Abstract
Oxidative DNA damage accumulation has been implicated in neurodegenerative diseases such as Alzheimer’s and Parkinson’s. The base excision repair (BER) pathway is a primary responder to oxidative DNA damage. Effects of loss of BER on normal brain function is a relatively nascent area of research that needs further exploration for better understanding of related brain diseases. Recently, we found that loss of a versatile DNA glycosylase endonuclease eight-like 1 (NEIL1) causes deficits in spatial memory retention using the Morris water maze test. Further we found that there is a significant loss of NEIL1 enzyme levels and its activity in postmortem Alzheimer’s disease brains. Based on the Allen Brain Atlas in situ hybridization data, the expression levels of Neil1 mRNA are higher in the olfactory bulb compared to other areas of the brain. Olfaction in mice is a central brain function that involves many central nervous system pathways. Here we studied the effect of complete loss of Neil1 gene on olfactory function. We explored olfactory function in mice with three different behavioral tests namely, olfactory sensitivity, performance and buried food tests. Neil1−/− mice performed poorly compared to wild-type mice in all three tests. Our data indicates that loss of Neil1 causes olfactory function deficits supporting our previous findings and that normal brain function requires robust DNA repair.
Introduction
Brain suffers from high levels of endogenous oxidative stress (Floyd and Hensley, 2002; Halliwell, 1992). One of the major macromolecules that are damaged due to reactive oxygen species is DNA. Proper repair of DNA is essential for normal brain function and behavior (Canugovi et al., 2013). The major pathway involved in repair of the oxidative DNA damage is BER (Wilson and Bohr, 2007). There are four major steps performed by specialized enzymes during BER that take place in the following order. DNA glycosylases recognize and remove DNA damage generating an abasic site, apurinic/apyrimidinic (AP) endonuclease processes abasic sites to generate a single nucleotide gap, DNA polymerase incorporates a nucleotide leaving a nick, and ligase seals the nick to complete the repair process. NEIL1 is a versatile DNA glycosylase as it not only recognizes a broad spectrum of DNA damage substrates but also participates in nucleotide excision repair and replication associated repair pathways in addition to BER (Grin and Zharkov, 2011). We have recently showed that loss of Neil1 causes spatial memory retention deficits in mice (Canugovi et al., 2012). We further showed that there is a significant loss of NEIL1 protein and its activity in postmortem Alzheimer’s patient brains (Canugovi et al., 2014).
Olfactory function includes the sense of differentiating various smells and their intensities. Although the olfactory bulb is mainly involved in olfaction, memory associated with each smell is a function that involves the majority of the brain. Within the brain, olfaction is performed by the rhinencephalon, one of the oldest brain regions. Rhinencephalon development is variable in different species. It is more prominent in rodents than in humans. This could potentially be due to an underestimation of olfactory functions in humans; as there is an increasing awareness of the importance of olfactory brain functions in association with diseases (Velayudhan and Lovestone, 2009). In humans, defects in olfaction are among the earliest symptoms of several neurological diseases including Alzheimer’s and Parkinson’s (Hawkes et al., 1999; Wattendorf et al., 2009; Wesson et al., 2010). Olfactory tests are being applied to test loss of normal brain function and brain related pathology assessment. These tests are further used to differentiate various neurological diseases that encompass loss of olfaction, along with other symptoms from those that do not (Doty, 2012; Doty, 2013; Doty et al., 1993). In rodents, loss of olfaction could affect their very livelihood, as they largely depend on smell to find their food source. Based on this behavior, several tests have been designed to study olfaction in rodents.
In this study, we used three different tests to investigate olfactory function in mice with a loss of Neil1 compared to wild type. Two of the three modules were designed to test the ability of mice to differentiate a favorable smell from an aversive smell (performance test), intensity of a specific smell (sensitivity test) and the third module tested the ability of mice to find buried food after starvation. We found that in all three tests, Neil1−/− mice performed worse than wild-type mice.
Materials and Methods
Mice
Neil1−/− mice were a generous gift from Steven Lloyd (Oregon Health and Science University, Portland, OR). The mice were maintained on a mixed background (C57 and 129). Neil1−/− mice and wild-type littermates were used for all experiments described below. Mice were maintained in a constant-temperature facility with a 12 hr. light/12 hr. dark cycles, with free access to food and water. All procedures were approved by the Animal Care and Use Committee of the National Institute on Aging Intramural Research Program.
