Abstract
Oxidative DNA damage is mainly repaired by base excision repair (BER). Previously, our lab showed that mice lacking the BER glycosylases Ogg1 or Neil1 recover more poorly from focal ischemic stroke than wild-type mice. Here, a mouse model was used to investigate whether loss of one of the two alleles of Xrcc1, which encodes a non-enzymatic scaffold protein required for BER, alters recovery from stroke. Ischemia and reperfusion caused higher brain damage and lower functional recovery in Xrcc1+/− mice than in wild-type mice. Additionally, a greater percentage of Xrcc1+/− mice died as a result of the stroke. Brain samples from human individuals who died of stroke and individuals who died of non-neurological causes were assayed for various steps of BER. Significant losses of thymine glycol incision, abasic endonuclease incision and single nucleotide incorporation activities were identified, as well as lower expression of XRCC1 and NEIL1 proteins in stroke brains compared to controls. Together, these results suggest that impaired BER is a risk factor in ischemic brain injury and contributes to its recovery.
Keywords: Base excision repair, Stroke, Xrcc1, Oxidative stress
1. Introduction
Neurons encounter periods of oxidative stress because of their high metabolic requirements needed to support electrical and synaptic activities. During ischemic stroke, excessive oxidative stress is generated increasing the burden on stress resistance mechanisms, including the repair of oxidatively damaged DNA. Thus, the integrity of repair systems that restore damaged DNA is critical for the survival and normal function of neurons. Oxidative DNA damage mainly includes modified bases, abasic sites and single strand breaks. The major pathway responsible for the repair of these forms of DNA damage is BER, and its related sub-pathway, single strand break repair (SSBR) (Canugovi, et al., 2013).
Enzymes in the BER pathway coordinately recognize and remove oxidative DNA lesions, such as 8-oxoguanine (8-oxo-dG). The first step of BER is mediated by a DNA glycosylase, such as 8-oxo-dG glycosylase 1 (OGG1), which releases the damaged base from the DNA backbone by cleaving the glycosidic bond. In the next step of BER, the phosphodiester bond in the DNA backbone is cleaved by an apurinic/apyrimidinic (AP) endonuclease, after which the DNA ends are processed. The missing nucleotide(s) are replaced by a gap-filling DNA polymerase, and the nicked DNA is sealed by a DNA ligase (Tomkinson, et al., 2006).
X-ray repair cross-complementing protein 1 (XRCC1) is a non-enzymatic scaffold protein that interacts with several BER enzymes, including DNA glycosylases (Campalans, et al., 2005,Marsin, et al., 2003), human AP endonuclease 1 (APE1) (Vidal, et al., 2001), DNA polymerase β (POLβ) (Caldecott, et al., 1996,Marintchev, et al., 2000), and DNA ligase IIIα (Mani, et al., 2007,Mortusewicz and Leonhardt, 2007). XRCC1 stimulates the activity of DNA glycosylases that repair oxidative and alkylated base lesions and stimulates the AP endonuclease and 3′-phosphodiesterase activities of APE1. XRCC1 also binds to and enhances DNA damage-specific binding of polynucleotide kinase/phosphatase (PNKP) (Mani, et al., 2007), accelerating repair of DNA SSBs (Whitehouse, et al., 2001). Furthermore, DNA ligase IIIα (LIG3α) is stabilized through a specific interaction between its C-terminal BRCT domain and the BRCT II domain of XRCC1 (Taylor, et al., 1998). XRCC1 also interacts with tyrosyl-DNA phosphodiesterase 1 (TDP1) and Aprataxin (APTX), which excise 3’ or 5’ end blocking groups from DNA, such as 3’-phosphate, 3’-tyrosyl and 5’-AMP residues (Caldecott, 2008,Wilson and Mattson, 2007). Mutations in TDP1 or APTX are linked to the neurodegenerative disorders spinocerebellar ataxia with axonal neuropathy (SCAN1) and ataxia-oculomotor apraxia 1 (AOA1), respectively (Date, et al., 2001,Moreira, et al., 2001,Takashima, et al., 2002). During direct SSBR, poly(ADP–ribose) polymerase (PARP) likely recognizes the single strand break and recruits XRCC1, which in-turn coordinates PNKP, POLβ and LIG3α to complete the repair process. Thus, biochemical and genetic studies suggest that XRCC1 plays crucial roles in BER/SSBR with potentially important roles in protecting neuronal cells against oxidative stress (Kulkarni, et al., 2008) and promoting neurogenesis during development (Lee, et al., 2009).
In mice, deletion of both alleles of Xrcc1 leads to early embryonic lethality (Tebbs, et al., 1999). In a comprehensive study, Xrcc1 heterozygous (Xrcc1+/−) mice did not show any signs of accelerated aging (McNeill, et al., 2011). Further, the brain histology and behavior were normal, but curiously, some Xrcc1+/− mice showed organ rupture. While motor coordination and overall activity were deemed normal in the Xrcc1+/− mice, it is noteworthy that a small number of animals exhibited behavioral features commonly associated with stroke, namely impaired lower leg movement or tilting of the head (McNeill, et al., 2011).
Stroke is a neurological disease that causes brain damage and injury due to acute oxidative stress, and is the second leading cause of neurological disability following dementia (http://www.world-heart-federation.org/cardiovascular-health/stroke/). While limiting the acute effects of stroke is of very high importance, the speedy recovery after stroke is just as important. A slow recovery is associated with long term neurological sequelae and is very costly for the individual and the society. Thus investigation of how the recovery after stroke can be improved is as important as identification of risk factors for stroke. The role of BER in antagonizing acute oxidative stress may be significant in stroke survivors. Therefore, understanding the role of BER and its components in response to stroke in human tissue and animal models is an important line of investigation. We previously showed that mice lacking either Ogg1 or Neil1, glycosylases that initiate BER of oxidative base lesions, recovered more poorly from stroke than the wild type (Canugovi, et al., 2012,Liu, et al., 2011). We here examine whether mice haploinsufficient for the BER/SSBR protein, XRCC1, are more affected by stroke induction.
Our results indicate that partial loss of the DNA repair protein, Xrcc1 renders brain cells vulnerable to oxidative stress after ischemic injury. We have also examined the relative activity levels of BER components in human stroke affected post-mortem and age-matched control brains. We find that, individual who died of stroke had a significant impairment in BER function. Together, our results underscore the importance of BER and further argue that BER genotypes should be evaluated as a risk factor for stroke and its recovery
2. Materials and methods
2.1. Mice
Xrcc1+/− and littermate wild-type mice (McNeill, et al., 2011) were used for all experiments. Five to twelve month-old male mice were used for the experiments. Mice were maintained in a constant-temperature facility with a 12 h light/12 h dark cycle, and free access to food and water. All procedures were approved by the Animal Care and Use Committee of the National Institute on Aging Intramural Research Program.
2.2. Rotarod
This test was used to analyze the motor function and endurance in mice. The rotarod machine (Med Associates) has nine different fixed speeds and an accelerating speed program. The parameters tested and recorded in each case were the number of falls in 5 min duration and the latency to the first fall. All mice were trained and tested for their motor ability at 2–20 rpm before middle cerebral artery occlusion/reperfusion (MCAO/R) procedure. After MCAO/R, the accelerating rotarod test was performed at the 24 h and 48 h time points.
2.3. Middle Cerebral Artery Occlusion and Reperfusion
The focal ischemia/reperfusion model was performed using the intraluminal suture technique described previously; the left middle cerebral artery was occluded for 1 h (Arumugam, et al., 2010). Body temperature was maintained at 37 °C using heating pads and a heatin g lamp throughout the surgical procedure and afterward until the mouse recovered from anesthesia. In a separate set of experiments anesthetized animals from all groups (three mice per group) underwent cerebral blood flow (CBF) measurements using a laser Doppler perfusion monitor. All CBF measurements were performed with the mouse fixed in a plastic frame with the probe placed in the region of cerebral cortex perfused by the middle cerebral artery (MCA).
2.4. Evaluation of Neurological Deficits and Infarct Volume
Neurological impairment was assessed using a five-point neurological deficit score (0, no neurological deficit; 1, failure to extend left paw; 2, circling to the left; 3, falling to the left; and 4, unable to walk spontaneously) (Bederson, et al., 1986) and were assessed without knowledge of mouse genotype. Differences between groups were analyzed by a nonparametric Kruskal–Wallis one-way ANOVA test at 5% significance level. At 48 h after the experimental stroke, the mice were euthanized and infarct area was evaluated by 2,3,5-triphenyltetrazolium chloride (TTC) staining as described previously (Arumugam, et al., 2010). For each brain section, the infarct area was determined by subtracting the area of the non-infarcted ipsilateral hemisphere from that of the intact contralateral hemisphere. The percentage of infarct volume was calculated by dividing the sum of the area from all sections of infarction by the total of that of contralateral hemisphere to avoid the influence of tissue edema (Swanson and Sharp, 1994). A nonparametric statistical test (Mann–Whitney test) and also a two-sample t-test were conducted to analyze the significance of the data.
