A novel role for ATF3 is identified in acinar-to–duct cell metaplasia during pancreatic injury. ATF3 targets transcriptional regulators that affect acinar cell fate and reduces the severity of pancreatic injury. However, by doing so, ATF3 may also increase the susceptibility for progression to pancreatic adenocarcinoma.
Abstract
Pancreatitis is a debilitating disease of the exocrine pancreas that, under chronic conditions, is a major susceptibility factor for pancreatic ductal adenocarcinoma (PDAC). Although down-regulation of genes that promote the mature acinar cell fate is required to reduce injury associated with pancreatitis, the factors that promote this repression are unknown. Activating transcription factor 3 (ATF3) is a key mediator of the unfolded protein response, a pathway rapidly activated during pancreatic insult. Using chromatin immunoprecipitation followed by next-generation sequencing, we show that ATF3 is bound to the transcriptional regulatory regions of >30% of differentially expressed genes during the initiation of pancreatitis. Of importance, ATF3-dependent regulation of these genes was observed only upon induction of pancreatitis, with pathways involved in inflammation, acinar cell differentiation, and cell junctions being specifically targeted. Characterizing expression of transcription factors that affect acinar cell differentiation suggested that acinar cells lacking ATF3 maintain a mature cell phenotype during pancreatitis, a finding supported by maintenance of junctional proteins and polarity markers. As a result, Atf3–/– pancreatic tissue displayed increased tissue damage and inflammatory cell infiltration at early time points during injury but, at later time points, showed reduced acinar-to–duct cell metaplasia. Thus our results reveal a critical role for ATF3 as a key regulator of the acinar cell transcriptional response during injury and may provide a link between chronic pancreatitis and PDAC.
INTRODUCTION
Pancreatitis involves fibrosis and inflammation of the exocrine pancreas, often the result of exposure to acute or chronic environmental stresses, including alcohol consumption, gall stone obstruction of the pancreatic duct, or hypersensitivity to pharmaceutical drugs (Yadav and Lowenfels, 2013; Roberts, 2015). Chronic and hereditary forms of pancreatitis are a significant susceptibility factor for pancreatic ductal adenocarcinoma (PDAC; Logsdon and Ji, 2009; Yadav and Lowenfels, 2013), likely due to prolonged loss of the acinar cell phenotype in these conditions (Grady et al., 1998; Shi et al., 2013). The initiating events in all forms of pancreatitis include intracellular activation of digestive enzymes (Grady et al., 1998), deregulation of Ca2+ homeostasis (Krüger et al., 2000), loss of acinar cell maturation factors (von Figura et al., 2014), and activation of interstitial fibroblasts to stellate cells (Erkan et al., 2012). The degree of activation of these events determines the severity of pancreatitis. Therefore understanding how they are transcriptionally regulated is of significant interest. In particular, several studies have shown rapid activation of the unfolded protein response (UPR) within minutes of initiating pancreatitis in rodent models (Kubisch et al., 2006; Kowalik et al., 2007).
The UPR and endoplasmic reticulum (ER) stress pathways have a dual role in cell physiology and pathology. The UPR is considered to be an immediate response to cell stress, typically activated by the presence of misfolded proteins (Wu and Kaufman, 2006; Ron and Walter, 2007). In this situation, the UPR works to mitigate damage caused by acute stress through reduction of protein load. Translation of new protein is significantly reduced (Ron and Walter, 2007), ubiquitination and degradation of existing protein is increased (Bernales, 2006), and the transcription profile is altered to promote the expression of proteins involved in these events (Bernales, 2006). However, chronic or recurrent activation of the UPR is detrimental, promoting cell apoptosis (Bernales, 2006) or alterations in gene expression that can enhance disease (Bernales, 2006; Kubisch et al., 2006). Therefore tight regulation of the UPR is required for an appropriate response to injury.
The UPR consists of three main signaling pathways that exhibit significant cross-talk (Bernales, 2006; Maas and Diehl, 2015). The PKR-like ER kinase (PERK) pathway is activated after dissociation of BiP/GRP78, leading to autoactivation, dimerization, and phosphorylation of eIF2α, a key member of the translation initiation complex (Harding et al., 2000; Ron et al., 2000). Phosphorylation of eIF2α reduces its ability to interact with this complex, promoting a global reduction in mRNA translation (Zhang et al., 2002). Activating transcription factor 4 (ATF4) is a direct downstream mediator of PERK (Bernales, 2006). The inositol-requiring enzyme 1 (IRE1) pathway is activated in a similar manner to PERK but directly targets and promotes splicing of X-box binding protein 1 (Xbp1) mRNA, leading to the expression of a potent transcription factor (sXBP1; Calfon et al., 2002; Lee et al., 2002), which promotes increased expression of numerous chaperones leading to protein degradation (Maas and Diehl, 2015). ATF6 is the third pathway to be activated and is believed to provide an adaptive response to long-term cell stress (Bernales, 2006; Wu et al., 2007). Targeted deletion of Ire1, Perk, Atf4, or Xbp1 within the pancreas leads to altered morphology, function, and cell survival, underscoring their important physiological relevance to acinar cell homeostasis (Zhang et al., 2002; Iida et al., 2007; Iwawaki et al., 2010; Hess et al., 2011).
The UPR also plays a role in the pathological response to pancreatic injury. During pancreatitis, the UPR is rapidly activated (Kubisch et al., 2006; Kowalik et al., 2007), suggesting a protective mechanism of action. However, prolonged periods of both increased and decreased UPR levels have been linked to more severe pancreatic injury (Alahari et al., 2011; Lugea et al., 2011; Sah et al., 2014). A better understanding of UPR mediators that are activated by pancreatic injury will provide clarity on these conflicting findings.
Activating transcription factor 3 (ATF3) is a mediator of the PERK/ATF4 pathway (Jiang et al., 2004) that, within the pancreas, is expressed only after pancreatitis is initiated (Kowalik et al., 2007). Rapid increases in ATF3 expression have been identified in several models of experimental pancreatitis (Allen-Jennings et al., 2001; Kowalik et al., 2007), yet it is unclear how this increase affects the pancreatic response to injury. ATF3 can function as a transcriptional repressor, targeting pancreatic and duodenal homeobox 1 (PDX1) expression and function during β-cell stress (Jang, 2011; Kim et al., 2011). However, a common event in acinar cells during pancreatitis is the repression of factors that stabilize a mature phenotype, including Mist1 (Kowalik et al., 2007), Nr5a2 (Masui et al., 2010), and Rbpjl (von Figura et al., 2014), and this loss of acinar cell maturity decreases the severity of injury (Krah et al., 2015; Martinelli et al., 2015).
The goal of this study was to characterize the transcriptional targets and roles of ATF3 in the cerulein-induced pancreatitis (CIP) model. Global chromatin immunoprecipitation sequencing (ChIP-seq) and RNA-sequencing (RNA-seq) suggest that ATF3 is enriched at more than one-third of all genes differentially expressed within 4 h of initiating CIP. Examination of differential RNA expression in mice carrying a targeted deletion of Atf3 (Atf3–/–) confirms a role for ATF3 in mediating the UPR and affecting cell differentiation. Acinar cells in Atf3–/– mice maintain their mature phenotype during CIP, correlating to increased tissue injury. However, Atf3–/– mice also show decreased acinar-to-ductal metaplasia (ADM) during the regenerative phases of CIP. These results suggest that ATF3 reduces the initial severity of pancreatic injury but leads to increased potential for events that promote PDAC.
RESULTS
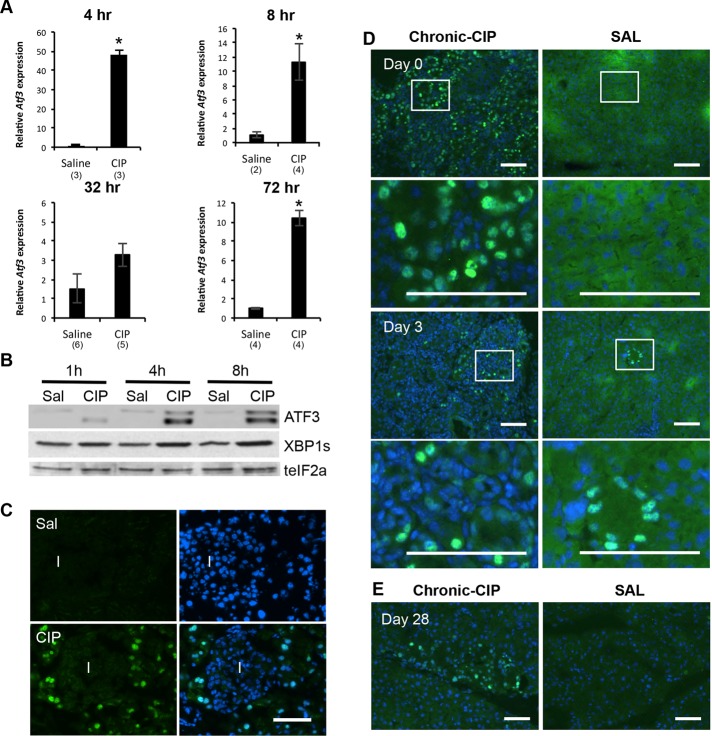
Previous studies examining gene expression during pancreatitis provided only limited information on ATF3 (Kubisch et al., 2006; Kowalik et al., 2007). Therefore we characterized temporal and spatial ATF3 expression in acute and recurrent models of CIP. During acute CIP, Atf3 was significantly increased as early as 1 h after initial cerulein injections, peaking 4 h into CIP (48-fold higher than Atf3 levels in control pancreatic tissue; Figure 1A). There was a transient decrease in Atf3 expression 32 h into CIP, but by 72 h, Atf3 levels remained significantly higher (10-fold) than in saline-treated pancreatic tissue. Western blot analysis confirmed increased ATF3 accumulation (Figure 1B), and immunofluorescence (IF) showed that ATF3 accumulation is specific to acinar cells (Figure 1C). Western blot analysis identified two bands for ATF3 (Figure 1C), and previous work suggested two isoforms exist for ATF3 that may have opposite roles in gene expression (i.e., activating vs. repressing functions; Chen et al., 1994). However, we cannot exclude the possibility that posttranslational modification of ATF3 accounts for bands of different mobility. Similar increases in expression were observed for other UPR mediators, including spliced (s) XBP1 (Figure 1B and Supplemental Figure S1) and Atf4 (unpublished data), consistent with previous studies demonstrating significant increases in Xbp1 splicing after pancreatic insult (Kubisch et al., 2006; Kubisch and Logsdon, 2007; Lugea et al., 2011). However, unlike ATF3, sXBP1 and ATF4 accumulation was observed in pancreatic tissue before CIP induction. Recurrent CIP treatment (Supplemental Figure S2) led to similar transient increases in ATF3 expression. ATF3 accumulation was apparent at the cerulein injection endpoint (day 0; Figure 1D). Three days after CIP treatment, ATF3 accumulation was still readily apparent, but by 28 d after cessation of CIP treatment, only small pockets of ATF3+ cells were observed (Figure 1E).