General testing procedure
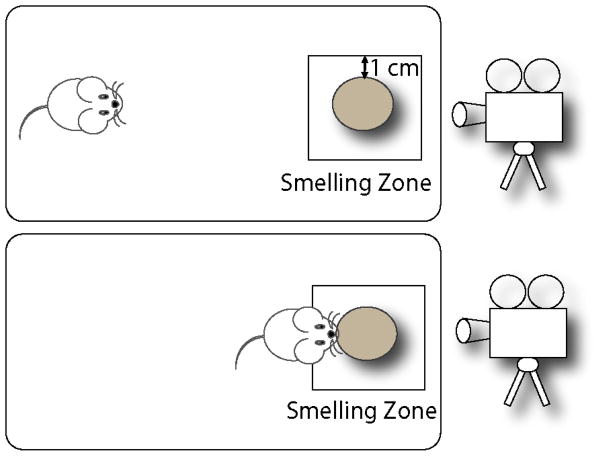
The protocols were adapted from Witt RM et al. (Witt et al., 2009). All mice were checked by a veterinarian for general health prior to olfactory testing. Each test was conducted in a clean new 8 H × 10 W × 18 L inch cage to avoid mixing of odors of different substances as well as mouse specific smells. All smells were sampled on a round filter paper (Whatman, diameter: 10 cm) in the cage arena (Fig 1). Time spent in smelling zone is defined as the time spent by the mouse within the area of smelling zone as depicted in figure 1. When the mouse enters its nose, paws or whole body near the odorant containing filter paper, it is considered as time spent in smelling zone. However bouts of sitting without smelling have been eliminated from data. In all the experiments described below, two researchers were involved in testing, one blind to subject genotypes and/or condition. The videos were started with mouse information, labeled with original tag numbers, without the genotype information and the odors were numbered without details for the blindfolding purposes. All tests were video recorded for data analysis at a later time.
Figure 1.
Top view of general smell test procedure. The smaller square is representative of the smelling zone. The rest of the area is considered as non-smelling zone. Each test is video-captured for analysis of the behavior data.
Olfactory sensitivity test
In this test multiple dilutions of a specific scent are used to test olfactory sensitivity between the two groups of mice (Tsukatani et al., 2003; Wysocki and Beauchamp, 1984). Each individual mouse was tested for various dilutions, one at a time in an empty cage container without bedding. Serial dilutions of cinnamon extracts (McCormick, Sparks, MD) (0.1, 0.01, 0.001, 0.0001, 0.00001 and 0 (water only)) were tested on all mice. The exploration of the scent by each mouse was recorded for 3 min. Total exploratory time and the time spent by the mouse near the scent-containing zone was calculated from the videos. The dose-dependent response is plotted for the two groups.
Olfactory performance test
This test is designed to measure the capacity to identify and differentiate scents (Wessling-Resnick, 2012). The scents were designated as attractive if the exploratory time is higher than water in the smelling zone, whereas the scent is designated as aversive if the exploratory time is less than water. If the exploratory time is equal to or comparable to water the scent is considered as a neutral smell. The scents tested were, peanut butter (J.M. Smucker Company, Orrville, OH) 10% w/v in mineral oil (attractive), vanillin (neutral) (McCormick, Sparks, MD) and 2-methylbutyricacid (2MBA) (Sigma-Aldrich, St. Louis, MO) (aversive). The exploration time was recorded for a total of 3 min. for all subjects and smells. Total and zonal exploratory time was counted from each video and the average of total and zonal exploratory times were plotted separately.
Buried food test
This test was designed based on a previous report (Lu et al., 2008; Yang and Crawley, 2009) to study the ability of mice to find buried food after 16 hours of starvation and dependent on their ability to smell the buried food. Each animal was tested in an individual cage with clean 3 inch high bedding and a fixed amount of food pellet at the bottom of the bedding. Prior to the experiment, a food pellet (peanut butter flavored cookie crisp, Nestle, Columbia, MD) used in this test was given to the animals as a feed to study the consumption relative to their regular diet pellet as well as to familiarize the smell of the food. During the test, each mouse was given a 15 min. period to find the buried food based on smell. All tests were video recorded and used later to calculate the latency of extracting the buried food under the bedding.
