A transposon event, resulting in partial suppression of a Chlamydomonas IFT46 null mutant, allowed the function of the N-terminus of IFT46 in flagellar assembly to be explored. The IFT46 N-terminus is not required for IFT complex assembly but is required for transport of outer arm dynein and its adaptor, ODA16, into the flagellum.
Abstract
Cilia are assembled via intraflagellar transport (IFT). The IFT machinery is composed of motors and multisubunit particles, termed IFT-A and IFT-B, that carry cargo into the cilium. Knowledge of how the IFT subunits interact with their cargo is of critical importance for understanding how the unique ciliary domain is established. We previously reported a Chlamydomonas mutant, ift46-1, that fails to express the IFT-B protein IFT46, has greatly reduced levels of other IFT-B proteins, and assembles only very short flagella. A spontaneous suppression of ift46-1 restored IFT-B levels and enabled growth of longer flagella, but the flagella lacked outer dynein arms. Here we show that the suppression is due to insertion of the transposon MRC1 into the ift46-1 allele, causing the expression of a fusion protein including the IFT46 C-terminal 240 amino acids. The IFT46 C-terminus can assemble into and stabilize IFT-B but does not support transport of outer arm dynein into flagella. ODA16, a cargo adaptor specific for outer arm dynein, also fails to be imported into the flagella in the absence of the IFT46 N-terminus. We conclude that the IFT46 N-terminus, ODA16, and outer arm dynein interact for IFT of the latter.
INTRODUCTION
Cilia and flagella (terms used interchangeably here) are microtubule-based organelles that extend from the cell surface into the environment. They are important for cell motility, for cells to sense their environment, and for signal transduction. Defects in ciliary structure or signaling cause a large number of human diseases, collectively called ciliopathies (Mitchison and Valente, 2017).
Ciliary assembly and signaling both depend on a highly conserved process known as intraflagellar transport (IFT; Kozminski et al., 1993; Rosenbaum and Witman, 2002; Lechtreck, 2015; Taschner and Lorentzen, 2016). IFT is the movement of large particles, termed IFT trains, from the base to the tip of the flagellum (anterograde IFT) and back to the base (retrograde IFT). Anterograde IFT is powered by kinesin motors (Kozminski et al., 1995; Snow et al., 2004; Verhey et al., 2011), while retrograde IFT is driven by cytoplasmic dynein 2/1b (Pazour et al., 1998, 1999; Porter et al., 1999; Hou and Witman, 2015). The trains are composed primarily of two complexes, IFT-A and IFT-B, the latter of which can be subdivided into IFT-B1 and IFT-B2 (Table 1) (Cole et al., 1998; Lechtreck, 2015; Taschner and Lorentzen, 2016). The trains transport structural components of the axoneme to the tip of the flagellum, where axonemal assembly occurs, and return the products of axonemal turnover to the cell body (Qin et al., 2004; Ishikawa et al., 2014; Lechtreck, 2015). IFT also transports signals and signaling proteins between cilia and the cell body in several pathways, including mating in Chlamydomonas (Wang et al., 2006) and Hedgehog, Wnt, transforming factor β, receptor tyrosine kinase, and Notch signaling in vertebrates (Mourão et al., 2016).
TABLE 1:
IFT-particle proteins.
| IFT-B | ||
|---|---|---|
| IFT-A | IFT-B1 | IFT-B2 |
| IFT144 | IFT88 | IFT172 |
| IFT140 | IFT81 | IFT80 |
| IFT139 | IFT74/72 | IFT57 |
| IFT122 | IFT70 | IFT54 |
| IFT121 | IFT56 | IFT38 |
| IFT43 | IFT52 | IFT20 |
| IFT46 | ||
| IFT27 | ||
| IFT25 | ||
| IFT22 | ||
With the exception of tubulin (Bhogaraju et al., 2013; Craft et al., 2015; Kubo et al., 2016), little is known about how IFT particles interact with their cargoes of axonemal precursors. Two highly conserved proteins that are providing a paradigm for such interactions are ODA16 and IFT46.
ODA16, the crystal structure of which was recently determined (Taschner et al., 2017), is proposed to be an IFT cargo adaptor. Chlamydomonas insertional mutants null for ODA16 assemble flagella, but these flagella have greatly reduced numbers of outer dynein arms (Ahmed and Mitchell, 2005). Because the outer dynein arms generate much of the force for flagellar bending, cells lacking these arms swim slowly. Outer arm dynein in the oda16 mutant is preassembled in the cell cytoplasm as in wild-type cells and is competent to bind to axonemes in vitro, and isolated oda16 axonemes are capable of binding outer arm dynein from wild-type cells, indicating that the underlying cause of the defect is failure to transport the dynein into the flagellum (Ahmed et al., 2008). ODA16 has a cellular localization similar to that of IFT proteins and interacts with IFT46, leading to the hypothesis that ODA16 functions as a cargo-specific adaptor that facilitates IFT of outer arm dynein (Ahmed et al., 2008; Taschner et al., 2017).
IFT46 is a subunit of IFT-B1, also known as the IFT-B core (Lucker et al., 2005; Taschner et al., 2016). IFT46 has no known domains or motifs; however, the N-terminal region (corresponding to Chlamydomonas aa 1–101) is much less well conserved than most of the rest of the protein (Hou et al., 2007; Taschner et al., 2017). Two Chlamydomonas insertional mutants null for IFT46 have been identified: in both ift46-1, previously described by us, and ift46-2, described by Lucker and colleagues, the cells assemble only very short nonmotile flagella (Hou et al., 2007; Lucker et al., 2010). In ift46-1, loss of IFT46 is correlated with a severe reduction in the levels of other IFT-B proteins, indicating that IFT46 is essential for stability of IFT-B and explaining the flagellar assembly defect (Hou et al., 2007). Importantly, we also identified a strain in which the ift46-1 mutation is suppressed. In this strain, termed Supift461, some cells assemble longer flagella when grown under stress conditions, but axonemes of these flagella have a severe deficiency of outer dynein arms, and the cells swim slowly. No IFT46 could be detected in Supift461 cells with the anti-IFT46 antibody then available. However, reverse transcription PCR showed that the 3′ end of the IFT46 gene is transcribed in Supift461 cells but not in ift46-1 cells (Hou et al., 2007). This raised the possibility that an intragenic mutation occurred in Supift461 that allows expression of the 3′ end of the IFT46 gene, which then leads to the suppression.
Here we identify the genomic basis for this suppression and demonstrate that the change results in expression of a fusion protein in which the N-terminal 104 amino acids of IFT46 are replaced by 10 amino acids derived from a sequence of an MRC1 retroposon that inserted into the ift46-1 allele. This protein assembles into and stabilizes IFT-B. We have recapitulated the suppression by transforming ift46-1 with a construct expressing a similarly truncated protein containing only IFT46 aa 106–344 that also stabilizes IFT-B and supports better flagellar growth under stress. Outer arm dynein in this strain is competent to bind to axonemes, but its transport into the flagellum is largely curtailed. We further find that the N-terminus of IFT46 is crucial for transport of ODA16 into the flagellum. The results establish a model for how an IFT-particle protein links to a major axonemal cargo to establish the unique ciliary protein composition. Finally, we explore the requirement for stress to enable flagellar assembly when IFT-B is defective.
RESULTS
A transposon in the Supift461 allele enables expression of a truncated IFT46 protein that supports flagellar assembly
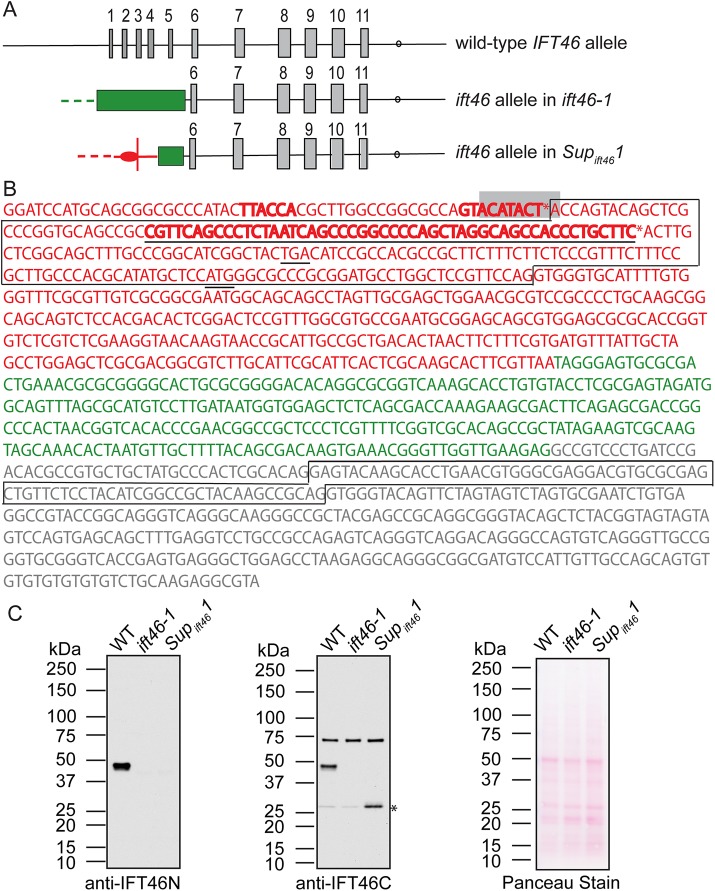
To determine the genomic basis for the suppression of ift46-1, we cloned and sequenced the ift46 alleles in ift46-1 and Supift461. In ift46-1, the NIT1 sequence originally used to create ift46-1 by insertional mutagenesis (Hou et al., 2007) was found inserted into the fifth intron of the IFT46 gene (Figure 1A). In Supift461, MRC1 (miniature retrotransposon of Chlamydomonas) sequence was found inserted into the exogenous NIT1 sequence (Figure 1A). To see whether this change at the genomic level caused the transcription of the 3′ end of the IFT46 gene, we cloned the 5′ end of the transcript by using 5′ rapid amplification of cDNA ends (RACE). Two cDNAs were identified that differed in their 5′ untranslated regions (UTRs); one clone was 78 base pairs longer than the other. The 5′UTR and first exon of these cDNAs are from MRC1, and the second, third, and subsequent exons are from the sixth, seventh, and subsequent exons, respectively, of the IFT46 gene (Figure 1, A and B). This result shows that the insertion of the transposon MRC1 into the ift46-1 allele in Supift461 causes expression of hybrid RNAs combining the MRC1 and IFT46 sequences.
