvement of multiple mitochondrial maintenance pathways in many of these diseases. In addition, these pathways are regulated in a highly coordinated manner. It has been demonstrated that oxidative stress will induce adaptive changes in UCPs, antioxidants, DNA repair and autophagy. Accordingly, the UCP2 knockout mouse shows a possible compensatory increase in SOD2 and mitochondrial glutathione levels [110]. Similarly, SOD2 heterozygous mice show a decreased respiratory control ratio and activation of the membrane permeability transition pore, perhaps indicating a compensatory increase in mitochondrial uncoupling and autophagy [111]. Further, increased oxidative stress due to CoQ10 deficiency leads to an adaptive increase in autophagy [112]. Additionally, it has recently been shown that a defect in mitochondrial protein maintenance can augment autophagy [113]. It follows that a decrease in ROS production will lead to a decrease in the mitochondrial maintenance pathways. This tight regulation of mitochondrial maintenance through ROS is a possible explanation for the disappointing results antioxidants have shown in some human trials. Going forward, assays for high throughput small molecule screens for mitophagy and mtDNA repair activators should be established and pharmaceuticals developed. With these achievements, a combinatorial treatment with drugs that simultaneously stimulate mitophagy, antioxidants and mtDNA repair could be viable options for diseases characterized by mitochondrial dysfunction.
Highlights.
Mitochondrial maintenance is essential for cellular and organismal function.
Maintenance includes ROS regulation, DNA repair, fusion-fission and mitophagy.
Loss of function of these pathways leads to disease.
Acknowledgments
We sincerely apologize to colleagues whose work we could not include due to space limitations. The research was supported entirely by the Intramural Research Program of the NIA/NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Prachar J. Mouse and human mitochondrial nucleoid--detailed structure in relation to function. Gen Physiol Biophys. 2010;29:160–174. [PubMed] [Google Scholar]
- 2.Becker T, et al. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci. 2012;37:85–91. doi: 10.1016/j.tibs.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole JC, et al. Multiplex analysis of mitochondrial DNA pathogenic and polymorphic sequence variants. Biol Chem. 2010;391:1115–1130. doi: 10.1515/BC.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunbar DR, et al. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc Natl Acad Sci U S A. 1995;92:6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haas RH, et al. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- 7.Taanman JW, et al. Analysis of mutant DNA polymerase gamma in patients with mitochondrial DNA depletion. Hum Mutat. 2009;30:248–254. doi: 10.1002/humu.20852. [DOI] [PubMed] [Google Scholar]
- 8.Manea EM, et al. Pearson syndrome in the neonatal period: two case reports and review of the literature. J Pediatr Hematol Oncol. 2009;31:947–951. doi: 10.1097/MPH.0b013e3181bbc4ef. [DOI] [PubMed] [Google Scholar]
- 9.Aure K, et al. Chronic progressive ophthalmoplegia with large-scale mtDNA rearrangement: can we predict progression? Brain. 2007;130:1516–1524. doi: 10.1093/brain/awm067. [DOI] [PubMed] [Google Scholar]
- 10.Lorenzoni PJ, et al. MERRF: Clinical features, muscle biopsy and molecular genetics in Brazilian patients. Mitochondrion. 2011;11:528–532. doi: 10.1016/j.mito.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Lorenzoni PJ, et al. MELAS: clinical features, muscle biopsy and molecular genetics. Arq Neuropsiquiatr. 2009;67:668–676. doi: 10.1590/s0004-282x2009000400018. [DOI] [PubMed] [Google Scholar]
- 12.Wu S, et al. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 2011;278:941–954. doi: 10.1111/j.1742-4658.2011.08010.x. [DOI] [PubMed] [Google Scholar]
- 13.Echtay KS, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 14.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths LM, et al. Dynamic compartmentalization of base excision repair proteins in response to nuclear and mitochondrial oxidative stress. Mol Cell Biol. 2009;29:794–807. doi: 10.1128/MCB.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellegrino MW, et al. Signaling the mitochondrial unfolded protein response. Biochim Biophys Acta. 2013;1833:410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osman C, et al. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen MB, Jasper H. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 2014;20:214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker BM, et al. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magen D, et al. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am J Hum Genet. 2008;83:30–42. doi: 10.1016/j.ajhg.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt IJ, Reyes A. Human mitochondrial DNA replication. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasiviswanathan R, et al. The interface of transcription and DNA replication in the mitochondria. Biochim Biophys Acta. 2012;1819:970–978. doi: 10.1016/j.bbagrm.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palin EJ, et al. Mesencephalic complex I deficiency does not correlate with parkinsonism in mitochondrial DNA maintenance disorders. Brain. 2013;136:2379–2392. doi: 10.1093/brain/awt160. [DOI] [PubMed] [Google Scholar]
- 25.Tang S, et al. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE)-like phenotype: an expanded clinical spectrum of POLG1 mutations. J Neurol. 2012;259:862–868. doi: 10.1007/s00415-011-6268-6. [DOI] [PubMed] [Google Scholar]