FIGURE 1:
ATF3 is elevated during acute and recurrent experimentally induced pancreatitis. (A) qRT-PCR analysis for Atf3 after saline or cerulein treatment 4–72 h after initiating treatment. *p < 0.05; n values are indicated on the graph. (B) Representative Western blot analysis for ATF3, spliced (s) XBP1, or total (t) eIF2α (as a loading control) 1–8 h after initial saline (Sal) or cerulein (CIP) treatment. (C) IF analysis for ATF3 at 4 h into CIP shows expression exclusively in acinar tissue. I, islet. Bar, 60 μm. IF for ATF3 in recurrent CIP (D) 1 d, 3 d, or (E) 28 d after cessation of treatment with cerulein or saline. Boxed areas are shown at higher magnification. In all instances, sections are counterstained with DAPI (blue). Bar, 36 μm.
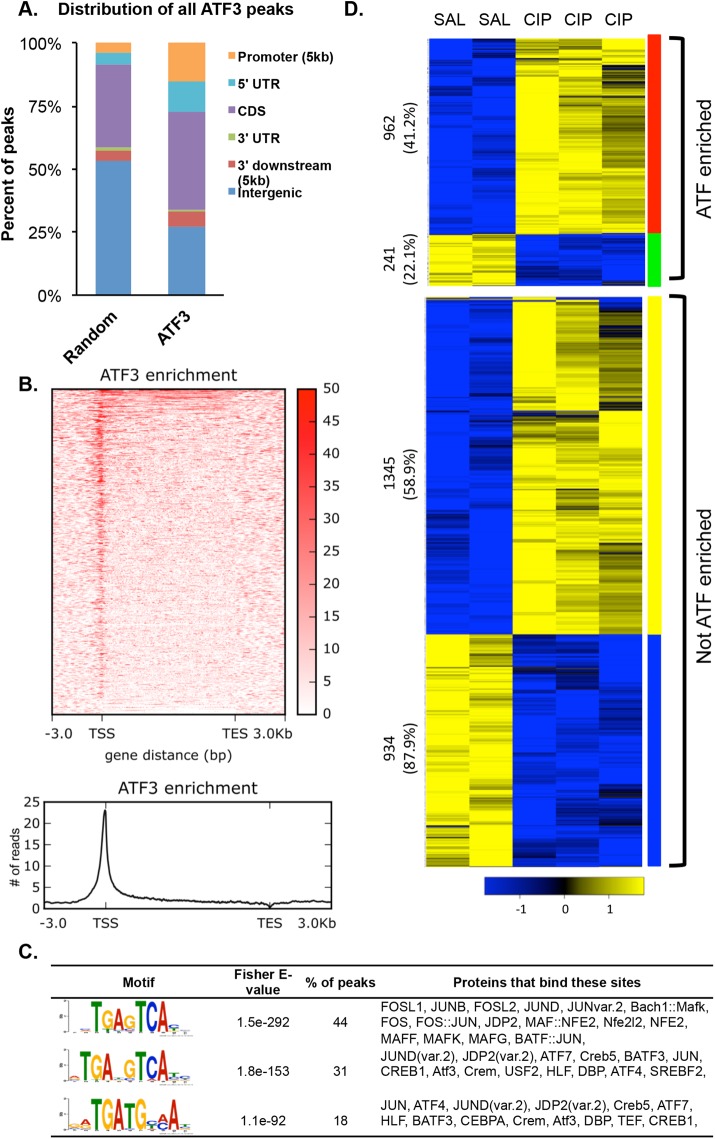
Because ATF3 is a transcription factor, we hypothesized that it would play an important role in determining gene expression patterns in response to CIP treatment. To delineate the targets of ATF3 during CIP, we performed ChIP-seq and RNA-seq for ATF3 on whole pancreatic tissue at a time consistent with peak ATF3 levels (i.e., 4 h into CIP). Because IF indicated acinar-specific localization of ATF3, we were not concerned about the inclusion of non–acinar cell populations. Peak calling identified 12,535 peaks of ATF3 enrichment over input controls (Table 1). This number of peaks is significantly lower (∼40%) than reported in previous studies on global ATF3 enrichment (Zhao et al., 2016) and could reflect the nature of the starting source, as we used whole tissue that expressed ATF3 for only 4 h versus cell cultures that constitutively expressed ATF3. Zhao et al. (2016) identified 19.4% of ATF3 enrichment peaks in promoter regions, whereas our study identified 45.9% of all ATF3 enrichment sites (5759) localized within promoters. This discrepancy could reflect the increased number of peaks in intragenic regions obtained in the previous study but also may reflect differences in defining promoter regions. Zhao et al. (2016) used regions ±2 kb surrounding the transcription start site (TSS) and identified >6000 genes enriched for ATF3. We used ±5 kb surrounding TSSs, which included 2880 sites within the 5’ UTR and coding sequence (CDS; Figure 2, A and B). Using this close association (±5 kb from a known TSS), we identified ATF3 enrichment sites associated with 3411 genes within 4 h of inducing CIP (Table 1). Of importance, both studies showed that the majority of ATF3 enrichment sites within promoter regions appear near the TSS, indicating a close association between ATF3 and annotated gene start sites.
TABLE 1:
ChIP-seq and RNA-seq comparisons of WT and Atf3–/– response to CIP.
| A. Changes in gene expression during CIP related to ATF3 enrichment | |||
|---|---|---|---|
| ChIPa | (+) RNASeq WTb | (+) RNAseq Atf3–/–b | |
| Peaks | 12,535 | ||
| Peaks ± 5 kb | 5759 (3411) | 1203 | 959 |
| Peaks ± 25 kb | 10,556 (6116) | ||
| Common (± 5 kb) | 860 | 860 | |
| Unique WT | 343 | ||
| Unique Atf3–/– | 99 | ||
| B. Changes in gene expression during CIP | |||
|---|---|---|---|
| Genotype | Total change | Increased expression | Decreased expression |
| WT | 3482 | 2263 | 1219 |
| Atf3–/– | 2621 | 1702 | 919 |
| Common | 2189 | 1482 | 707 |
| Unique to WT | 1293 | 781 | 512 |
| Unique to Atf3–/– | 432 | 220 | 212 |
aATF3 enrichment peaks were called using MACS2 with a cutoff p of 0.0001.
bRNA-seq gene expression changes were derived with the DESeq2 and an adjusted p value of 0.01.
FIGURE 2:
ChIP-seq analysis for ATF3 targets 4 h into CIP. (A) Annotation of all called peaks (12,535) for ATF3 ChIP-seq at 4 h into cerulein treatment compared with 12,535 random locations. (B) Heat map showing ATF3 enrichment (input subtracted) at all mm10 RefSeq genes. ATF3 is generally localized near the TSSs (TES, transcriptional end site). (C) Motif analysis identifying consensus sequences found in peaks located within 5 kb of a TSS, along with the percentage of peaks in which these motifs appear and the potential proteins that bind these motifs. (D) Gene expression analysis of pancreatic tissue treated for 4 h with saline (SAL) or cerulein (CIP), showing increased (yellow) or decreased (blue) expression was cross-referenced to ChIP-seq analysis showing ATF3 enrichment. Numbers indicate the total number of genes in each group, with the percentage in brackets reflecting the proportion of ATF3-enriched genes that increase or decrease.
Motif analysis identified three consensus sites that appear in a significant number of ATF3-enriched regions. These motifs are found in 44% (5’ TGAG/CTCA 3’), 31% (5’ TGAT/CG/CTCA 3’), and 18% (5’ TGATGC/TA/CA 3’) of all ATF3-enriched sites (Figure 2C) and have been reported as target sites for ATF3 enrichment in an unrelated cell type by Zhao et al. (2016), confirming the validity of our ChIP-seq data set. No other motifs were found in a significant number of ATF3 enrichment sites. These findings suggest a significant role for ATF3 in regulating the acinar cell transcriptome during the immediate response to pancreatitis.