FIGURE 1:
A transposon inserted into the ift46-1 allele allows expression of the IFT46 C-terminus. (A) Diagram of the IFT46 gene in wild-type, ift46-1, and Supift461 as revealed by sequencing of genomic DNA and cDNAs. Exons are shown as tall boxes and introns as lines; the polyadenylation site is indicated by a circle. The ift46-1 allele was generated by insertion of the NIT1 gene (green box) into intron 5 of IFT46. The Supift461 allele resulted from insertion of the MRC1 transposon (red) into the exogenous NIT1 gene in ift46-1. After transcription and splicing, a hybrid RNA is formed in which the 5′ UTR (oval) and first exon are from MRC1 and subsequent exons are from IFT46 exons 6, 7, 8, 9, 10, and 11 (see B). Dashes indicate regions not sequenced. (B) Sequence of the genomic region where the transposon MRC1 (red) is inserted into the exogenous NIT1 gene (green) just upstream of exon 6 in the IFT46 gene (gray) in Supift461. Sequence found in cDNAs is boxed. The 5′ RACE yielded cDNAs with two different start sites (asterisks); one is immediately downstream of predicted bacterial promoter regions (bold), one of which is the known bacterial transcription initiation factor RpoD binding site (gray box); the other is immediately downstream of a predicted eukaryotic promoter (bold, underlined). A predicted start codon and a stop codon just upstream of the predicted start codon are underlined. (C) Western blots of whole-cell lysates of the indicated strains probed with anti-IFT46N (left) or anti-IFT46C (middle). A duplicate blot was stained with Ponceau S (right) to demonstrate equal loading of the lanes. The wild-type IFT46 protein at ∼48 kDa was recognized by both antibodies. A band at ∼28 kDa, the predicted mass for IFT46T1, was recognized strongly by anti-IFT46C in Supift461 cells only (asterisk), showing that a truncated IFT46 is expressed in this strain. A faint band at ∼28 kDa was sometimes detected in wild-type and ift46-1 whole-cell lysates; these bands may represent nonspecific cross-reactivity with the antibody or they may be the products of alternative transcription initiation. The anti-IFT46C antibody also detected a band just below 75 kDa in all three samples; this band was not detected in the flagellar sample (Figure 2A) and was not detected by the anti-IFT46N antibody in the whole-cell lysate and therefore likely is nonspecific.
The complete MRC1 transposon includes a sequence of 420 base pairs bracketed by 600–base pair long terminal repeats (LTRs) (Kim et al., 2006). In Supift461, MRC1 is inserted in the opposite orientation to the IFT46 gene, with one of its LTRs adjacent to the NIT1 and IFT46 genes. Sequence analysis predicted a bacterial promoter and a known bacterial transcription factor RpoD binding site (rpoD15) at the correct positions relative to the identified transcription start point for the longer RNA, and a eukaryotic promoter just upstream of the transcription initiation site for the shorter RNA (Figure 1B).
The hybrid RNAs from Supift461 are predicted to encode a fusion protein (which we term IFT46T1) in which the first 10 amino acids are derived from the MRC1 sequence, followed by 240 amino acids corresponding to IFT46 aa 105–344. The published IFT46 antibody (anti-IFT46N) was raised against the first 19 amino acids of IFT46 and did not detect IFT46 sequence in Supift461 (Figure 1C, left) (Hou et al., 2007). To determine whether IFT46T1 is expressed in Supift461, we generated a guinea pig polyclonal antibody (anti-IFT46C) to IFT46 aa 105–344. In Western blots of whole-cell lysates, the anti-IFT46C antibody reacted strongly with IFT46 at ∼48 kDa in wild type and with a band of ∼28 kDa, which is the predicted size for IFT46T1, in Supift461 (Figure 1C, center). Therefore IFT46T1 is expressed in Supift461.
IFT46T1 is assembled into IFT-B and stabilizes it
To determine whether IFT46T1 enters flagella, we probed Western blots of isolated flagella with anti-IFT46C (Figure 2A, left). IFT46T1 was detected in flagella of Supift461 at a level similar to that of IFT46 in wild-type flagella.
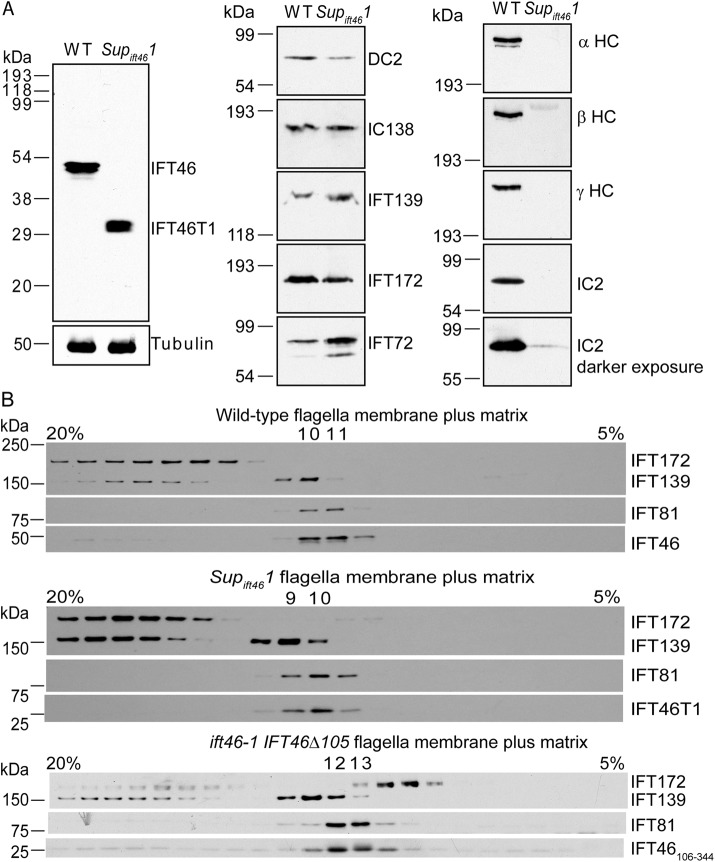
FIGURE 2:
IFT46T1 and IFT46106-344 incorporate into the IFT-B1 subcomplex in Supift461 and ift46-1 IFT46Δ105 flagella, respectively. (A) Western blots of isolated flagella of wild type and Supift461. Left, a blot probed with the anti-IFT46C antibody, which specifically recognizes IFT46 at ∼48 kDa in the wild-type sample (WT) and IFT46T1 at ∼30 kDa in the Supift461 sample. Tubulin served as a loading control. Center and right, blots of the same samples probed for the indicated proteins. Inner arm dynein intermediate chain IC138 is present at near-normal levels in Supift461 flagella but outer arm dynein α, β, and γ heavy chains and intermediate chain IC2 are missing or greatly reduced. DC2, an outer arm dynein docking-complex protein that is transported into flagella independently of outer arm dynein (Takada and Kamiya, 1994; Wakabayashi et al., 2001), is slightly reduced in Supift461 flagella; this is expected for flagella lacking the outer dynein arm (Wakabayashi et al., 2001). (B) IFT46T1 and IFT46106-344 comigrate with the IFT-B1 subcomplex. Flagellar membrane-plus-matrix fractions from wild-type (top), Supift461 (middle), or ift46-1 IFT46Δ105 (bottom) cells were further fractionated on 5–20% sucrose gradients, and the fractions were analyzed by Western blotting using antibodies to the indicated IFT proteins. IFT-B1 proteins migrated together as shown by IFT81 and IFT46 in the wild-type sample. Like their wild-type counterpart, both IFT46T1 and IFT46106-344 comigrated with the IFT-B1 protein IFT81.
We previously showed that other IFT-B proteins, which are largely destabilized in ift46-1 cell bodies, are restored to near-normal levels in Supift461 whole-cell lysates (Hou et al., 2007). To determine whether the expression of IFT46T1 resulted in normal levels of other IFT proteins in Supift461 flagella, we probed Western blots of isolated flagella with antibodies to several IFT-A and IFT-B proteins (Figure 2A, center). The IFT-B protein IFT72 was elevated relative to wild type. In contrast, the IFT-B protein IFT172 was slightly decreased. IFT172 can function independently of the IFT particle (Pedersen et al. 2005), and its level often trends differently from levels of other IFT-B proteins (Hou et al., 2007; Brown et al., 2015). IFT-A, as represented by IFT139, was elevated relative to wild type. Therefore expression of IFT46T1 results in slight changes in levels of IFT proteins in flagella of Supift461.
Importantly, the Supift461 flagella contained normal levels of the inner arm dynein intermediate chain IC138, but antibodies failed to detect the α, β, or γ outer arm dynein heavy chains and detected only a trace of the outer arm dynein intermediate chain IC2, even at long exposure times (Figure 2A, center and right). These results confirm our previous conclusions, based on electron microscopy (Hou et al., 2007), that the outer dynein arms are drastically and specifically reduced in Supift461 axonemes.
To investigate whether IFT46T1 in the Supift461 flagella is associated normally with other IFT-B proteins, we analyzed the flagellar detergent-soluble membrane-plus-matrix fraction by sucrose density gradient centrifugation followed by Western blotting. Under our experimental conditions, proteins of the wild-type IFT-B1 subcomplex remained associated with one another, as indicated by comigration of IFT46 and IFT81, but were separate from IFT172 and IFT-A proteins as represented by IFT139 (Figure 2B). In fractions from Supift461 flagella, IFT46T1 similarly comigrated with IFT81 but not with IFT172 or IFT139. Therefore IFT46T1 is incorporated into the IFT-B1 subcomplex.
Taken together, results at the genomic, mRNA, and protein levels show that, in Supift461, the insertion of an MRC1 transposon into the ift46-1 allele causes a hybrid RNA to be expressed and translated into an N-terminally truncated version of IFT46 that we term IFT46T1. Like its wild-type counterpart, IFT46T1 is incorporated into IFT-B1 and enters flagella, allowing some flagellar assembly, including incorporation of inner dynein arms. However, IFT46T1 does not support assembly of outer dynein arms into flagella.
Expression of IFT46 aa 106–344 is sufficient for partial rescue of ift46-1
These results indicate that the C-terminal two-thirds of IFT46 is sufficient to stabilize IFT-B and support some flagellar assembly in Supift461. However, because IFT46T1 is expressed from an unusual exogenous promoter and is a fusion protein in which the first 10 amino acids are derived from the MRC1 sequence, it was unclear whether part of the phenotype of Supift461 might be due to abnormal expression of IFT46T1 or to its MRC1-derived amino acids. To clarify this, we cotransformed ift46-1, which is null for IFT46 and is nonmotile, with a drug-resistance gene and a construct designed to express a Met plus IFT46 aa 106–344 (termed IFT46106-344) from the endogenous IFT46 promoter. About 10% of the resulting transformants contained slow-swimming cells when grown in 96-well plates. Western blotting of 11 of the partially motile cell lines revealed that they all expressed a protein of appropriate size that is detected by our anti-IFT46C antibody but is not detected in ift46-1 (Figure 3); this protein thus must be IFT46106-344. Southern blotting of DNA from the partially motile transformants confirmed that they were the products of independent transformation events (Supplemental Figure S1). Thus the C-terminal 239 amino acids of IFT46 are sufficient to partially rescue the ift46-1 phenotype.