- 26.Agostino A, et al. Mutations of ANT1, Twinkle, and POLG1 in sporadic progressive external ophthalmoplegia (PEO) Neurology. 2003;60:1354–1356. doi: 10.1212/01.wnl.0000056088.09408.3c. [DOI] [PubMed] [Google Scholar]
- 27.Bestwick ML, Shadel GS. Accessorizing the human mitochondrial transcription machinery. Trends Biochem Sci. 2013;38:283–291. doi: 10.1016/j.tibs.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falkenberg M, et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 29.Shutt TE, et al. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci U S A. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasarman F, et al. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotig A. Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta. 2011;1807:1198–1205. doi: 10.1016/j.bbabio.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Smirnova E, et al. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cipolat S, et al. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson JD. The molecular structure and contact relationships of cell membranes. Prog Biophys Mol Biol. 1960;10:343–418. [PubMed] [Google Scholar]
- 37.Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 38.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizzuto R, et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 40.Lahiri S, et al. A Conserved Endoplasmic Reticulum Membrane Protein Complex (EMC) Facilitates Phospholipid Transfer from the ER to Mitochondria. PLoS Biol. 2014;12:e1001969. doi: 10.1371/journal.pbio.1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugiura A, et al. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 42.Dolman NJ, et al. Stable Golgi-mitochondria complexes and formation of Golgi Ca(2+) gradients in pancreatic acinar cells. J Biol Chem. 2005;280:15794–15799. doi: 10.1074/jbc.M412694200. [DOI] [PubMed] [Google Scholar]
- 43.Elbaz-Alon Y, et al. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Soubannier V, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 45.Gilkerson RW, et al. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod. 2000;15(Suppl 2):148–159. doi: 10.1093/humrep/15.suppl_2.148. [DOI] [PubMed] [Google Scholar]
- 47.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vevea JD, et al. Inheritance of the fittest mitochondria in yeast. Trends Cell Biol. 2014;24:53–60. doi: 10.1016/j.tcb.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang EF, et al. Defective Mitophagy in XPA via PARP-1 Hyperactivation and NAD(+)/SIRT1 Reduction. Cell. 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mai S, et al. Decreased expression of Drp1 and Fis1 mediates mitochondrial elongation in senescent cells and enhances resistance to oxidative stress through PINK1. J Cell Sci. 2010;123:917–926. doi: 10.1242/jcs.059246. [DOI] [PubMed] [Google Scholar]
- 51.Yu-Wai-Man P, et al. Multi-system neurological disease is common in patients with OPA1 mutations. Brain. 2010;133:771–786. doi: 10.1093/brain/awq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muglia M, et al. Clinical and genetic study of a large Charcot-Marie-Tooth type 2A family from southern Italy. Neurology. 2001;56:100–103. doi: 10.1212/wnl.56.1.100. [DOI] [PubMed] [Google Scholar]
- 53.Calvo J, et al. Genotype-phenotype correlations in Charcot-Marie-Tooth disease type 2 caused by mitofusin 2 mutations. Arch Neurol. 2009;66:1511–1516. doi: 10.1001/archneurol.2009.284. [DOI] [PubMed] [Google Scholar]
- 54.Camaschella C. Treating iron overload. N Engl J Med. 2013;368:2325–2327. doi: 10.1056/NEJMcibr1304338. [DOI] [PubMed] [Google Scholar]
- 55.Pandolfo M, Pastore A. The pathogenesis of Friedreich ataxia and the structure and function of frataxin. J Neurol. 2009;256(Suppl 1):9–17. doi: 10.1007/s00415-009-1003-2. [DOI] [PubMed] [Google Scholar]
- 56.Yee C, et al. The Intrinsic Apoptosis Pathway Mediates the Pro-Longevity Response to Mitochondrial ROS in C. elegans. Cell. 2014;157:897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 58.Mattiasson G, et al. Uncoupling protein-2 prevents neuronal death and diminishes brain dysfunction after stroke and brain trauma. Nat Med. 2003;9:1062–1068. doi: 10.1038/nm903. [DOI] [PubMed] [Google Scholar]
- 59.Marzouki N, et al. Vitamin E deficiency ataxia with (744 del A) mutation on alpha-TTP gene: genetic and clinical peculiarities in Moroccan patients. Eur J Med Genet. 2005;48:21–28. doi: 10.1016/j.ejmg.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 60.Wang J, Hegele RA. Microsomal triglyceride transfer protein (MTP) gene mutations in Canadian subjects with abetalipoproteinemia. Hum Mutat. 2000;15:294–295. doi: 10.1002/(SICI)1098-1004(200003)15:3<294::AID-HUMU14>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 61.Kimura H, Kummerow FA. The effect of alpha-tocopherol on essential fatty acid oxidation in liver mitochondria from vitamin E-deficient chicks. Arch Biochem Biophys. 1963;102:86–91. doi: 10.1016/0003-9861(63)90323-0. [DOI] [PubMed] [Google Scholar]
- 62.Taly AB, et al. Wilson disease: description of 282 patients evaluated over 3 decades. Medicine (Baltimore) 2007;86:112–121. doi: 10.1097/MD.0b013e318045a00e. [DOI] [PubMed] [Google Scholar]
- 63.Ristoff E, et al. Long-term clinical outcome in patients with glutathione synthetase deficiency. J Pediatr. 2001;139:79–84. doi: 10.1067/mpd.2001.114480. [DOI] [PubMed] [Google Scholar]