2.5. Immunoblot analysis
Mice were euthanized by cervical dislocation, and the brain was removed from the skull and the cerebral cortical tissue was removed from both hemispheres and rapidly frozen. Protein was extracted by homogenizing the samples in RIPA buffer with protease inhibitor (Pierce). The samples were left on ice for 30 min before centrifugation at 10,000 g at 4°C for a further 30 min. Supernatant w as removed and aliquoted before storage at −80°C. Protein concentration was determined using a bicinchoninic acid (BCA) assay using bovine serum albumin standards (Pierce). Proteins in 200 μg of total protein lysate were separated on a mini-protean®TGX™ gradient (4–15%) gel (Bio-Rad) at 100V for 120 min. Proteins in the gel were transferred electrophoretically to a Novex® PVDF 0.2 μm membrane (life science) at 4°C at 30V. Membranes were blocke d in 5% milk protein for 60 min at room temperature followed by overnight incubation with primary antibody (1:1000 dilutions) at 4°C. Primary antibodies used were: anti-Xrcc1 (Cell Signaling, Cat # 2735S); anti-NEIL1 (Santa Cruz, SC-134547); anti-DNA ligase III (BD Biosciences, Cat # 611876); anti-Caspase-3 (Cell Signaling, Cat #9662); and anti-β-actin (Santa Cruz, SC-1616). After washing the membrane, it was incubated in the presence of secondary (1:10000) for 120 min at room temperature. Secondary HRP conjugate was detected by chemiluminescence (Super Signal West Femto, Pierce) on a chemidoc imager XRS+™ (Bio-Rad). Images were analyzed and quantitated using ImageLab (V3.0) (Bio-Rad) with protein of interest signal normalized to -β-Actin signal.
2.6. TUNEL Staining, Microscopy, and Quantification
To evaluate apoptosis, TUNEL staining was performed on brain sections using a Deadend kit (Promega). Multiple sections from individual brain samples were layered on a glass slide before the TUNEL assay; brain sections were fixed in 4% (vol/vol) paraformaldehyde and permeablized with 0.2% Triton X-100 and proteinase K as per manufacturer’s instructions. After staining, the sections were mounted in hardset Vectashield medium containing DAPI. Additional staining was done with hematoxylin–eosin to locate the boundary of the ischemic penumbra (Borlongan, et al., 2000). The sections were photographed using a fluorescence microscope equipped with AxioVision software. Four wild type and four Xrcc1+/− brains were analyzed without knowledge of the genotype of the mice. A minimum of four microscopic fields within the ischemic penumbra of each section were photographed. TUNEL+ cells were counted in a double-blinded manner in each of these areas. An average of all of the TUNEL+ cells from these areas counted was taken from each brain section (Khan, et al., 2009). Data were analyzed by Student’s two-tailed t-test.
2.7. Human brain samples and source
Human autopsy brain samples stored within 24 hours after death (n=18) were obtained from the Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center, Los Angeles, CA 90073 which is sponsored by NINDS/NIMH, National Multiple Sclerosis Society, and Department of Veterans Affairs. All samples were taken from the parietal cortex region. Nine samples were from stroke victims and nine samples were from individuals whose death was attributed to non-neurological causes. Patient data are shown in Table 1.
Table 1.
Details of tissue specimen obtained from the Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center, Los Angeles, CA 90073 which is sponsored by NINDS/NIMH, National Multiple Sclerosis Society, and Department of Veterans Affairs.
| Sample | PMI | Background | Age | Sex | Area | Cause of Death |
|---|---|---|---|---|---|---|
| 2528 | 19.8 | Stroke | 76 | F | Parietal Cortex | Stroke |
| 3968 | 13.6 | Stroke | 81 | F | Parietal Cortex | Stroke |
| 3741 | 25.3 | Stroke | 76 | F | Parietal Cortex | Stroke |
| 3614 | 14.0 | Stroke | 84 | M | Parietal Cortex | Stroke |
| 4321 | 12.3 | Stroke | 85 | M | Parietal Cortex | Stroke |
| 4513 | 15.6 | Stroke | 74 | M | Parietal Cortex | Stroke |
| 4377 | 9.0 | Stroke | 87 | F | Parietal Cortex | Stroke |
| 4238 | 13.2 | Stroke | 84 | M | Parietal Cortex | Stroke |
| 3739 | 12.0 | Stroke | 62 | F | Parietal Cortex | Stroke |
| 3504 | 11.0 | Non-neurological | 80 | M | Parietal Cortex | Renal Cancer |
| 3531 | 13.6 | Non-neurological | 74 | M | Parietal Cortex | Lung Cancer |
| 3535 | 14.0 | Non-neurological | 81 | M | Parietal Cortex | Non-Hodgkin Lymphoma |
| 3565 | 11.0 | Non-neurological | 76 | M | Parietal Cortex | Cardiomyopathy |
| 3590 | 11.5 | Non-neurological | 75 | M | Parietal Cortex | Coronary Heart Disease |
| 3602 | 13.3 | Non-neurological | 66 | M | Parietal Cortex | Bone and Liver metastasis |
| 3603 | 12.0 | Non-neurological | 74 | F | Parietal Cortex | Pancreas Cancer |
| 4064 | 15.4 | Non-neurological | 75 | F | Parietal Cortex | Liver Cancer |
| 4135 | 12.6 | Non-neurological | 57 | M | Parietal Cortex | Colon and Liver Cancer |
2.8. Oligonucleotide substrates
All PAGE purified oligonucleotides were ordered from IDT (San Diego, CA). The oligonucleotide sequences and location of the specific lesions can be found in Table 2.
Table 2.
Names and sequences of oligonucleotides used in the study
| Name | Sequence |
|---|---|
| TG-51mer | 5’-TAATAATAACAATTGAATGTCT(TG)CACAGCCACTTTCCACACAGACATCAT-3’ 3’-ATTATTATTGTTAACTTACAGA(A)GTGTCGGTGAAAGGTGTGTCTGTAGTA-5’ |
| THF-91mer | 5’-TAATTAATGCTTGTAGGACATAATAATAACAATTGAATGTCT(F)CACA … 3’-ATTAATTACGAACATCCTGTATTATTATTGTTAACTTACAGA(C)GTGT … … GCCACTTTCCACACAGACATCATAACAAAAAATTTCCACCAAAC-3’ … CGGTGAAAGGTGTGTCTGTAGTATTGTTTTTTAAAGGTGGTTTG-5’ |
| GAP-91mer | 5’-TAATTAATGCTTGTAGGACATAATAATAACAATTGAATGTCTGCACA … 3’-ATTAATTACGAACATCCTGTATTATTATTGTTAACTTACAGA( )GTGT … … GCCACTTTCCACACAGACATCATAACAAAAAATTTCCACCAAAC-3’ … CGGTGAAAGGTGTGTCTGTAGTATTGTTTTTTAAAGGTGGTTTG-5’ |
TG containing thymine glycol; THF = containing a tetrahydrofuran abasic site analog; GAP =containing a single nucleotide gap.
2.9. Preparation of brain tissue lysates
Human and mouse tissue lysates were prepared as described previously (Weissman, et al., 2007). Briefly, brain tissue lysates were prepared by homogenizing with a dounce homogenizer in buffer (20 mM Hepes, pH 7.5, 50 mM KCl, 2 mM EGTA and complete protease inhibitor (Roche). Lysates were centrifuged at 800 × g for 10 min to remove large cell debris. The resulting lysates were resuspended (2 mg/mL) in 20 mM Hepes (pH 7.0), 150 mM KCl, 2 mM EGTA, 1% (wt/vol) CHAPSO (Sigma-Aldrich), and protease inhibitor mixture and incubated at 4 °C for 1 h wit h end-over-end rotation. The lysates were centrifuged at 100,000 × g for 1 h, and the supernatants were collected. The samples were flash frozen in liquid nitrogen and stored at −80 °C unti l use. Protein concentration was determined using the BCA assay kit (Pierce).