FIGURE 3:
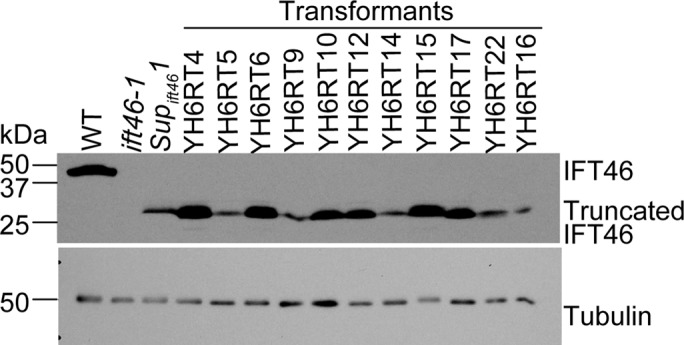
Expression of IFT46106-344 partially rescues the ift46-1 phenotype. Cells of ift46-1 were cotransformed with a gene that expresses IFT46106-344 and, on a separate plasmid, a drug-resistance gene that served as a selectable marker. Among the resulting transformants, ∼10% were partially rescued; they mimicked the Supift461 phenotype in that some cells could assemble longer flagella and swim slowly when stressed by growing in 96-well plates. The panel shows a Western blot of cell lysates from the motile transformants and wild-type, ift46-1, and Supift461 cells probed with the anti-IFT46C antibody; all of the partially suppressed strains expressed the truncated IFT46 protein, albeit at different levels. Strain YH6RT4 was chosen for further analysis and renamed ift46-1 IFT46Δ105. A blot of the same samples diluted 1:20 was probed with an anti-tubulin antibody as a loading control. Supplemental Figure S1 provides evidence that the motile transformants resulted from independent transformation events. In a control experiment, 200 cell lines obtained by transformation with the drug-resistance gene alone were examined and all found to be nonmotile.
To more rigorously examine the ability of IFT46106-344 to carry out IFT46 function, we chose the cell line YH6RT4 (hereafter ift46-1 IFT46Δ105) for detailed analysis, because it expresses IFT46106-344 at a level similar to that of IFT46 in wild type (Figure 3). Like Supift461, these cells have very short flagella when aerated; however, they are much more readily induced to form longer flagella when stressed. Western blotting of whole-cell lysates showed that IFT complex A and B proteins are present above normal levels in these cells (Figure 4A), suggesting that IFT46106-344 is assembled into IFT-B1 and stabilizes the complex. The increased expression of both IFT-A and IFT-B proteins may reflect a response by the cell to try to remedy a defect in IFT (see IFT is reduced in ift46-1 IFT46Δ105 flagella).
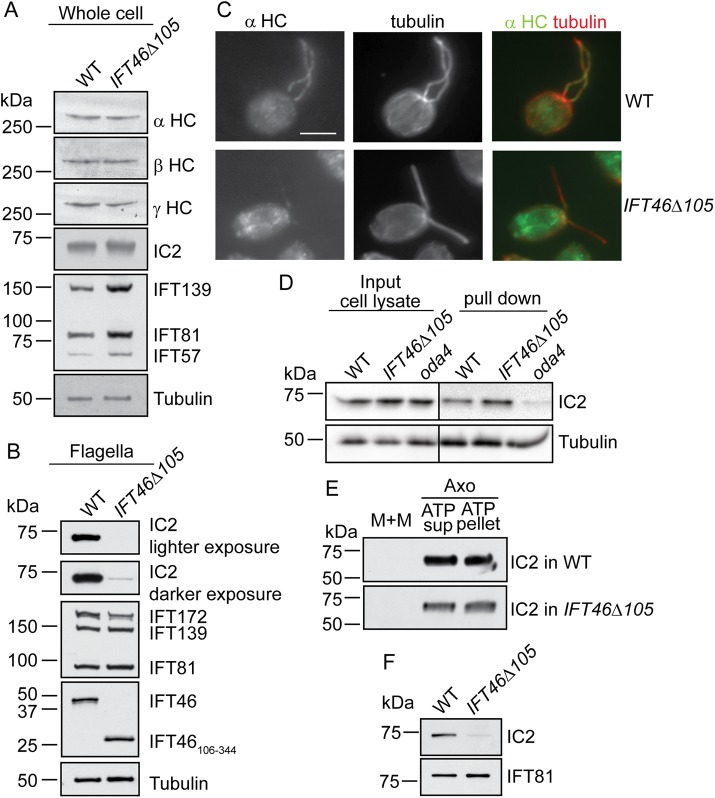
FIGURE 4:
ift46-1 IFT46Δ105 cells have impaired transport of outer arm dynein into their flagella. (A) Western blots of wild-type and ift46-1 IFT46Δ105 whole-cell lysates indicate that levels of outer arm dynein subunits are normal in ift46-1 IFT46Δ105. Levels of IFT-A (IFT139) and IFT-B (IFT81, IFT57) proteins are above normal in these cells. A separate blot using 1/200 of each sample was analyzed for the tubulin loading control. Here and in the following figures, ift46-1 IFT46Δ105 is abbreviated IFT46Δ105. (B) Western blots of wild-type and ift46-1 IFT46Δ105 flagella revealed that the latter contain greatly reduced amounts of outer arm dynein as represented by IC2. IFT139 and IFT81 levels are normal. For the tubulin control, the amount of protein loaded was 1/100 that loaded for detection of the other proteins. (C) Immunofluorescence images of wild-type and ift46-1 IFT46Δ105 cells doubly labeled with antibodies to α-tubulin and the α heavy chain of outer arm dynein. The outer arm dynein signal in ift46-1 IFT46Δ105 flagella was greatly reduced compared with that in wild-type flagella. In the ift46-1 IFT46Δ105 cell shown, most of the outer arm dynein label was near the flagellar base, but in general, any labeling was distributed evenly along the flagella in ift46-1 IFT46Δ105 cells. Scale bar: 5 µm. (D) Outer arm dynein preassembled in the ift46-1 IFT46Δ105 cell body is competent to bind to axonemes. Lysates from wild-type and ift46-1 IFT46Δ105 cells were incubated with ift46-1 IFT46Δ105 axonemes, and the axonemes were then separated from unbound protein by centrifugation and assayed for bound IC2. The Western blot of the cell lysate (input cell lysate) confirmed that the wild-type and ift46-1 IFT46Δ105 cells contained about the same amount of IC2. The Western blot of the axonemes (pull down) showed that outer arm dynein from wild-type and ift46-1 IFT46Δ105 cytoplasm is equally competent to bind to axonemes. An oda4 cell lysate served as a control for nonspecific binding to the axonemes. Cells of oda4 lack the β heavy chain of outer arm dynein but have normal levels of the other outer arm dynein subunits in their cytoplasm; because the β heavy chain is missing, the dynein cannot bind specifically to the axoneme (Ahmed et al., 2008). For the tubulin control, the amount of protein loaded was 1/14 that loaded for detection of IC2. (E) Western blots of wild-type and ift46-1 IFT46Δ105 flagellar fractions probed with an anti-IC2 antibody. The flagella were fractionated into membrane-plus-matrix (M+M) and axoneme (Axo) fractions via detergent treatment. The axoneme fraction was further divided into ATP-soluble (ATP sup) and ATP-insoluble (ATP pellet) fractions. The trace amount of IC2 in the ift46-1 IFT46Δ105 flagella is associated with the axoneme, not the matrix. IC2 was distributed similarly between the two axonemal fractions in wild-type and ift46-1 IFT46Δ105, suggesting that its attachment to the axoneme is normal in ift46-1 IFT46Δ105. For samples from the same strain, protein from an equal number of flagella was loaded in each lane; more protein was loaded for the ift46-1 IFT46Δ105 samples than for the wild-type samples in order to detect the very small amount of IC2 present in the former. (F) Western blots of membrane-plus-matrix fractions from regenerating flagella of wild-type and ift46-1 IFT46Δ105 cells probed with antibodies to IC2 and IFT81 (loading control). IC2 was greatly reduced in the ift46-1 IFT46Δ105 sample. Cells were deflagellated by pH shock and were allowed to regenerate new flagella by bringing the pH back to the pre-deflagellation level. The regenerating flagella were then collected when about half-grown (4.0 ± 1.3 µm for wild-type flagella; 3.8 ± 0.8 µm for ift46-1 IFT46Δ105 flagella; n = 100, p = 0.08).
To confirm that IFT46106-344 is assembled normally into IFT-B1, we checked the distribution of the truncated protein in sucrose gradient fractions of ift46-1 IFT46Δ105 flagellar membrane-plus-matrix and found that IFT46106-344 cosedimented normally with IFT81 (Figure 2B). We also used label-free quantitative mass spectrometry (MS) to determine whether IFT46 and IFT46106-344 were incorporated into IFT-B1 in the same molar ratio in wild type and ift46-1 IFT46Δ105, respectively. The wild-type or ift46-1 IFT46Δ105 flagellar membrane-plus-matrix was fractionated on sucrose gradients, and the peak fractions of IFT46 or IFT46106-344 were concentrated and used for the analysis. The intensities of the top three common precursors in the MS spectra for IFT46/IFT46106-344 and IFT81 (the internal standard) were used to calculate, for each protein, the fold change between the wild-type and ift46-1 IFT46Δ105 samples. The fold change for IFT46/IFT46106-344 was 0.8, while that for IFT81was 0.6. We conclude that a normal molar ration of IFT46106-344 is incorporated into IFT-B1.
Levels of outer arm dynein proteins are normal in the whole cell but greatly reduced in the flagella of ift46-1 IFT46Δ105
We previously found that IC2 is present in the Supift461 cell body at normal levels (Hou et al., 2007); similarly, in this study, we found by Western blot analysis that outer arm dynein heavy and intermediate chains were present at normal levels in whole-cell lysates of ift46-1 IFT46Δ105 (Figure 4A). Therefore truncation of the IFT46 N-terminus has no effect on the level of expression of outer arm dynein proteins.
In wild-type Chlamydomonas, ∼90% of the total cellular pool of IFT proteins is in the cell body; only ∼10% is in the flagella (Ahmed et al., 2008; Lechtreck et al., 2009). In contrast, outer arm dynein proteins are about evenly distributed between the cell body and the flagella (Ahmed et al., 2008). To determine how truncation of the IFT46 N-terminus affects localization of these proteins to the flagella, we compared the protein content of isolated flagella from wild type and ift46-1 IFT46Δ105 by Western blotting (Figure 4B). IFT81 (IFT-B) and IFT139 (IFT-A) protein levels were about the same as in wild-type flagella; IFT172 was slightly reduced. However, the amount of the outer arm dynein intermediate chain IC2 was greatly decreased in ift46-1 IFT46Δ105 flagella. Immunofluorescence labeling of wild-type and ift46-1 IFT46Δ105 cells with an antibody to the outer arm dynein α heavy chain confirmed a drastic reduction in the fluorescence signal for outer dynein arms in ift46-1 IFT46Δ105 flagella (Figure 4C). Therefore, despite being expressed at normal levels in the cytoplasm, outer arm dynein is not properly localized to the flagella of ift46-1 IFT46Δ105 cells. The loss of outer dynein arms undoubtedly accounts for the slow-swimming phenotype of the ift46-1 IFT46Δ105 cells.