- 64.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 65.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 66.Kaina B, et al. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst) 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 68.Robertson AB, et al. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cleaver JE, et al. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 70.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gredilla R, et al. Mitochondrial DNA repair and association with aging--an update. Exp Gerontol. 2010;45:478–488. doi: 10.1016/j.exger.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Souza-Pinto NC, et al. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009;8:704–719. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LeDoux SP, et al. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 74.Lakshmipathy U, Campbell C. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res. 1999;27:1198–1204. doi: 10.1093/nar/27.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sage JM, et al. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J Biol Chem. 2010;285:18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dmitrieva NI, et al. Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA. Am J Physiol Regul Integr Comp Physiol. 2011;301:R632–R640. doi: 10.1152/ajpregu.00853.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kornblum C, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ronchi D, et al. Mutations in DNA2 link progressive myopathy to mitochondrial DNA instability. Am J Hum Genet. 2013;92:293–300. doi: 10.1016/j.ajhg.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.el-Khamisy SF, Caldecott KW. DNA single-strand break repair and spinocerebellar ataxia with axonal neuropathy-1. Neuroscience. 2007;145:1260–1266. doi: 10.1016/j.neuroscience.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 80.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 81.Scheibye-Knudsen M, et al. Mitochondrial deficiency in Cockayne syndrome. Mech Ageing Dev. 2013;134:275–283. doi: 10.1016/j.mad.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 83.DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132:785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Das BB, et al. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci U S A. 2010;107:19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sykora P, et al. Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc Natl Acad Sci U S A. 2011;108:7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kamenisch Y, et al. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aamann MD, et al. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valentin-Vega YA, et al. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scheibye-Knudsen M, et al. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rubinsztein DC, et al. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 96.Ding WX, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen Y, Dorn GW. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cullup T, et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet. 2013;45:83–87. doi: 10.1038/ng.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao H, et al. Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J Cell Biol. 2013;200:731–741. doi: 10.1083/jcb.201211014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martinez TN, Greenamyre JT. Toxin models of mitochondrial dysfunction in Parkinson’s disease. Antioxid Redox Signal. 2012;16:920–934. doi: 10.1089/ars.2011.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Allen GF, et al. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maron BJ, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301:1253–1259. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van der Ploeg AT, Reuser AJ. Pompe’s disease. Lancet. 2008;372:1342–1353. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 105.Schlotawa L, et al. SUMF1 mutations affecting stability and activity of formylglycine generating enzyme predict clinical outcome in multiple sulfatase deficiency. Eur J Hum Genet. 2011;19:253–261. doi: 10.1038/ejhg.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scheibye-Knudsen M, et al. A novel diagnostic tool reveals mitochondrial pathology in human diseases and aging. Aging (Albany NY) 2013;5:192–208. doi: 10.18632/aging.100546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garone C, et al. Clinical and genetic spectrum of mitochondrial neurogastrointestinal encephalomyopathy. Brain. 2011;134:3326–3332. doi: 10.1093/brain/awr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Graf WD, et al. Phenotypic heterogeneity in families with the myoclonic epilepsy and ragged-red fiber disease point mutation in mitochondrial DNA. Ann Neurol. 1993;33:640–645. doi: 10.1002/ana.410330613. [DOI] [PubMed] [Google Scholar]
- 109.Virgilio R, et al. Mitochondrial DNA G8363A mutation in the tRNA Lys gene: clinical, biochemical and pathological study. J Neurol Sci. 2009;281:85–92. doi: 10.1016/j.jns.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 110.de BF, et al. Resistance to cerebral ischemic injury in UCP2 knockout mice: evidence for a role of UCP2 as a regulator of mitochondrial glutathione levels. J Neurochem. 2004;89:1283–1292. doi: 10.1111/j.1471-4159.2004.02432.x. [DOI] [PubMed] [Google Scholar]
- 111.Williams MD, et al. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J Biol Chem. 1998;273:28510–28515. doi: 10.1074/jbc.273.43.28510. [DOI] [PubMed] [Google Scholar]
- 112.Rodriguez-Hernandez A, et al. Coenzyme Q deficiency triggers mitochondria degradation by mitophagy. Autophagy. 2009;5:19–32. doi: 10.4161/auto.5.1.7174. [DOI] [PubMed] [Google Scholar]
- 113.Takeda K, et al. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc Natl Acad Sci U S A. 2010;107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]