Gene Ontology (GO) analysis was performed using the 3411 genes enriched for ATF3 and identified a number of broad GO categories, including regulation of intracellular signal transduction, protein phosphorylation, and cell junction organization. GO analysis also indicated that the response to unfolded proteins was directly targeted by ATF3 (highlighted in blue; Supplemental Figure S3A and Supplemental Table S2). Several additional GO terms, including the response to topologically incorrect proteins and response to ER stress, supported the involvement of ATF3 in responding to cell stress (highlighted in red; Supplemental Figure S3A). GO analysis also revealed significant ATF3 enrichment at gene networks involved in catabolic processes, cell junction assembly, and cell organization. The same gene set was used for PANTHER and KEGG pathway analysis (Supplemental Figure S3, B and C, and Supplemental Table S3). Genes enriched for ATF3 were identified in pathways known to be involved in cerulein-induced pancreatic injury (cholecystokinin [CCK] signaling [ Kowalik et al., 2007] and epidermal growth factor [EGF] receptor and mitogen-activated protein kinase signaling [ Ardito et al., 2012]) and pancreatic cancer (Shi et al., 2013), as well as those involved in endocytosis, pancreatic cell differentiation, including Notch (Gomez et al., 2004) and Wnt signaling (Morris et al., 2010), tight junctions, and adherens junctions (Collares-Buzato et al., 2004). Several of these pathways were also identified by Zhao et al (2016; Supplemental Tables S2 and S9).
To determine whether ATF3 enrichment corresponded to changes in gene expression after CIP treatment, we cross-referenced the ChIP-seq data to RNA-seq, comparing gene expression in pancreatic tissue from wild-type mice treated for 4 h with cerulein or saline (Figure 2D and Supplemental Figure S4A). Based on RNA-seq analysis alone, 3482 genes were differentially expressed 4 h into cerulein treatment (adjusted p < 0.01). Of these genes, 2263 were significantly increased and 1219 genes were significantly decreased (Table 1 and Supplemental Figure S4A). In total, >34% (1203) of genes differentially expressed during CIP were also enriched for ATF3 at this 4-h time point, confirming an important role for ATF3 in mediating acinar cell transcription during CIP. Although ATF3 has been described as a transcriptional repressor, the presence of ATF3 at a gene did not exclusively correlate with decreased gene expression during CIP (Figure 2D). Of all genes with significantly increased expression during CIP, 41.2% are enriched for ATF3, and 22.1% of all genes with significantly decreased expression are enriched for ATF3. Similarly, no correlation to gene expression (i.e., increased or decreased) was identified for where ATF3 enrichment occurred or the type of consensus sites found within the ATF3 enrichment site (unpublished data).
GO overexpression analysis on the 1203 ATF3 genes that are differentially expressed 4 h into CIP treatment identified 146 pathways with greater than twofold enrichment (based on p < 0.01; Supplemental Table S4). This suggests a widespread effect of ATF3 on acinar cell function in response to CIP. Pathway analysis via PANTHER/KEGG analysis (Supplemental Tables S5 and S6) again identified pathways linked to the UPR, cell organization, cerulein treatment (CCK and MAPK signaling), and pathways involved in affecting pancreatitis severity, such as oxidative stress (Sah et al., 2014) and platelet-derived growth factor signaling (Aghdassi et al., 2011), or implicated in acinar-to–duct cell metaplasia (RAS (Logsdon and Lu, 2016), EGF (Ardito et al., 2012), and Wnt signaling (Morris et al., 2010)).
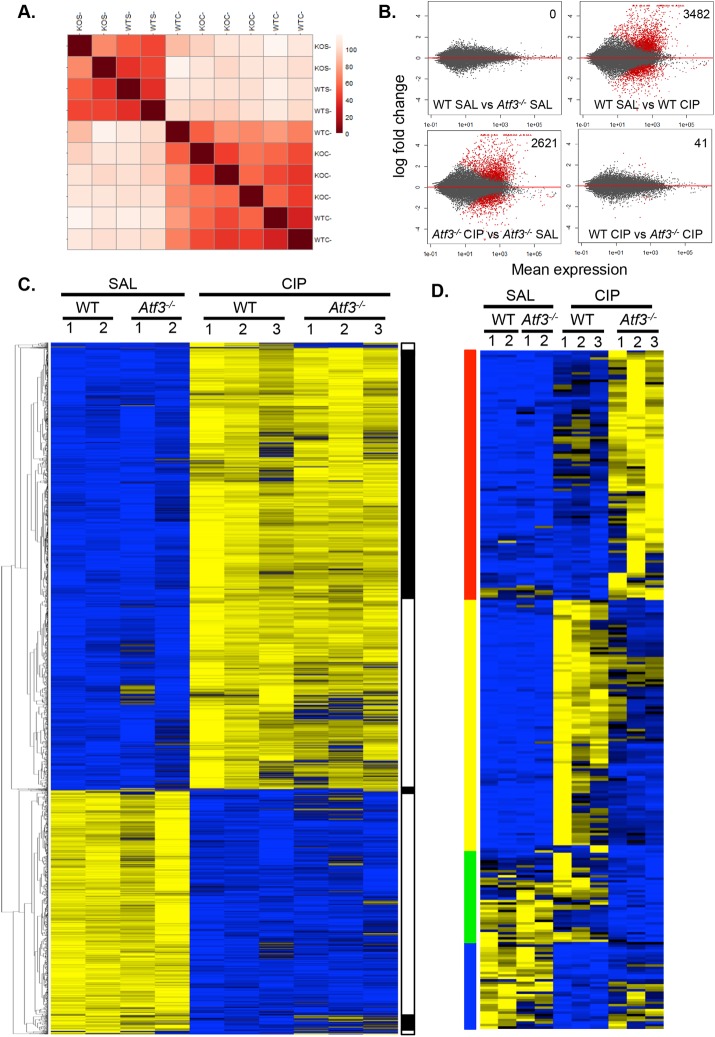
To determine the expression changes that are dependent on ATF3 during CIP, we compared changes in pancreatic mRNA accumulation between WT and Atf3–/– mice after 4 h of cerulein treatment (Figure 3). Sample-to-sample distance clustering showed a distinct segregation of RNA expression patterns based on saline versus cerulein treatment (Figure 3A). Saline-treated Atf3–/– pancreatic tissue showed no significant differences in gene expression compared with saline-treated WT tissue (adjusted p < 0.01; Figure 3B), suggesting that ATF3 has little contribution to the normal physiology and function of the pancreas. This finding is expected because ATF3 is not expressed under conditions of normal pancreatic homeostasis. After CIP treatment, the expression of 3482 genes (adjusted p < 0.01) was significantly altered in WT pancreatic tissue relative to saline-treated tissue (Table 1 and Figure 3B). Of these 3482 genes, only 2189 (62.9%) were also differentially expressed between saline- and CIP-treated Atf3–/– tissues. An additional 432 genes were altered in cerulein-treated Atf3–/– mice that were not changed in WT mice after CIP. This suggests that the significant alterations in CIP gene expression profiles in the absence of ATF3 may be due to activating and repressive effects of ATF3. Of the 1293 genes differentially expressed in only WT tissue, 298 (23%) were identified as enriched for ATF3 based on ChIP-seq analysis, suggesting both direct and indirect transcriptional effects of ATF3 on the acinar cell response to CIP.
FIGURE 3:
Lack of ATF3 alters the molecular response to CIP. (A) Distance correlation heat map comparing global gene expression in male WT and Atf3–/– mice treated with saline (SAL; n = 2) or cerulein (CIP; n = 3). (B) MA plots for global gene expression of the four different conditions in the RNA-seq; significantly differentially expressed genes (p-adj < 0.01) are indicated in red. Number of differentially expressed genes is indicated in the top right. (C) Heat maps comparing gene expression differences between WT and Atf3–/– pancreatic tissue 4 h into saline or cerulein treatment identified seven ill-defined gene clusters based on similar changes in expression (indicated in black and white). (D) Heat maps of genes determined as differentially expressed between WT and Atf3–/– cerulein-treated pancreatic tissue (p < 0.01) identified 274 genes grouped into four clusters based on expression changes, including those that increased only in Atf3–/– (red) or WT tissue (yellow), or decreased only in Atf3–/– (green) or WT tissue (blue) See Supplemental Table S10 for gene lists.
GO analysis of biological processes associated with differentially expressed genes of CIP-treated WT and Atf3–/– mice confirmed differences in the way WT and Atf3–/– mice responded to CIP (Supplemental Table S7) and suggested that the response to stress, posttranscriptional regulation of gene expression, regulation of apoptosis signaling, and cell junction organization may all be affected by the loss of ATF3. Analysis of both KEGG and PANTHER pathways (Supplemental Figure S4, A and B, and Supplemental Tables S8 and S9) showed marked differences between WT and Atf3–/– tissue in the response to CIP—specifically, in pathways linked to inflammation, EGF signaling, adherens junctions, and Wnt signaling. Of interest, generating a heat map for all genes differentially expressed between saline and cerulein treatments in either WT or Atf3–/– mice (Figure 3C; 3914 genes in total based on an adjusted p < 0.01) revealed gene clusters in which expression was increased or decreased after CIP. Whereas some differences between genotypes could be identified based on expression changes, more-defined clusters of genes were not readily apparent. Therefore we focused on genes that show differential expression specific between cerulein-treated WT and Atf3–/– mice. Using an adjusted p < 0.01, we identified only 41 differentially expressed genes (Figure 3B), suggesting a minimal effect of ATF3 on the pancreatic response to cerulein. Because very few genes were seen to be differentially expressed at an adjusted p < 0.01 (chosen due to the higher variability between cerulein and saline conditions), we reduced the stringency of our analysis to a nonadjusted p < 0.01 to encompass more potentially relevant genes. This direct comparison between ATF3 CIP versus WT CIP samples identified 274 genes that require ATF3 for appropriate changes in expression (Figure 3D and Supplemental Table S10). These genes could be clustered into four distinct groups: genes increased or decreased in only WT tissue, and genes increased or decreased in only Atf3–/– tissue. Several genes linked to the UPR (including Ddit3, Hsp5a, and Fgf21) showed higher expression in the absence of ATF3 after CIP, indicating that ATF3 may suppress or limit this pathway. Conversely, a number of genes linked to acinar cell differentiation, including Sox9 and Krt19, were increased in only WT mice during CIP. These findings support a role for ATF3 in affecting acinar cell differentiation.