2.10. Incision assays
Incision assays were performed as described previously (Weissman, et al., 2007). Briefly, incision reactions contained 100 fmol of 32P-labeled duplex oligonucleotide with a specific modification. Samples were diluted in 10 mM HEPES–KOH (pH 7.4) containing 100 mM KCl. Reactions (10 μl) contained 25 mM HEPES–KOH (pH 7.4), 25 mM KCl, 0.1 mg/ml BSA, 5 mM MgCl2, 10% glycerol, 0.05% Triton X-100 and different amount of protein. The reactions were incubated with whole tissue lysates at 32 °C. The incubation times and amount of tissue lysate varied for each type of activity assay. While assaying for abasic site analog tetrahydrofuran cleavage, we incubated with 2 μg of whole tissue lysates for 5 minutes, whereas 60 μg of whole tissue lysates was incubated overnight when assaying for thymine glycol incision in the same lysates. The reactions were terminated by the addition of 5 mg/mL proteinase K and 10% SDS and incubated at 37 °C for 30 minutes follo wed by the addition of formamide-containing buffer and continued incubation at 95 °C for 5 minutes. These samples were resolved on a denaturing 15% polyacrylamide gel containing 7M urea then exposed to a phosphorimager screen. The gels were visualized by the Typhoon Phosphorimager (Molecular Dynamics, GE Healthcare, Ramsey, MN, USA). The images were analyzed using ImageQuant 5.2 software (GE Healthcare, Pittsburgh, PA, USA). The percent incision was calculated by dividing the product band intensity by the total intensity in the lane.
2.11. Incorporation assay
Single-nucleotide gap-filling reaction was performed as described previously (Canugovi, et al., 2014) using an unlabeled DNA oligo substrate with a single gap in the middle of the substrate (Table 2). Samples were diluted in 10 mM Tris-HCl (pH 7.4) containing 100 mM KCl to achieve similar protein concentrations in each of the independent samples. Reactions (10 μl) contained 50 mM Tris-HCl (pH 7.4), 50 mM KCl, 1 mM dithiothreitol (DTT), 5 mM MgCl2, 5% glycerol, 5 mM deoxycytidinetriphosphate (dCTP) (Roche Applied Sciences, Indianapolis, IN, USA), 1 pmol of duplex gap oligonucleotide, 4 μCi of 32P-dCTP (GE Healthcare, Pittsburgh, PA, USA), and 40 μg protein. Reactions were incubated at 37 °C for 3 hours and terminated by the addition of 1 μL of 0.5M ethylenediaminetetraacetic acid (EDTA) and formamide dye followed by the heating at 95 °C for 5 minutes. Samples were resolved on a polyacrylamide gel as described above.
2.12. Immunodetection of XRCC1 and NEIL1 in postmortem human brain tissue
Multiple sections from a single brain were layered on a glass slide, fixed in 4% (vol/vol) formaldehyde for 20 min, and air dried overnight at 4 °C. Slides wer e rehydrated, microwaved in 0.1 M citrate buffer (pH 6) for 10 min, and gradually allowed to cool to room temperature. Slides were washed twice in PBS and blocked for 1 h with 5% goat serum in PBS. Antibodies were diluted (1:200) in 5% goat serum in PBS. Tissue sections were incubated with primary antibodies for overnight at 4 °C, washed three times with PBS and incubated with diluted secondary antibody for 90 min at room temperature. Samples were rinsed and mounted in Vector shield hard set with DAPI. Sections were photographed using a fluorescence microscope equipped with AxioVision software. The primary antibodies used were rabbit Anti-Xrcc1 (Cell Signaling, Cat # 2735S) and rabbit Anti-Neil1 (Santa Cruz, SC-134547). Alexa Fluor 488 goat anti-rabbit IgG was used as the secondary antibody (Molecular Probe, USA).
2.13. Hexokinase Assay
Human brain tissue lysates were prepared and hexokinase activities in the brain tissue were determined as per manufacturer instruction (Hexokinase Colorimetric Assay Kit, Sigma Cat # MAK091).
2.14. Statistics
When comparing the means (or medians) of two independent samples, a two-sample Student's t-test was used (or the nonparametric Mann–Whitney test). When comparing the means (or medians) among a number of independent samples, a one-way analysis of variance (ANOVA) was used (or the nonparametric Kruskal–Wallis test). In the experiments where each animal was measured on three occasions (pre-stroke and at 24 and 48 h after stroke), a randomized complete block design was applied to account for the repeated measures on the animals (this is equivalent to a two-sample ANOVA with no interaction term). Appropriate post hoc approaches, as mentioned in the results section were conducted to control the false positive error rate. A 5% significance level is used throughout. Graphpad Prism version 5 and Statistical Aanalysis System (SAS) programming were used to calculate statistics in various experiments.
For analyzing human data, the results are reported as mean ± standard error. Each assay was performed at least twice (on 9 independent biological samples per set). The differences among human control and stroke samples were analyzed by the Student's t-test, and a p < 0.05 was considered statistically significant.
3. Results
3.1. Xrcc1 +/− mice exhibit greater brain damage, increased mortality, and poorer functional recovery after ischemic stroke
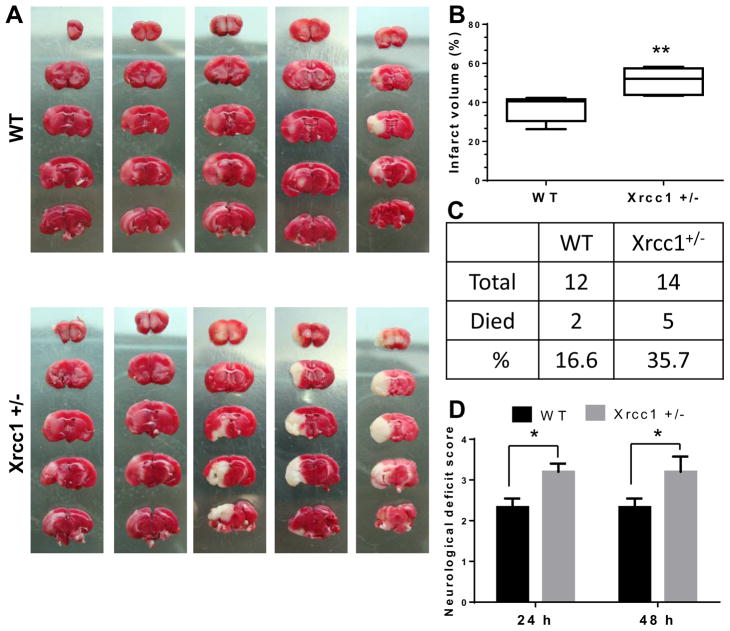
To study the role of Xrcc1 and BER in brain cells subjected to severe stress, we performed middle cerebral artery occlusion/reperfusion (MCAO/R) on 12 wild type (WT) and 14 Xrcc1 +/− mice at 12 months of age. Post-stroke mortality rate, infarct volume, and neurological deficits were measured in each case. The infarct volume was measured in brains of mice that were alive at 48 h after stroke by TTC staining, and neurological deficits were assessed for these mice as described in the materials and methods section (n = 6 each). The infarct volume was significantly higher in Xrcc1 +/− mice compared with WT mice (37.2 ± 2.7% for WT and 50.9 ± 3.1% for Xrcc1 +/−, p<0.01) as determined by the Mann–Whitney test (Fig. 1A and B). The observed mortality rate was about 2.2-fold higher in Xrcc1 +/− mice (35.7%) compared to WT mice (16.6%) after ischemic stroke, although it did not reach statistical significance (Fig. 1C). Furthermore, the neurological deficit score was increased significantly in Xrcc1 +/− mice relative to WT mice (p<0.05) at both 24 and 48 h post-stroke (Fig. 1D).
Figure 1.
Infarct volume, mortality rate, and neurological deficit after middle cerebral artery occlusion/ reperfusion (MCAO/R) in WT and Xrcc1 +/− mice. (A) Mouse brain sections stained with 2,3,5-triphenyltetrazolium chloride (TTC) after 48 h of MCAO/R. (B) Quantification of the infarct volume shown in A. The data are presented as a box plot diagram, which represents the minimum to maximum distribution of the average percentage of infarct volume ± SEM. (C) Percentage of death in mice within 48 h of the MCAO/R procedure. (D) Graphic of the assigned neurological deficit score at 24 and 48 h after the MCAO/R procedure. Data are presented as means ± SEM. Statistical analysis was performed by one-way ANOVA followed by Bonferroni’s post-hoc test for pairwise comparisons, *p<0.05, **p<0.01.