The low level of outer arm dynein in ift46-1 IFT46Δ105 flagella is due to a defect in IFT
Outer arm dynein is preassembled in the cell cytoplasm before it is transported into the flagellum (Fowkes and Mitchell, 1998), and it must undergo a poorly understood maturation step to be able to attach to its binding site on the outer doublet microtubules (Desai et al., 2015). To investigate whether ift46-1 IFT46Δ105 outer arm dynein in the cytoplasmic pool is preassembled, mature, and competent to bind to axonemes, we performed an in vitro binding assay. Cell lysates from wild-type and ift46-1 IFT46Δ105 cells were incubated with ift46-1 IFT46Δ105 axonemes, the axonemes were then separated from unbound proteins by centrifugation, and binding of outer arm dynein to the axonemes was assessed by Western blotting using an anti-IC2 antibody as a probe (Figure 4D). Similar amounts of IC2 from wild-type and ift46-1 IFT46Δ105 whole-cell lysates bound to the axonemes, indicating that outer arm dynein in the ift46-1 IFT46Δ105 cell cytoplasm is fully preassembled and competent to bind to axonemes. This result also shows that the ift46-1 IFT46Δ105 axoneme is capable of binding outer arm dynein. This is consistent with the previous finding that outer arm dynein in the original ift46 mutant is fully preassembled and competent to bind to axonemes (Ahmed et al., 2008).
In our analyses of ift46-1 IFT46Δ105 flagella, we detected small amounts of IC2 (Figure 4B). To determine whether this represented outer arm dynein bound to the axoneme or simply present in the flagellar matrix, possibly associated with IFT particles, flagellar samples were fractionated by detergent treatment into membrane-plus-matrix and axonemal fractions. The axonemal fraction was further treated with ATP to yield an ATP-soluble fraction and an ATP-insoluble fraction. The former contains residual IFT particles not removed during the demembranation step (Hou et al., 2004) and some outer arm dynein that is solubilized by ATP treatment (Goodenough and Heuser, 1984; Hou et al., 2004). Western blotting of the fractions (Figure 4E) revealed 1) that the outer arm dynein in the ift46-1 IFT46Δ105 flagella is associated with the axoneme, not the flagellar matrix; and 2) that it has the same distribution in the ATP-treated fractions as does outer arm dynein in the wild-type fractions, indicating that its attachment to the axoneme is normal.
We also considered the possibility that outer dynein arms are transported normally by IFT into the flagella of ift46-1 IFT46Δ105 but not released from the IFT particle and so are returned directly to the cell body rather than being incorporated into the axoneme. This is unlikely to be the case because the above experiment showed that the small amount of IC2 in the ift46-1 IFT46Δ105 flagellum is located in the axoneme, not the membrane-plus-matrix fraction, where we would expect it to be if the truncation of IFT46 did not prevent IFT of outer arm dynein but did block release of outer arm dynein from the IFT particle. Nevertheless, to further test this hypothesis, we compared the membrane-plus-matrix fractions from half-length regenerating flagella of wild-type and ift46-1 IFT46Δ105 cells. We used regenerating flagella because, although the number of IFT particles traversing the flagella is more or less independent of flagellar length (Dentler, 2005; Engel et al., 2009), the loading of axonemal precursors (as represented by DRC4, a subunit of the nexin-dynein regulatory complex) onto IFT particles increases ∼10-fold in such flagella (Wren et al., 2013). Western blotting revealed that there was much less IC2 in the ift46-1 IFT46Δ105 membrane-plus-matrix fraction than in the wild-type membrane-plus-matrix fraction (Figure 4F). These results strongly suggest that loading of outer arm dynein onto IFT particles is severely reduced in the absence of the IFT46 N-terminus.
In summary, ift46-1 IFT46Δ105 cells have normal amounts of outer arm dynein, and the dynein is competent to bind to ift46-1 IFT46Δ105 axonemes in vitro. The few arms that are present in ift46-1 IFT46Δ105 flagella appear to be bound to the axoneme normally. Given that the only known genetic defect in ift46-1 IFT46Δ105 cells is in IFT46, we conclude that the reduction in outer dynein arms in ift46-1 IFT46Δ105 flagella is a result of the cells being unable to move preassembled, assembly-competent outer arm dynein efficiently into the flagella due to truncation of the N-terminus of IFT46.
ODA16 is not transported normally into ift46-1 IFT46Δ105 flagella
ODA16, a 49-kDa WD-repeat protein, is required for assembly of a full complement of outer dynein arms into the axoneme (Ahmed and Mitchell, 2005; Ahmed et al., 2008). Its transport into the flagellum is dependent on IFT and occurs even in the absence of outer arm dynein, suggesting that it interacts directly with the IFT machinery, an interaction confirmed by coimmunoprecipitation experiments. Yeast two-hybrid experiments and pull-down assays using recombinantly produced proteins have shown that ODA16 interacts directly with IFT46 in vitro (Ahmed et al., 2008) and that this interaction is mediated by the IFT46 N-terminus (Taschner et al., 2017). To determine whether loss of the N-terminus of IFT46 affects the transport of ODA16 into flagella, we used Western blotting to compare ODA16 levels in whole-cell lysates and isolated flagella of wild-type and ift46-1 IFT46Δ105 cells. As is the case for IFT-particle proteins, the majority of ODA16 in wild-type cells is in the cell body, while much less is in the flagella (Ahmed et al., 2008). ODA16 was present in ift46-1 IFT46Δ105 cells at a normal level (Figure 5A), but was dramatically reduced in ift46-1 IFT46Δ105 flagella (Figure 5B). Therefore the IFT46 N-terminus is crucial for the IFT-mediated import of ODA16 into flagella. The small amount of ODA16 that does access the ift46-1 IFT46Δ105 flagellum may be carried in by low-affinity binding to sites still present in IFT46106-344. Alternatively, it may be diffusing in, because the flagellar transition zone, which functions as a selective barrier controlling entry of proteins into the flagellum, is permeable to soluble proteins smaller than ∼50 kDa (Kee et al., 2012; Breslow et al., 2013; Awata et al., 2014). In any case, our results provide in vivo evidence for the interaction of IFT46 with ODA16 and suggest that elimination of this interaction is the underlying cause for loss of the outer arms from the axonemes of ift46-1 IFT46Δ105 cells.
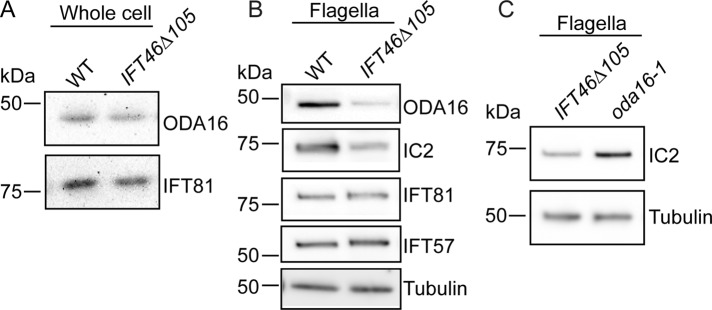
FIGURE 5:
Cells of ift46-1 IFT46Δ105 have impaired transport of ODA16 into their flagella, which contain even less outer arm dynein than oda16-1 flagella. (A) Western blots of wild-type and ift46-1 IFT46Δ105 whole-cell lysates probed with anti-ODA16 and anti-IFT81 antibodies. The ODA16 level in the ift46-1 IFT46Δ105 whole-cell lysate is normal. (B) Western blot of isolated flagella from wild-type and ift46-1 IFT46Δ105 cells probed with the indicated antibodies. ODA16 is greatly reduced in the ift46-1 IFT46Δ105 flagella. Levels of IC2 also are reduced, while levels of the IFT-B1 proteins IFT81 and IFT57 are normal. For the tubulin controls, the sample loadings were 1/100 those for the other proteins. (C) Western blots of ift46-1 IFT46Δ105 and oda16-1 flagella probed with anti-IC2. ift46-1 IFT46Δ105 flagella contain even less outer arm dynein than oda16-1 flagella. The samples were diluted 1/1000 for the tubulin loading controls.
In the mutant oda16-1, which is null for ODA16, the flagellar level of outer arm dynein is only ∼10% that of wild type ((Ahmed and Mitchell, 2005). If IFT of outer arm dynein is mediated solely by ODA16, the flagella of oda16-1 should have levels of outer arm dynein at least as low as those of ift46-1 IFT46Δ105. To test this, we compared the levels of outer arm dynein in the flagella of the two strains by Western blotting (Figure 5C). In fact, ift46-1 IFT46Δ105 flagella contained much less outer arm dynein than those of oda16-1 flagella. This suggests that either the N-terminus of IFT46 is capable of binding some outer arm dynein, even in the absence of ODA16, or outer arm dynein can enter flagella in an IFT-independent manner (e.g., by diffusion; see Discussion) more readily in oda16-1 than in ift46-1 IFT46Δ105.
Flagellar assembly requires both stress and a threshold level of truncated IFT46
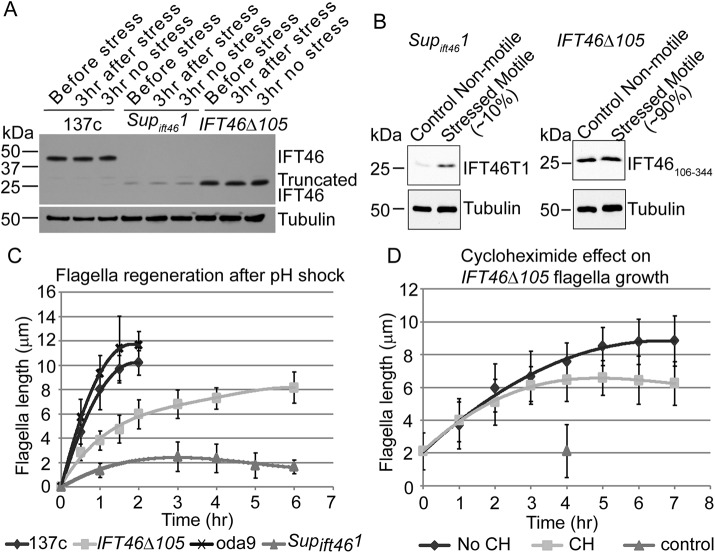
Cells of Supift461 are aflagellate or have very short flagella when grown with aeration, but a small percentage of cells will grow longer flagella of various lengths when stressed by hypoxia (Hou et al., 2007). Cells of ift46-1 IFT46Δ105 similarly are aflagellate or have very short flagella when grown with aeration, but are much more readily induced to form flagella under a variety of stress conditions. To better understand the basis for this phenotypic difference, we compared the levels of truncated IFT46 in these strains under two different treatments for inducing stress.