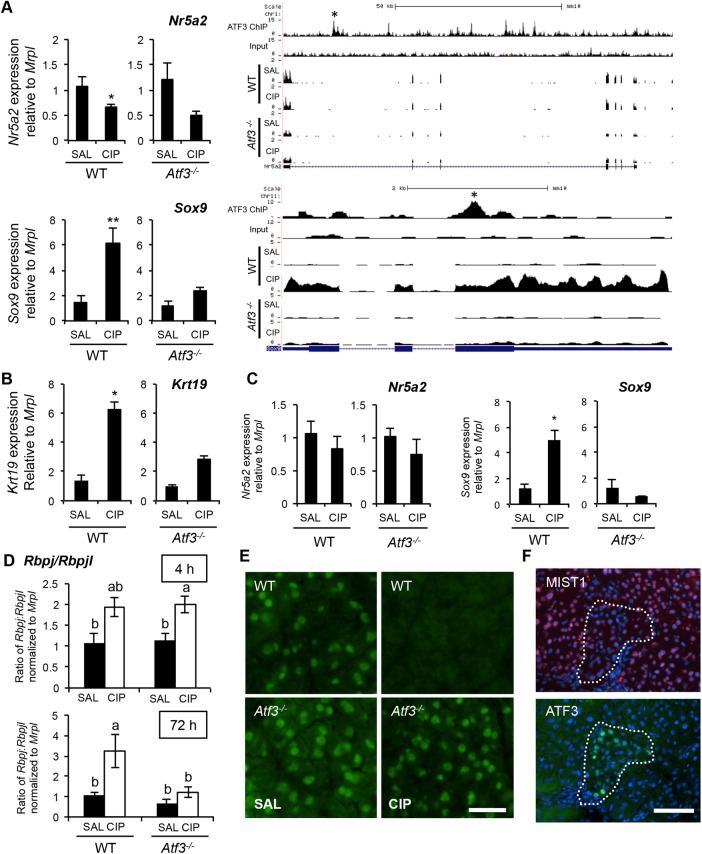
In support of a role for ATF3 in affecting acinar cell differentiation during CIP, ChIP-seq of WT pancreatic tissue revealed ATF3 enrichment at both acinar cell maturity (Nr5a2, Mist1) and progenitor (Sox9, Rbpj) marker genes (Figure 4A and Supplemental Figure S7B; data not shown for Rbpj). Thus we compared the expression of both between WT and Atf3–/– tissue during CIP (Figure 4A). Comparison of gene expression by quantitative reverse transcription-PCR (qRT-PCR) showed a significant decrease in Nr5a2 expression in WT tissue 4 h into CIP (Figure 4A), which did not reach significance in Atf3–/– mice, although a distinct trend was observed. In agreement with the RNA-seq data, significant differences in Sox9 were observed only in WT tissue (Figure 4A), suggesting that ATF3 is required for up-regulation of factors that promote acinar cell dedifferentiation. Examination of other markers of ADM identified a similar increase in Krt19 (Figure 4B), but not Pdx1 (Supplemental Figure S5A), only in WT tissue. By 72 h into CIP, a time consistent with pancreatic regeneration, both Sox9 (Figure 4C) and Pdx1 (Supplemental Figure S5B) expression are increased only in WT mice, whereas Nr5a2 showed no difference in expression between saline- and CIP-treated tissue from either genotype. We next assessed Rbpj and Rbpjl, which are cofactors for Ptf1a and are expressed in progenitor and mature acinar cells, respectively (Masui et al., 2010). Rbpj and Rbpjl were quantified relative to Mrpl, an internal control gene that does not change during pancreatitis, and then presented as a ratio of Rbpj to Rbpjl. The ratio of Rbpj to Rbpjl, indicating a switch from mature to progenitor cells, was significantly increased 4 h into CIP for both genotypes but only in WT mice 72 h after initiating CIP (Figure 4D), supporting a requirement for ATF3 in promoting acinar cell dedifferentiation.
FIGURE 4:
Atf3–/– mice show maintained expression of differentiation factors during CIP.
(A) qRT-PCR analysis and ChIP- and RNA-sequencing gene tracks for Nr5a2 or Sox9 4 h into saline (SAL) or cerulein (CIP) treatment in wild-type (WT; n = 4 [SAL] or 5 [CIP]) and Atf3–/– tissue (n = 6; *p < 0.05; **p < 0.01). Called peaks of ATF3 enrichment are marked by asterisks. Similar qRT-PCR analysis for Krt19 at 4 h (B) or Sox9 and Nr5a2 at 72 h (C) after initiating CIP. n = 4 or 5; *p < 0.05. (D) Ratio of Rbpj:Rbpjl (relative to Mrpl) based on qRT-PCR analysis 4 or 72 h after initiating CIP. Letters indicate statistically different values (n = 4 or 5; p < 0.05). (E) Representative immunofluorescence analysis for MIST1 in saline or CIP-treated wild type (WT) and Atf3–/– mice 4 h into cerulein treatment show maintained MIST1 expression only in Atf3–/– tissue. Bar, 20 μm. (F) Representative IF for MIST1 or ATF3 on serial sections from pancreatic tissue 28 d after chronic CIP injections. Pockets of ATF3-positive/MIST1-negative cells are outlines by the dotted line. Cell nuclei are stained by DAPI. Bars, 70 μm.
During pancreatitis, acinar cell dedifferentiation involves down-regulation of factors that promote a mature acinar cell phenotype, including the transcription factor MIST1 (Karki et al., 2015). We previously showed that MIST1 is required for complete acinar cell maturation (Pin et al., 2001), and others showed the requirement for Mist1 repression during ADM after CIP (Karki et al., 2015). To determine whether ATF3 plays a role in regulating MIST1 expression, we examined the expression of MIST1 in WT and Atf3–/– tissue after CIP. IF analysis (Figure 4E) confirmed MIST1 protein accumulation in WT tissue is lost 4 h into cerulein treatment and is present at lower levels at 8 h after initial cerulein injection (Supplemental Figure S6). Similar analysis in Atf3–/– tissue showed maintained MIST1 accumulation at both time points (Figure 4E and Supplemental Figure S6), suggesting that ATF3 is required for reduced MIST1 accumulation observed during CIP (Karki et al., 2015). Further confirmation for a role of ATF3 in reducing MIST1 accumulation was observed through IF analysis of serial sections from pancreatic tissue 28 d after cessation of recurrent CIP treatment. These data showed limited MIST1 accumulation where pockets of ATF3 expression were maintained (Figure 4F), indicating a prolonged role for ATF3 in repressing Mist1 expression after CIP.
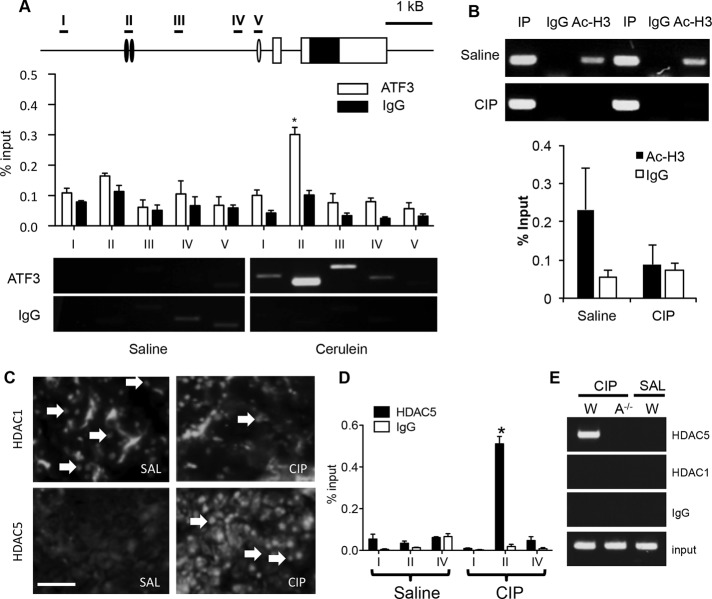
To illustrate a direct transcriptional role for ATF3 in CIP, we next examined ATF3’s ability to directly regulate the Mist1 gene. Mist1 is an attractive target for further analysis since CIP leads to decreased cell differentiation, and MIST1 is an overall regulator of the mature acinar cell phenotype (Pin et al., 2001). Both ChIP-seq and targeted ChIP-PCR confirmed increased ATF3 enrichment upstream of the Mist1 TSS, suggesting that ATF3 directly targets Mist1 early in CIP. ATF3 enrichment was consistently observed ∼2500 base pairs upstream of the Mist1 TSS 4 h into CIP treatment (Figure 5A), whereas saline-treated tissue showed no such enrichment, which is expected given the negligible level of ATF3 accumulation in the absence of CIP. Examination of the sequence 2500 base pairs upstream of the Mist1 TSS identified two putative ATF3-binding sites that are conserved across multiple species (Supplemental Figure S7).