3.2. Impaired motor function in Xrcc1+/− mice after ischemic stroke
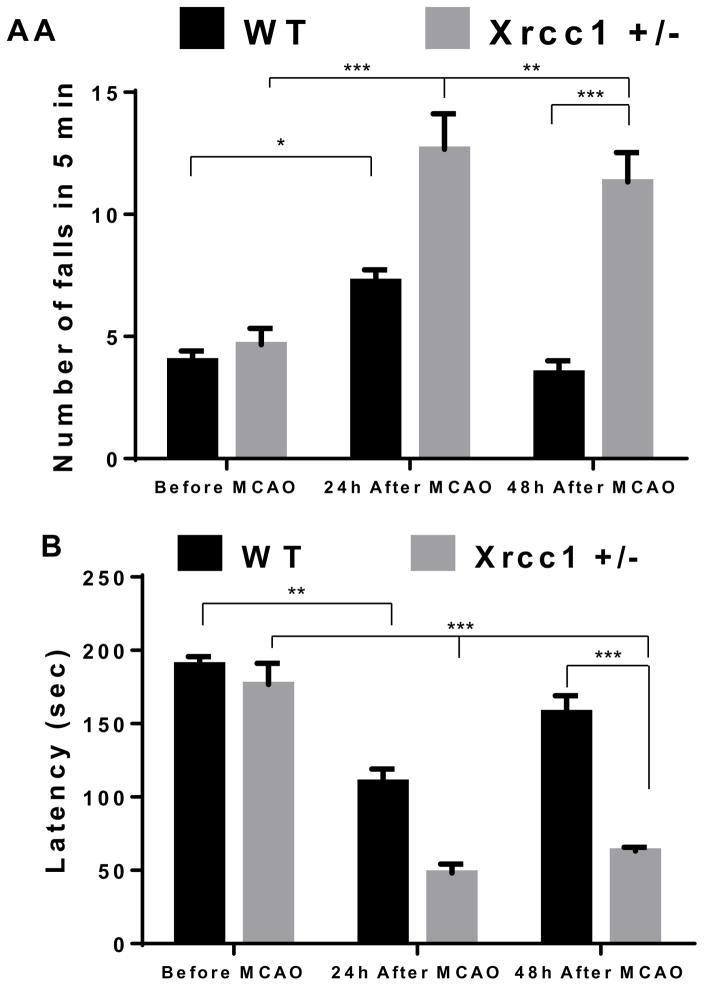
Stroke patients are prone to partial or complete loss of motor function, clinically known as paralysis. This is mainly attributed to the degeneration of cortical motor neurons (and/or their axons in their subcortical course) caused by mechanisms involving excess oxidative stress, DNA damage, and apoptosis (Broughton, et al., 2009,Niizuma, et al., 2009). To study the effect of partial loss of Xrcc1 on motor function after ischemic stroke, rotarod tests were performed at 24 and 48 h after stroke in WT (n = 4) and Xrcc1 +/− (n = 4, age =5–6 months) mice. Prior to stroke, Xrcc1+/− mice did not show any difference on the rotarod test relative to WT mice (Fig. 2), consistent with the previous findings (McNeill, et al., 2011). However, at both the 24 and 48 h time points after stroke, the measured number of falls during the rotarod tests was significantly higher in Xrcc1 +/− mice compared to WT mice (Fig. 2A). Indeed, our studies indicate that WT mice appear to recover completely as assessed by number of falls post-stroke by 48 h, whereas Xrcc1 +/− mice still exhibit profound deficits at that time point. A second parameter, latency to the first fall, which measures motor coordination and grip strength, was also significantly reduced and showed an impaired rate of recovery in Xrcc1 +/− mice after MCAO/R (Fig. 2B). These data suggest that the partial loss of Xrcc1 in mice exacerbates stroke-induced motor dysfunction.
Figure 2.
Motor dysfunction in Xrcc1 +/− and WT mice before and after middle cerebral artery occlusion/ reperfusion (MCAO/R). (A) Average number of falls and (B) average latency of first fall measured during rotarod test. Data indicate average ± SEM, where the sample size is WT, n = 4 and Xrcc1 +/− n = 3. A two-way ANOVA followed by Bonferroni’s correction as well as LSD was used to analyze the data. Asterisks denote the various p-values as follows: *, p≤0.024; **p≤0.001; *** p≤0.0001..
3.3. Partial loss of Xrcc1 results in enhanced apoptosis in brain cells of mice subjected to stroke
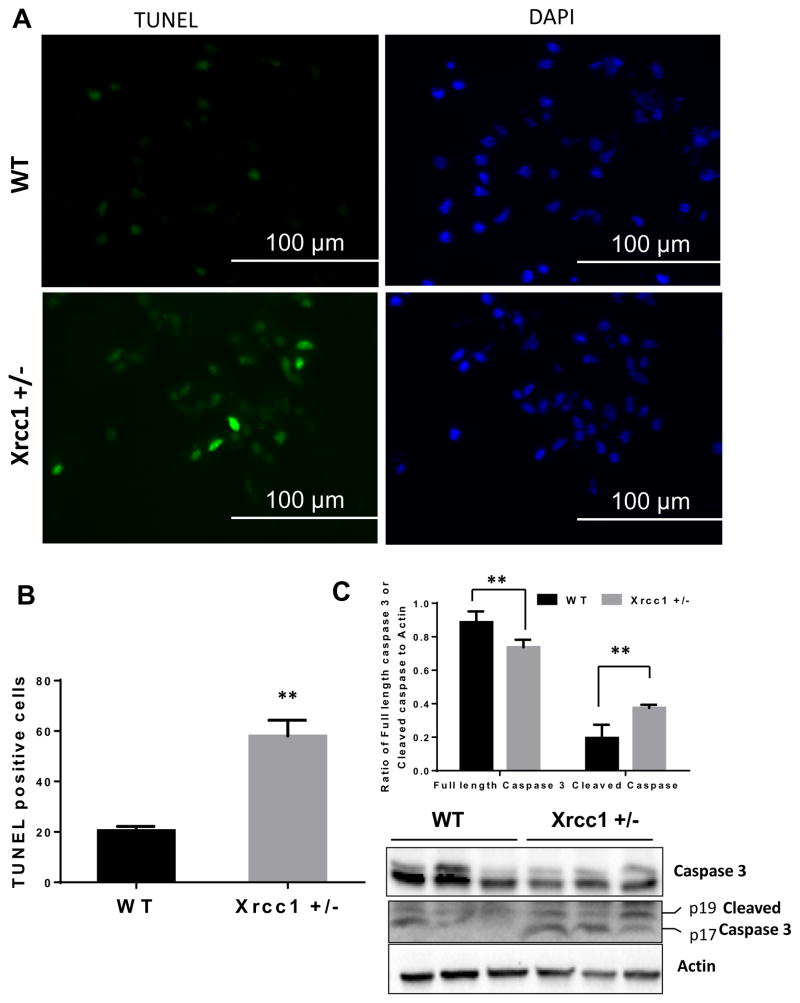
Previous data show that Xrcc1 haploinsufficiency does not cause adverse phenotypic changes under normal conditions, in terms of life expectancy, weight, body fat composition, organ function, motor coordination, brain integrity or memory, bone marrow cellularity, blood cell composition or morphology, and chromosome status (McNeill, et al., 2011). However, these mice were sensitive to the exogenous genotoxin azoxymethane, an alkylating agent that causes DNA damage (McNeill, et al., 2011). Here, we used stroke to induce oxidative stress, which could in turn generate increased levels of DNA lesions that trigger apoptosis in cells. We determined the impact of the reduced DNA repair capacity in Xrcc1 +/− mice on the vulnerability of neurons in the ischemic penumbra to apoptosis. A terminal deoxynucleotidyl transferase (TdT) dUTP nick end-labeling (TUNEL) assay was performed on brain sections from Xrcc1+/− and WT mice (n=4) at 48 h after focal ischemia/reperfusion. Data were analyzed by a two-tailed Student's t-test, and revealed that there were significantly more TUNEL+ cells in the ischemic penumbra of Xrcc1 +/− mice compared with WT mice (Fig. 3A and B). Consistently, protein markers of apoptosis including cleaved caspase-3 (sum of p17 and p19 fragments) were increased and full length caspase 3 was decreased in the brain lysates of Xrcc1 +/− mice compared with WT mice (Fig. 3C), indicating that partial loss of Xrcc1 renders brain cells vulnerable to apoptosis following a stroke. There was no observable increase in TUNEL+ cells in the brain tissue from the nonischemic (contralateral) cerebral hemispheres of Xrcc1 +/− mice in comparison to WT mice, as there were no TUNEL+ cells in either sample (Fig. S1A).Moreover, there was no difference in the level of cleaved caspase 3 in the brain tissue from the nonischemic (contralateral) cerebral hemispheres of WT and Xrcc1 +/− mice (data not shown).
Figure 3.
Apoptosis in Xrcc1 +/− mouse brains after middle cerebral artery occlusion/ reperfusion (MCAO/R). (A) Representative picture of a mouse brain section after TUNEL staining with FITC-labeled dUTP and terminal deoxynucleotidyl transferase enzyme in WT and Xrcc1 +/− mouse brains after MCAO/R. (B) Quantification of TUNEL+ nuclei in WT and Xrcc1 +/− mice. An average total TUNEL+ cells count was derived from at least six brain section areas using 4 WT and 3 Xrcc1 +/− mice is shown. Data are mean ± SEM. *p<0.05, **p<0.01. Data were analyzed by two-tailed Student's t-test. (C) Levels of the full length and cleaved caspase-3 in whole brain lysates of ipsilateral area of the stroked brains. The graph indicates the quantitation of full length caspase 3 and cleaved caspase 3 (sum of p19 + p17) levels shown in the blot normalized to the actin levels.