In the first regimen, mid–log phase cells were concentrated eightfold during the day, and aeration was then stopped. This treatment does not induce flagellar growth in Supift461 cells but does induce slow growth of flagella of uniform length in ift46-1 IFT46Δ105 cells (see Figure 6D). Western blotting showed that, before application of stress, wild-type and ift46-1 IFT46Δ105 cells had similar amounts of IFT46 or IFT46106-344, respectively, whereas the level of IFT46T1 was much less in Supift461 cells (Figure 6A). The level of IFT46 in wild-type cells or of truncated IFT46 in Supift461 or ift46-1 IFT46Δ105 cells did not noticeably increase as a result of the stress. These results suggest that the ift46-1 IFT46Δ105 cells are more readily induced to assemble flagella because they have cytoplasmic levels of truncated IFT46 above a required threshold, whereas the lower amount of truncated IFT46 in Supift461 cells is below that threshold and inadequate to support flagellar assembly.
FIGURE 6:
Truncated IFT46 levels correlate with ability to assemble flagella in Supift461 and ift46-1 IFT46Δ105 cells, and flagellar assembly is slow in ift46-1 IFT46Δ105 cells. (A) Western blot showing that the levels of IFT46 and truncated IFT46 did not change when the cells were stressed by concentration during the day and then maintained for 3 h without aeration. Under these conditions, ift46-1 IFT46Δ105 cells grew longer flagella, but Supift461 cells did not. Before stress, the amount of IFT46T1 in Supift461 cells was much less than the amounts of IFT46 in wild-type cells or of IFT46106-344 in ift46-1 IFT46Δ105 cells; there was little or no change in these amounts at the end of the 3-h stress period. A separate blot using 1/5 of the same samples was probed with an anti-tubulin antibody as a loading control. (B) Western blot comparing the levels of IFT46T1 in Supift461 cells and of IFT46106-344 in ift46-1 IFT46Δ105 cells after cultures were kept overnight with aeration (Control) or without aeration (Stressed); the stress condition induced flagellar formation in ∼10% of Supift461 cells and 90% of ift46-1 IFT46Δ105 cells. The lack of aeration caused a substantial increase in the level of truncated IFT46 in Supift461 but not in ift46-1 IFT46Δ105. For the tubulin controls, the sample loadings were 1/20th of those used for the detection of truncated IFT46. (C) Flagellar regeneration curves after pH shock. Wild-type and oda9 cells regenerated full-length flagella within 2 h, ift46-1 IFT46Δ105 cells assembled flagella more slowly, and Supift461 cells very slowly formed only very short flagella. (D) Flagellar assembly curves for ift46-1 IFT46Δ105 cells induced to form flagella by concentration followed by maintenance without aeration in the presence or absence of 10 µg/ml cycloheximide (CH). Control cells were not concentrated and were aerated without interruption in the absence of cycloheximide. For C and D, one flagellum from each of 50 cells was measured and the mean value was calculated for each data point; nonflagellated cells were not included. Error bars show SDs.
In the second regimen, which is the most optimal that we have found for inducing flagellar growth in Supift461 cells, cells were grown to mid–log phase with aeration, and aeration was then stopped overnight. The next morning, ∼10% of Supift461 cells and 90% of ift46-1 IFT46Δ105 cells had flagella of approximately normal length and were swimming. Western blot analysis revealed that the level of IFT46106-344 in the ift46-1 IFT46Δ105 cells did not change under the stress conditions (Figure 6B). In contrast, IFT46T1 in the Supift461 cells was significantly increased by these stress conditions, although its level did not reach that of IFT46106-344 (unpublished data). These findings suggest that the level of IFT46T1 in Supift461 cells had to increase above a threshold level for the cells to grow flagella. The results support the hypothesis that, compared with ift46-1 IFT46Δ105 cells, Supift461 cells are less readily induced to grow flagella because they have lower levels of truncated IFT46.
Finally, the experiments shown in Figure 6, A and B, together indicate that a level of IFT46106-344 in ift46-1 IFT46Δ105 cells that is similar to that of IFT46 in wild type and adequate to support flagellar assembly under stress conditions is not sufficient to support flagellar assembly in the absence of stress. In the presence of adequate amounts of truncated IFT46, stress itself enables flagellar assembly through a still unknown mechanism.
ift46-1 IFT46Δ105 cells have abnormal flagellar assembly kinetics
To determine how the absence of the IFT46 N-terminus affects the rate of flagellar assembly, we subjected cells to pH shock, which causes abscission of any flagella present, thus resetting flagellar length to zero. We then centrifuged the cells, resuspended them in fresh medium, and subsequently incubated them in a beaker with light but no aeration. Under these conditions, wild-type cells rapidly formed new flagella with kinetics similar to that typically observed for wild-type strains (Rosenbaum et al., 1969) (Figure 6C). Supift461 cells reformed only very short flagella, consistent with the poor ability of these cells to assemble flagella under most conditions. Cells of ift46-1 IFT46Δ105 formed new flagella, but the initial rapid rate of growth was ∼63% that of wild type. The flagella then transitioned to a slow elongation phase similar to that of wild type (Rosenbaum et al., 1969; Wood et al., 2012), but this transition occurred at a shorter flagellar length than in wild type, and the flagella never reached an average length greater than about two-thirds that of wild-type flagella. Because ift46-1 cells rescued with the wild-type IFT46 gene have normal flagellar regeneration kinetics, and strain YH6RT10 generated by rescue of ift46-1 in an independent transformation event with the construct expressing IFT46106-344 has regeneration kinetics similar to that of ift46-1 IFT46Δ105 (Supplemental Figure S2), the ift46-1 IFT46Δ105 flagellar regeneration phenotype is not due to some other mutation. Its slow initial rate of flagellar assembly also is not due to the absence of outer dynein arms on the ift46-1 IFT46Δ105 doublet microtubules or on IFT particles: cells of oda9, which lack the outer dynein arm intermediate chain IC1 (Kamiya, 1988; Wilkerson et al., 1995) and thus do not preassemble outer arms in the cell body (Fowkes and Mitchell, 1998), do not transport them into their flagella as IFT cargo during flagellar regeneration (Supplemental Figure S3), and do not incorporate them into their axonemes (Kamiya, 1988; Wilkerson et al., 1995), had initial rapid regeneration kinetics similar to that of wild-type cells (Figure 6C). Therefore ift46-1 IFT46Δ105 cells, despite having a level of IFT46106-344 similar to the levels of IFT46 in wild type (Figure 6A), have abnormally slow flagellar growth, even after induction of flagellar assembly.
IFT is reduced in ift46-1 IFT46Δ105 flagella
The slower flagellar regeneration kinetics in ift46-1 IFT46Δ105 cells could be the result of a defect in IFT caused by the absence of the IFT46 N-terminus. To investigate this possibility, we used differential interference contrast (DIC) microscopy to compare IFT velocity and frequency in wild-type versus ift46-1 IFT46Δ105 cells. Because shorter flagella exhibit slower IFT speeds (Engel et al., 2009), our measurements were carried out on growing wild-type or ift46-1 IFT46Δ105 flagella that were ∼8 µm long. Anterograde velocity and frequency in ift46-1 IFT46Δ105 were ∼70% of their values in wild type; retrograde velocity and frequency were respectively ∼60% and 70% of their wild-type values (Figure 7A). Figure 7, B and C, shows the distributions of anterograde and retrograde IFT velocities for wild-type and ift46-1 IFT46Δ105 cells. The distributions of anterograde velocities fell into similarly sharp peaks in both strains, whereas the retrograde velocities were much more widely distributed, suggesting that retrograde trains are more heterogeneous in size or number of motors than are anterograde trains; similar differences between the distributions of anterograde and retrograde velocities have been reported previously (Dentler, 2005). The decrease in anterograde velocity coupled with the decrease in anterograde frequency in ift46-1 IFT46Δ105 could account for the reduction in the initial rate of flagellar assembly that we observed in this strain (Figure 6C).
FIGURE 7:
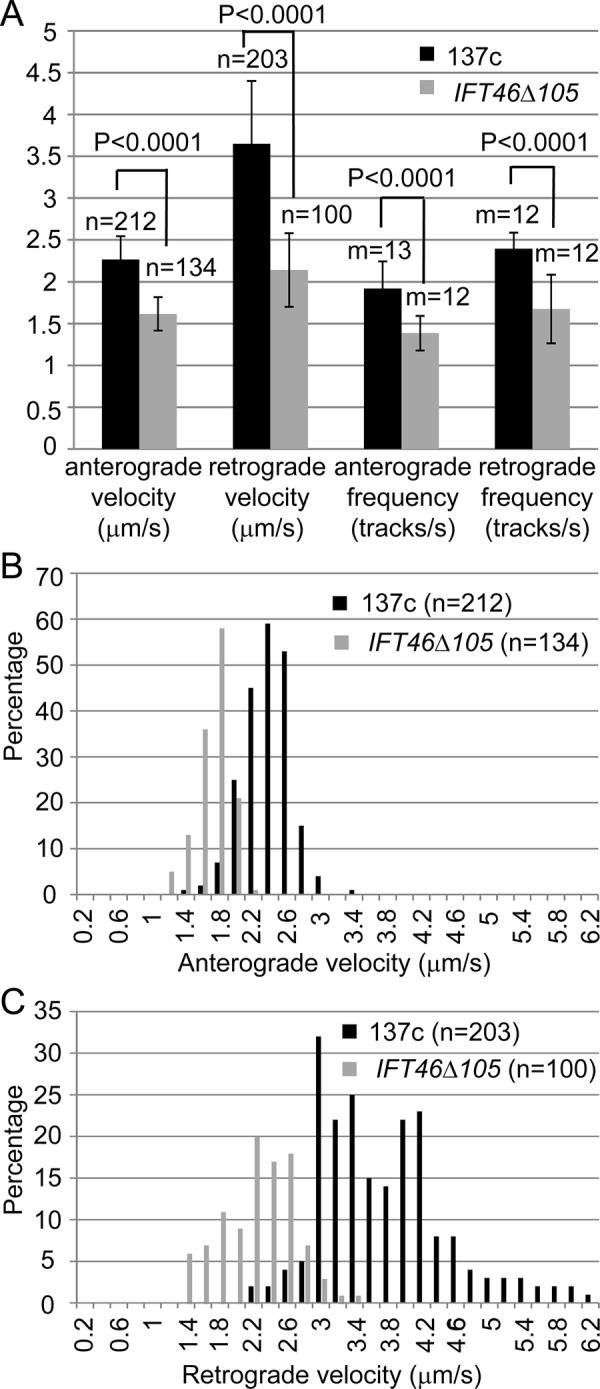
IFT is impaired in ift46-1 IFT46Δ105 cells. (A) ift46-1 IFT46Δ105 cells have lower IFT velocities and frequencies than wild-type cells. In both strains, measurements were carried out on flagella that were ∼8 µm long, and the mean values were plotted. Error bars show SDs. (B, C) Distributions of anterograde (B) and retrograde (C) IFT velocities in wild-type and ift46-1 IFT46Δ105 flagella. In flagella of both strains, anterograde velocities distributed in a sharp peak, whereas retrograde velocities spread more widely. n, number of IFT particles analyzed; m, number of flagella analyzed.