FIGURE 5:
ATF3 represses Mist1 expression in part through recruitment of HDAC5. (A) ChIP-qPCR for ATF3 enrichment along the Mist1 gene shows enrichment specifically at region II (–2500 base pairs relative to the Mist1 TSS) only after induction of CIP (4 h). Representative ChIP-PCR shows ATF3 enrichment at the Mist1 promoter. (B) ChIP-PCR and ChIP-qPCR reveals decreased acetylated H3 (Ac-H3) at the Mist1 promoter 4 h into CIP. Two different saline and CIP samples are shown for ChIP-PCR. (C) IF shows increased nuclear accumulation of HDAC5 at 4 h into CIP (arrows) with no change in HDAC1 accumulation. Bar, 70 μm. (D) ChIP-qPCR for HDAC5 shows enrichment ∼2.5 kb upstream of the Mist1 TSS only after 4 h of CIP treatment. (E) HDAC5 requires ATF3 for recruitment to the Mist1 gene (A–/– = Atf3–/–). *p < 0.05, n = 3 in all cases.
ATF3 is known to repress genes through multiple mechanisms, including the recruitment of repressive histone deacetylases (HDACs) to gene promoters (Li et al., 2010; Zhao et al., 2016). ATF3 recruitment of HDAC1 underlies inhibition of inflammatory gene transcription (Gilchrist et al., 2006; Li et al., 2010), and ATF3 can act as a corepressor in conjunction with HDAC6 (Li et al., 2016). Several HDACs are expressed in pancreatic tissue, and our previous microarray data suggested that HDAC5 might be specifically increased in pancreatic tissue in response to CIP (Kowalik et al., 2007). It is possible that ATF3 recruits HDACs to the Mist1 gene after CIP. In support of this theory, ChIP–quantitative PCR (qPCR) for acetylated H3 showed decreased enrichment of acetylated H3 across the Mist1 promoter after CIP treatment, suggesting ATF3 represses Mist1 expression through histone deacetylation (Figure 5B). To determine which HDACs may contribute to ATF3-mediated repression, we examined the accumulation of several HDACs during CIP treatment. IF analysis 4 h into CIP showed consistent increases in nuclear accumulation for HDAC5 (Figure 5C) and decreased HDAC3 accumulation (unpublished data), with HDAC1 accumulating to a similar extent in both saline- and cerulein-treated mice (Figure 5C). ChIP analysis for HDAC5 revealed enhanced enrichment at the ATF3-binding site within the Mist1 promoter only in CIP-treated chromatin samples, whereas ChIP for HDAC1 showed no enrichment (Figure 5D). HDAC5 recruitment was dependent on ATF3, as no such recruitment was observed in Atf3–/– tissue (Figure 5E).
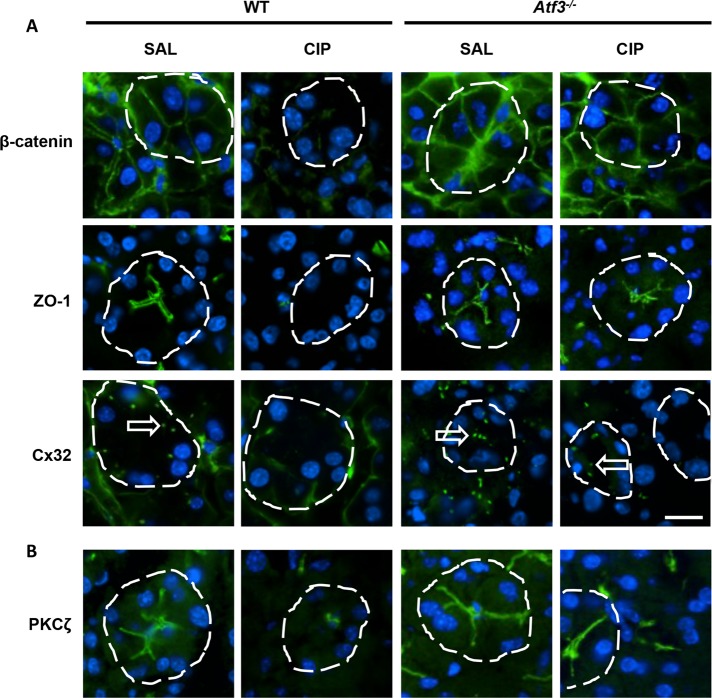
Altering genes that affect ADM in Atf3–/– acinar cells is consistent with the ChIP-seq and RNA-seq analyses, which suggested involvement of ATF3 in regulating genes involved in cell–cell interactions (adherens junctions, tight junctions, etc.). Loss of the mature acinar phenotype, including disruption of cell junctions, is an early event in the progression of pancreatitis (Shi et al., 2013). Therefore we examined accumulation of cell junctional complexes as an indicator of acinar cell maturity (Figure 6). IF for markers of tight junctions (ZO-1), adherens junctions (β-catenin), and gap junctions (Cx32) all showed a reduction in accumulation within 4 h of initial cerulein injections in WT mice (Figure 6A), consistent with previous studies (Lerch et al, 1997; Frossard et al, 2003; Schmitt et al, 2004). Conversely, Atf3–/– acinar cells maintained accumulation of these junctional markers 4 h (Figure 6A) into CIP, suggesting that the mature acinar cell phenotype is maintained in these animals. IF analysis for atypical PKCζ, a marker of cell polarity that regulates junctional formation (Storz, 2015), also revealed maintained accumulation only in Atf3–/– tissue during CIP (Figure 6B). Cell–cell junction organization and cell junction assembly were identified in our GO analysis as targets of ATF3 regulation (Supplemental Table S2). Whereas ChIP-seq showed enrichment for ATF3 at genes encoding Cx32 and ZO-1 (unpublished data), the genes encoding ZO-1, β-catenin, and Cx32 were not significantly reduced during CIP based on RNA-seq data. This suggests that either factors involved in stabilization or targeting of ZO-1, β-catenin, and Cx32 are affected by ATF3 or changes in gene expression occur at later time than 4 h after CIP. However, loss of accumulation for ZO-1, β-catenin, and Cx32 is consistent with previous work and suggests that multiple mechanisms exist to reduce protein levels.
FIGURE 6:
Atf3–/– acinar cells maintain cell junction complexes during CIP. IF for markers of (A) adherens junctions (β-catenin), tight junctions (ZO-1), and gap junctions (Cx32) or (B) cell polarity (PKCζ). Expression and localization is maintained in Atf3–/– acini 4 h into CIP treatment, contrasting the loss of accumulation observed in CIP-treated WT mice. Individual acini are delineated by a dashed line and Cx32 plaques indicated by arrows. Bar, 14 µm.
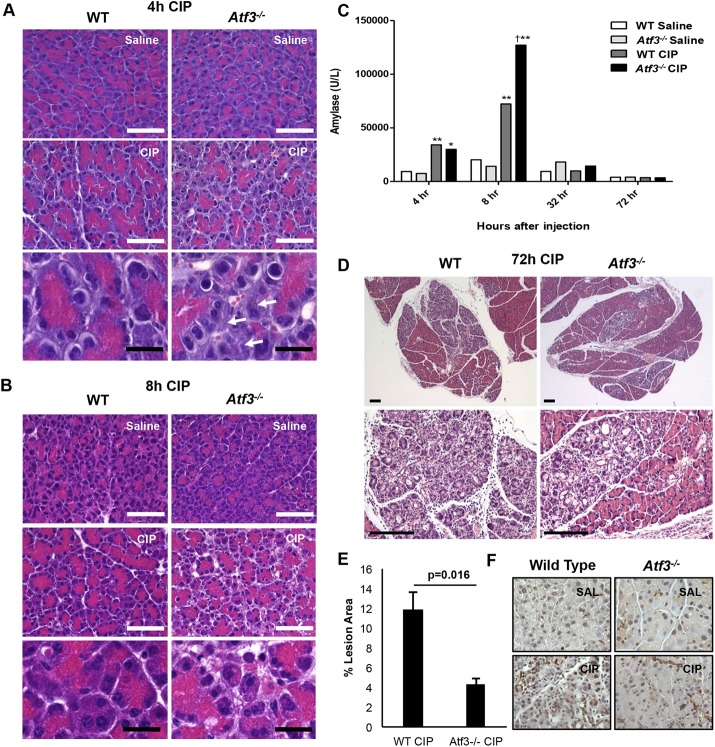
Acinar cell dedifferentiation is believed to be an important step in limiting damage to pancreatic injury. Therefore, on the basis of the foregoing findings, we predicted that Atf3–/– mice would show increased damage to CIP treatment as they maintain the differentiated acinar cell phenotype during injury. Examination of CIP severity through histological analysis confirmed that Atf3–/– mice are more sensitive to cerulein treatment. At both 4 h (Figure 7A) and 8 h (Figure 7B) into cerulein treatment, Atf3–/– mice showed a markedly worse response to CIP, with increased necrosis and edema readily observed relative to WT mice (unpublished data). Histological grading confirmed increased leukocyte infiltration at these time points, and qRT-PCR revealed increased levels of the proinflammatory cytokine interleukin 6 in Atf3–/– animals (Supplemental Figure S8). Elevated serum amylase levels were also observed in Atf3–/– mice 8 h into treatment, consistent with increased tissue damage (Figure 7C). However, histological analysis revealed dramatically different results 72 h into CIP treatment, when WT tissue showed significantly more areas of regeneration, characterized by fibrosis, inflammation (Figure 7, D and E), and ADM based on histology and SOX9 IHC (Figure 7F). Combined, these results suggest that ATF3 protects against pancreatic injury by, in part, targeting Mist1 for repression by HDACs, thereby leading to loss of the mature acinar cell phenotype.