3.4. Post stroke brain samples from Xrcc1 +/− mice show loss of DNA ligase III expression and reduced DNA repair capacity
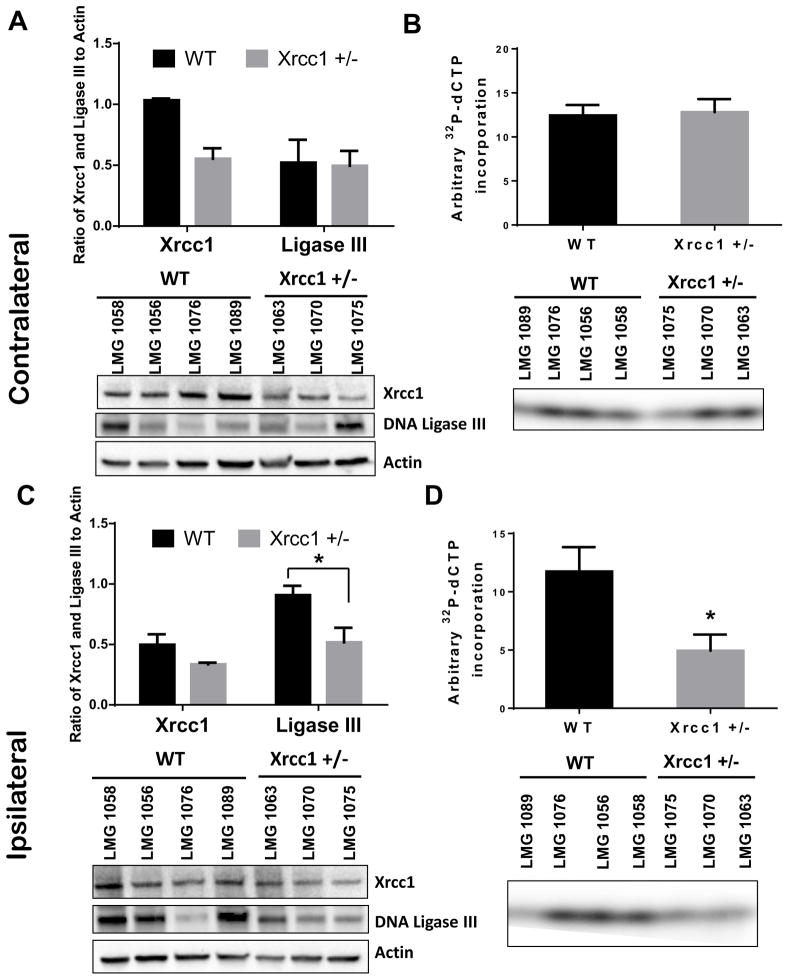
To further characterize the above observation, whole cell lysates were prepared from ipsilateral (ischemic) and contralateral (non-ischemic) cerebral cortex of WT and Xrcc1 +/− mice 48 h after MCAO/R, and the lysates were subsequently analyzed for the expression of Xrcc1 and DNA Lig3 by immunoblotting. In addition, BER synthesis and ligation was measured in prepared lysates using a 91-mer double-strand substrate containing a single nucleotide gap and radiolabeled dCTP nucleotides. For the contralateral side, immunoblotting revealed half the signal for Xrcc1, whereas there was no change in Lig3 levels in brain tissue from the non-ischemic cerebral hemispheres of Xrcc1 +/− mice (Fig. 4A). Furthermore, levels of BER activity were similar in tissue from the contralateral cerebral hemispheres of WT and Xrcc1 +/− mice (Fig. 4B and Fig. S1B). On the ipsilateral side, Xrcc1 and Lig3 levels were lower in Xrcc1 +/− mice relative to WT mice (Fig. 4C). Moreover, BER activity in whole cell lysates of cerebral cortex tissue in the ischemic penumbra of Xrcc1 +/− mice was significantly reduced compared to WT mice (58% lower in Xrcc1 +/− mice than WT mice, Fig. 4D and Fig. S1B).
Figure 4.
Expression of Xrcc1 & DNA Ligase III and whole BER activity in Xrcc1 +/− mouse brains after middle cerebral artery occlusion/ reperfusion (MCAO/R). (A) A representative immunoblot of Xrcc1 and Ligase III levels in whole brain lysates of contralateral area of the stroked brains. The graph indicates the quantitation of Xrcc1 and ligase III levels shown in the blot normalized to the actin levels. Data were averaged from 4 or 3 mice and at least two independent experiments. (B) Relative dCTP incorporation and ligation into a 91-mer in whole brain lysates of contralateral area of the stroked brains. Data indicate average ± SEM (*p<0.05 using a two-tailed Student's t-test with a sample size of n = 4 for WT and n = 3 for Xrcc1 +/−). (C) A representative immunoblot of Xrcc1 and Ligase III levels in whole brain lysates of ipsilateral area of the brains. The graph indicates the quantitation of Xrcc1 and ligase III levels shown in the blot normalized to the actin levels. Data were represented from at least two experiments. (D) Relative dCTP incorporation and ligation into a 91-mer in whole brain lysates of ipsilateral area of the brains. Data indicate average ± SEM.
Together, these results suggest that the slightly reduced protein level of Xrcc1 has no effect on overall Lig3 levels or BER activity in the nonischemic brain region (contralateral part). Conversely, the overall protein level of Xrcc1 appears to be more important in the induction/stability of Lig3 and BER capacity in the brain cells (ipsilateral part) that are challenged with acute ischemic stroke.
3.5. Impaired BER activities in postmortem human brain tissue samples from individuals who died of stroke
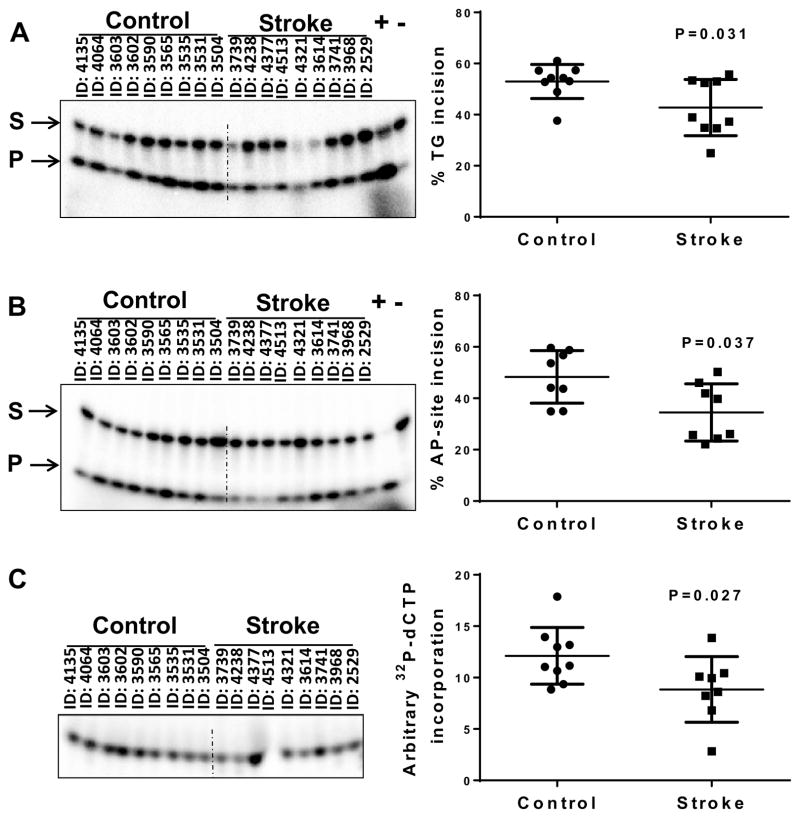
The data presented so far indicate the importance of BER capacity in recovering from focal ischemic stroke in mice. We wondered if altered BER activities might be seen in human stroke patients. Although population studies predicted that loss or modifications of BER players such as OGG1 or XRCC1 might enhance vulnerability to stroke (Dutra, et al., 2006,Mahabir, et al., 2007,Shyu, et al., 2012), detailed biochemical analysis has not been reported. Hence, we determined the capacity of specific BER steps in extracts prepared from human post-mortem cerebral cortical tissue samples from individuals who had died from stroke as well as individuals who had died of non-neurological causes (Table 1). DNA glycosylase activity was assayed by the incision of a radiolabeled DNA oligonucleotide substrate containing a single lesion, thymine glycol, and an oligonucleotide containing the abasic site analog THF was used to measure AP site incision activity. Incision activity was calculated as the amount of radioactivity in the band corresponding to the damage-specific cleavage product over the total radioactivity in the lane. Interestingly, lysates from the stroke affected brain regions showed significantly reduced thymine glycol incision (42.76 ± 11.01 % vs 52.97 ± 6.67 %, p-value 0.031) and AP endonuclease activity (34.51 ± 11.14 % versus 48.31 ± 10.24 %, p-value 0.037) (Fig. 5A and 5B). We have also checked the excision of 5-hydroxy uracil (5-OHU), the preferred substrate for NEIL1 but the activity was found too low for quantitative analysis (data not shown). We then assayed for whole BER activity, as described above. Whole BER activity was 27% lower in stroke samples than in control samples (P = 0.027) (Fig. 5C).