Stress-induced flagella formation does not require new protein synthesis
To try to understand the mechanism by which stress induces flagellar assembly in ift46-1 IFT46Δ105 cells, we investigated whether the induction requires new protein synthesis. Cells were grown to log phase with aeration and then concentrated 8- to 10-fold and thereafter kept in the light without aeration. Immediately after concentration, the cells were divided into two aliquots, and cycloheximide, which in Chlamydomonas blocks protein synthesis within 1–2 min (Rosenbaum et al., 1969), was added to one of the aliquots. Samples were then taken at 1-h intervals for measurement of flagellar length. In the absence of cycloheximide, ift46-1 IFT46Δ105 cells grew new flagella with slow kinetics (Figure 6D); the flagellar growth curve was similar to the curve for this strain in the experiment shown in Figure 6C. In the presence of cycloheximide, new flagella were formed at a rate that initially was identical to that of the cells without cycloheximide, and growth continued for several hours before leveling off as the flagella reached a length of ∼6 µm. Growth of flagella of wild-type cells in cycloheximide similarly levels off at ∼6 µm (Rosenbaum et al., 1969); this is believed to reflect exhaustion of the cellular pool of flagellar precursors. Therefore, consistent with our results for outer arm dynein (Figure 4A), ift46-1 IFT46Δ105 cells likely have a cellular pool of flagellar precursors similar to that of wild-type cells. We conclude that new protein synthesis is not required for the induction of flagellar growth by stress in ift46-1 IFT46Δ105 cells.
DISCUSSION
Genome remodeling by transposon MRC1 under environmental stress and selective pressure restored evolutionary fitness
Our data show that the spontaneous suppression of the ift46-1 mutant was caused by transposition of the mobile element MRC1 (Kim et al., 2006). The original suppressor was identified in an ift46-1 culture (Hou et al., 2007) that had been grown to stationary phase with aeration and then left for more than 1 wk without aeration, during which time the nonmotile cells sank to the bottom of the flask and likely experienced hypoxia, especially during the dark cycle. It is possible that the stress activated the transposition (Wheeler, 2013); it certainly would have resulted in a strong selection for cells that could swim toward the air:liquid interface.
Although it is known that insertion of transposable elements can activate downstream genes by initiating transcription within the transposon (Charlier et al., 1982; Ciampi et al., 1982), this is the first time that MRC1 was shown to activate gene expression via a similar mechanism. In this case, the insertion of the MRC1 sequence enabled flagellar assembly by providing both gene regulatory elements and a protein-coding sequence that resulted in partial restoration of IFT. Such transposon-mediated restructuring of the genome has been proposed to play an important role in the evolution of plants and algae in response to environmental change (Kim et al., 2006). The current study provides an excellent laboratory example of this.
The level of truncated IFT46 transcript in Supift461 cells is much lower than that of the full-length transcript in wild-type cells (Figure 8 in Hou et al., 2007), suggesting that transcription initiated by the MRC1 LTR sequence is much weaker than that initiated by the endogenous IFT46 promoter. Consistent with this, the level of IFT46T1 protein in Supift461 cells is much less than the level of IFT46 in wild-type cells or of IFT46106-344 in ift46-1 IFT46Δ105 cells. A promoter is predicted upstream of each of the two transcription start sites that we identified in the MRC1 LTR sequence. The promoter that gives rise to the shorter transcript is a eukaryotic polymerase II promoter. A predicted binding site for the transcription factor snail is upstream of this promoter region; however, there is no Chlamydomonas orthologue of snail. The promoter that gives rise to the longer transcript has bacterial promoter elements at positions −31 and −9 and a predicted RpoD binding site at −7, all correctly positioned relative to the transcription initiation site. RpoD is a member of the sigma 70 family of proteins that target RNA polymerase to the transcription start site in prokaryotes and plastids (Paget and Helmann, 2003). Chlamydomonas has only one sigma 70 protein, RPOD (Carter et al., 2004; Bohne et al. 2006; Kawazoe et al., 2012), which is encoded by a nuclear gene but believed to function in the chloroplast. It would be of interest to determine whether RPOD is involved in the initiation of transcription of the ift46 allele in the Supift461 nucleus.
The C-terminus of IFT46 stabilizes IFT-B, while the N-terminus mediates IFT of outer arm dynein
Expression of IFT46106-344 in ift46-1 cells recapitulated the suppression observed in Supift461 cells, confirming that the suppression in the latter is due to expression of an N-terminally truncated IFT46. IFT46106-344 stabilizes the other IFT-B1 proteins and is incorporated at the normal molar ratio into IFT-B1; as a result, ift46-1 IFT46Δ105 cells are able to form flagella. This is consistent with previous findings that the C-terminus of IFT46 interacts with the C-terminus of IFT52 and that the assembly of an IFT46/IFT52 subcomplex is a prerequisite for the assembly of IFT-B1 (Lucker et al., 2010; Taschner et al., 2014).
However, cells expressing IFT46106-344, like Supift461 cells, still have greatly reduced numbers of outer dynein arms in their flagella. We determined that this is due to reduced ability of IFT to transport outer dynein arms into the flagella. This transport is mediated by ODA16 acting as a cargo adaptor (Ahmed and Mitchell, 2005, Ahmed et al., 2008). Indeed, we found that the level of ODA16 is greatly reduced in ift46-1 IFT46Δ105 flagella, which is consistent with recent studies demonstrating that the N-terminal 147 amino acids of IFT46 interact with ODA16 in vitro and may contain the entire ODA16-binding domain (Taschner et al., 2017). Thus loss of ODA16 binding by IFT46 explains the reduction in outer arm dynein in flagella of ift46-1 IFT46Δ105. Interestingly, ift46-1 IFT46Δ105 flagella contain even less outer arm dynein than oda16-1 flagella, raising the possibility that the N-terminus of IFT46 is involved in limited outer arm dynein transport in an ODA16-independent manner.
How the very small amount of outer arm dynein present even in ift46-1 IFT46Δ105 flagella gets there is still unknown, but there are several possibilities. It may be carried in by the small amount of ODA16 that still enters the flagellum, perhaps by binding with low affinity to sites between IFT46 aa 106–147. It may diffuse in; although preassembled outer arm dynein is larger than the apparent size limit for diffusion through the flagellar transition zone (Kee et al., 2012; Breslow et al., 2013; Awata et al., 2014), the barrier may not be absolutely impermeable to large molecules (Lin et al., 2013). Finally, a small amount of dynein may enter the flagellum through a pathway that bypasses IFT and the transition zone barrier—for example, through the basal body lumen.
Stress enables flagellar assembly in IFT mutants with partially repaired IFT-B complexes
Although IFT46106-344 stabilizes IFT-B in the cytoplasm, and cells expressing this truncated IFT46 can assemble longer flagella than Supift461 cells, they assemble longer flagella only under stress (hypoxic) conditions. When recombinant His6-tagged full-length IFT46 or IFT46 lacking the first 25 amino acids were electroporated into ift46-2 cells, the cells’ flagellar assembly defect was rescued, but similar electroporation of recombinant His6-tagged IFT46 lacking the first 50 or 100 amino acids did not rescue the cells (Lucker et al., 2010). In these experiments, the cells were agitated every few minutes following electroporation, so they probably were not stressed sufficiently to support rescue by the more severely truncated proteins.
In addition to our initial report on stress-induced suppression of ift46-1 (Hou et al., 2007) and the findings reported here, there have been other reports of cells with defects in IFT-B proteins and impaired ciliary assembly that is partially corrected under stress conditions. In Chlamydomonas, the IFT74 null mutant ift74-2 has greatly reduced levels of IFT-B proteins in its cytoplasm and severely impaired flagellar assembly, even in the absence of aeration (Brown et al., 2015). However, in the mutant ift74-1, expression of a truncated form of IFT74 lacking the first 196 amino acids supports IFT-B assembly, but the cells still have severely impaired flagellar assembly unless they are grown without aeration, in which case the cells form half-length flagella and become motile.
In Tetrahymena, an IFT52 knockout mutant lacking cilia was observed to undergo frequent spontaneous partial suppression resulting in sparse short cilia usually lacking the central pair of microtubules; these cilia grew longer and acquired a “9+2” axoneme under hypoxic conditions (Brown et al., 2003). Further analysis revealed that the conditional suppression involved gene reorganization and alternative splicing leading to expression of a modified IFT52 that lacks seven of the original amino acids but has 43 additional amino acids (Dave et al., 2016). In Chlamydomonas, the lack of IFT52 destabilizes IFT-B (Richey and Qin, 2012), so it is possible that the original Tetrahymena IFT52 knockout mutant had a defect in IFT-B assembly or stability that was repaired by expression of the modified IFT52.
In none of these cases is the mechanism by which hypoxia promotes ciliary assembly clear. However, in at least two of the cases, there was expression of a partially functional IFT-B protein that stabilizes the IFT-B complex in the cytoplasm but does not restore full function to the complex. Stress then enables flagellar assembly to proceed. This is not due to a mechanism that bypasses IFT, because it does not occur in null mutants lacking IFT-B. Rather, it likely involves further repair or stabilization of the partially restored IFT-B. This does not involve new protein synthesis, because we found that stress can induce ift46-1 IFT46Δ105 cells to assemble half-length flagella in the presence of cycloheximide. The mechanism could involve posttranslational modification of IFT-B itself or of a chaperone that then interacts with IFT-B, repairing IFT-B’s ability to assemble into IFT trains or bind cargo. It also could involve the way IFT particles interact with the flagellar transition zone. When a Chlamydomonas mutant null for the transition zone protein CEP290 is grown with aeration, most cells have extremely short flagella, but when grown in the absence of aeration, many cells form longer flagella, indicating a connection between stress and transition zone function (Craige et al., 2010).
A similar mechanism to restore IFT may be operating in Caenorhabditis elegans. The cilia of hypomorphic IFT mutants, which are severely truncated in young adults, become longer as the worms age. This partial restoration of ciliary length is dependent upon IFT and requires the heat shock factor HSF-1 and probably the HSF1-regulated chaperone Hsp90 (Cornils et al., 2016).
IFT defects likely explain the abnormal flagellar assembly kinetics of ift46-1 IFT46Δ105 cells
Although stressed ift46-1 IFT46Δ105 cells assemble new flagella following deflagellation, the initial rapid rate of assembly is slower than normal. This is not due to the lack of incorporation of outer dynein arms into the axoneme, the absence of outer arm dynein as cargo on IFT trains, a mutation at a second site, or a lack of flagellar precursors. It appears to be due to a residual defect in IFT. Anterograde IFT frequency is slower in ift46-1 IFT46Δ105 cells than in wild-type cells; retrograde IFT frequency is slower by about the same amount, likely reflecting the reduced frequency with which anterograde trains reach the flagellar tip. Anterograde and retrograde velocity also are reduced; this might occur if the lower frequency of entry into the flagellum results in larger IFT trains, as trains with larger apparent size move more slowly (Iomini et al., 2001; Schmidts, Hou, et al., 2015). The combination of lower IFT frequency and velocity would be expected to slow delivery of axonemal precursors to the flagellar tip, resulting in a reduction in the flagellar assembly rate and shorter flagella (Marshall and Rosenbaum, 2001; Ishikawa and Marshall, 2011).