Data Analysis
The following set of rules was followed the analysis of data from the videos: 1. Two researchers were blinded for the genotype of the mouse video being analyzed. 2. The latency of zonal exploration smelling the scent was counted only when the mouse examined the filter paper from within 1 cm of its location (smell zone, Fig 1). 3. Bouts of sitting near or on the filter paper were excluded from counting. 4. Each mouse was observed for total exploration time within the 3 min. test-period to check if the mouse behavior in an open field of the cage area is comparable between groups.
Cardiac perfusion, sectioning and staining
Approximately ten months old mice were deeply anesthetized by isoflurane inhalation. The animals were cardiac perfused with 0.1M phosphate buffered saline followed by 4% paraformaldehyde. Animals were decapitated and brains were immediately removed. The brains were placed in 4% paraformaldehyde for 48 h, followed by equilibration in 30% sucrose. Tissue was sectioned coronally (40 μm) on a freezing microtome (Thermo-Fisher) and stored at −20°C in cryoprotectant solution. All animal procedures were done in accordance with the National Institute of Health Animal Care and Use Committee.
Immunohistochemistry
For accessing the olfactory bulb morphology and histology, a 1:6 series of free-floating coronal sections (40 μm) was washed 3x in TBS buffer for 10 min at room temperature. After thorough washing, the sections were blocked with TBS++ (3% Donkey Serum, 0.05M TBS, 0.5% Triton-X 100) for 30 min at room temperature and incubated with the neuronal marker mouse anti-NeuN (1:100 Millipore, CA) and the astrocyte marker rabbit anti-GFAP (1:500, EnCor Biotechnology, FL) for 72 h at 4°C. After rinses with TBS and blocking in TBS++, sections were co-incubated with donkey anti-mouse Cy™3 (1:250, Jackson ImmunoResearch, PA) and donkey anti-rabbit Cy™5 dyes (1:250, Jackson ImmunoResearch, PA) for 3 h at room temperature. After rinses with TBS sections were mount in the Pro Long®Gold antifade reagent with DAPI (Life technologies, OR). Fluorescent signals were imaged with a Axiovert 200M Zeiss microscope (Carl Zeiss Microimaging, NY) equipped with the software AxioVision 4.8.3.0. Mosaics and z-stacked images were used to determine the morphology of the olfactory bulb. For general histological analysis the sections of various areas of the brain are stained with hematoxylin and eosin dyes.
Statistical significance
The data presented here is a mean ± standard error of exploratory time in different experimental conditions. In addition to standard t-tests and analysis of variance, a linear mixed-effects regression model to analyze the repeated-measures data presented in Figure 2 was conducted. The model accounts for the repeated measure through a random effect for mouse in the model. This was followed by a Bonferroni’s posthoc test to get adjusted p-values. A non-parametric statistical hypothesis test (Wilcoxon Rank Sum test) was conducted to test the significance of latency difference between groups during the buried food test. the Fishers exact test was used to compare the frequencies of finding food with in the 15 minute test window. The data were considered significant when the p-value was less than 0.05.
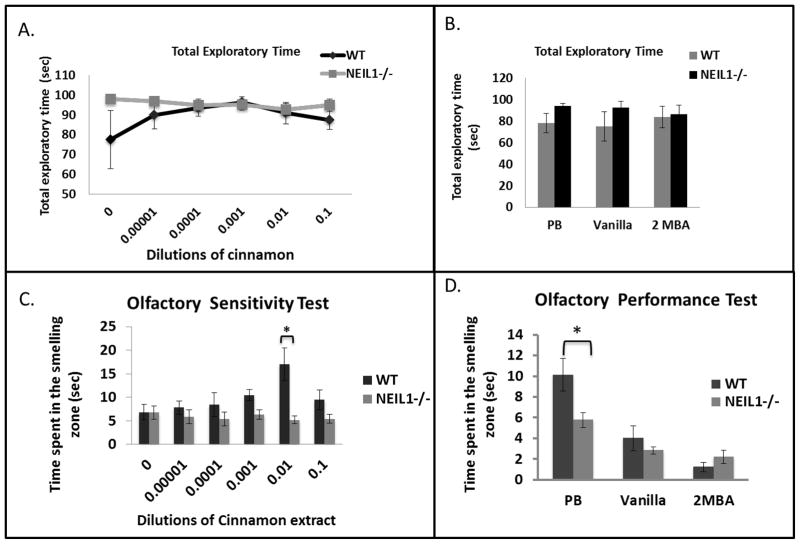
Figure 2.