Figure 5.
BER activities in control and stroke human brain samples. (A) Whole tissue lysates from control and stroke postmortem brains were assessed for thymine glycol incision activity using oligonucleotide sequence (Table 2). (B) Whole tissue lysates from control and stroke postmortem brains were assessed for abasic site (tetrahydrofuran [THF]) cleavage using oligonucleotide sequence (Table 2). (C) Whole tissue lysates from control and stroke postmortem brains were assessed for deoxycytidinetriphosphate (dCTP) incorporation and ligation activity using oligonucleotide sequence (Table 2). The data are represented by mean ± SD, n = 9.
The postmortem intervals (PMIs) of the samples used in this study averaged 14.9 hours in stroke patients and 12.7 hours in control brains. To eliminate the possibility of loss of activity due to stroke itself or storage conditions we performed enzymatic assays on one of the constitutively expressed enzymes in brain, hexokinase. We found no significant differences in the protein’s activity in the sample groups (Fig. S2). Moreover, we plotted the PMI against each of the assays for all samples to determine if there was any correlation in activity to the length of PMI. No correlation was found between length of PMI and repair activities (Fig. S3). These results are consistent with our prior study where a similar analysis had been done (Canugovi, et al., 2014).
3.6. Reduced expression of XRCC1, NEIL1 and DNA LIG3 proteins in human brain samples derived from individuals who died of stroke
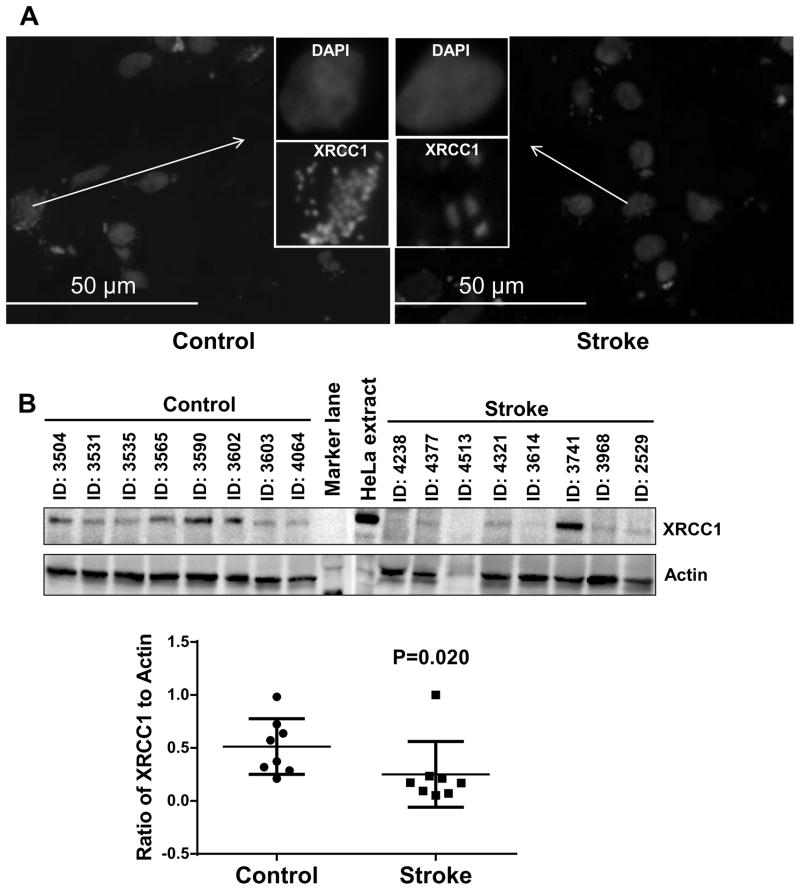
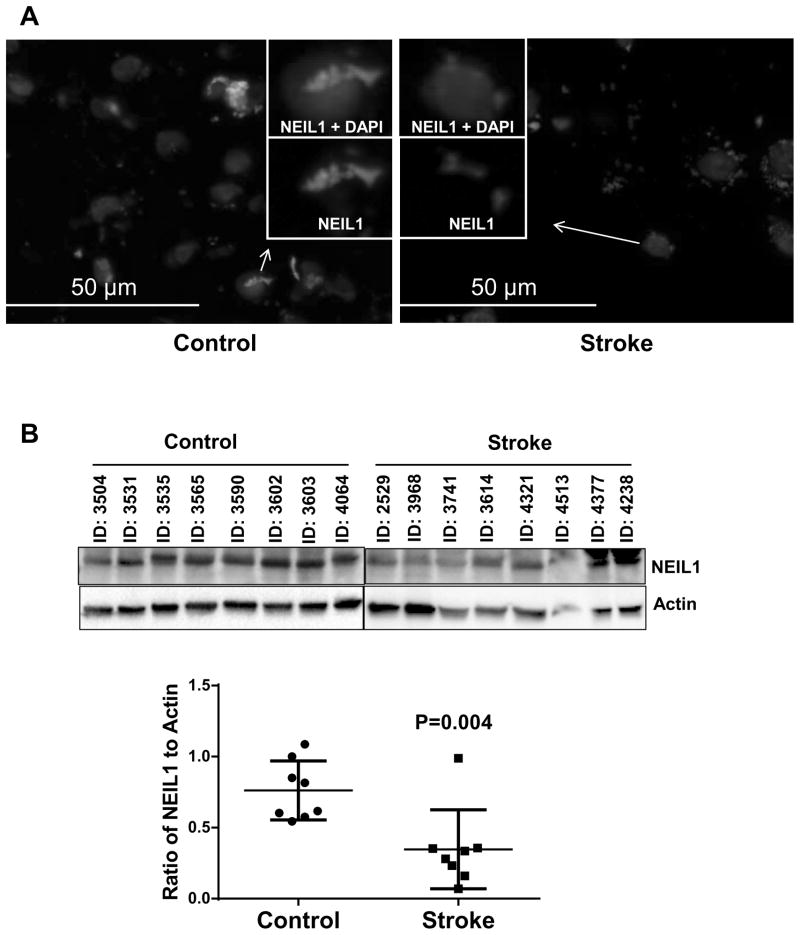
We wondered whether the loss of the BER activities described above related to a loss of protein expression. Since NEIL1 plays an important role and serves as back-up glycosylase (Hazra, et al., 2002,Katafuchi, et al., 2004,Takao, et al., 2002) in the initial steps of BER. DNA LIG3 and XRCC1 plays a crucial role in later steps of BER, it was of interest to study the expression of these three proteins in stroke affected human brains compared to controls. To examine this, we utilized immunoblotting and immunofluorescence methods. In both cases, we found significantly lower XRCC1 expression (51% lower, P = 0.020) in the human stoke brain samples compared to control brain samples (Fig. 6A and B) and lower expression of DNA LIG3 in the human stroke patient brain samples compared to control brain samples (Fig. S4). Similarly, we also found a significant loss of NEIL1, a major DNA glycosylase that removes oxidative damage from DNA (54% lower, P=0.004, Fig. 7A and B). Together, these data suggest that there may be significantly less BER activity in stroke postmortem brain samples.
Figure 6.
XRCC1 expression in control and stroke human brain samples. (A) Representative image of Immunofluorescence staining of XRCC1 in control and stroke human brain samples, n=8 (Insert) image of a single cell is enlarged to show the expression of XRCC1. (B) A representative immunoblot of XRCC1 levels in control and stroke human brain samples. The graph indicates the quantitation of XRCC1 levels shown in the blot normalized to the actin levels. The data are represented by mean ± SD, n = 8.
Figure 7.
NEIL1 expression in control and stroke human brain samples. (A) Representative image of Immunofluorescence staining of NEIL1 in control and stroke human brain samples, n=8 (Insert) image of a single cell is enlarged to show the expression of NEIL1. (B) A representative immunoblot of NEIL1 levels in control and stroke human brain samples. The graph indicates the quantitation of NEIL1 levels shown in the blot normalized to the actin levels. The data are represented by mean ± SD, n = 8.