FIGURE 7:
Atf3–/– mice show a differential response to cerulein-induced pancreatitis. H&E histology at 4 h (A) and 8 h (B) into CIP support more damage in the Atf3–/– pancreas based on increased inflammatory cells, necrosis, and apoptosis. Bars, 160 µm (white), 25 µm (black). (C) Serum amylase levels show increased levels in both WT and Atf3–/– tissue 4 and 8 h after initial cerulein treatment; *, p < 0.05 relative to WT saline; **, p < 0.01 relative to WT saline; †, p < 0.05 relative to WT CIP. Representative H&E histology 72 h into CIP (D) shows more damage in the presence of ATF3, as quantified by significantly increased lesion area (E). Bars, 167 µm (small), 150 µm (large). p value represents a two-tailed unpaired t test. (F) Representative IHC images for SOX9 expression show increased SOX9 accumulation in WT tissue relative to Atf3–/– pancreatic tissue. Bars, 100 μm.
DISCUSSION
During pancreatic injury, the acinar cell differentiation program is repressed, with cells taking on a more progenitor-like state in an effort to reduce the amount of tissue damage associated with injury. Whereas several studies highlighted the importance of acinar cell maturation factors in reducing ADM (von Figura et al., 2014; Karki et al., 2015; Martinelli et al., 2015), it is unclear how these factors are repressed during acute pancreatic injury. In this study, we identified ATF3 as an important mediator of gene expression early in CIP, including repressing the differentiated acinar molecular profile. The absence of ATF3 is correlated with maintained expression of differentiated acinar cell gene expression and increased pancreatic damage during early injury. During tissue regeneration, lack of ATF3 results in decreased expression of ADM-promoting factors and a resultant reduction of ADM. On a mechanistic level, ATF3 was enriched at multiple genes that either promote or repress acinar cell differentiation and was required for recruiting HDAC5 to the Mist1 gene, illustrating a direct role for ATF3 in regulating the acinar cell phenotype.
Previous work by several laboratories has shown a rapid onset of ATF3 expression in the pancreas after injury (Hartman et al., 2004; Jiang et al., 2004), likely as part of the UPR/ER stress response pathway. The UPR has a physiological role in the pancreas, likely due to the high level of protein production within acinar cells. Deletion of Perk, Ire1, or Xbp1 has significant consequences on pancreatic tissue (Zhang et al., 2002; Iida et al., 2007; Iwawaki et al., 2010; Hess et al., 2011). However, unlike other key mediators of the UPR in the pancreas, ATF3 expression is limited to injured tissue, and the loss of Atf3 has no observable effect on pancreatic biology or gene expression in the absence of injury. This suggests that ATF3 may distinguish UPR functions between normal acinar cell physiology (maintenance of protein-folding capacity) and pathology (general repression of protein translation and alterations in gene transcription).
One of the earliest responses to experimental models of pancreatitis is the rapid repression of the acinar differentiation program. Repression of the acinar cell phenotype through decreased expression of acinar differentiation factors (Mist1, Rbpjl, Nr5a2) helps reduce tissue damage (Masui et al., 2010; von Figura et al., 2014). We showed a clear role for ATF3 in repression of the acinar phenotype through enrichment of ATF3 at the promoters of the foregoing genes 4 h after injury and, in the case of Mist1, a role for ATF3 in the recruitment of repressive HDACs to the gene. Furthermore, loss of ATF3 expression is correlated to maintained MIST1 expression, and stabilization of the differentiated acinar cell profile evidenced with maintenance of junctional complexes (ZO-1, β catenin, and Cx32) and polarity markers (PKCζ). We show that the maintenance of these factors is correlated with increased damage at early time points in the pancreatitis response, which is consistent with previous findings that maintained MIST1 was correlated with increased severity of CIP (Karki et al., 2015). Surprisingly, none of these genes (Mist1, Cx32, ZO1, and β catenin) was identified by RNA-seq to be significantly decreased in either wild-type or Atf3–/– mice during CIP. However, all of these factors have been identified as repressed by previous studies (Lerch et al., 1997; Frossard et al., 2003; Schmitt et al., 2004; Karki et al., 2015), and each showed a trend toward repression in WT tissue (unpublished data). Therefore we suggest that the differences are likely due to the short time after induction of injury that we examined RNA expression (4 h) compared with these other studies and indicate that multiple transcriptional and posttranslational mechanisms (i.e., protein degradation) may combine to regulate the expression of these factors.
Conversely, ATF3 was also enriched at the promoters of genes that promote acinar cell dedifferentiation, including Sox9, Pdx, and Rbpj, which are all increased in expression during pancreatic injury. Furthermore, Atf3–/– tissue shows reduced expression of these factors during injury, illustrating that ATF3 activates and represses genes regulating the ADM process during CIP. Previous work in pancreatic β-cells showed that ATF3 promotes loss of the mature β-cell phenotype by inhibiting both PDX1 expression and activity (Jang, 2011; Kim et al., 2011), and our ChIP-seq data showed enrichment of ATF3 at the Pdx1 gene (unpublished data). However, this enrichment did not correlate with decreased Pdx1 expression, suggesting that ATF3 may be involved in targeting transcriptional complexes to the genome but has different effects based on the transcriptional complex to which it is associated. Whereas ATF3 interacts with a variety of transcription factors and recruits multiple HDACs (this study; Li et al., 2010; Zhao et al., 2016), it can also interact with NFκB (Kwon et al., 2015), CEBP (Jang et al., 2012), PDX1 (Jang, 2011), and other bZIP transcription factors (Thompson et al., 2009). This promiscuity suggests a multifaceted role for ATF3 in the transcriptional profile of the cell. Our findings that ATF3 is enriched within 5 kb of >30% of all the genes that show greater than twofold increase in expression indicates that ATF3 is a crucial mediator of gene transcription during injury, affecting multiple cellular pathways in addition to its effects on acinar cell differentiation.
Although loss of the acinar cell phenotype may initially be protective, ATF3-mediated ADM could promote development of PDAC because enhanced ADM leads to increased sensitivity to oncogenic KRAS induction of PDAC (Logsdon and Ji, 2009; Krah et al., 2011; Shi et al., 2013). Combined with the knowledge that maintained expression of MIST1 prevents KRAS-driven PanIN formation (Shi, 2009), we postulate that ATF3-mediated repression of Mist1 may promote susceptibility to PDAC. Analysis of our RNA-seq data indicates loss of ATF3 affects many cancer initiating pathways, including EGF receptor signaling, MAPK signaling, and the Ras pathway. Therefore repression of ATF3 during pancreatitis may reduce ADM and decrease susceptibility to PDAC. We are generating Atf3–/– mice in an oncogenic KRAS background to test this possibility.
In conclusion, we identified ATF3 as an important transcriptional regulator of pancreatic acinar cells during the response to cerulein-induced pancreatic injury. Our data show that ATF3 regulates multiple gene networks and cellular functions, including a primary role in promoting the loss of the acinar cell phenotype through activating and repressive transcriptional mechanisms. Specifically, we show that ATF3 is required for HDAC recruitment to repress transcription at the promoter for the acinar cell maturation factor, MIST1. Although promoting a more progenitor-like cell phenotype in response to injury will limit the amount of tissue damage, it is unclear whether long-term activation of ATF3 will allow for a greater sensitivity to factors that promote PDAC.
MATERIALS AND METHODS
Animals and induction of cerulein-induced pancreatitis
Mouse experiments followed guidelines in protocol 2008-116 approved by the Animal Care and Use Committee at the University of Western Ontario. Pancreatitis was induced in 2- to 4-mo-old C57Bl6 mice and congenic mice containing homozygous deletion of Atf3 (Atf3–/–; Hartman et al., 2004). Atf3–/– mice were kindly provided by Tsonwin Hai, The Ohio State University. Mice were given up to eight hourly intraperitoneal injections of cerulein (Sigma-Aldrich, St. Louis, MO; 50 µg/kg of body weight), a cholecystokinin analogue, to induce mild edematous, acute pancreatitis, as previously described (Johnson et al., 2009). Animals were killed 1–72 h after initiation of pancreatitis. Blood was obtained through cardiac puncture for assessment of serum amylase as described (Kowalik et al., 2007).
Protein isolation and Western blotting
Pancreatic protein was isolated as described (Kowalik et al., 2007) and quantified using a Bio-Rad protein assay (Bio-Rad, Hercules, CA). Isolated protein was resolved by SDS–PAGE and transferred to polyvinylidene fluoride membrane (Bio-Rad). Western blot analysis was carried out as described (Fazio et al., 2011) using antibodies specific for ATF3 (Santa Cruz Biotechnology, Dallas, TX), sXBP1 (BioLegend, San Diego, CA), MIST1 (Pin et al, 2000), and total ERK (Cell Signaling Technology, Danvers, MA). Blots were visualized using the VersaDoc Imaging System with Quantity One 1-D Analysis software (Bio-Rad).
RNA isolation, qRT-PCR, and RNA-seq
RNA was isolated from mouse pancreatic tissue using TRIZOL (Invitrogen, Carlsbad, CA) as previously described (Johnson et al., 2009). cDNA was synthesized using ImPromII Reverse transcriptase (Sigma-Aldrich). PCR was carried out using GoTaq flexi DNA polymerase (Sigma-Aldrich) and gene-specific primer sequences (Supplemental Table S1). qRT-PCR was carried out using the GoTaq PCR Mastermix system (Promega, Madison, WI) using an ABI Prism 7900HT Sequence Detection System and corresponding SDS 2.2.1 software (Applied Biosystems, Foster City, CA). The quality and quantity of RNA samples were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Total RNA was used to construct a stranded, rRNA-depleted RNA library, followed by next generation sequencing using the Illumina HiSeq 2500 with 100–base pair paired-end reads (Centre for Applied Genomics, Toronto, Canada). The complete RNA-seq data can be found at Gene Expression Omnibus (GEO) accession (pending).