Summary of olfactory test data. A) Total exploratory time of mice during cinnamon exposure. Data are average total exploratory time ± standard error. Each experiment is repeated with n=9 Neil1−/− and n=7 wild-type mice. B) Total exploratory time of mice with peanut butter (PB), Vanilla and 2-methylbutyric acid (2MBA). C) Effect of increasing concentrations of cinnamon extracts on olfactory sensitivity. Data are average zonal exploratory time ± standard error. Each experiment was repeated with n=9 Neil1−/− and n=7 wild-type mice. ‘0’ is water only control. The * symbol indicates significance of data indicating p values 0.02 and 0.005, respectively, at 0.001 and 0.01 cinnamon extract dilutions. D) Loss of Neil1 decreases the ability to identify specific smells. Data are average zonal exploratory time ± standard error. Each experiment is repeated with n=9 Neil1−/− and n=7 wild-type mice. PB is peanut butter and 2MBA is 2-methylbutyric acid. * indicates significance of the data and represents a p-value of 0.016.
Results
Loss of NEIL1 is accompanied by olfactory defects in mice: We have tested olfactory function in wild type and Neil1−/− mice utilizing three different tests that measure recognition, sensitivity, and differentiation of various smells. During the olfactory sensitivity test, we exposed each mouse to various dilutions of cinnamon extract as described in materials and methods. We have first measured the total time each mouse spent in exploring the entire cage area to make sure that there was no significant difference in the mouse behavior and exploratory activity. We found that wild type and Neil1−/− mouse groups were equally active. Although there seem to be a difference in exploration time during the water and peanut butter exposure the data is not statistically significant (Fig 2A and 2B). The amount of time the mouse spent examining the smell on the filter paper (exploratory time) was plotted against the dilution factor. There was a trend towards increased average exploratory time for the wild-type animal group with 0.001 and 0.01 dilutions of cinnamon concentration in the smelling zone, but the 0.01 dilution differed significantly with all other dilutions except with 0.1 dilution (WT, 0.01 ≠ 0 (p=0.00151), 0.01 ≠ 0.00001 (p=0.00691), 0.01 ≠ 0.0001 (p=0.01695), 0.01 (p=0.06727) is marginally significantly different from 0.1). Whereas the Neil1−/− mouse groups’ average exploration time within the smelling zone remained unchanged at all doses of cinnamon (Fig. 2C). At 0.01 dilution there was a significant difference (p=0.00027) between the average zonal exploratory time of the WT and Neil1−/− groups. This result indicates that the Neil1−/− mice are not sensitive to the intensity of the smell presented to them.
A clearer test for olfaction is to study if the mice are able to differentiate various smells, as this indicates an essential survival instinct such as recognizing a consumable food from aversive toxic material. We exposed all animals to three different smells: peanut butter (food material/attractive), vanillin (non-essential/neutral) and 2MBA (pungent/aversive). There was a significant difference in the average exploration time between the two groups for the peanut butter scent (wild type=10 seconds and Neil1−/− 5 seconds) while no difference was found for either vanillin or 2MBA. Neil1−/− mice explored the peanut butter smell significantly less (p=0.016) than wild-type mice indicating that there is an olfactory deficit in Neil1−/− mice (Fig. 2D). Interestingly, there was a trend in the Neil1−/− mice exploring the 2MBA scent, as they spent exploring this scent for relatively longer period of time. However, the test was not sensitive enough to show whether Neil1−/− mice explored the aversive scent more significantly than wild-type mice due to their inability of differentiating an aversive smell. We therefore decided to test the animals more rigorously regarding food-related smells using a buried food test.