4. Discussion
In this study, we provide evidence that effective and efficient BER is essential for speedy recovery from stroke. Specifically, we found that heterozygous Xrcc1 mice recover poorly from stroke as compared to wild type littermates. Importantly, reduced DNA repair capacity in heterozygous Xrcc1 mice was sufficient to trigger higher cellular death in the ischemic penumbra. These observations underscore the importance of robust BER for recovery of mice from stroke. To extend our analysis to human subjects, we investigated BER capacity in stroke-affected human post-mortem brain samples and compared them with human brain autopsy samples from individuals who had died of non-neurological causes. The results indicate that stroke is associated with a significant impairment of general BER function and reduced expression of XRCC1 and NEIL1. These findings suggest that effective BER is likely important for recovery from stroke.
XRCC1 is a non-enzymatic scaffold protein that plays critical roles in BER and SSBR. In mouse models, loss or dysfunction of both alleles of Xrcc1 causes death at embryonic stages E6.5–7.5, due to abnormal development of the embryonic endoderm (Tebbs, et al., 1999). In addition, published data links functional defects in XRCC1 with neurodegenerative diseases and cancer (Caldecott, 2008,Ladiges, 2006,Wilson, et al., 2011). Therefore, heterozygous and other hypomorphic Xrcc1 mouse strains are a useful system for investigating the in vivo biological role(s) of XRCC1.
Initial characterization of Xrcc1+/− heterozygous mice demonstrated that the phenotypic consequences of copy number reduction of Xrcc1 are relatively minor in the absence of exogenous stressors (McNeill, et al., 2011). Mice with one functional allele of Xrcc1 have no prominent neurological defects during development and display normal cognitive ability as adult mice. Minor neurological defects have been noted in some adult Xrcc1+/− animals and a few a animals exhibited behavioral features commonly associated with stroke (McNeill, et al., 2011). Furthermore, specific inactivation of both alleles of Xrcc1 in the nervous system of adult mice contributes to neuropathology including loss of cerebellar interneurons, abnormal hippocampal neurons, progressive mild ataxia and episodic spasms (Lee, et al., 2009). These data support the idea that Xrcc1 plays an important role in neurons in immature and mature mice. However, ectopic expression of a small amount (<10% the normal level) of wild type Xrcc1 fully complements the developmental defect in Xrcc1 null embryos and reverses sensitivity to DNA damaging agents in Xrcc1 null fibroblasts (Tebbs, et al., 1999). Therefore, it is reasonable to propose that under some conditions, one functional allele of Xrcc1 is adequate for normal functions of the cell, while under other conditions, likely involving stress, higher levels of Xrcc1 expression may be required to prevent such damage. The data presented in this paper are fully consistent with this hypothesis.
Two human population studies revealed that polymorphisms in BER genes APE1 and XRCC1 had no association with large artery atherosclerotic (LAA) stroke (Dutra, et al., 2006,Mahabir, et al., 2007). A second population study involving cigarette smoking and non-smoking groups showed that polymorphisms of ERCC2 and XPD were associated with LAA stroke risk, and expression of both OGG1 and ERCC2 polymorphisms concurrently elevated the risk of LAA stroke; additionally, this risk was elevated with cigarette smoking in all groups (Shyu, et al., 2012). Although there is no association of APE/ref-1, XRCC1, OGG1 gene polymorphisms with stroke by themselves, combinations of these three polymorphisms increased the risk for LAA stroke in smokers, indicating the importance of efficient BER in an increased oxidative stress environment (Canugovi, et al., 2013).
It has been shown that Xrcc1 is one of the BER proteins that is upregulated with transient ischemia as part of cellular stress resistance mechanisms (Li, et al., 2007). However, we did not observe this as seen in western blots in Fig. 4, compare WT ipsilateral to contralateral. Additionally, we found that at 24 and 48 h after the stroke, mice having a single Xrcc1 gene copy exhibited significantly poorer performance in the rotarod test assessing motor function than to wild type mice. This indicates that higher levels of Xrcc1 are essential for resistance to adverse effects caused by ischemic stroke and for a recovery after stroke.
The interpretation of above results is complicated by the fact that XRCC1 is required for the stability of Lig3, a DNA repair protein expressed in both mitochondrial and nuclear compartments whose function overlaps with, but is not redundant to, that of DNA ligases I and IV. Although XRCC1 and Lig3 play apparently independent roles in response to DNA damage in the nuclear and mitochondrial compartments, respectively (Lakshmipathy and Campbell, 2000), on the biochemical level, XRCC1 is required to stabilize and ensure the enzymatic competence of DNA Lig3 (Simsek, et al., 2011a,Simsek, et al., 2011b). In addition, XRCC1 interacts with and stimulates the function of other DNA repair proteins and pathways, supporting the hypothesis that XRCC1 increases the efficiency of the BER process, possibly more so when cells are challenged with oxidative stress.
Our data suggest that the partial loss of the important DNA repair protein, Xrcc1 renders brain cells susceptible to ischemic stroke and its robust action is critical in the survival of cells under conditions of severe oxidative stress. However, other factors in operation in stroke apart from oxidative stress may also contribute to the overall response. This is in agreement with previous studies showing that BER is a major pathway for the repair of oxidatively modified DNA bases in neurons (Harrison, et al., 2005,Karahalil, et al., 2002). With regards to brain damage from stroke, our findings suggest that neurons that are better able to repair oxidative DNA lesions have an increased chance of survival, consistent with the previous studies of BER activity in mouse models of ischemic stroke (Canugovi, et al., 2012,Liu, et al., 2011). In a recent human study, it has been shown that there is little plasticity both in terms of neurogenesis or genomic rearrangements in human cortical neurons and the neurons having effective DNA repair system survived long-term after stroke (Huttner, et al., 2014). Since BER is important for mice to recover from focal ischemic stroke, we measured BER activities in brain specimens from patients that died of stroke or normal controls who died from non-neurological causes. . Our findings show that XRCC1 and NEIL1 protein levels were significantly lower in brains of stroke patients than in controls. Our findings are consistent with earlier studies where we have shown that a lower level of BER protein is associated with neurological diseases (Canugovi, et al., 2014,Weissman, et al., 2007). These findings, along with our prior publications (Canugovi, et al., 2012,Liu, et al., 2011) provide compelling evidence that BER capacity matters with respect to one’s ability to recover from an acute ischemic event. Furthermore, as we enter the era of personalize medicine an individual’s collective BER genotype may be useful to identify individuals as risk of poor recover from ischemic events.
Collectively, these results show that even a haploinsufficiency, and not a complete knockout, of a DNA repair gene is detrimental to brain cell survival after stroke. Together our data suggest that effective and efficient BER plays significant role in protecting cells from stroke-induced adverse effects.
Supplementary Material
(A) Apoptosis in Xrcc1 +/− mouse contralateral brain sections after middle cerebral artery occlusion/ reperfusion (MCAO/R). Representative picture of a mouse brain section after TUNEL staining with FITC-labeled dUTP and terminal deoxynucleotidyl transferase enzyme in WT and Xrcc1 +/− mice. There is no TUNEL staining signal (green). (B) Whole gel is shown for whole BER activity in WT and Xrcc1 +/− mouse brains after middle cerebral artery occlusion/ reperfusion (MCAO/R).
Hexokinase activity in whole tissue lysate of in control and stroke human brain samples to determine if enzyme activity was affected because of postmortem interval in control and stroke brain sample.
Postmortem intervals (PMIs) were plotted against the respective repair activities to determine if a correlation existed between the enzyme activity and postmortem interval in both Stroke (A–B) and Control (C–D) brain whole tissue lysate. Panels A and C report thymine glycol incision and AP endonuclease 1 activities while panels B and D display whole BER activity.
DNA ligase III expression in control and stroke human brain samples. A representative immunoblot of DNA ligase III levels in control and stroke human brain samples is shown. Samples were prepared as described in the materials and methods.
Acknowledgments
This research was supported entirely by the Intramural Research Program of the National Institutes of Health and National Institute on Aging (AG 000723-07). We would like to thank Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center, Los Angeles, CA 90073 which is sponsored by NINDS/NIMH, National Multiple Sclerosis Society, and Department of Veterans Affairs for the human postmortem brain samples used in this study. Additionally, we would like to thank Drs. Beverly A. Baptiste and Raghavendra A. Shamanna for critically reading the manuscript.