RNA-seq reads were aligned to the mouse mm10 genome (mm10_v6) with Gencode M9 annotations using STARv2.4.2a (Dobin et al., 2013); notable options included –outFilterScoreMinOverLread 0.55, –outFilterMatchNminOverLread 0.55. STARv2.4.2a was also used to generate .wig files for visualization in the UCSC Genome Browser. Aligned reads were sorted with samtools (Li et al., 2009; Li, 2011), and gene counts were done with HTSeq (Anders et al., 2015) using the htseq-count function and Gencode M9 annotations. Differential expression analysis was derived with the DESeq2 (Love et al., 2014) package. All differential expression data used for downstream analysis were cut off at an adjusted p value of 0.01 (p-adj < 0.01). GO (Ashburner et al., 2000; The Gene Ontology Consortium, 2015) terms and pathways were derived via PANTHER (Thomas et al., 2003; Mi and Thomas, 2009; Mi et al., 2017), whereas KEGG pathways were derived from DAVID (Huang et al., 2009a,b).
ChIP PCR, ChIP-seq, and bioinformatics analysis
Chromatin was isolated from pancreatic tissue of mice treated for 4 h with cerulein as described (Fazio et al., 2011; Mehmood et al., 2014). Purified antibodies against ATF3 (1:500, Santa Cruz Biotechnology), HDAC1 (1/250; Active Motif, Carlsbad, CA), HDAC5 (Cell Signaling Technology), and acetylated H3 (Millipore, Etobicoke, Canada) were used for ChIP and qPCR performed using the GoTaq PCR Mastermix system. Average Ct values for individual ChIP and immunoglobulin G controls were expressed as a percentage of starting chromatin samples (input). ATF3 ChIP was followed by next-generation sequencing (ChIP-seq). Chromatin was pooled from whole pancreatic tissue of three mice, and ChIP was carried out as described (Fazio et al., 2011; Mehmood et al., 2014). Enriched pooled DNA fragments from ChIP reactions from three biological replicates per group were sequenced using the Illumina 2.0 genome analyzer (Beijing Genomics Institute, Hong Kong). The complete ChIP-seq data can be found at GEO accession GSE60250.
ChIP-seq reads were aligned to mouse genome mm10 using Bowtie2 (Langmead and Salzberg, 2012) with default settings. Peaks were called using MACS2 (Zhang et al., 2008) with p = 0.0001. Custom Perl scripts were used to generate .wig files for visualization in the UCSC Genome Browser and to annotate peaks with gene features using Gencode M9 annotations. Heat maps and density profiles were made using deepTools (Ramirez et al., 2014). Random positions were generated using Bedtools shuffle (Quinlan and Hall, 2010). DNA sequence motifs were identified using MEME-ChIP (Machanick and Bailey, 2011) with the -nmeme option increased to 5000: motifs were discovered using MEME and then passed to Centrimo to identify centrally enriched motifs and to TOMTOM to identify putative binding proteins from the Jaspar Core (2016) vertebrate database (Mathelier et al., 2016).
Histological analysis and serum amylase assay
Mouse pancreata were immediately frozen in cryomatrix and sectioned at 6 µm. Cryostat sections were processed for IF as described (Fazio et al., 2005). For paraffin expression, mouse pancreata were fixed in 4% Formalin overnight at 4°C and washed with phosphate-buffered saline several times before paraffin embedding, followed by sectioning at 5 µm.
Paraffin sections of mouse pancreatic tissue were stained with hematoxylin and eosin (H&E) and imaged using light microscopy. Sections from WT and Atf3–/– mice 72 h after initiation of CIP were assayed for the presence of lesions and ADM. Total lesion area was calculated as a percentage of the total pancreatic tissue area. For both lesion and ADM analysis, three different sections were assayed from each individual pancreas to produce an average value. Primary antibodies used for IF included those specific for MIST1 (1:500), ATF3 (1:500), β-catenin (1:2000; Sigma-Aldrich), ZO-1 (1:100; Chemicon, Temecula, CA), Cx32 (1:100; Chemicon), PKCζ (1/500; Sigma-Aldrich), HDAC1 (1/250; Active Motif), HDAC3 (1:500; Abcam, Cambridge, MA), and HDAC5 (1/250; Cell Signaling Technology), and SOX9 expression (1:500; Abcam) was determined by IHC. Secondary antibodies included anti-mouse fluorescein isothiocyanate (FITC) and anti-rabbit FITC (1:250, Jackson ImmunoResearch, West Grove, PA). Tissue sections were counterstained with eosin upon completion of immunohistochemical staining or 4′,6-diamidino-2-phenylindole (DAPI) after IF.
Statistical analysis
Differences between experimental groups were analyzed using two-way analysis of variance, followed by a Bonferroni post hoc test. All qRT-PCR results were analyzed using unpaired t tests, with the exception of the Rbpj and Rbpjl comparisons. Significance is considered as p < 0.05.
Supplementary Material
Acknowledgments
We thank Fred Dick, Gabriel DiMattia, and Martin Fernandez-Zapico for thoughtful insight and suggestions on the study. E.F. was supported by a Canadian Institutes of Health Research/Canadian Digestive Health Foundation Doctoral Award and an Ontario Graduate Scholarship, C.Y. was supported by a Canadian Institutes of Health Research Canada Graduate Scholarship, J.T. was supported by an Ontario Graduate Scholarship, and R.M., M.L., and W.M. were supported by Epigenetics Fellowships from the Children’s Health Research Institute. The work would not have been possible without an operating grant from the Canadian Institute of Health Research and funds from the Children’s Health Foundation, the Lawson Health Research Institute, and the Cancer Research and Technology Transfer Program. This work was made possible by the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET; www.sharcnet.ca) and Compute/Calcul Canada.
Abbreviations used:
- ADM
acinar-to-ductal metaplasia
- ATF
activating transcription factor
- ChIP
chromatin immunoprecipitation
- CIP
cerulein-induced pancreatitis
- HDAC
histone deacetylase
- PDAC
pancreatic ductal adenocarcinoma
- TES
transcriptional end site
- TSS
transcriptional start site.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-04-0254) on July 12, 2017.
REFERENCES
- Aghdassi AA, Mayerle J, Christochowitz S, Weiss FU, Sendler M, Lerch MM. Animal models for investigating chronic pancreatitis. Fibrogenesis Tissue Repair. 2011;4:26. doi: 10.1186/1755-1536-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alahari S, Mehmood R, Johnson CL, Pin CL. The absence of MIST1 leads to increased ethanol sensitivity and decreased activity of the unfolded protein response in mouse pancreatic acinar cells. PLoS One. 2011;6:e28863. doi: 10.1371/journal.pone.0028863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen-Jennings AE, Hartman MG, Kociba GJ, Hai T. The roles of ATF3 in glucose homeostasis. A transgenic mouse model with liver dysfunction and defects in endocrine pancreas. J Biol Chem. 2001;276:29507–29514. doi: 10.1074/jbc.M100986200. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardito CM, Gruner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, Delgiorno KE, Carpenter ES, Halbrook CJ, Hall JC, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S. Intracellular signaling by the unfolded protein response. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chen BPC, Liang G, Whelan J, Hai T. ATF3 and ATF3ΔZip. Transcriptional repression versus activation by alternatively spliced isoforms. J Biol Chem. 1994;269:15819–15826. [PubMed] [Google Scholar]
- Collares-Buzato CB, Carvalho CPF, Furtado AG, Boschero AC. Upregulation of the expression of tight and adherens junction-associated proteins during maturation of neonatal pancreatic islets in vitro. J Mol Histol. 2004;35:811–822. doi: 10.1007/s10735-004-1746-0. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkan M, Adler G, Apte MV, Bachem MG, Buchholz M, Detlefsen S, Esposito I, Friess H, Gress TM, Habisch HJ, et al. StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut. 2012;61:172–178. doi: 10.1136/gutjnl-2011-301220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazio EN, Dimattia GE, Chadi SA, Kernohan KD, Pin CL. Stanniocalcin 2 alters PERK signalling and reduces cellular injury during cerulein induced pancreatitis in mice. BMC Cell Biol. 2011;12:17. doi: 10.1186/1471-2121-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazio EN, Everest M, Colman R, Wang R, Pin CL. Altered Glut-2 accumulation and beta-cell function in mice lacking the exocrine-specific transcription factor, Mist1. J Endocrinol. 2005;187:407–418. doi: 10.1677/joe.1.06376. [DOI] [PubMed] [Google Scholar]
- Frossard JL, Rubbia-Brandt L, Wallig MA, Benathan M, Ott T, Morel P, Hadengue A, Suter S, Willecke K, Chanson M. Severe acute pancreatitis and reduced acinar cell apoptosis in the exocrine pancreas of mice deficient for the Cx32 gene. Gastroenterology. 2003;124:481–493. doi: 10.1053/gast.2003.50052. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;44:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- Gomez G, Englander EW, Wang G, Greeley GH. Increased expression of hypoxia-inducible factor-1alpha, p48, and the Notch signaling cascade during acute pancreatitis in mice. Pancreas. 2004;28:58–64. doi: 10.1097/00006676-200401000-00009. [DOI] [PubMed] [Google Scholar]