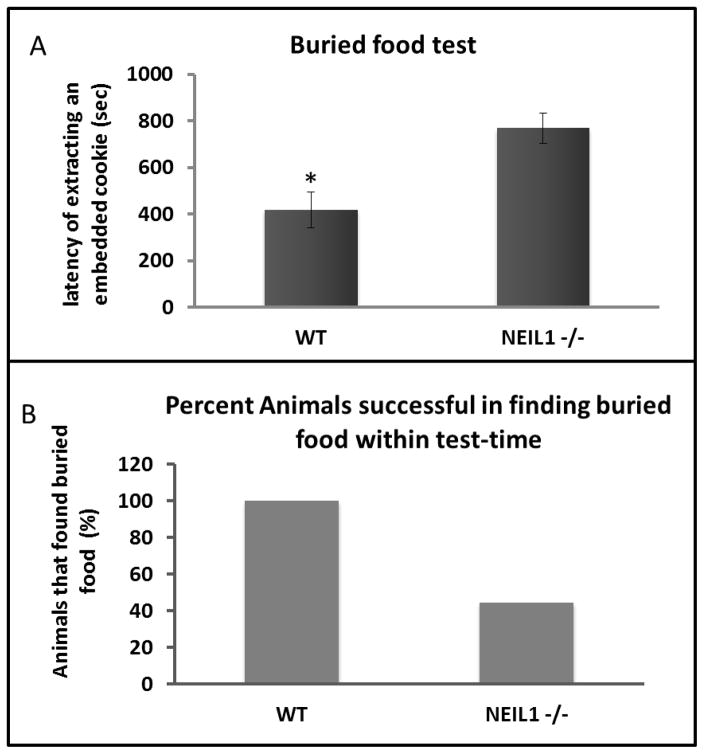
The buried food test is designed to observe olfactory differences between mouse groups by a more rigorous method as this includes a 16hr. solid food deprivation as a pre-condition. The mice were provided with water during this period. Prior to the starvation, we familiarized all the animals in the test with peanut butter cookies and also tested the consumption of this new food between the groups. Both wild type and Neil1−/− mice consumed the cookies equally well. Three peanut butter cookies were placed at the bottom of bedding and the mouse needed to extract the cookie and eat it within the stipulated period of 15 min. The average latency of finding the cookie was significantly longer in Neil1−/− mice (wild type=418 seconds and Neil1−/− =769 seconds) (p=0.0074) (Fig. 3A). More significantly, several mice from the Neil1−/− group (5 out of 9 mice tested) never extracted the cookie within the 15 min. time limit (Fig. 3B). The frequencies of finding food between groups were subjected to Fisher’s exact test of significance and the observed p-value, 0.0294 indicates that the Neil1−/− mice were unable to find food after a long period of food deprivation due to an olfactory deficit.
Figure 3.
Buried-food test. A) Data are average latency in extracting buried food ± standard error. Each experiment is repeated with n=9 Neil1−/− and n=8 wild-type mice. B) Percentage animals that was successful in finding the buried food with the test-time of 900 seconds.
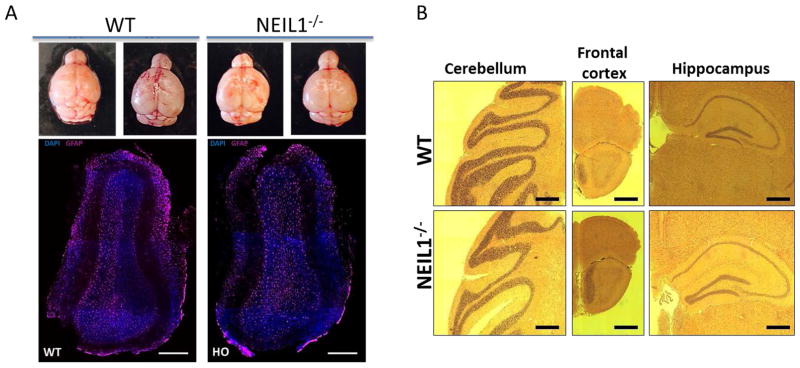
Morphological/histological parameter remain unchanged in Neil1−/− mice compared to wild type: We wondered if there is a major change in the olfactory bulb size or basic histology between the Neil1−/− and wild type mouse groups. As shown in the top panel of Fig. 4A, the general size and shape of the olfactory bulb are similar between the mouse groups. Further we assessed the astrocyte staining in the olfactory bulb and found no significant difference between the mouse groups (Fig. 4A bottom panel). Histology by hematoxylin and eosin staining of the frontal cortex, hippocampus, and cerebellum did not reveal any pathological changes (Fig. 4B).
Figure 4.
Histology and immunohistochemistry of the WT and NEIL1−/− mouse brains (A) Top panel represents images of the brains isolated from Neil1+/+ and Neil1−/− 10 months old mice. Bottom panel shows the images (coronal sections) of the adult olfactory bulb stained with DAPI and astrocyte’s marker GFAP (Scale bars, 300 μm). (B) Hematoxylin and eosin staining of various regions from WT and NEIL1−/− mouse brains (Scale bars, 500 μm).