Footnotes
Disclosure statement
Authors have no conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arumugam TV, Phillips TM, Cheng A, Morrell CH, Mattson MP, Wan R. Age and energy intake interact to modify cell stress pathways and stroke outcome. Annals of neurology. 2010;67(1):41–52. doi: 10.1002/ana.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bederson JB, Pitts LH, Tsuji M, Nishimura MC, Davis RL, Bartkowski H. Rat middle cerebral artery occlusion: evaluation of the model and development of a neurologic examination. Stroke; a journal of cerebral circulation. 1986;17(3):472–6. doi: 10.1161/01.str.17.3.472. [DOI] [PubMed] [Google Scholar]
- Borlongan CV, Yamamoto M, Takei N, Kumazaki M, Ungsuparkorn C, Hida H, Sanberg PR, Nishino H. Glial cell survival is enhanced during melatonin-induced neuroprotection against cerebral ischemia. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2000;14(10):1307–17. doi: 10.1096/fj.14.10.1307. [DOI] [PubMed] [Google Scholar]
- Broughton BR, Reutens DC, Sobey CG. Apoptotic mechanisms after cerebral ischemia. Stroke; a journal of cerebral circulation. 2009;40(5):e331–9. doi: 10.1161/STROKEAHA.108.531632. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nature reviews Genetics. 2008;9(8):619–31. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular 'nick-sensor' in vitro. Nucleic acids research. 1996;24(22):4387–94. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A, Marsin S, Nakabeppu Y, O'Connor TR, Boiteux S, Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA repair. 2005;4(7):826–35. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Canugovi C, Misiak M, Ferrarelli LK, Croteau DL, Bohr VA. The role of DNA repair in brain related disease pathology. DNA repair. 2013;12(8):578–87. doi: 10.1016/j.dnarep.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C, Shamanna RA, Croteau DL, Bohr VA. Base excision DNA repair levels in mitochondrial lysates of Alzheimer's disease. Neurobiology of aging. 2014;35(6):1293–300. doi: 10.1016/j.neurobiolaging.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C, Yoon JS, Feldman NH, Croteau DL, Mattson MP, Bohr VA. Endonuclease VIII-like 1 (NEIL1) promotes short-term spatial memory retention and protects from ischemic stroke-induced brain dysfunction and death in mice. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(37):14948–53. doi: 10.1073/pnas.1204156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nature genetics. 2001;29(2):184–8. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- Dutra AV, Lin HF, Juo SH, Mohrenweiser H, Sen S, Grewal RP. Analysis of the XRCC1 gene as a modifier of the cerebral response in ischemic stroke. BMC medical genetics. 2006;7:78. doi: 10.1186/1471-2350-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JF, Hollensworth SB, Spitz DR, Copeland WC, Wilson GL, LeDoux SP. Oxidative stress-induced apoptosis in neurons correlates with mitochondrial DNA base excision repair pathway imbalance. Nucleic acids research. 2005;33(14):4660–71. doi: 10.1093/nar/gki759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(6):3523–8. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner HB, Bergmann O, Salehpour M, Racz A, Tatarishvili J, Lindgren E, Csonka T, Csiba L, Hortobagyi T, Mehes G, Englund E, Solnestam BW, Zdunek S, Scharenberg C, Strom L, Stahl P, Sigurgeirsson B, Dahl A, Schwab S, Possnert G, Bernard S, Kokaia Z, Lindvall O, Lundeberg J, Frisen J. The age and genomic integrity of neurons after cortical stroke in humans. Nature neuroscience. 2014;17(6):801–3. doi: 10.1038/nn.3706. [DOI] [PubMed] [Google Scholar]
- Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2002;16(14):1895–902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. The Journal of biological chemistry. 2004;279(14):14464–71. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- Khan M, Im YB, Shunmugavel A, Gilg AG, Dhindsa RK, Singh AK, Singh I. Administration of S-nitrosoglutathione after traumatic brain injury protects the neurovascular unit and reduces secondary injury in a rat model of controlled cortical impact. Journal of neuroinflammation. 2009;6:32. doi: 10.1186/1742-2094-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A, McNeill DR, Gleichmann M, Mattson MP, Wilson DM., 3rd XRCC1 protects against the lethality of induced oxidative DNA damage in nondividing neural cells. Nucleic acids research. 2008;36(15):5111–21. doi: 10.1093/nar/gkn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladiges WC. Mouse models of XRCC1 DNA repair polymorphisms and cancer. Oncogene. 2006;25(11):1612–9. doi: 10.1038/sj.onc.1209370. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic acids research. 2000;28(20):3880–6. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Katyal S, Li Y, El-Khamisy SF, Russell HR, Caldecott KW, McKinnon PJ. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nature neuroscience. 2009;12(8):973–80. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wu H, Yang S, Chen D. Ischemic preconditioning induces XRCC1, DNA polymerase-beta, and DNA ligase III and correlates with enhanced base excision repair. DNA repair. 2007;6(9):1297–306. doi: 10.1016/j.dnarep.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Liu D, Croteau DL, Souza-Pinto N, Pitta M, Tian J, Wu C, Jiang H, Mustafa K, Keijzers G, Bohr VA, Mattson MP. Evidence that OGG1 glycosylase protects neurons against oxidative DNA damage and cell death under ischemic conditions. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31(2):680–92. doi: 10.1038/jcbfm.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahabir S, Abnet CC, Qiao YL, Ratnasinghe LD, Dawsey SM, Dong ZW, Taylor PR, Mark SD. A prospective study of polymorphisms of DNA repair genes XRCC1, XPD23 and APE/ref-1 and risk of stroke in Linxian, China. Journal of epidemiology and community health. 2007;61(8):737–41. doi: 10.1136/jech.2006.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Fanta M, Karimi-Busheri F, Silver E, Virgen CA, Caldecott KW, Cass CE, Weinfeld M. XRCC1 stimulates polynucleotide kinase by enhancing its damage discrimination and displacement from DNA repair intermediates. The Journal of biological chemistry. 2007;282(38):28004–13. doi: 10.1074/jbc.M704867200. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Robertson A, Dimitriadis EK, Prasad R, Wilson SH, Mullen GP. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic acids research. 2000;28(10):2049–59. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. The Journal of biological chemistry. 2003;278(45):44068–74. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- McNeill DR, Lin PC, Miller MG, Pistell PJ, de Souza-Pinto NC, Fishbein KW, Spencer RG, Liu Y, Pettan-Brewer C, Ladiges WC, Wilson DM., 3rd XRCC1 haploinsufficiency in mice has little effect on aging, but adversely modifies exposure-dependent susceptibility. Nucleic acids research. 2011;39(18):7992–8004. doi: 10.1093/nar/gkr280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nature genetics. 2001;29(2):189–93. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O, Leonhardt H. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC molecular biology. 2007;8:81. doi: 10.1186/1471-2199-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niizuma K, Endo H, Chan PH. Oxidative stress and mitochondrial dysfunction as determinants of ischemic neuronal death and survival. Journal of neurochemistry. 2009;109(Suppl 1):133–8. doi: 10.1111/j.1471-4159.2009.05897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu HY, Shieh JC, Ji-Ho L, Wang HW, Cheng CW. Polymorphisms of DNA repair pathway genes and cigarette smoking in relation to susceptibility to large artery atherosclerotic stroke among ethnic Chinese in Taiwan. Journal of atherosclerosis and thrombosis. 2012;19(4):316–25. doi: 10.5551/jat.10967. [DOI] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS genetics. 2011a;7(6):e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011b;471(7337):245–8. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RA, Sharp FR. Infarct measurement methodology. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1994;14(4):697–8. doi: 10.1038/jcbfm.1994.88. [DOI] [PubMed] [Google Scholar]
- Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. The Journal of biological chemistry. 2002;277(44):42205–13. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nature genetics. 2002;32(2):267–72. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- Taylor RM, Wickstead B, Cronin S, Caldecott KW. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Current biology : CB. 1998;8(15):877–80. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Developmental biology. 1999;208(2):513–29. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chemical reviews. 2006;106(2):687–99. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. The EMBO journal. 2001;20(22):6530–9. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic acids research. 2007;35(16):5545–55. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104(1):107–17. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutation research. 2011;711(1–2):100–12. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Mattson MP. Neurodegeneration: nicked to death. Current biology : CB. 2007;17(2):R55–8. doi: 10.1016/j.cub.2006.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Apoptosis in Xrcc1 +/− mouse contralateral brain sections after middle cerebral artery occlusion/ reperfusion (MCAO/R). Representative picture of a mouse brain section after TUNEL staining with FITC-labeled dUTP and terminal deoxynucleotidyl transferase enzyme in WT and Xrcc1 +/− mice. There is no TUNEL staining signal (green). (B) Whole gel is shown for whole BER activity in WT and Xrcc1 +/− mouse brains after middle cerebral artery occlusion/ reperfusion (MCAO/R).
Hexokinase activity in whole tissue lysate of in control and stroke human brain samples to determine if enzyme activity was affected because of postmortem interval in control and stroke brain sample.
Postmortem intervals (PMIs) were plotted against the respective repair activities to determine if a correlation existed between the enzyme activity and postmortem interval in both Stroke (A–B) and Control (C–D) brain whole tissue lysate. Panels A and C report thymine glycol incision and AP endonuclease 1 activities while panels B and D display whole BER activity.
DNA ligase III expression in control and stroke human brain samples. A representative immunoblot of DNA ligase III levels in control and stroke human brain samples is shown. Samples were prepared as described in the materials and methods.