It is not clear why the frequency with which IFT trains enter the flagellum is reduced in ift46-1 IFT46Δ105 cells. The IFT frequency presumably is not reduced as a result of the inability to load outer arm dynein onto IFT trains, because oda9 cells do not load outer arm dynein onto their IFT trains yet regenerate flagella at nearly normal initial assembly rates. The reduction in IFT frequency may be due to an impairment in the way the partially restored IFT-B1 assembles with IFT-B2, IFT-A, or the IFT motors; the way it interacts with IFT cargo; the way it docks on the transition fibers during IFT train assembly (Deane et al., 2001; Brown et al., 2015); or the way it interacts with the IFT quality control machinery that regulates entry of trains into the flagellum (Dentler, 2005; Craige et al., 2010). Future studies should be able to distinguish between these possibilities.
MATERIALS AND METHODS
Cells and culture conditions
Chlamydomonas reinhardtii strain 137c (nit1, nit2, mt+) from the Chlamydomonas Resource Center (University of Minnesota, St. Paul, MN) was used as a wild-type control. Additional strains used included oda4 (Okagaki and Kamiya, 1986); oda9 (Kamiya, 1988); oda16-1 (Ahmed and Mitchell, 2005); and ift46-1, 6R29, and Supift461 (Hou et al., 2007). Partially rescued strains were generated by cotransforming ift46-1 cells with linearized plasmid pSP124S (Lumbreras et al., 1998) and the truncated ift46 gene plasmid (see Creation of DNA constructs for Chlamydomonas transformation); one of these strains (YH6RT4) was renamed ift46-1 IFT46Δ105.
Cells were grown on a 14:10 h light:dark cycle in M (Sager and Granick, 1953) medium I altered to have 0.0022 M KH2PO4 and 0.00171 M K2HPO4. Normally, cells were aerated with 5% CO2 and 95% air (Witman, 1986). Two modified culture conditions were used to stress cells. Under the first, cultures were inoculated with 105 cells/ml on day 0 and grown with aeration. On day 2, cells were concentrated 8- to 10-fold in the morning and then maintained without aeration. This regimen induces ift46-1 IFT46Δ105 cells but not Supift461 cells to grow flagella slowly during the remainder of the day. Under the second growth condition, cultures were inoculated at 105 cells/ml on day 0 and grown with aeration. On the evening of day 2, aeration was stopped, and the cells were left overnight without aeration. The next morning, some of the Supift461 cells and most of the ift46-1 IFT46Δ105 cells had longer flagella. Experiments involving immunofluorescence microscopy and isolation of flagella were done using cells grown under the second growth condition. Analysis of IFT was done using cells grown under the first growth condition.
DNA isolation and analysis
Genomic DNA was isolated from Chlamydomonas cells as described by Pazour et al. (1998). The DNA was cut by PstI, and DNA gel electrophoresis was done using standard procedures (Sambrook et al., 1987). Southern blotting was performed using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science, Penzberg, Germany); instead of using the kit’s hybridization buffer, we used Church buffer (7% SDS, 1 mM EDTA, 0.25 M Na2HPO4, pH 7.2; Church and Gilbert, 1984) and hybridized at 65°C. The 1.2-kb and 1.5-kb PstI fragments cut from the cloned 4.8-kb IFT46 gene (Hou et al., 2007) were used as templates for probes. For detection of the truncated IFT46 gene by PCR, the primer pair IFT46-1 and IFT46-10 was used; all primers are detailed in Supplemental Table S1. Primer pair Fus3 and Fus4, specific for the plus mating-type gene FUS1 (Ferris et al., 1996), was used as a control to confirm that DNA was present.
Cloning of ift46 alleles from ift46-1 and Supift461 and 5′ RACE of ift46 transcript in Supift461
For cloning the ift46 allele from ift46-1, ift46-1 genomic DNA was cut by PstI and self-ligated. Primer pair IFT46-3 and IFT46-10 was used to amplify the region where exogeneous DNA was integrated during the insertional mutagenesis that created ift46-1. The PCR product was cloned and sequenced.
To clone the ift46 allele from Supift461, we used the Vectorette PCR system (Sigma-Aldrich, St. Louis, MO). Supift461 genomic DNA was cut by BamH1. Primer IFT46-34 was used in the first-round PCR, and primer IFT46-10 was used for the nested PCR. The nested PCR product was cloned and sequenced.
For cloning the 5′ end of the ift46 transcripts in Supift461, total RNA was extracted from Supift461 cells using TRIzol LS Reagent (Thermo Fisher Scientific, Waltham, MA). The 5′ RACE then was carried out on the total RNA according to the standard protocol (Anonymous, 2005) with some modifications. Specifically, the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA) was used to produce total cDNA from total RNA. Poly A was added to the end of the cDNAs. Primer IFT46-4 was then used together with the (dT)17-adaptor primer to amplify the ift46 cDNA by PCR. The ift46 cDNA was further amplified by nested PCR using primers IFT46-26 and the adaptor primer. The PCR product was cloned and sequenced.
Sequence analysis
The bacterial promoter elements and the bacterial transcription factor binding site rpoD15 were identified using BPROM (prediction of bacterial promoters: http://linux1.softberry.com/berry.phtml?topic = bprom&group = programs&subgroup = gfindb). The eukaryotic polymerase II promoter was identified using the Neural Network Promoter Prediction tool (www.fruitfly.org/seq_tools/promoter.html). Putative transcription factor binding sites for snail were identified by means of the sequence analysis tool at http://consite.genereg.net/cgi-bin/consite.
Creation of DNA constructs for Chlamydomonas transformation
For generation of the truncated IFT46 gene for rescue experiments, a 1.4-kb NcoI fragment that contains the 5′ end of the IFT46 gene was removed from the plasmid that contains the 4.8-kb full-length IFT46 gene (Hou et al., 2007) and cloned into the NcoI site of the pGEM-T Easy Vector (Promega, Madison, WI). To mutagenize the 1.4-kb NcoI fragment, we used primers IFT46-30 (which encodes the first amino acid from the IFT46 gene and the neighboring upstream vector sequence) and IFT46-33 (which starts with sequence encoding IFT46 amino acid 106 and the following amino acids) to amplify the vector sequence and a portion of the 5′ end of the IFT46 gene that lacks the sequence encoding aa 2–105. The ends of the PCR products were polished by T4 polymerase and T4 kinase. The modified PCR product was self-ligated to yield circular plasmids and sequenced to verify that no additional mutations had been introduced into the coding region by the PCR. The truncated NcoI fragment (0.4 kb) was then inserted back into the IFT46 gene from which the 1.4-kb NcoI fragment had been removed to yield a construct that can express IFT46 aa 1 and 106–344.
Transformation
ift46-1 cells were cotransformed with linearized plasmid pSP124S (obtained from the Chlamydomonas Resource Center: http://chlamycollection.org/plasmid/psp124s-ble-cassette) and the construct that contains the truncated IFT46 gene using the glass-bead method of Kindle (1990). The cells were then mixed in TAP medium (Gorman and Levine, 1965) containing 0.5% agar and layered over TAP-1.5% agar containing 10 µg/ml zeocin. Zeocin-resistant transformants were picked and grown in 96-well plates in M medium.
Antibodies
The antibodies used are listed in Supplemental Table S2. For generation of an antibody to the IFT46 C-terminus, PCR primer pair IFT46-29 and IFT46-14 was used to amplify a 0.7-kb sequence encoding IFT46 aa 105–344 from a cloned IFT46 cDNA (Hou et al., 2007). The PCR product was digested with BamHI and inserted into the BamHI site of pMAL-cR1 (New England Biolabs, Beverly, MA). Expression of this construct in Escherichia coli produced a protein in which IFT46 aa 105–344 were fused to maltose-binding protein. The fusion protein was purified by amylase affinity chromatography, and antibodies were raised against the fusion protein in guinea pigs (Covance Research Products, Denver, PA).
Protein biochemistry
Preparation of Chlamydomonas whole-cell extracts, PAGE, and Western blotting were performed as described by Pazour et al. (1999). The signals on blots were detected using x-ray film or a FluorChem Q from Cell Biosciences (Santa Clara, CA). Image contrast was adjusted using Adobe Photoshop (Adobe Systems, San Jose, CA). Preparation of Chlamydomonas flagella and the further fractionation of the flagella into membrane-plus-matrix and axonemes were performed as described by Witman (1986), except that HMDEK buffer (30 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM dithiothreitol, 0.5 mM EGTA, 25 mM KCl) was used instead of HMDEKP buffer. The demembranated axonemal fraction was incubated with HMDEK buffer containing 10 mM ATP magnesium salt for 10 min on ice and then further fractionated into an ATP-soluble fraction and an ATP-insoluble axoneme fraction using the same centrifugation condition as was used to prepare the whole axoneme fraction (Witman, 1986). Sucrose gradient analysis was as described by Hou et al. (2004). The outer dynein arm binding assay was performed as described by Ahmed et al. (2008).
Flagellar regeneration experiments
After deflagellation by pH shock (Lefebvre, 1995), cells were concentrated and resuspended in the original volume of fresh M medium without aeration. The concentration step and/or subsequent incubation without aeration are necessary to induce flagellar growth in ift46-1 IFT46Δ105 cells; pH shock alone does not accomplish this (unpublished data). Aliquots were removed at various times after pH shock and mixed with an equal volume of 2% glutaraldehyde to fix the cells. Cells were then imaged by phase-contrast optics and photographed using the microscope system described in the next subsection. ImageJ (http://rsbweb.nih.gov/ij) was used to measure ∼50 flagella per sample. One flagellum per cell was measured. For determination of whether stress-induced flagellar assembly required protein synthesis, cycloheximide (final concentration 10 μg/ml) was added to the sample either immediately before or after cell concentration.
Immunofluorescence microscopy
Cells were fixed and stained by the alternate protocol of Cole et al. (1998) using Alexa Fluor 488– or 568–conjugated secondary antibodies (Molecular Probes, Eugene, OR); images were acquired with an AxioCam camera, AxioVision 3.1 software, and an Axioskop 2 plus microscope (Zeiss, Thornwood, NY). DIC imaging of IFT and the subsequent analysis of IFT were done as described by Craige et al. (2010).
Label-free quantitation by MS
Flagellar membrane-plus-matrix fractions from wild-type and ift46-1 IFT46Δ105 cells were further fractionated on 5–20% sucrose gradients (Hou et al., 2004). The peak fractions for IFT46/IFT46106-344 were determined by Western blotting using anti-IFT46C antibody. The peak fractions were combined and concentrated using Amicon Ultra-4 centrifugal filters with 10,000 molecular weight cutoff (Millipore Corporation, Billerica, MA). Proteins were loaded on an SDS–polyacrylamide gel and run briefly for 3–5 min until the dye front was ∼2–3 mm into the gel. The gel regions containing protein were excised and analyzed three times by MS (Proteomics and Mass Spectrometry Facility, University of Massachusetts Medical School). The MS data were analyzed by Scaffold 3.6.2 software (Proteome Software, Portland, OR). Fold changes were calculated using the top three common precursor intensities with normalization.