Discussion
In this study we investigated the olfactory capability of wild type and NEIL1−/− mice. We employed three well-established olfactory tests that explored three different parameters of smelling efficiency. We tested smelling sensitivity to different intensities of a specific scent (dilutions of cinnamon extract) and whether the mice could distinguish separate odors such as attractive (peanut butter), neutral (vanillin extract) and aversive (2MBA). Finally, we tested the mice for their ability to find buried food after a period of starvation. We found that in all three cases the Neil1−/− group performed significantly worse than the wild-type mouse group suggesting that loss of Neil1 affects their olfactory senses.
Three major DNA glycosylases remove oxidatively modified bases from DNA. They are 8-oxoguanine DNA glycosylase (OGG1), endonuclease three like-1 (NTH1) and NEIL1 (Ide and Kotera, 2004). When their expression levels were compared based on in situ hybridization data from the Allen Brain Atlas, Nth1 and Neil1 expression levels were at least 2-fold greater in olfactory areas compared to other areas of the brain (Canugovi et al., 2013). Consequently, we sought to determine if loss of this gene might impact olfactory function in Neil1−/− mice. We found that the loss of Neil1 has a profound effect on olfaction in these mice. For instance, 5 out of 9 mice tested were unable to find the buried food by the end of the 15 min. observation time, while all the wild-type mice were able to find the buried food within the test period (Fig 3B). Further, the average latency of finding the buried food was significantly higher in the Neil1−/− mouse group compared to wild type (Fig 3A). Interestingly, during sensitivity test there was a steady increase in average zonal exploratory time when presented with increasing concentration of cinnamon extracts in wild-type group. However, Neil1−/− mice did not show any dose dependent response in this test (Fig 2C). At the highest concentration of cinnamon extract exposure (indicated by 0.1, Fig 2C) the wild-type mice did not appear to follow the trend (p=0.06), thus we believe that the sensitivity of the test had reached its limit. Further Neil1−/− mice could not efficiently identify food-related smells relative to wild-type mice during exposure to the peanut butter smell (Fig 2D). Based on the statistical analysis, time spent by Neil1−/− and WT mice in the smelling zone differ for Peanut Butter (p=0.01187). Among Neil1−/− mice, the time spent in smelling zone during exposure to 2MBA is significantly different than peanut butter (p=0.01089). Peanut butter and vanilla are only marginally significantly (p=0.05383) different. Whereas among WT mice 2MBA is significantly different than peanut butter (p=0.00000) and vanilla is significantly different than peanut butter (p=0.00007). Overall, these results suggest that Neil1−/− mice are acting similar to WT mice. It is very well possible that lower sensitivity in smelling capacity is responsible for the difference between groups with Peanut butter rather than identification issue. For the olfactory behavior tests to be valid, all mice must be equally active and explore the arena of the cage. In every case, we measured the total exploratory time in mice (Fig 2A and 2B) to show that in general both groups were similarly exploring the whole arena. We found that though the mouse groups were equally active, the zonal explorations (near the scent) changed significantly. However, we did not found any major differences in the size, or morphology of the olfactory bulb between the two groups (Fig. 4). Similarly, the histology by hematoxylin and eosin staining of various regions of the brain did not show significant changes (Fig. 4) that could explain the differences in olfactory regulated behavior. The mechanistic reason for the change in smelling pattern is yet to be uncovered.
Olfactory function involves the combined action of the peripheral and central nervous systems. The peripheral olfactory sensory neurons consist of a number of odorant receptors whose axons converge on the olfactory bulb of the central nervous system to form a glomerular map that reflect odorant receptor identity. The mitral cells form the connection between the olfactory bulb and the olfactory cortex which is linked to the limbic system (Shepherd, 1972). Loss of olfactory function can occur due to severe head trauma, cancer, or neurological diseases (Doty et al., 1997). However, one of the biggest risk factors for loss of olfactory function is aging (Conley et al., 2003). Aging is associated with loss of DNA repair as well as neurological problems (Vyjayanti et al., 2012). Hence this study is an advance in understanding the possible role of DNA repair in normal brain function. Further, this is the first example of a DNA repair deficient mouse showing a deficit in olfactory behavior.