- Grady T, Mah’Moud M, Otani T, Rhee S, Lerch MM, Gorelick FS. Zymogen proteolysis within the pancreatic acinar cell is associated with cellular injury. Am J Physiol Gastrointest Liver Physiol. 1998;275:G1010–G1017. doi: 10.1152/ajpgi.1998.275.5.G1010. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Hartman MG, Lu D, Kim ML, Kociba GJ, Shukri T, Buteau J, Wang X, Frankel WL, Guttridge D, Prentki M, et al. Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol Cell Biol. 2004;24:5721–5732. doi: 10.1128/MCB.24.13.5721-5732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DA, Humphrey SE, Ishibashi J, Damsz B, Lee A-H, Glimcher LH, Konieczny SF. Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology. 2011;141:1463–1472. doi: 10.1053/j.gastro.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Li Y, McGrath BC, Frank A, Cavener DR. PERK eIF2 alpha kinase is required to regulate the viability of the exocrine pancreas in mice. BMC Cell Biol. 2007;8:38. doi: 10.1186/1471-2121-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K. IRE1α disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS One. 2010;5:e13052. doi: 10.1371/journal.pone.0013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK. ATF3 represses PDX-1 expression in pancreatic β-cells. Biochem. Biophys Res Commun. 2011;412:385–390. doi: 10.1016/j.bbrc.2011.07.108. [DOI] [PubMed] [Google Scholar]
- Jang MK, Kim CH, Seong JK, Jung MH. ATF3 inhibits adipocyte differentiation of 3T3-L1 cells. Biochem Biophys Res Commun. 2012;421:38–43. doi: 10.1016/j.bbrc.2012.03.104. [DOI] [PubMed] [Google Scholar]
- Jiang H-Y, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, Wang X, Ron D, Cavener DR, Wek RC. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol Cell Biol. 2004;24:1365–1377. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Weston JY, Chadi SA, Fazio EN, Huff MW, Kharitonenkov A, Köester A, Pin CL. Fibroblast growth factor 21 reduces the severity of cerulein-induced pancreatitis in mice. Gastroenterology. 2009;137:1795–1804. doi: 10.1053/j.gastro.2009.07.064. [DOI] [PubMed] [Google Scholar]
- Karki A, Humphrey SE, Steele RE, Hess DA, Taparowsky EJ, Konieczny SF. Silencing Mist1 gene expression is essential for recovery from acute pancreatitis. PLoS One. 2015;10:e0145724. doi: 10.1371/journal.pone.0145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W-H, Jang MK, Kim CH, Shin HK, Jung MH. ATF3 inhibits PDX-1-stimulated transactivation. Biochem Biophys Res Commun. 2011;414:681–687. doi: 10.1016/j.bbrc.2011.09.132. [DOI] [PubMed] [Google Scholar]
- Kowalik AS, Johnson CL, Chadi SA, Weston JY, Fazio EN, Pin CL. Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1123–G1132. doi: 10.1152/ajpgi.00512.2006. [DOI] [PubMed] [Google Scholar]
- Krah NM, De La O JP, Swift GH, Hoang CQ, Willet SG, Chen Pan F, Cash GM, Bronner MP, Wright CV, MacDonald RJ, Murtaugh LC. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. Elife. 2015;4:e07125. doi: 10.7554/eLife.07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger B, Albrecht E, Lerch MM. The role of intracellular calcium signaling in premature protease activation and the onset of pancreatitis. Am J Pathol. 2000;157:43–50. doi: 10.1016/S0002-9440(10)64515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubisch CH, Logsdon CD. Secretagogues differentially activate endoplasmic reticulum stress responses in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1804–G1812. doi: 10.1152/ajpgi.00078.2007. [DOI] [PubMed] [Google Scholar]
- Kubisch CH, Sans MD, Arumugam T, Ernst SA, Williams JA, Logsdon CD. Early activation of endoplasmic reticulum stress is associated with arginine-induced acute pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2006;291:G238–G245. doi: 10.1152/ajpgi.00471.2005. [DOI] [PubMed] [Google Scholar]
- Kwon J-W, Kwon H-K, Shin H-J, Choi Y-M, Anwar MA, Choi S. Activating transcription factor 3 represses inflammatory responses by binding to the p65 subunit of NF-κB. Sci Rep. 2015;5:14470. doi: 10.1038/srep14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch MM, Lutz MP, Weidenbach H, Müller-Pillasch F, Gress TM, Leser J, Adler G. Dissociation and reassembly of adherens junctions during experimental acute pancreatitis. Gastroenterology. 1997;113:1355–1366. doi: 10.1053/gast.1997.v113.pm9322531. [DOI] [PubMed] [Google Scholar]
- Li C, Zhou Y, Loberg A, Tahara SM, Malik P, Kalra VK. Activated transcription factor 3 in association with histone deacetylase 6 negatively regulates microRNA 199a2 transcription by chromatin remodeling and reduces endothelin-1 expression. Mol Cell Biol. 2016;36:2838–2854. doi: 10.1128/MCB.00345-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-F, Cheng C-F, Liao W-J, Lin H, Yang R-B. ATF3-mediated epigenetic regulation protects against acute kidney injury. J Am Soc Nephrol. 2010;21:1003–1013. doi: 10.1681/ASN.2009070690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon CD, Ji B. Ras activity in acinar cells links chronic pancreatitis and pancreatic cancer. Clin Gastroenterol Hepatol. 2009;7:S40–S43. doi: 10.1016/j.cgh.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon CD, Lu W. The significance of Ras activity in pancreatic cancer initiation. Int J Biol Sci. 2016;12:338–346. doi: 10.7150/ijbs.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugea A, Tischler D, Nguyen J, Gong J, Gukovksy I, French SW, Gorelick FS, Pandol SJ. Adaptive unfolded protein response attenuates alcohol-induced pancreatic damage. Gastroenterology. 2011;140:987–997. doi: 10.1053/j.gastro.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas NL, Diehl JA. Molecular pathways: the PERKs and pitfalls of targeting the unfolded protein response in cancer. Clin Cancer Res. 2015;21:675–679. doi: 10.1158/1078-0432.CCR-13-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli P, Madriles F, Cañamero M, Pau ECS, Pozo ND, Guerra C, Real FX. The acinar regulator Gata6 suppresses KrasG12V-driven pancreatic tumorigenesis in mice. Gut. 2015;65:476–486. doi: 10.1136/gutjnl-2014-308042. [DOI] [PubMed] [Google Scholar]
- Masui T, Swift GH, Deering T, Shen C, Coats WS, Long Q, Elsässer H, Magnuson MA, MacDonald RJ. Replacement of Rbpj with Rbpjl in the ptf1 complex controls the final maturation of pancreatic acinar cells. Gastroenterology. 2010;139:270–280. doi: 10.1053/j.gastro.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas A, Chen C, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood R, Varga G, Mohanty SQ, Laing SW, Lu Y, Johnson CL, Kharitonenkov A, Pin CL. Epigenetic reprogramming in Mist1−/− mice predicts the molecular response to cerulein-induced pancreatitis. PLoS One. 2014;9:e84182. doi: 10.1371/journal.pone.0084182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Thomas P. Protein networks and pathway analysis. Methods Mol Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JP, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin CL, Bonvissuto AC, Konieczny SF. Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. Anat Rec. 2000;259:157–167. doi: 10.1002/(SICI)1097-0185(20000601)259:2<157::AID-AR6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Pin CL, Rukstalis JM, Johnson C, Konieczny SF. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol. 2001;155:519–530. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GH. Pancreatitis. J Contin Educ Top Issues. 2015;17:4–11. [Google Scholar]
- Ron D, Bertolotti A, Zhang Y, Hendershot LM, Harding HP. Dynamic interaction of BiP and ER stress transducers in the unfolded-proteinresponse. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Sah RP, Garg SK, Dixit AK, Dudeja V, Dawra RK, Saluja AK. Endoplasmic reticulum stress is chronically activated in chronic pancreatitis. J Biol Chem. 2014;289:27551–27561. doi: 10.1074/jbc.M113.528174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M, Klonowski-Stumpe H, Eckert M, Lüthen R, Häussinger D. Disruption of paracellular sealing is an early event in acute caerulein-pancreatitis. Pancreas. 2004;28:181–190. doi: 10.1097/00006676-200403000-00010. [DOI] [PubMed] [Google Scholar]
- Shi G. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology. 2009;136:1368–1378. doi: 10.1053/j.gastro.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, DiRenzo D, Qu C, Barney D, Miley D, Konieczny SF. Maintenance of acinar cell organization is critical to preventing Kras-induced acinar-ductal metaplasia. Oncogene. 2013;32:1950–1958. doi: 10.1038/onc.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P. Targeting protein kinase C subtypes in pancreatic cancer. Expert Rev Anticancer Ther. 2015;15:433–438. doi: 10.1586/14737140.2015.1003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MR, Xu D, Williams BRG. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl) 2009;87:1053–1060. doi: 10.1007/s00109-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Figura G, Morris JP, Wright CVE, Hebrok M. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut. 2014;63:656–664. doi: 10.1136/gutjnl-2012-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD-Y, Kaufman RJ. ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252–1261. doi: 10.1053/j.gastro.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Li X, Guo M, Yu J, Yan C. The common stress responsive transcription factor ATF3 binds genomic sites enriched with p300 and H3K27ac for transcriptional regulation. BMC Genomics. 2016;17:335. doi: 10.1186/s12864-016-2664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.