Supplementary Material
Acknowledgments
We thank D. Cole (University of Idaho), H. Qin (Texas A&M University), W. Sale (Emory University School of Medicine), M. Wirschell (University of Mississippi Medical Center), D. Mitchell (SUNY Upstate Medical University), and J. Rosenbaum (Yale University) for antibodies; B. Craige (University of Massachusetts Medical School) and J. Brown (Salem State University) for help with DIC microscopy; and J. Leszyk at the University of Massachusetts Medical School Proteomics and Mass Spectrometry Facility for his expert assistance with MS. This work was supported by National Institutes of Health grants R37 GM030626 and R35 GM122574 to G.B.W. and by the Robert W. Booth Endowment at the University of Massachusetts Medical School (G.B.W.).
Abbreviations used:
- DIC
differential interference contrast
- IFT
intraflagellar transport
- LTR
long terminal repeat
- MS
mass spectrometry
- RACE
rapid amplification of cDNA ends
- UTR
untranslated region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-03-0172) on July 12, 2017.
REFERENCES
Boldface names denote co–first authors.
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed NT, Mitchell DR. ODA16p, a Chlamydomonas flagellar protein needed for dynein assembly. Mol Biol Cell. 2005;16:5004–5012. doi: 10.1091/mbc.E05-07-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous. Rapid amplification of 5′ complementary DNA ends (5′ RACE) Nat Methods. 2005;2:629–630. doi: 10.1038/nmeth0805-629. [DOI] [PubMed] [Google Scholar]
- Awata J, Takada S, Standley C, Lechtreck KF, Bellve KD, Pazour GJ, Fogarty KE, Witman GB. NPHP4 controls ciliary trafficking of membrane proteins and large soluble proteins at the transition zone. J Cell Sci. 2014;127:4714–4727. doi: 10.1242/jcs.155275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, et al. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne AV, Irihimovitch V, Weihe A, Stern DB. Chlamydomonas reinhardtii encodes a single sigma70-like factor which likely functions in chloroplast transcription. Curr Genet. 2006;49:333–340. doi: 10.1007/s00294-006-0060-7. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Cochran DA, Craige B, Kubo T, Witman GB. Assembly of IFT trains at the ciliary base depends on IFT74. Curr Biol. 2015;25:1583–1593. doi: 10.1016/j.cub.2015.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Fine NA, Pandiyan G, Thazhath R, Gaertig J. Hypoxia regulates assembly of cilia in suppressors of Tetrahymena lacking an intraflagellar transport subunit gene. Mol Biol Cell. 2003;14:3192–3207. doi: 10.1091/mbc.E03-03-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ML, Smith AC, Kobayashi H, Purton S, Herrin DL. Structure, circadian regulation and bioinformatic analysis of the unique sigma factor gene in Chlamydomonas reinhardtii. Photosynth Res. 2004;82:339–349. doi: 10.1007/s11120-004-4213-6. [DOI] [PubMed] [Google Scholar]
- Charlier D, Piette J, Glansdorff N. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res. 1982;10:5935–5948. doi: 10.1093/nar/10.19.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi MS, Schmid MB, Roth JR. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc Natl Acad Sci USA. 1982;79:5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils A, Maurya AK, Tereshko L, Kennedy J, Brear AG, Prahlad V, Blacque OE, Sengupta P. Structural and functional recovery of sensory cilia in C. elegans IFT mutants upon aging. PLoS Genet. 2016;12:e1006325. doi: 10.1371/journal.pgen.1006325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft JM, Harris JA, Hyman S, Kner P, Lechtreck KF. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J Cell Biol. 2015;208:223–237. doi: 10.1083/jcb.201409036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave D, Pandiyan G, Wloga D, Gaertig J. Unusual intragenic suppression of an IFT52 gene disruption links hypoxia to the intraflagellar transport in Tetrahymena thermophila. bioRxiv. 2016 doi: https://doi.org/10.1101/044420. [Google Scholar]
- Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- Dentler W. Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J Cell Biol. 2005;170:649–659. doi: 10.1083/jcb.200412021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PB, Freshour JR, Mitchell DR. Chlamydomonas axonemal dynein assembly locus ODA8 encodes a conserved flagellar protein needed for cytoplasmic maturation of outer dynein arm complexes. Cytoskeleton (Hoboken) 2015;72:16–28. doi: 10.1002/cm.21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Ludington WB, Marshall WF. Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J Cell Biol. 2009;187:81–89. doi: 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Woessner JP, Goodenough UW. A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol Biol Cell. 1996;7:1235–1248. doi: 10.1091/mbc.7.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowkes ME, Mitchell DR. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol Biol Cell. 1998;9:2337–2347. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Heuser J. Structural comparison of purified dynein proteins with in situ dynein arms. J Mol Biol. 1984;180:1083–1118. doi: 10.1016/0022-2836(84)90272-9. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Pazour GJ, Witman GB. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol Biol Cell. 2004;15:4382–4394. doi: 10.1091/mbc.E04-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Witman GB. Dynein and intraflagellar transport. Exp Cell Res. 2015;334:26–34. doi: 10.1016/j.yexcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Babaev-Khaimov V, Sassaroli M, Piperno G. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J Cell Biol. 2001;153:13–24. doi: 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ide T, Yagi T, Jiang X, Hirono M, Sasaki H, Yanagisawa H, Wemmer KA, Stainier DY, Qin H, et al. TTC26/DYF13 is an intraflagellar transport protein required for transport of motility-related proteins into flagella. Elife. 2014;3:e01566. doi: 10.7554/eLife.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe R, Mahan KM, Venghaus BE, Carter ML, Herrin DL. Circadian regulation of chloroplast transcription in Chlamydomonas is accompanied by little or no fluctuation in RPOD levels or core RNAP activity. Mol Biol Rep. 2012;39:10565–10571. doi: 10.1007/s11033-012-1942-z. [DOI] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Kustu S, Inwood W. Natural history of transposition in the green alga Chlamydomonas reinhardtii: use of the AMT4 locus as an experimental system. Genetics. 2006;173:2005–2019. doi: 10.1534/genetics.106.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Brown JM, Bellve K, Craige B, Craft JM, Fogarty K, Lechtreck KF, Witman GB. Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J Cell Sci. 2016;129:2106–2119. doi: 10.1242/jcs.187120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF. IFT-cargo interactions and protein transport in cilia. Trends Biochem Sci. 2015;40:765–778. doi: 10.1016/j.tibs.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, Witman GB. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre PA. Flagellar amputation and regeneration in Chlamydomonas. Methods Cell Biol. 1995;47:3–7. doi: 10.1016/s0091-679x(08)60782-7. [DOI] [PubMed] [Google Scholar]
- Lin YC, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue T, Rohatgi R, Inoue T. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat Chem Biol. 2013;9:437–443. doi: 10.1038/nchembio.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker BF, Behal RH, Qin H, Siron LC, Taggart WD, Rosenbaum JL, Cole DG. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J Biol Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- Lucker BF, Miller MS, Dziedzic SA, Blackmarr PT, Cole DG. Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46. J Biol Chem. 2010;285:21508–21518. doi: 10.1074/jbc.M110.106997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Stevens DR, Purton S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998;14:441–447. [Google Scholar]
- Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison HM, Valente EM. Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- Mourão A, Christensen ST, Lorentzen E. The intraflagellar transport machinery in ciliary signaling. Curr Opin Struct Biol. 2016;41:98–108. doi: 10.1016/j.sbi.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Kamiya R. Microtubule sliding in mutant Chlamydomonas axonemes devoid of outer or inner dynein arms. J Cell Biol. 1986;103:1895–1902. doi: 10.1083/jcb.103.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Miller MS, Geimer S, Leitch JM, Rosenbaum JL, Cole DG. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr Biol. 2005;15:262–266. doi: 10.1016/j.cub.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Porter ME, Bower R, Knott JA, Byrd P, Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richey EA, Qin H. Dissecting the sequential assembly and localization of intraflagellar transport particle complex B in Chlamydomonas. PLoS One. 2012;7:e43118. doi: 10.1371/journal.pone.0043118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas: the use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies with Chlamydomonas reinhardi. Ann NY Acad Sci. 1953;56:831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1987. [Google Scholar]
- Schmidts M, Hou Y, Cortes CR, Mans DA, Huber C, Boldt K, Patel M, van Reeuwijk J, Plaza JM, van Beersum SE, et al. TCTEX1D2 mutations underlie Jeune asphyxiating thoracic dystrophy with impaired retrograde intraflagellar transport. Nat Commun. 2015;6:7074. doi: 10.1038/ncomms8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, Brust-Mascher I, Scholey JM. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Takada S, Kamiya R. Functional reconstitution of Chlamydomonas outer dynein arms from alpha-beta and gamma subunits: requirement of a third factor. J Cell Biol. 1994;126:737–745. doi: 10.1083/jcb.126.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Kotsis F, Braeuer P, Kuehn EW, Lorentzen E. Crystal structures of IFT70/52 and IFT52/46 provide insight into intraflagellar transport B core complex assembly. J Cell Biol. 2014;207:269–282. doi: 10.1083/jcb.201408002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E. The intraflagellar transport machinery. Cold Spring Harb Perspect Biol. 2016;8:a028092. doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Mourao A, Awasthi M, Basquin J, Lorentzen E. Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46. J Biol Chem. 2017;292:7462–7473. doi: 10.1074/jbc.M117.780155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Weber K, Mourao A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 2016;35:773–790. doi: 10.15252/embj.201593164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Dishinger J, Kee HL. Kinesin motors and primary cilia. Biochem Soc Trans. 2011;39:1120–1125. doi: 10.1042/BST0391120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Takada S, Witman GB, Kamiya R. Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil Cytoskeleton. 2001;48:277–286. doi: 10.1002/cm.1015. [DOI] [PubMed] [Google Scholar]
- Wang Q, Pan J, Snell WJ. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell. 2006;125:549–562. doi: 10.1016/j.cell.2006.02.044. [DOI] [PubMed] [Google Scholar]
- Wheeler BS. Small RNAs, big impact: small RNA pathways in transposon control and their effect on the host stress response. Chromosome Res. 2013;21:587–600. doi: 10.1007/s10577-013-9394-4. [DOI] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Koutoulis A, Pazour GJ, Witman GB. The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J Cell Biol. 1995;129:169–178. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Wood CR, Wang Z, Diener D, Zones JM, Rosenbaum J, Umen JG. IFT proteins accumulate during cell division and localize to the cleavage furrow in Chlamydomonas. PLoS One. 2012;7:e30729. doi: 10.1371/journal.pone.0030729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, Lechtreck KF. A differential cargo-loading model of ciliary length regulation by IFT. Curr Biol. 2013;23:2463–2471. doi: 10.1016/j.cub.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.