Olfaction is not simply a function of the olfactory bulb area alone but also of other parts of the brain (Rolls, 2013). For instance, to memorize a specific smell and associated stimuli, a first experience smelling that substance is necessary for re-identifying that smell. Repeated smelling of specific scents lead to the permanent memory of the smell indicating that long-term potentiation is essential for smell-related memory (Czerniawska et al., 2013). We recently showed that normal brain function is disrupted in the absence of NEIL1 in mice by Morris water maze test probe trials (Canugovi et al., 2012). The Neil1−/− mice were able to acquire spatial memory similar to wild type however they were unable to retain the acquired memory (Canugovi et al., 2012). This deficit in memory retention may also contribute to the olfactory deficits, as the mice were unable to extract buried food (that they were previously familiarized with) despite the need to find food. Olfactory memory loss is associated with neurological problems (Wattendorf et al., 2009; Wesson et al., 2010). Olfactory deficit is one of the earliest symptoms of Alzheimer’s. A trend toward this deficit has been seen in a precursor version of this disease, mild cognitive impairment(Stamps et al., 2013). Our group and others have reported loss of DNA repair previously in mild cognitive impairment as well as late stages of Alzheimer’s (Bucholtz and Demuth, 2013; Mantha et al., 2013; Weissman et al., 2007). We have more recently found that there is a significant loss of NEIL1 protein levels and activity in both whole cell and mitochondrial extracts of Alzheimer’s postmortem brains further supporting the importance of NEIL1 function in brain-related diseases (Canugovi et al., 2014). These findings cumulatively suggest that NEIL1 may be playing a more important role in normal brain function than previously thought.
HighLights.
Olfaction in mice is a central brain function that involves many central nervous system pathways
We studies olfaction in mice deficient for an important DNA base excision repair enzyme NEIL1, and found a marked defect in olfaction.
Robust DNA repair is required for normal brain function
Acknowledgments
Funding for this work is entirely provided by National Institute on Aging. We thank Dr. R. Stephen Lloyd for the gift of NEIL1 knockout mice. We greatly appreciate Christopher Morrell, NIA, for his help and advice with the statistical analysis of our data. We would like to thank Dr. Baptiste Beverly and Dr. Somnath Ghosh for reading and revising the manuscript prior to submission.
Footnotes
All Authors Declare No Financial Interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bucholtz N, Demuth I. DNA-repair in mild cognitive impairment and Alzheimer’s disease. DNA Repair (Amst) 2013;12:811–816. doi: 10.1016/j.dnarep.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Canugovi C, Misiak M, Ferrarelli LK, Croteau DL, Bohr VA. The role of DNA repair in brain related disease pathology. DNA Repair (Amst) 2013;12:578–587. doi: 10.1016/j.dnarep.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C, Shamanna RA, Croteau DL, Bohr VA. Base excision DNA repair levels in mitochondrial lysates of Alzheimer’s disease. Neurobiol Aging. 2014 doi: 10.1016/j.neurobiolaging.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C, Yoon JS, Feldman NH, Croteau DL, Mattson MP, Bohr VA. Endonuclease VIII-like 1 (NEIL1) promotes short-term spatial memory retention and protects from ischemic stroke-induced brain dysfunction and death in mice. Proc Natl Acad Sci U S A. 2012;109:14948–14953. doi: 10.1073/pnas.1204156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley DB, Robinson AM, Shinners MJ, Kern RC. Age-related olfactory dysfunction: cellular and molecular characterization in the rat. Am J Rhinol. 2003;17:169–175. [PubMed] [Google Scholar]
- Czerniawska E, Zegardlo E, Wojciechowski J. Memories evoked by odors stimulating the olfactory nerve versus odors stimulating both the olfactory and trigeminal nerves: possible qualitative differences? Percept Mot Skills 117:1290–1298. 2013 doi: 10.2466/24.27.pms.117x15z5. [DOI] [PubMed] [Google Scholar]
- Doty RL. Olfaction in Parkinson’s disease and related disorders. Neurobiol Dis. 2012;46:527–552. doi: 10.1016/j.nbd.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty RL. The Scientist Magazine. 2013. [Google Scholar]
- Doty RL, Golbe LI, McKeown DA, Stern MB, Lehrach CM, Crawford D. Olfactory testing differentiates between progressive supranuclear palsy and idiopathic Parkinson’s disease. Neurology. 1993;43:962–965. doi: 10.1212/wnl.43.5.962. [DOI] [PubMed] [Google Scholar]
- Doty RL, Yousem DM, Pham LT, Kreshak AA, Geckle R, Lee WW. Olfactory dysfunction in patients with head trauma. Arch Neurol. 1997;54:1131–1140. doi: 10.1001/archneur.1997.00550210061014. [DOI] [PubMed] [Google Scholar]
- Floyd RA, Hensley K. Oxidative stress in brain aging. Implications for therapeutics of neurodegenerative diseases. Neurobiol Aging. 2002;23:795–807. doi: 10.1016/s0197-4580(02)00019-2. [DOI] [PubMed] [Google Scholar]
- Grin IR, Zharkov DO. Eukaryotic endonuclease VIII-like proteins: new components of the base excision DNA repair system 1. Biochemistry (Mosc) 2011;76:80–93. doi: 10.1134/s000629791101010x. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Reactive oxygen species and the central nervous system. J Neurochem. 1992;59:1609–1623. doi: 10.1111/j.1471-4159.1992.tb10990.x. [DOI] [PubMed] [Google Scholar]
- Hawkes CH, Shephard BC, Daniel SE. Is Parkinson’s disease a primary olfactory disorder? QJM 92:473–480. 1999 doi: 10.1093/qjmed/92.8.473. [DOI] [PubMed] [Google Scholar]
- Ide H, Kotera M. Human DNA glycosylases involved in the repair of oxidatively damaged DNA 1. Biol Pharm Bull. 2004;27:480–485. doi: 10.1248/bpb.27.480. [DOI] [PubMed] [Google Scholar]
- Lu DC, Zhang H, Zador Z, Verkman AS. Impaired olfaction in mice lacking aquaporin-4 water channels. FASEB J. 2008;22:3216–3223. doi: 10.1096/fj.07-104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantha AK, Sarkar B, Tell G. A short review on the implications of base excision repair pathway for neurons: Relevance to neurodegenerative diseases. Mitochondrion. 2013 doi: 10.1016/j.mito.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Rolls ET. Limbic systems for emotion and for memory, but no single limbic system. Cortex. 2013 doi: 10.1016/j.cortex.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. Synaptic organization of the mammalian olfactory bulb. Physiol Rev. 1972;52:864–917. doi: 10.1152/physrev.1972.52.4.864. [DOI] [PubMed] [Google Scholar]
- Stamps JJ, Bartoshuk LM, Heilman KM. A brief olfactory test for Alzheimer’s disease. J Neurol Sci. 2013;333:19–24. doi: 10.1016/j.jns.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukatani T, Miwa T, Furukawa M, Costanzo RM. Detection thresholds for phenyl ethyl alcohol using serial dilutions in different solvents. Chem Senses. 2003;28:25–32. doi: 10.1093/chemse/28.1.25. [DOI] [PubMed] [Google Scholar]
- Velayudhan L, Lovestone S. Smell identification test as a treatment response marker in patients with Alzheimer disease receiving donepezil. J Clin Psychopharmacol. 2009;29:387–390. doi: 10.1097/JCP.0b013e3181aba5a5. [DOI] [PubMed] [Google Scholar]
- Vyjayanti VN, Swain U, Rao KS. Age-Related Decline in DNA Polymerase beta Activity in Rat Brain and Tissues 2. Neurochem Res. 2012 doi: 10.1007/s11064-011-0694-9. [DOI] [PubMed] [Google Scholar]
- Wattendorf E, Welge-Lussen A, Fiedler K, Bilecen D, Wolfensberger M, Fuhr P, Hummel T, Westermann B. Olfactory impairment predicts brain atrophy in Parkinson’s disease. J Neurosci. 2009;29:15410–15413. doi: 10.1523/JNEUROSCI.1909-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessling-Resnick VRKaM. Influence of Iron Deficiency on Olfactory Behavior in Weanling Rats. Journal of Behavioral and Brain Science. 2012;2:167–175. doi: 10.4236/jbbs.2012.22020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesson DW, Levy E, Nixon RA, Wilson DA. Olfactory dysfunction correlates with amyloid-beta burden in an Alzheimer’s disease mouse model. J Neurosci. 2010;30:505–514. doi: 10.1523/JNEUROSCI.4622-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, III, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Witt RM, Galligan MM, Despinoy JR, Segal R. Olfactory behavioral testing in the adult mouse. J Vis Exp. 2009 doi: 10.3791/949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki CJ, Beauchamp GK. Ability to smell androstenone is genetically determined. Proc Natl Acad Sci U S A. 1984;81:4899–4902. doi: 10.1073/pnas.81.15.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Crawley JN. Simple behavioral assessment of mouse olfaction. Curr Protoc Neurosci. 2009;Chapter 8(Unit 8):24. doi: 10.1002/0471142301.ns0824s48. [DOI] [PMC free article] [PubMed] [Google Scholar]